Abstract
The role of myeloid differentiation factor 88 (MyD88) in malignant tumors is largely unknown. Therefore, in this study, we aimed to examine the function and underlying mechanism of MyD88 in colorectal carcinoma in vitro using SW480 and HCT116 cell lines and in vivo using a nude mouse model. SW480 and HCT116 cells were infected with a lentiviral-based effective MyD88 siRNA virus. CCK-8 and colony formation assay were used to assess cell proliferation. Transwell and scratch assays were used to test the migration of colorectal cancer cells, and the Transwell assay was further used to analyze the invasiveness of colorectal cancer cells. Western blotting was performed to analyze the underlying mechanism of MyD88 regulation. In vitro experiments demonstrated that silencing MyD88 in SW480 and HCT116 cells markedly suppressed growth and invasion. Furthermore, MyD88 knockdown affected the MyD88-NF-κB/AP-1 signaling pathways in SW480 and HCT116 cells. In vivo, MyD88 knockdown inhibited tumor growth in a HCT116 cell subcutaneous nude model. We found that knockdown of the MyD88 gene can affect proliferation, invasion, and migration of colorectal cancer cells. We further verified that MyD88 knockdown can reduce the activity of NF-κB and AP-1 pathways. These results show that MyD88 gene plays an important role in promoting colorectal cancer, and thus can be exploited as a potential diagnostic and prognostic biomarker for colorectal cancer.
Keywords: myeloid differentiation factor 88, colorectal cancer, proliferation, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, with the second highest incidence in men, the third in women, and the third and fourth in cancer-related deaths among men and women (1). Despite recent improvements in screening strategies and the development of more effective CRC treatments, the prognosis for advanced CRC remains poor (2). Many risk factors, including viral and bacterial infections, alcohol, tobacco smoke, aging, ulcerative colitis, and a sedentary lifestyle, as well as genetic mutations may be associated with CRC (3). There are three types of genetic aberrations, namely chromosomal instability (CIN), microsatellite instability (MSI), and CpG island methylator phenotype (CIMP), involved in the pathogenesis of CRC (4).
Myeloid differentiation factor 88 (MyD88) is an essential adaptor molecule for IL-1 and Toll-like receptor (TLR) signaling (5). TLRs are a family of pattern recognition receptors that identify various pathogen-associated molecular patterns (PAMPs), molecules derived from various pathogens, and activate host innate immune defense against pathogen invasion. MyD88 signaling plays a predominant role in mediating systemic and cardiac cytokine responses in the survival of activated CD4+ T cells to promote tumor cell proliferation, invasion, metastasis and are correlated with the prognosis of HCC patient-mediated inflammatory pathway injury and neurodegenerative tissue injury (6-9). MyD88 expression is an adverse prognostic factor in ovarian cancer and is essential in adenovirus keratitis (10,11). MyD88 is a therapeutic target for inflammatory lung diseases (12). It also contributes to ocular surface homeostasis (13). However, the role of MyD88 in CRC and the mode of action following its expression remain unknown.
Previous findings showed MyD88 expression in cancer tissue and adjacent normal colorectal tissues of patients with CRC; however, the expression levels were significantly higher in the cancer tissues than in the adjacent tissues (14). The MyD88 expression level was correlated with the clinical stage, T stage, M stage and lymph node metastasis, and the survival rate of patients with CRC and higher MyD88 expression was significantly lower than that of the patients with CRC and lower MyD88 expression.
The aim of the present study was to determine the role of MyD88 in CRC. The MyD88 gene was knocked down to dissect its functional role in CRC cells. In addition, the mechanism of MyD88 knockdown, which causes change in the related signal pathway, was explored. The findings showed that MyD88 is a crucial factor affecting CRC progression.
Materials and methods
MyD88 siRNA synthesis and transfection
siRNA target sequences were identified on the human MyD88 sequence. According to the siRNA design guidelines, DNA template oligonucleotides corresponding to three different siRNA sequences (siRNA-1, siRNA-2 and siRNA-3) were designed as follows: siRNA-1: GCC TAT CGC TGT TCT TGA A, siRNA-2: GAC TGA TTC CTA TTA AAT A, siRNA-3: CAGCGAGCTAATTGAGAAA. These siRNA and negative control (NC) sequences were produced by Genechem Co. Ltd.. The SW480 and HCT116 cells were cultured in a medium with 10% FBS. When these CRC cells were at approximately 90% confluency, the NC, siRNA-1, siRNA-2 and siRNA-3 sequences were transfected into the cells, using Lipofectamine 3000 (Invitrogen), according to the manufacturer's instructions.
RNA preparation and quantitative PCR amplification. RT-qPCR was used to test mRNA expression of the MyD88 gene. Total RNA was extracted from CRC cell lines, using the Qiagen RNeasy kit (Qiagen Bioinformatics) according to the manufacturer's instructions, and quantified using UV260/280 nm to an absorption ratio of z1.8. An RT Reagent kit (Takara Bio Lnc.) was used for reverse transcription of RNA into cDNA. The primers (BioSune Biotechnology Co., Ltd.) MyD88 F: GGC TGC TCT CAA CAT GCG A, R: CTG TGT CCG CAC GTT CAA GA and GAPDH F: GAA GGT GAA GGT CGG AGT C, R: GAA GAT GGT GAT GGG ATT TC, were designed to amplify cDNA with SYBR Premix EX Taq kit (Takara Bio Lnc.). PCR conditions were 95°C for 2 min, 95°C for 15 sec, and 60°C for 30 sec for 40 cycles. The relative amount of MyD88 mRNA was normalized to that of GAPDH. The 2−ΔΔCq method was used to calculate this as Livak and Schmittgen had reported (15).
Construction of lentiviral vectors
In order to establish better stable transfection for later experiments, we constructed lentiviral-based siRNA targeting MyD88 vector. The primers, designed by BioSune Biotechnology Co., Ltd. contained two restriction sites, AgeI (underlined) and EcoRI (in bold) (siRNA1 F: CCGGGC CTA TCG CTG TTC TTG AAT TCA AGA GAT TCAA GAA CAG CGA TAG GCT TTT TTG GTA CC, R: AATTGG TAC CAA AAA AGC CTA TCG CTG TTC TTG AAT CTC TTG AAT TCA AGA ACA GCG ATA GGC; siRNA2 F: CCGGGA CTG ATT CCT ATT AAA TAT TCA AGA GAT ATT TAA TAG GAA TCA GTC TTT TTT GGT ACC, R: AAT TGG TAC CAA AAA AGA CTG ATT CCT ATT AAA TAT CTC TTG AAT ATT TAA TAG GAA TCA GTC; and siRNA3 F: CCGGCA GCG AGC TAA TTG AGA AAT TCA AGA GAT TTC TCA ATT AGC TCG CTG TTT TTT GGT ACC, R: AAT TGG TAC CAA AAA ACA GCG AGC TAA TTG AGA AAT CTC TTG AAT TTC TCA ATT AGC TCG CTG). The primers were designed to anneal and form double-stranded DNA. pLKO.1-EGFP-Puro (Fenghui Bio) was used as an empty vector for purification and recovery (Tiangen) after double digestion with AgeI and EcoRI (Thermo Fisher Scientific, Inc.). The purified linear empty vector and the double-stranded DNA formed by annealing were then ligated with T4 DNA Ligase (Invitrogen) and transformed, using stabl3, cloned, sequenced, and then subjected to plasmid extraction (Tiangen) for the next experiment.
Lentiviral infection and stable cell line screening
The SW480 and HCT116 cell lines were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). The two cell lines were cultured and maintained in DMEM (Gibco BRL) with 10% fetal bovine serum (FBS), and incubated at 37°C and 5% CO2. The validated MyD88 knockdown plasmid and empty vector were co-transfected with pMD2.G and psPAX2 (Fenghui Bio) into 293T cells, and the supernatant was filtered after 2 days of culture. HCT116 and SW480 cells were infected with the supernatant, and after 2 weeks of screening with puromycin, the protein was extracted for verification. Stably expressed cells were selected for subsequent experiments.
Western blot analysis
Western and IP cell lysis buffer (Beyotime), containing 1% PMSF (Amresco), was used to cleave proteins for 30 min on ice. After 30 min, the lysed cells were centrifuged at 12,000 × g for 10 min at 4°C to extract the supernatant for protein quantification using the BCA Protein Assay kit (Thermo Fisher Scientific, Inc.), and boiled for 10 min by adding 5X SDS. The total amount of protein (50 µg) was added to the prepared 12% SDS-PAGE gels for electrophoretic separation and transferred to 0.45 µm PVDF membranes (Amersham Hybond, GE Healthcare). The membranes were then blocked with 1% albumin from bovine serum (Amresco) for 2 h. Next, the membranes were incubated overnight with diluted MyD88 (1:1,000, AF5195; Affinity), GAPDH (1:1,000, ab181602; Abcam), NF-κB p65 (1:1,000, AF5006; Affinity), p-NF-κB p65 (1:1,000, #3033; Cell Signaling), c-jun (1:1,000, bs-0670R; Bioss), and p-c-jun (1:1,000, bs-3172R; Bioss) antibodies on a shaker at 4°C. The membranes were washed for 10 min three times with TBS-T (0.1% Tween-20) at room temperature, incubated in goat anti-rabbit IgG H&L (HRP) (1:2,000, ab7090; Abcam) for 1 h, and then washed. Subsequently the membranes were exposed to enhanced chemiluminescence substrate detection solution (Lulong Biotech).
Colony formation assay
Cells (500/well) were seeded in a 6-cm plate, and cultured for 14 days in a culture medium, containing 10% FBS. The culture solution was discarded and cells were infiltrated with methanol for 10 min, stained with crystal violet, washed with water, dried and counted.
Cell proliferation assay
CCK-8 (Dojindo) was used to detect cell proliferation. Cells were seeded, at densities of 1,000 cells/well, in a 96-well plate, then incubated at 37°C, 5% CO2. After 24, 48, 72, and 96 h, the culture solution was discarded and 100 µl of 10% CCK-8 serum-free medium was added. Cell proliferation was estimated using a microplate reader (Bio-Tek) after 1 h of incubation.
Cell migration and invasion assay
In the migration experiment, 4×104 CRC cells, in serum-free medium, were seeded into the upper chamber of a Transwell insert (8-mm pore size; Corning Inc.), and a medium with 20% FBS was added in the lower chamber as a chemoattractant. In the invasion experiment, Matrigel (BD Bioscience) was coated on the upper chamber seeded with 9×104 cells, and the lower chamber contained 20% FBS medium. After incubation at 37°C, 5% CO2 for 48 h, the Transwell chamber was taken out and the medium in the well was discarded and washed with calcium-free PBS. The cells were, then, fixed with methanol for 30 min and stained with 0.1% crystal violet for 20 min. The upper unmigrated cells were gently wiped off with a cotton swab, and counted under microscope.
Wound healing assay for CRC cell migration
The cells were seeded in a 6-well plate. After the cells had grown to 100% confluency, the cell layer was scratched with a 20 µl pipette tip and the medium containing 10% FBS was replaced with a serum-free medium. Images of the cells were captured at 0, 24, 48, and 72 h.
Animal experiments
The experiment performed with animals was approved by the Institutional Animal Care and Use Committee at the Fujian Medical University. Ten five-week-old BALB/c nude mice raised at the Animal Experimental Center of Fujian Medical University were used in this study. HCT116-pLKO.1 or HCT116-pLKO.1-sh3 cells (2×106), in serum-free DMEM medium, were injected into the bilateral subcutaneous (HCT116-pLKO.1 cells on one side and HCT116-pLKO.1-sh3 cells on the other side) of 10 mice. The manipulation was stable, rapid and gentle to reduce discomfort in mice. During tumor growth, the tumor volume was measured every four days using the formula: length x width 2/2. and the correlation function graph was plotted. After 6 weeks of tumor growth the nude mice were euthanized by carbon dioxide (100% CO2 gas replacement rate at 10-30% container volume/min), and the tumors were removed after the heartbeat and respiratory arrest of nude mice, photographed and immediately weighed. The tumors were soaked in formalin for immunohistochemical analysis.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tumor tissue specimens from the mice. After dewaxing, hydration, and antigen retrieval, the remaining experimental procedures were performed in accordance with the UltraSensitiveTM SP (mouse/rabbit) IHC kit instructions. Finally, the degree of staining of the slices was observed under the microscope after DAB staining, hematoxylin counterstaining and neutral resin sealing.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software. The data were expressed as the means ± standard deviation (SD) and analyzed by one-way ANOVA or Student's t-test. Differences with P-values <0.05 were considered to be statistically significant.
Results
Expression of MyD88 mRNA and protein after siRNA transfection
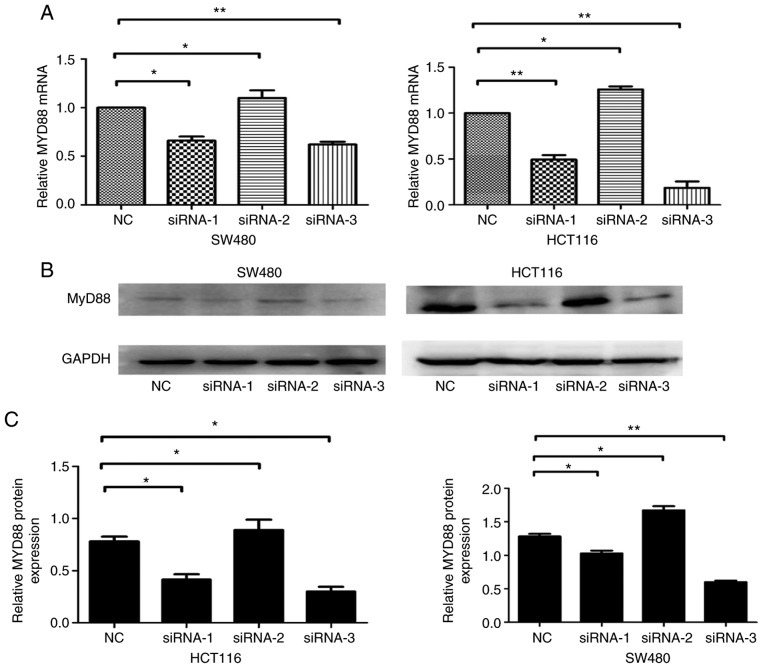
An RNAi-based method was employed to silence MyD88 and detect the effects of knocking down the gene. NC, siRNA-1, siRNA-2 and siRNA-3 sequences were transfected into SW480 and HCT116 cells. The MyD88 mRNA and protein levels, determined 48 h after transfection using RT-qPCR and western blot analysis, were shown to decrease in the siRNA-transfected SW480 and HCT116 cells. Compared with NCsiRNA and siRNA-2, which had no effect on MyD88 mRNA and protein expression levels, siRNA-1 and siRNA-3 sequences inhibited MyD88 mRNA and protein expression levels (Fig. 1A-C). Since siRNA-1 and siRNA-3 were the most effective siRNA, and these sequences were selected to silence MyD88 in this study.
Figure 1.
Knockdown of MyD88 in the SW480 and HCT116 cells. (A) Total RNA from SW480 and HCT116 cells after transfection using NC, siRNA-1, siRNA-2 and siRNA-3 sequences were assessed by RT-qPCR using MyD88 primers. GAPDH was amplified for an internal control. The siRNA-1 and siRNA-3 sequences resulted in higher suppression of the levels of MyD88 mRNA. (B) MyD88 protein in the SW480 and HCT116 cells after transfection using NC, siRNA-1, siRNA-2 and siRNA-3 sequences were detected by western blotting. GAPDH protein was used an internal control. (C) Semi-quantitative analysis showed that MyD88 protein levels in siRNA-1 and siRNA-3 cells were markedly lower compared with NC and siRNA-2 cells. The mRNA expression of MyD88 in SW480 NC group and HCT116 NC group cells were taken as 1. Error bars represent mean ± SEM, representative of three experiments, *P<0.05, **P<0.01.
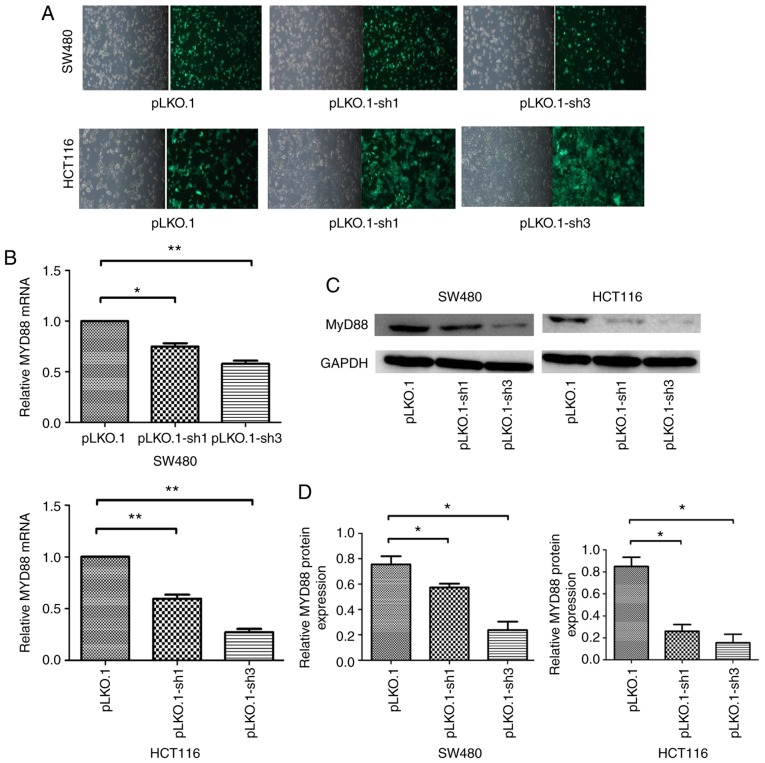
Expression of MyD88 mRNA and protein after infection with pLKO.1-sh1 and pLKO.1-sh3 lentiviral vectors
The NC, siRNA-1 and siRNA-3 sequences were constructed into lentiviral vectors and packaged as lentiviruses carrying NC, siRNA-1 and siRNA-2 sequences named pLKO.1, pLKO.1-sh1 and pLKO.1-sh3, respectively. Infection efficiency was quantified by counting cells under a fluorescence microscope 48 h after infection, because the pLKO.1 vector can express green fluorescent protein in the SW480 and HCT116 cells, which had >95% efficiency (Fig. 2A). Quantitative PCR and western blot analyses showed that MyD88 mRNA and protein levels were markedly inhibited after infection in SW480 and HCT116 cells. Compared with the pLKO.1 group, the pLKO.1-sh1 and pLKO.1-sh3 groups revealed relatively lower MyD88 mRNA expression levels in SW480 and HCT116 cells (Fig. 2B). Consistent with mRNA expression, western blotting showed inhibition of MyD88 protein expression in the pLKO.1-sh1 and pLKO.1-sh3 groups compared with pLKO.1 group in SW480 and HCT116 cells (Fig. 2C and D). To generate stably transfected pLKO.1, the pLKO.1-sh1 and pLKO.1-sh3 cells, the culture medium was replaced with a selection medium containing 2 µg/ml puromycin (Sigma) for two weeks. Stably transfected CRC cells were cultured using medium containing 0.5 µg/ml puromycin. These cells were then used for subsequent experiments.
Figure 2.
Downregulation of MyD88 mRNA and protein expression by lentivirus-mediated MyD88 siRNA in SW480 and HCT116 cells. (A) pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 lentivirus infected the SW480 and HCT116 cells. After 72 h infection, the green fluorescent protein (GFP) as demonstrated by fluorescence microscopy. (B) The mRNA and (C) protein expression was analyzed using RT-qPCR and western blot analysis after infection with lentivirus in the SW480 and HCT116 cells. GAPDH was used as an internal control. (D) Semi-quantitative analysis showed that the level of MyD88 protein were signifi-cantly inhibited by pLKO.1-sh1 and pLKO.1-sh3 in the SW480 and HCT116 cells. The mRNA expression of MyD88 in SW480 pLKO.1 group and HCT116 pLKO.1 group cells were taken as 1. Error bars represent mean ± SEM, representative of three experiments. *P<0.05, **P<0.01.
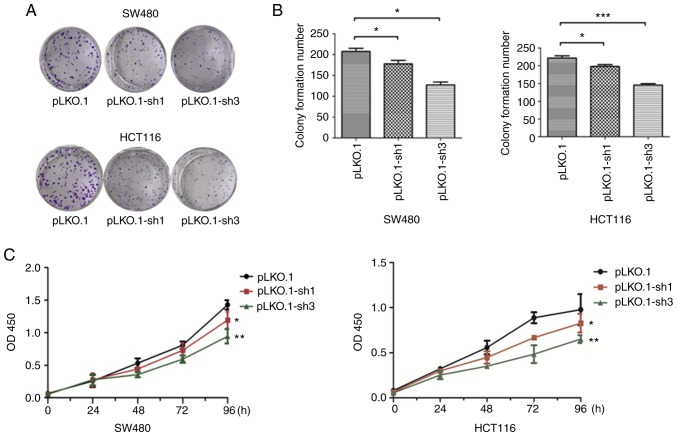
Knockdown of MyD88 reduces the proliferation of SW480 and HCT116 cells
To verify whether the expression of MyD88 can affect tumor proliferation, stably transfected pLKO.1, the pLKO.1-sh1 and pLKO.1-sh3 CRC cells were used. Cell proliferation was estimated using colony formation and CCK-8 assays. The colony formation assay showed a decrease (P<0.05) in the number of colonies in the pLKO.1-sh1 and pLKO.1-sh3 groups, while there was no significant difference in the colony numbers in the pLKO.1-sh1 and pLKO.1-sh3 groups (P>0.05), when compared to the pLKO.1 group in the SW480 and HCT116 cells (Fig. 3A and B). CCK-8 experiment results showed that compared to the LKO.1 group, cell proliferation of the pLKO.1-sh1 and pLKO.1-sh3 groups, which were similar, was lower in the SW480 and HCT116 cells (P<0.05) (Fig. 3C). The cell viabilities of the pLKO.1-sh1 and pLKO.1-sh3 groups were decreased when compared with the pLKO.1 group in both colorectal carcinoma cell lines. These results indicate that silencing MyD88 contributes to the inhibition of CRC cell proliferation.
Figure 3.
Effects of siMyD88 on the viability of pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 in the SW480 and HCT116 cells. (A and B) Downregulation of MyD88 reduced the colony number in colony formation assays in the SW480 and HCT116 cells. (C) CCK-8 assays revealing that MyD88 knock-down inhibits the proliferation of SW480 and HCT116 cells. Error bars represent mean ± SEM, representative of three experiments. *P<0.05, **P<0.01, ***P<0.001.
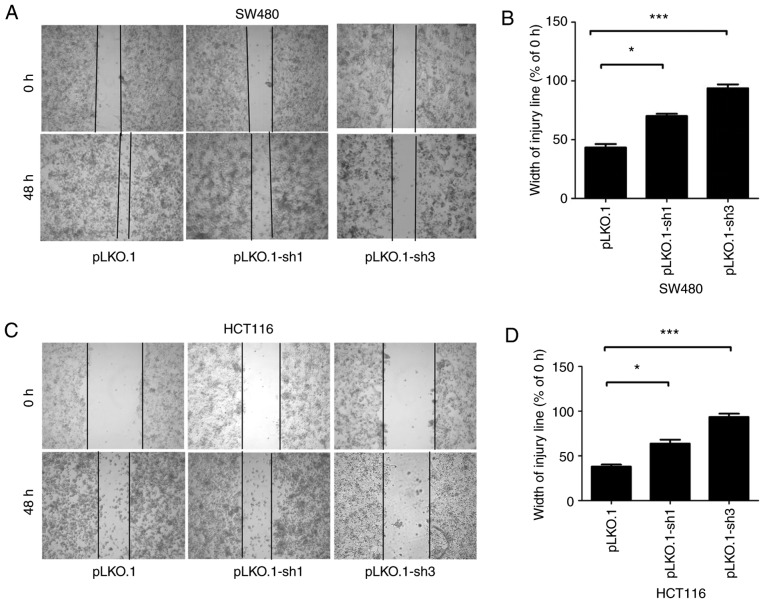
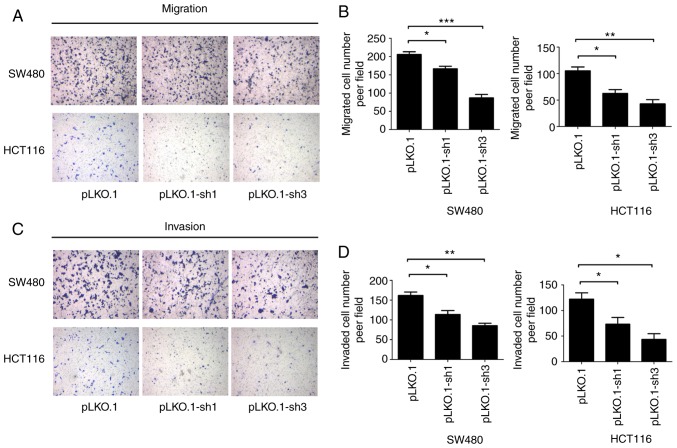
MyD88 knockdown attenuates migration and invasion of colorectal carcinoma cells
To determine the ability of MyD88 knockdown in colorectal carcinoma cells to suppress migration, the Transwell assay and scratch test were used to verify the migration ability of SW480 and HCT116 cells harboring MyD88 knockdown. Similar to migration, the invasiveness of the SW480 and HCT116 cells was tested using the Transwell assay. Wound healing/scratch was used to confirm changes in cell migration by observing the extent of wound closure. In the pLKO.1-sh1 and pLKO.1-sh3 groups (P>0.05), the healing speed of the scratches were lower than in the pLKO.1 group in SW480 and HCT116 cells (P<0.05) (Fig. 4A-D). To further verify the results of the migration, we used the Transwell experiment. The number of colorectal cells in the pLKO.1 group that migrated through the Transwell polycarbonate filter was markedly higher than that of cells in the pLKO.1-sh1 group (P<0.05), which was similar to the number of cells in the pLKO.1-sh3 group in the SW480 and HCT116 cells (P>0.05) (Fig. 5A and B). The results showed that knockdown of MyD88 significantly suppressed cell migration ability compared with the NC group. In vitro cell invasion showed that the number of cells penetrating the basement membrane was significantly higher in the pLKO.1 group than in the pLKO.1-sh1 and pLKO.1-sh3 groups (P>0.05) in the SW480 and HCT116 cells (Fig. 5C and D).
Figure 4.
Effects of siMyD88 on the migration of pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 in the SW480 and HCT116 cells. (A) In the SW480 cells, representative images of pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 cell wound healing and microscopic observations were photographed 0 and 48 h after scratching the cell surface. (B) The width of the scratch was analyzed using a histogram in the SW480 cells. (C) In the HCT116 cells, representative images of pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 cells wound healing and microscopic observations were photographed 0 and 48 h after scratching the cell surface. (D) The width of the scratch was analyzed using a histogram in the HCT116 cells. Error bars represent mean ± SEM, representative of three experiments. *P<0.05, ***P<0.001.
Figure 5.
Effects of siMyD88 on SW480 and HCT116 cells migration and invasion. (A) Migration ability of pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 in the SW480 and HCT116 cells. (B) Number of migrated cells in the pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 groups in the SW480 and HCT116 cells. (C) Invasion ability of pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 in the SW480 and HCT116 cells. (D) Number of invaded cells in the pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 groups in the SW480 and HCT116 cells. Error bars represent mean ± SEM, representative of three experiments. *P<0.05, **P<0.01, ***P<0.001.
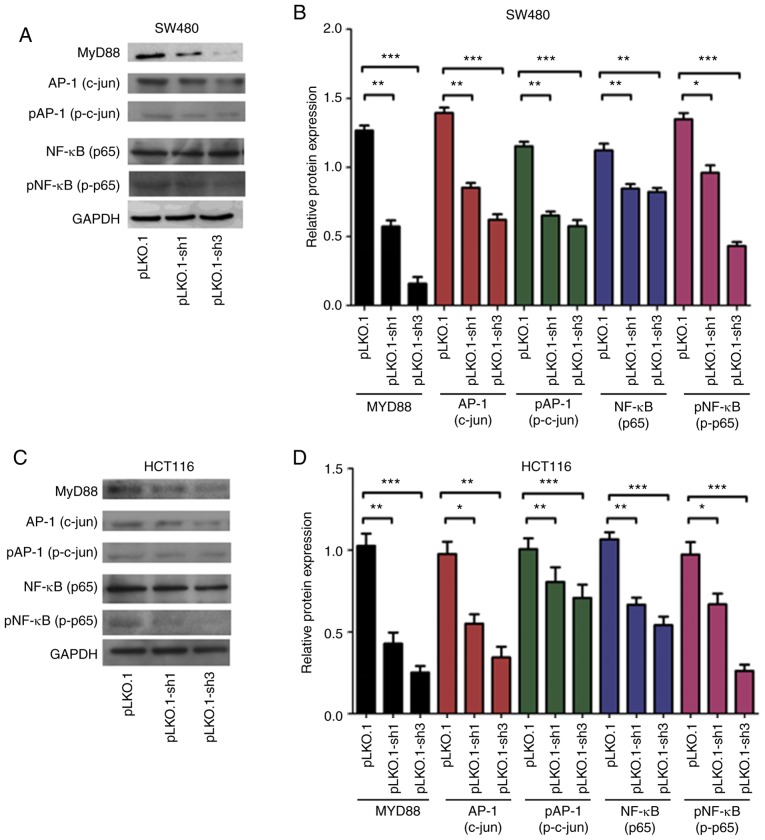
MyD88 affects NF-κB and AP-1 signaling pathways
In the above experiments, we confirmed that MyD88 can affect the biological behavior of CRC cells, but the underlying mechanisms remain unknown. To investigate the mechanism by which MyD88 silencing leads to a decrease in proliferation and invasion, we evaluated NF-κB (p65), p-NF-κB (p-p65), AP-1 (c-jun) and p-AP-1 (pc-Jun) proteins; these proteins are critical for the growth and invasion of cancer cells, and are very important for NF-κB and AP-1 signaling pathways. Western blot analysis showed that NF-κB (p65), p-NF-κB (p-p65), AP-1 (c-jun) and p-AP-1 (p-c-jun) protein levels in the pLKO.1 group cells increased compared with those in the pLKO.1-sh1 and pLKO.1-sh3 groups (P<0.05) in the SW480 and HCT116 cells (Fig. 6A-D). The results indicated that MYD88 suppressed the expression of related signaling pathway proteins.
Figure 6.
Western blot analysis revealed that the silencing of MyD88 markedly inhibited the expression NF-κB (p65), p-NF-κB (p-p65), AP-1 (c-jun) and p-AP-1 (p-c-jun) protein in the SW480 and HCT116 cells. (A) The protein electropherogram showed the protein expression situation of NF-κB (p65), p-NF-κB (p-p65), AP-1 (c-jun), p-AP-1 (p-c-jun) and MyD88 in pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 groups in the SW480 cells. (B) The protein expression of the pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 groups was analyzed using a histogram in the SW480 cells. (C) An electropherogram was used to show the protein expression of NF-κB (p65), p-NF-κB (p-p65), AP-1 (c-jun), p-AP-1 (p-c-jun) and MyD88 in pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 groups in the HCT116 cells. (D) The protein expression of the pLKO.1, pLKO.1-sh1 and pLKO.1-sh3 groups was analyzed using a histogram in the HCT116 cells. Error bars represent mean ± SEM, representative of three experiments. *P<0.05, **P<0.01, ***P<0.001.
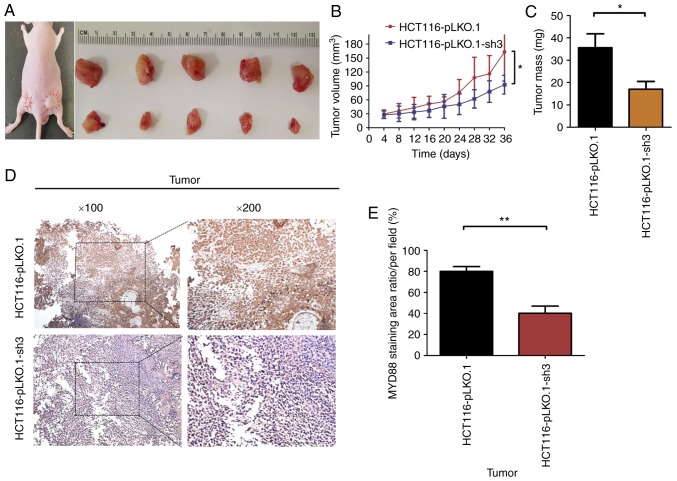
Knockdown of MyD88 suppresses tumor growth in BALB/c nude mice with subcutaneous xenograft tumors
To verify whether tumor growth in vivo was the same as in vitro, the HCT116-pLKO.1 or HCT116-pLKO.1-sh3 cells were subcutaneously xenografted into 5-week-old BALB/c nude mice (5 mice/group). At 6 weeks of subcutaneous tumor growth, nude mice were all successfully established in a subcutaneous mouse model (Fig. 7A). At autopsy, none of the mice had any ascites or liver, lung or lymph node metastasis. The results showed that the growth rate of subcutaneous tumors in the HCT116-pLKO.1-sh3 group was significantly lower than that in the HCT116-pLKO.1 group (P<0.05) (Fig. 7B). In addition, the tumor weight of the HCT116-pLKO.1-sh3 group was markedly lower than that of the HCT116-pLKO.1 group (P<0.05) (Fig. 7C). Since MyD88 protein expression was inhibited in HCT116-pLKO.1-sh3 cells, immunohistochemistry was used to analyze the expression levels of MyD88 in subcutaneous xenograft tumors. Compared with the HCT116-pLKO.1 group, MyD88 expression levels were markedly inhibited in the HCT116-pLKO.1-sh3 group (P<0.05) (Fig. 7D and E).
Figure 7.
Suppression of tumor growth by silencing the MyD88 gene in the HCT116 cells subcutaneous xenografts. Three groups (pLKO.1 and pLKO.1-sh3 groups) were subcutaneous xenografted onto nude mice. (A) The tumor growth of pLKO.1-sh3 group was significantly inhibited compared with pLKO.1 group. (B) The size of the primary tumors was measured every 4 days. Mice were sacrificed after 5 weeks. (C) The weights of tumors were weighed. Significant differences were identified between the pLKO.1 and pLKO.1-sh3 groups. (D) Immunohistochemical analysis of the expression of MyD88 in HCT116 cell subcutaneous xenograft tumors. Staining showed expression of MyD88 (×100 and ×200) in pLKO.1 and pLKO.1-sh3 groups. (E) Quantitative evaluation of the expression of MyD88. Bars show the MyD88 staining area ration/per field. Marked differences between the pLKO.1 and pLKO.1-sh3 groups were identified. *P<0.05, **P<0.01.
Discussion
MyD88 plays a central role in innate immune response. It regulates NF-κB signaling activity (16) and the MAPKs-c-jun (AP-1) signaling pathway (17). MyD88 and MyD88-related signaling have been shown to be involved in the progression and carcinogenesis of cancer-associated cells, with both intrinsic and extrinsic inflammation. In addition, detection of abnormal MyD88 expression is used to predict the prognosis of various human cancers (e.g., lymph, liver) (18). Hence, MyD88 can be considered a potential carcinogenic marker for related research. It will have a positive impact on the pathogenic factors and treatment of CRC.
In the current study, we successfully knocked down the MyD88 gene and verified the knockdown effect of the protein using cell fluorescence, RT-qPCR and western blot analysis. We determined whether MyD88 gene was able to affect the biological behavior of CRC cells, and subsequently confirmed the ability of MyD88 knockdown to inhibit cell proliferation and invasion in vivo using relevant cell function tests. The results suggest that knockdown MyD88 can suppress proliferation, invasion, and migration of CRC. These findings agree with those of previous studies which showed that, the SNP of MyD88 was associated with poor survival of Caucasian patients with CRC (19). Ikebe et al (20) reported that siMyD88 can decrease the invasiveness and migration ability of pancreatic cancer cells. Wang et al (21) indicated that a high MyD88 expression is associated with liver metastasis and poor prognosis in CRC patients. We also found that MyD88 expression was high in CRC tissues and was associated with poor survival in colorectal carcinoma patients (14).
Similarly, we explored whether knockdown of MyD88 gene had the same effect in vitro. We used subcutaneous injection of colon cancer cells to conduct subcutaneous tumorigenesis experiments, confirming that knockdown of MyD88 gene in mice can also inhibit tumor growth. This finding is in agreement with previous studies, which considered that MyD88 increased the risk of developing colorectal neoplasm (22,23). Our findings indicate that MyD88 can promote the growth of CRC. Thus, knockdown of MyD88 gene can inhibit tumor growth, invasion, migration and other related behavioral changes.
However, the specific mechanism by which MyD88 induces biological behavior in CRC is unclear. Previous findings showed that MyD88 can protect the intestine from tumorigenesis via IL-18/MyD88 signaling as compared to colitis-associated cancer animal models of IL-18−/−, IL-18R1−/−, and MyD88−/− mice, as well as wild-type mice (17). By contrast, MyD88 can promote the malignant transformation of colitis to colorectal cancer through the TLR/MyD88 pathway in the MyD88-knockout mice after LPS treatment (24). To date, it remains unclear how and why MyD88 possesses these dual functions in CRC, and the underlying mechanisms require further investigation.
As previously shown, MyD88 signaling plays a role in regulating the growth of normal epithelial and cancer cells in the gut. The activated form of NF-kB is upregulated and functionally correlated with many tumors, modulating proliferation and invasion. AP-1 and, specifically, c-Jun, affect tumor cell proliferation, migration and invasion (25). Recent studies have shown that mitogen-activated protein kinases (MAPKs) and nuclear factor-κB (NF-κB) signaling pathways are two key factors leading to the development of lipopolysaccharide (LPS)-induced acute lung injury (ALI) (26), osteoclastogenesis (27), neuroinflammation (28), atherosclerosis (29), tumorigenesis in prostate carcinoma and glioblastoma (25), and activation of human dendritic cells (30). Under the premise of MyD88-induced tumor bioethology, we investigated whether knockdown MyD88 induced changes in NF-κB (p65) and AP-1 (MAPKs-c-jun) proteins. In knockdown of MyD88 stably transfected cell lines, we found that p65, phosphorylated p65, c-jun, and phosphorylated c-jun protein expression resulted in different degrees of decline. This demonstrates that knockdown of MyD88 gene can affect MyD88-mediated activation of the NF-κB (p65) and AP-1 (MAPKs-c-jun) pathways, thereby effectively retarding the aggressive transformation of the tumor.
In summary, MyD88 could be an independent factor to explore its role in colon cancer, which can be used as a factor in the pathogenesis of colon cancer, and deserves further development as a potential diagnostic and prognostic biomarker. We found that knocking down the MyD88 gene affects proliferation, invasion, and migration of CRC cells, and can reduce the activity of NF-κB and AP-1 pathways. These results show that the MyD88 gene plays an important role in promoting CRC and may be exploited as a diagnostic and prognostic biomarker for CRC.
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81702424 and 81872364), the Fujian Provincial Health Department Young and Middle-aged Talents Training Project (grant no. 2018-ZQN-46), the Joint Funds for the Innovation of Science and Technology, Fujian Province (grant no. 2017Y9092), the Project of Science and Technology Research Program in Fujian Province (grant no. 2016B044), the Fujian Provincial Natural Science Foundation (grant no. 2018J05127), and the National Clinical Key Specialty Construction Project (General Surgery) of China.
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JY, GZ, YH and WZ conceived and designed the study. GZ, ZC and CL performed the experiments. SY and JY interpreted the results. GZ and ZC wrote the paper, and all authors contributed to writing. All authors read and approved the final manuscript.
Ethics approval and consent to pariticipate
The experiment performed with animals was approved by the Institutional Animal Care and Use Committee at the Fujian Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Testa U, Pelosi E, Castelli G. Colorectal cancer: Genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med Sci (Basel) 2018;6:E31. doi: 10.3390/medsci6020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaghoubi N, Soltani A, Ghazvini K, Hassanian SM, Hashemy SI. PD-1/PD-L1 blockade as a novel treatment for colorectal cancer. Biomed Pharmacother. 2019;110:312–318. doi: 10.1016/j.biopha.2018.11.105. [DOI] [PubMed] [Google Scholar]
- 4.Marmol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:E197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lingel A, Ehlers E, Wang Q, Cao M, Wood C, Lin R, Zhang L. Kaposi's sarcoma-associated herpesvirus reduces cellular myeloid differentiation primary-response gene 88 (MyD88) expression via modulation of its RNA. J Virol. 2016;90:180–188. doi: 10.1128/JVI.02342-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y, Zou L, Chen C, Li D, Chao W. Role of cardiac- and myeloid-MyD88 signaling in endotoxin shock: A study with tissue-specific deletion models. Anesthesiology. 2014;121:1258–1269. doi: 10.1097/ALN.0000000000000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Li M, Hung CY, Sinha M, Lee LM, Wiesner DL, LeBert V, Lerksuthirat T, Galles K, Suresh M, et al. MyD88 shapes vaccine immunity by extrinsically regulating survival of CD4+ T cells during the contraction phase. PLoS Pathog. 2016;12:e1005787. doi: 10.1371/journal.ppat.1005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Yin Y, Tang J, Xie Y, Han Z, Zhang X, Liu Q, Qin X, Huang X, Sun B. Long non-coding RNA Myd88 promotes growth and metastasis in hepatocellular carcinoma via regulating Myd88 expression through H3K27 modification. Cell Death Dis. 2017;8:e3124. doi: 10.1038/cddis.2017.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Syeda S, Patel AK, Lee T, Hackam AS. Reduced photoreceptor death and improved retinal function during retinal degeneration in mice lacking innate immunity adaptor protein MyD88. Exp Neurol. 2015;267:1–12. doi: 10.1016/j.expneurol.2015.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d'Adhemar CJ, Spillane CD, Gallagher MF, Bates M, Costello KM, Barry-O'Crowley J, Haley K, Kernan N, Murphy C, Smyth PC, et al. The MyD88+ phenotype is an adverse prognostic factor in epithelial ovarian cancer. PLoS One. 2014;9:e100816. doi: 10.1371/journal.pone.0100816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Ramke M, Chintakuntlawar AV, Lee JY, Rajaiya J, Chodosh J. Role of MyD88 in adenovirus keratitis. Immunol Cell Biol. 2017;95:108–116. doi: 10.1038/icb.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Padova F, Quesniaux VFJ, Ryffel B. MyD88 as a therapeutic target for inflammatory lung diseases. Expert Opin Ther Targets. 2018;22:401–408. doi: 10.1080/14728222.2018.1464139. [DOI] [PubMed] [Google Scholar]
- 13.Reins RY, Courson J, Lema C, Redfern RL. MyD88 contribution to ocular surface homeostasis. PLoS One. 2017;12:e0182153. doi: 10.1371/journal.pone.0182153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu G, Zheng W, Huang Y, Hua J, Yang S, Ye J. Expression of MyD88 in cancer tissue of patients with colorectal cancer and clinical significancer. Journal of Jilin University (Medicine Edition) 2018;44:1047–1051. [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Yan F, Guan J, Peng Y, Zheng X. MyD88 NEDDylation negatively regulates MyD88-dependent NF-κB signaling through antagonizing its ubiquitination. Biochem Biophys Res Commun. 2017;482:632–637. doi: 10.1016/j.bbrc.2016.11.084. [DOI] [PubMed] [Google Scholar]
- 17.Tanishima M, Takashima S, Honda A, Yasuda D, Tanikawa T, Ishii S, MaruYama T. Identification of optineurin as an interleukin-1 receptor-associated kinase 1-binding protein and its role in regulation of MyD88-dependent signaling. J Biol Chem. 2017;292:17250–17257. doi: 10.1074/jbc.M117.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Yu K, Zhang X, Yu S. Dual functional roles of the MyD88 signaling in colorectal cancer development. Biomed Pharmacother. 2018;107:177–184. doi: 10.1016/j.biopha.2018.07.139. [DOI] [PubMed] [Google Scholar]
- 19.Klimosch SN, Försti A, Eckert J, Knezevic J, Bevier M, von Schönfels W, Heits N, Walter J, Hinz S, Lascorz J, et al. Functional TLR5 genetic variants affect human colorectal cancer survival. Cancer Res. 2013;73:7232–7242. doi: 10.1158/0008-5472.CAN-13-1746. [DOI] [PubMed] [Google Scholar]
- 20.Ikebe M, Kitaura Y, Nakamura M, Tanaka H, Yamasaki A, Nagai S, Wada J, Yanai K, Koga K, Sato N, et al. Lipopolysaccharide (LPS) increases the invasive ability of pancreatic cancer cells through the TLR4/MyD88 signaling pathway. J Surg Oncol. 2009;100:725–731. doi: 10.1002/jso.21392. [DOI] [PubMed] [Google Scholar]
- 21.Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, Bando Y, Kudo E, Shimada M, Sano T. High expression of toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer. 2010;102:908–915. doi: 10.1038/sj.bjc.6605558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 23.Chang CM, Chia VM, Gunter MJ, Zanetti KA, Ryan BM, Goodman JE, Harris CC, Weissfeld J, Huang WY, Chanock S, et al. Innate immunity gene polymorphisms and the risk of colorectal neoplasia. Carcinogenesis. 2013;34:2512–2520. doi: 10.1093/carcin/bgt228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiechl G, Bauer B, Fuss I, Lang SA, Moser C, Ruemmele P, Rose-John S, Neurath MF, Geissler EK, Schlitt HJ, et al. Tumor development in murine ulcerative colitis depends on MyD88 signaling of colonic F4/80+CD11b(high)Gr1(low) macrophages. J Clin Invest. 2011;121:1692–1708. doi: 10.1172/JCI42540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galardi S, Mercatelli N, Farace MG, Ciafre SA. NF-κB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 2011;39:3892–3902. doi: 10.1093/nar/gkr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Yang X, Liu T, Guan M, Feng X, Dong W, Chu X, Liu J, Tian X, Ci X, et al. Kaempferol regulates MAPKs and NF-κB signaling pathways to attenuate LPS-induced acute lung injury in mice. Int Immunopharmacol. 2012;14:209–216. doi: 10.1016/j.intimp.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Park JW, Yoon HJ, Kang WY, Cho S, Seong SJ, Lee HW, Yoon YR, Kim HJ. G protein-coupled receptor 84 controls osteoclastogenesis through inhibition of NF-κB and MAPK signaling pathways. J Cell Physiol. 2018;233:1481–1489. doi: 10.1002/jcp.26035. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Fang W, Fan S, Liao W, Xiong Y, Liao S, Li Y, Xiao S, Liu J. Neurotropin inhibits neuroinflammation via suppressing NF-κB and MAPKs signaling pathways in lipopoly-saccharide-stimulated BV2 cells. J Pharmacol Sci. 2018;136:242–248. doi: 10.1016/j.jphs.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Pan JX. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-κB signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:322–328. [PubMed] [Google Scholar]
- 30.Parola C, Salogni L, Vaira X, Scutera S, Somma P, Salvi V, Musso T, Tabbia G, Bardessono M, Pasquali C, et al. Selective activation of human dendritic cells by OM-85 through a NF-κB and MAPK dependent pathway. PLoS One. 2013;8:e82867. doi: 10.1371/journal.pone.0082867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.