Abstract
To verify whether amyloid precursor protein (APP) affects the migration and invasion of breast cancer cell lines, and to understand its underlying mechanisms, epithelial-mesenchymal transition (EMT), the mitogen-activated protein kinase (MAPK) signaling pathway and the matrix metalloproteinase (MMP) family were investigated in MDA-MB-231, MCF-7 and BT474 human breast cancer cells. Breast cancer cell lines were transfected with plasmids containing APP coding sequences (pEGFP-n1-APP) and APP short hairpin RNA (pENTR APP shRNA). APP overexpres-sion efficiency, knockout efficiency and the expression levels of related genes were tested using reverse transcription-quantitative PCR (RT-qPCR) and western blot analyses. The effects of APP and mitogen-activated protein kinase kinase (MEK) inhibitor on cell migration and invasion were examined using Transwell assays. The results demonstrated that APP was significantly upregulated in the pEGFP-n1-APP group (P<0.05), and significantly downregulated in the pENTR APP shRNA group (P<0.05), compared with the control group. APP overexpression increased the migratory and invasive ability of human breast cancer cells (P<0.05), whereas APP silencing significantly inhibited cell migration and invasion (P<0.05). RT-qPCR and western blot analysis results suggested that APP overexpression significantly increased the expression of MMP-9, MMP-2, MMP-3, N-cadherin and vimentin (P<0.05). In addition, the enhanced expression of APP markedly affected the phosphorylation of mitogen-activated protein kinase kinase kinase 11 (MLK3), mitogen-activated protein kinase kinase 4 (MEK4) and mitogen-activated protein kinase 10 (JNK3; P<0.05). Additionally, APP overexpression had no effect on the total expression levels of MLK3, MEK4, and JNK3; however, APP overexpression significantly decreased the expression levels of E-cadherin and cytokeratin (P<0.05). Conversely, APP silencing had the opposite effects. When cells were treated with the MEK inhibitor PD0325901, the expression of APP was not altered, nor was the expression levels of MEK and its upstream signaling molecules. Taken together, the present findings suggested that APP could affect the migration and invasion of human breast cancer cells by mediating the activation of the MAPK signaling pathway, thereby promoting the EMT process.
Keywords: amyloid precursor protein, breast cancer, mitogen-activated protein kinase, migration, invasion
Introduction
Breast cancer is a serious threat to human health (1) and a major cause of morbidity and mortality among women, accounting for ~23% of female malignancies (2,3). With the development of medical technology and the prevalence of precancerous screening technology, breast cancer morbidity and mortality rates have declined in developed countries, but incidence and mortality are rising worldwide (4-6). Currently, the main treatment methods for breast cancer include surgical resection, radiotherapy and chemotherapy, as well as immunotherapy (7). However, due to its high metastatic rate, strong invasiveness and recurrence, the prognosis of patients with breast cancer remains poor (7,8).
Accumulating evidence suggested that breast cancer metastasis can occur early and has significant organ specificity (9,10), which may explain the high expression levels of adhesion molecules, such as vascular cell adhesion protein 1 (11), in addition to a series of genes associated with breast cancer metastasis. Breast cancer cells expressing the epidermal growth factor receptor oncogene possess neurotropic properties, and commonly metastasize to the central nervous system (12). A recent study demonstrated a significant correlation between the low expression level of breast cancer type 1 susceptibility protein in the tumor tissues of patients with breast cancer and brain metastasis, but the specific mechanism remains to be fully elucidated (13). In a previous study, investigation of circular RNA (circRNA) expression by high-throughput sequencing identified numerous circRNAs as potential molecular markers for brain metastasis in breast cancer (14). Among those, the interaction between hsa_circ_0001944 and microRNA-509 was found to be the most promising (14). In addition to brain metastasis, a subset of breast cancer cells expressing C-X-C chemokine receptor (CXCR2) tend to metastasize to lung tissues rich in stromal cell-derived factor 1 (SDF1) (15). Although the role of the SDF1/CXCR4 axis in breast cancer metastasis has not been fully elucidated, the results from a previous meta-analysis suggested that the SDF1/CXCR4 protein expression level is a reliable prognostic biomarker for breast cancer (16). Bone and liver metastases (17,18) also play a critical role in breast cancer. In vivo, it was confirmed that interleukin-1 was a mediator of bone metastasis in breast cancer cells (19). Although certain biomarkers of breast cancer can predict organ specificity, the mechanism of cancer metastasis in complex living systems is not fully understood. Epithelial-mesenchymal transition (EMT) has been considered to be responsible for tumor metastasis (20,21). Several studies identified that tumor cells display decreased epithelial properties and enhanced mesenchymal properties, associated with reduced expression of epithelial-related markers, including cytokeratin and E-cadherin, and increased expression of mesenchymal-related markers, such as vimentin, N-cadherin and matrix metalloproteinases (MMPs). This is accompanied by activation of the mitogen-activated protein kinase (MAPK) signaling pathway (22-24). Amyloid precursor protein (APP) has a long extracellular domain, a transmembrane domain and a shorter intracellular domain, and is widely expressed in most cells (25). Early studies have reported that β-APP is formed via the hydrolysis of extracellular fragments accumulated in the brain tissue, affecting the structure and function of the nervous system, and is one of the factors associated with the pathogenesis of Alzheimer's disease (26-29). In addition, tumor cells can promote hematogenous metastasis by increasing the expression of APP (30). In addition, the intracellular domain of APP can interact with adaptor proteins, activating downstream intracellular signaling molecules, thereby affecting the physiological functions of nerves and immune cells, influencing cell migration and invasion (31).
APP is upregulated in a variety of cancer types (32,33). A previous study found that APP is an independent predictor of poor prognosis in non-intraluminal breast cancer and an important risk factor for breast cancer distal lymph node metastasis (34). In the present study, immunohistochemistry and in vitro experiments were performed to examine the association between APP expression in breast cancer and clinical symptoms in patients with breast cancer. The present results suggested that APP was positively correlated with the expression of androgen receptor (AR) and Ki-67. In vitro experiments from the present study demonstrated that the bioactive androgen dihydrotestosterone induced APP mRNA transcription in a dose- and time-dependent manner, while hydroxyflutamide, an AR blocking agent, effectively inhibited this process. Moreover, the proliferative activity of breast cancer cells is associated with the expression levels of APP (35). However, little is known on the role of APP in breast cancer progression. In the present study, the effects of APP on the migration and invasion of breast cancer cells were investigated using APP overexpression and knockdown cell lines. The present results provides theoretical support for the development of APP as a novel therapeutic targets for the management of breast cancer.
Materials and methods
Cell lines
MDA-MB-231, MCF-7, MCF-10, BT549 and BT474 breast cancer cell lines were obtained from the Shanghai Institute of Life Sciences Cell Bank and cultured according to the manufacturer's instructions.
Related reagents
DMEM and FBS were purchased from Gibco (Thermo Fisher Scientific, Inc.). The empty plasmid pEGFP-n1-APP (cat. no. 69924) and pENTR APP short hairpin (sh)RNA (cat. no. 30135) plasmids were supplied by Addgene Inc. The transfection reagent polyetherimide (PEI; cat. no. 03880) was supplied by Sigma-Aldrich (Merck KGaA). PrimeScript RT reagent kit (Takara Bio, Inc.) and One Step SYBR-Green PrimeScript RT-PCR kit II (Takara Bio, Inc.) kits were used for reverse transcription (RT) and quantitative-PCR (q-PCR), respectively. Transwell chambers and Matrigel were purchased from BD Biosciences. Rabbit anti-human APP (1:2,000 for western blot analysis; 1:300 for immunohistochemistry; cat. no. 2452S), mouse anti-human E-cadherin (1:2,000; cat. no. 14472), mouse anti-human N-cadherin (1:2,000; cat. no. 14215), mouse anti-human cytokeratin (1:2,000; cat. no. 4545), mouse anti-human vimentin (1:2,000; cat. no. 49636), mouse anti-human MMP-9 (1:2,000; cat. no. 3852), rabbit anti-human MMP-2 (1:2,000; cat. no. 4022), rabbit anti-human MMP-3 (1:2,000; cat. no. 14351) and rabbit anti-human mitogen-activated protein kinase kinase kinase 11 (MLK3) primary antibodies (1:2,000; cat. no. 2817) were purchased from Cell Signaling Technology, Inc. Rabbit anti-human MEK4 (1:2,000; cat. no. ab33912), rabbit anti-human phosphorylated (p)-MEK4 (1:2,000; cat. no. ab131353), rabbit anti-human p-MLK3 (1:2,000; cat. no. ab191530), rabbit anti-human JNK3 (1:2,000; cat. no. ab126591), rabbit anti-human p-JNK3 (1:2,000; cat. no. ab124956) and rabbit anti-human β-actin primary antibodies (1:4,000; cat. no. ab179467), as well as horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:5,000; cat. no. ab6721) and goat anti-mouse (1:3,500; cat. no. ab6789) secondary antibodies were purchased from Abcam. TRIzol® reagent was obtained from Thermo Fisher Scientific, Inc. qPCR primers were synthesized by Shanghai Biotech.
Cell culture
MDA-MB-231, MCF-7 and BT474 cells were cultured in DMEM containing 10% FBS and 1% streptomycin mixture, and then placed in a humidified atmosphere with 5% CO2 at 37°C. Cell passaging was conducted using 0.25% trypsin + EDTA.
Human breast carcinoma tissues and immunohistochemistry
A total of eight female patients with breast cancer (age, 37-62 years) underwent clinical and histopathological diagnosis at the First Affiliated Hospital of Xiamen University between January and December 2018. All patients included in the study had clinical TNM stage III or IV breast cancer, and had not been treated with radiotherapy or chemotherapy prior to surgery. Written informed consent was obtained from each patient. The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Xiamen University. All immunohistochemical samples were retrieved from the Department of Pathology of the First Affiliated Hospital of Xiamen University. Immunohistochemical staining was performed on tissues. Samples were soaked in 10% neutral buffered formalin at 4°C and embedded in paraffin. Subsequently, the paraffined slices (thickness, 5 µm) were dewaxed and rehydrated, then placed on glass slides. Antigen retrieval was performed in a pressure cooker for 3 min at 120°C. Slices were then incubated with primary antibody (1:500) at 4°C overnight, followed by incubation with corresponding secondary antibodies for 2 h at room temperature. After washing in PBS, the slices were added with diaminobenzidine, stained with 10% hematoxylin for 10 sec at room temperature and sealed with neutral gum. Sections were visualized and photographed using an optical microscope (Leica Microsystems GmbH; magnification, ×200). Results were quantified using Image Pro Plus software (version 6.0; Media Cybernetics).
Plasmid transfection
MDA-MB-231, MCF-7 and BT474 cells in the logarithmic growth phase were trypsinized and counted, and then seeded into a 6-well plate at a density of 5×105 cells/well. To determine the effects of APP overexpression, five groups were defined as follows: i) Untransfected cells (PEI but no plasmid); ii) control plasmid (PEI with 2 µg control pEGFP plasmid); iii) control shRNA (PEI with 2 µg control pENTR plasmid); iv) APP overexpression (PEI with 2 µg pEGFP-n1-APP plasmid); and v) APP shRNA (PEI with 2 µg pENTR APP shRNA plasmid). For the subsequent experiments, cells were transfected and categorized into three groups as follows: i) Control (PEI but no plasmid); ii) overexpression (PEI with 2 µg pEGFP-n1-APP plasmid); and iii) silencing (PEI with 2 µg pENTR APP shRNA plasmid) groups. The transfected cells were used in subsequent experiments after 48 h.
Drug treatment
MDA-MB-231 cells were cultured in six-well plates at a density of 5×105 cells/well, and then treated with the MEK inhibitor PD0325901 (Sigma-Aldrich; Merck KGaA; 1 nM) for 48 h at 37°C. When drug treatment and transfection were combined, MDA-MB-231 cells were cultured in six-well plates at a density of 5×105 cells/well, and then transfected with APP overexpressing plasmid for 24 h, followed by treatment with the MEK inhibitor PD0325901 (1 nM) for 48 h.
Migration assay
MDA-MB-231, MCF-7 and BT474 cells in the logarithmic growth phase were trypsinized and counted, then seeded in a 6-well plate at a density of 5×105 cells per well. After reaching complete adherence and ~60% confluence, the cells were transfected with the corresponding plasmids. After 48 h of incubation, the cells were digested and counted, resuspended in serum-free medium, and inoculated into the upper Transwell chamber at a density of 1×104 cells/well. The lower chamber was filled with 500 µl complete medium containing 10% FBS in a 24-well plate. After the upper and lower chambers were assembled, they were placed in a cell culture incubator for 4 h, and the upper chamber was removed. The Transwell inserts were scraped with cotton swabs to remove residual cells. Cells on the lower surface of the well were stained with 0.1% crystal violet solution for 30 min at room temperature and visualized using an optical microscope (Leica Microsystems GmbH; magnification, ×100).
Invasion assay
The matrix collagen solution (8 mg/ml) was diluted 8X with serum-free medium. The diluted Matrigel (50 µl) was then added to the upper chamber of the Transwell insert in advance, and placed in a cell culture incubator for a minimum of 1 h at 37°C. After treating the cells as described in the migration assay, cells resuspended in the serum-free medium were seeded at a density of 1×104 cells/well into the upper chamber of the Transwell insert, where the Matrigel solution had been previously added at room temperature and then put into culture incubator at 37°C for 30 min. The lower chamber was filled with 500 µl complete medium containing 10% FBS. After the upper and lower chambers were assembled, they were placed in a cell culture incubator for 12 h at 37°C. Next, the cells were treated as described in the migration assay section. Stained cells were photographed using an optical microscope (Leica Microsystems GmbH; magnification, ×100) and counted in five randomly selected fields.
Gap closure assay
A total of 1×106 MDA-MB-231 cells were seeded into a 6-well plate, on which the UV-sterilized polydimethylsiloxane blocks (1 mm ×2 cm) were placed before. The culture medium was supplemented with 10% FBS. When the cells reached a confluence of 90%, the blocks were removed and the width was photographed using an optical microscope (Leica Microsystems GmbH; magnification, ×40) and measured at 24, 48 and 72 h. The gap closure values were calculated as follows: Percentage of gap closure= 1 -(widtht/width0) ×100% (36).
RT-qPCR
Total RNA was extracted from MDA-MB-231, MCF-7 and BT474 cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) at 4°C, according to the manufacturer's instructions. A ratio of A260/A280 was considered to be acceptable within the range of 1.8-2.0. The RT reaction was performed as follows: Initial incubation at 37°C for 5 min, followed by sequential steps at 42°C for 15 min, at 85°C for 10 sec and at 4°C for 60 min. β-actin was selected as the reference gene. For the q-PCR, the thermocycling conditions were as follows: Pre-denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 sec and of annealing at 60°C for 15 sec. The 2−ΔΔCq method (37) was used to quantify gene expression. The primer sequences used were as follows: β-actin forward, 5′-CCT CGC CTT TGC CGA TCC-3′ and reverse, 5′-GGA TCT TCA TGA GGT AGT CAG TC-3′; MMP-9 forward, 5′-AAT CTC ACC GAC AGG CAG CT-3′ and reverse, 5′-CCA AAC TGG ATG ACG ATG TC-3′; APP forward, 5′-TCT CGT TCC TGA CAA GTG CAA-3′ and reverse, 5′-GCA AGT TGG TAC TCT TCT CAC TG-3′; cytokeratin forward, 5′-ACC AAG TTT GAG ACG GAA CAG-3′ and reverse, 5′-CCC TCA GCG TAC TGA TTT CCT-3′; vimentin forward, 5′-GCC CTA GAC GAA CTG GGT C-3′ and reverse, 5′-GGC TGC AAC TGC CTA ATG AG-3′; MMP-2 forward, 5′-TAC AGG ATC ATT GGC TAC ACA CC-3′ and reverse, 5′-GGT CAC ATC GCT CCA GAC T-3′; MMP-3 forward, 5′-CTG GAC TCC GAC ACT CT G GA-3′ and reverse, 5′-CAG GAA AGG TTC TGA AGT G AC C-3′; E-cadherin forward, 5′-CGA GAG CTA CAC GTT CAC GG-3′ and reverse, 5′-GGG TGT CGA GGG AAA AAT AGG-3′; N-cadherin forward, 5′-TCA GGC GTC TGT AGA GGC TT-3′ and reverse, 5′-ATG CAC ATC CTT CGA TAA GA C TG-3′.
Western blot analysis
The cells were lysed with RIPA buffer (Beyotime Institute of Biotechnology) to extract the total protein from each group. Protein lysates were centrifuged at 12,000 × g for 15 min at 4°C and the protein concentration was determined using a bicinchoninic acid assay. Samples (30 µg per lane) were isolated using 8-10% SDS-PAGE and transferred to PVDF membranes, followed by blocking with 5% skimmed milk at room temperature for 2 h. Target proteins were detected with specific antibodies overnight at 4°C and observed on Bio-Rad Gel Imager using HRP-conjugated secondary antibodies (1:5,000; incubated for 3 h at room temperature) and enhanced chemiluminescence reagents (Western Lightning, Inc.). The bands were quantified using Quantity One 4.6 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data analysis were performed using SPSS 21.0 (IBM Corp.). Graphs were drawn using GraphPad Prism 6.0 (GraphPad Software, Inc.). Multivariate mean comparisons were performed using one-way ANOVA with Dunnett's test. Student's t-test was used to analyze the difference between two groups. All experimental data were obtained from ≥3 independent experiments. Data are the presented as mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
APP expression in human breast cancer tissue samples and verification of transfection efficiency following APP overexpression and knockdown in breast cancer cells
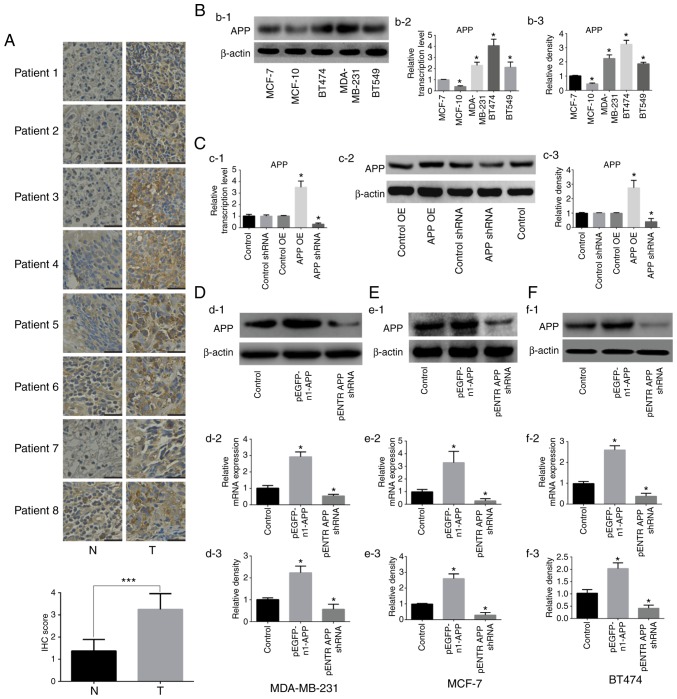
The expression of APP was detected by immunohistochemistry in human breast cancer tissues. The results shown in Fig. 1A indicated a significant upregulation of APP in breast cancer tissues compared with adjacent tissues (P<0.001). Subsequently, the expression levels of APP in five breast cancer cell lines were examined (Fig. 1B), and three cell lines (MDA-MB-231, MCF-7 and BT474) showing a moderate expression of APP were selected for further experiments. APP overexpression and knockdown were then performed on these selected cell lines.
Figure 1.
Expression of APP in human breast cancer tissues and effects of APP overexpression and knockdown in breast cancer cell lines. (A) APP expression in human breast cancer tissues by immunohistochemistry and statistical analysis. APP-positive staining was indicated by a brown color. Magnification, ×200. Scale bar, 100 µm. (Bb-1) Representative protein expression bands of APP in different human breast cancer cell lines. (Bb-2) RT-qPCR results of mRNA expression levels of APP. (Bb-3) Densitometric analysis of the protein expression level of APP. (Cc-1) Empty plasmids have no effect on the mRNA expression level of APP in MDA-MB-231 cells. (Cc-2) Protein expression of APP and (Cc-3) statistical results. (Dd-1) Representative images of APP protein expression by western blot analysis following APP overexpression and knockdown in MDA-MB-231 breast cancer cells. (Dd-2) APP mRNA expression in MDA-MB-231 breast cancer cells using RT-qPCR. (Dd-3) Statistical analysis of the protein expression level of APP. (Ee-1) Protein expression of APP by western blot analysis following APP overexpression and silencing in MCF-7 breast cancer cells. (Ee-2) APP mRNA expression using RT-qPCR in MCF-7 breast cancer cells. (Ee-3) Statistical analysis of the protein expression level of APP. (Ff-1) APP protein expression was analyzed by western blot analysis following APP overexpression and knockdown in BT474 breast cancer cells. (Ff-2) RT-qPCR results of the mRNA expression levels of APP in BT474 breast cancer cells. (Ff-3) Statistical analysis of the protein expression level of APP. Results are representative of three independent experiments. Data are presented as the mean ± SD. *P<0.05 vs. corresponding control. ***P<0.001. APP, amyloid precursor protein; OE, overexpression; PEI, polyetherimide; N, adjacent normal tissue; T, tumor tissue; RT-qPCR, reverse transcription-quantitative PCR; shRNA, short hairpin RNA.
Following transfection with the corresponding plasmids, it was found that the empty plasmids had no effects on the expression of APP in MDA-MB-231 cell line, therefore cells treated with PEI alone were used as the control in the subsequent experiments (Fig. 1C). The expression levels of APP in MDA-MB-231, MCF-7 and BT474 cells were detected by RT-qPCR and western blot analysis. The mRNA and protein expression levels of APP in cells transfected with the pEGFP-n1-APP plasmid were significantly increased compared with control cells (Fig. 1D-F; P<0.05). APP mRNA and protein expression levels in pENTR APP shRNA cells were significantly decreased compared with control cells (P<0.05). The present results suggested that the transfection experiments were successful.
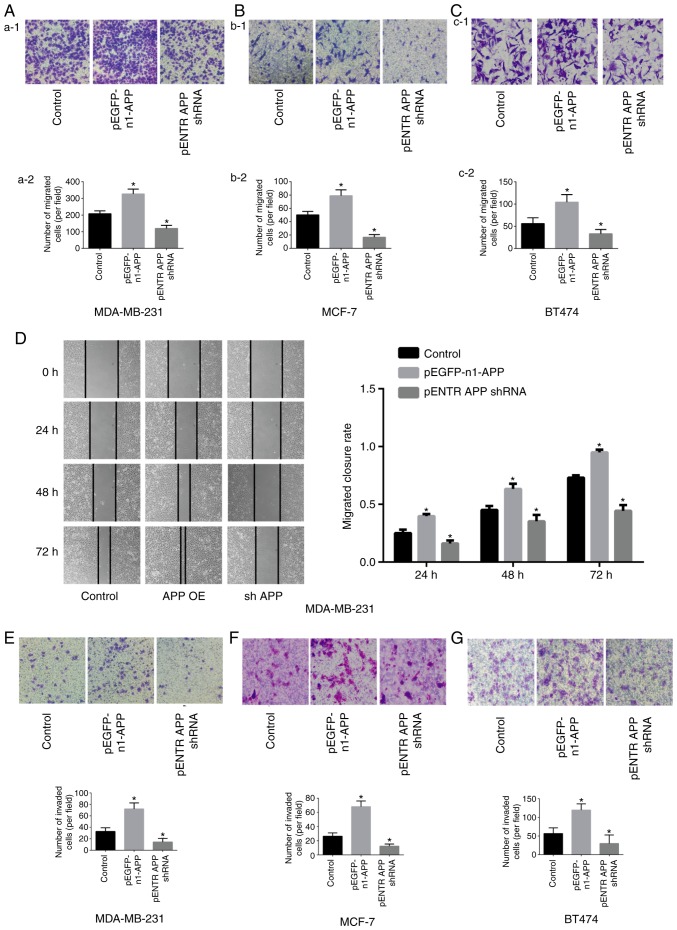
APP promotes the migratory and invasive ability of breast cancer cells
The results of the Transwell assay showed that the migratory (Fig. 2A-C) and invasive (Fig. 2E-G) abilities of pEGFP-n1-APP-transfected cells were significantly enhanced compared with the control group (P<0.05), whereas pENTR APP shRNA-transfected cells exhibited significantly impaired migratory and invasive abilities compared with the control group (P<0.05). The results of the gap closure assay revealed that APP overexpression significantly promoted gap closure in MDA-MB-231 cells, while APP knockdown inhibited gap closure (Fig. 2D; P<0.05).
Figure 2.
Effects of APP overexpression and silencing on the migration of MDA-MB-231, MCF-7 and BT474 breast cancer cells. Staining images from the Transwell assay. The effects of APP overexpression and silencing were investigated in (Aa-1) MDA-MB-231 cells and (Aa-2) quantified, in (Bb-1) MCF-7 cells and (Bb-2) quantified, and in (Cc-1) BT474 breast cancer cells and (Cc-2) quantified. Magnification in Aa-1 and Bb-1, ×100. Scale bar, 200 µm. Magnification in Cc-1, ×400. Scale bar, 50 µm. (D) Representative gap closure assay images and statistical analysis, showing the effects of APP overexpression and silencing on the cell migration of MDA-MB-231 cells. Magnification, ×40. Scale bar, 250 µm. Representative images of the effects of APP overexpression and silencing on the invasive ability of breast cancer (E) MDA-MB-231, (F) MCF-7 and (G) BT474 cells (magnification, ×100; Scale bar, 200 µm) and the statistical results of the number of invading cells. Five representative fields were randomly imaged and quantified for each well in cell migration and invasion assays. Data are presented as the mean ± SD from three independent experiments. *P<0.05 vs. control. APP, amyloid precursor protein; shRNA, short hairpin RNA; OE, overexpression.
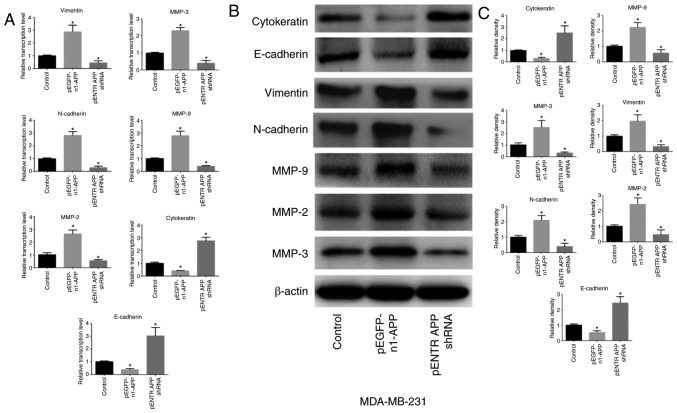
APP regulates the expression of EMT-related genes in breast cancer
APP overexpression significantly upregulated the expression levels of the mesenchymal-related genes MMP-9, MMP-2, MMP-3, N-cadherin and vimentin in MDA-MB-231 cells (P<0.05), whereas the expression levels of the epithelial-related genes E-cadherin and cytokeratin were significantly attenuated (Fig. 3; P<0.05). APP knockdown exhibited the opposite effects (P<0.05).
Figure 3.
Effects of APP overexpression and silencing on the expression levels of EMT-related factors in MDA-MB-231 breast cancer cells. (A) mRNA expression levels of EMT-related genes were detected using RT-qPCR following APP overexpression and silencing in MDA-MB-231 cells. (B) Representative western blotting bands of the protein expression of EMT-related factors. (C) Protein expression levels of EMT-related factors were detected by western blot analysis following APP overexpression and silencing in MDA-MB-231 cells. All experiments were performed in triplicate. Data are presented as the mean ± SD. *P<0.05 vs. control. APP, amyloid precursor protein; EMT, epithelial-to-mesenchymal transition; RT-qPCR, reverse transcription-quantitative PCR; MMP, matrix metalloproteinase; shRNA, short hairpin RNA.
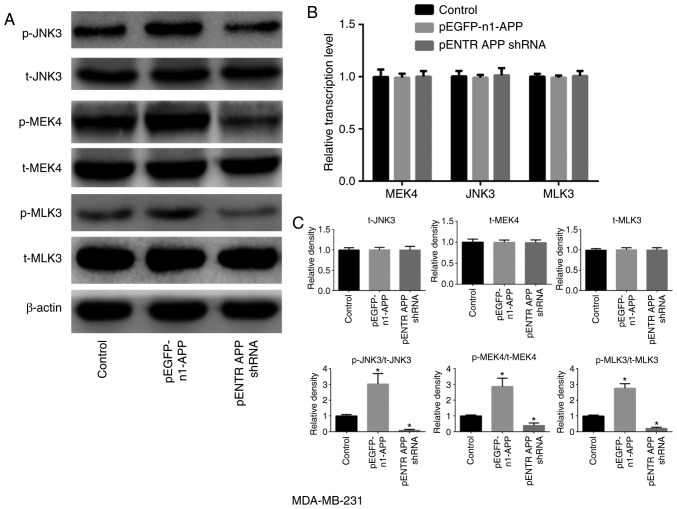
APP is involved in the expression and phosphorylation of MAPK signaling pathway-related molecules in breast cancer
APP overexpression notably increased the phosphorylation of MLK3, MEK4 and JNK3 in breast cancer MDA-MB-231 cells compared with the control group (Fig. 4; P<0.05). However, no significant differences were observed in the expression levels of total MLK3, MEK4 and JNK3 in any of the groups compared with the control. However, the phosphorylation levels of MLK3, MEK4 and JNK3 were significantly decreased following APP silencing (P<0.05).
Figure 4.
Effects of APP overexpression and silencing on the expression and phosphorylation levels of MAPK signaling pathway-related factors in MDA-MB-231 breast cancer cells. (A) Protein expression and phosphorylation levels of MAPK signaling pathway-related factors following APP overexpression and silencing. (B) mRNA expression level of MAPK signaling pathway-related molecules by RT-qPCR following APP overexpression and silencing. (C) Statistical analysis of the western blot bands. APP overexpression significantly increased the phosphorylation levels of MLK3, MEK4 and JNK3 in MDA-MB-231 breast cancer cells, when compared with the control groups. All experiments were performed in triplicate. Data are presented as the mean ± SD. *P<0.05 vs. control. APP, amyloid precursor protein; MLK3, mixed lineage kinase 3; MEK4, mitogen-activated protein kinase kinase 4; t-, total; p-, phosphorylated; shRNA, short hairpin RNA; MAPK, mitogen-activated protein kinase.
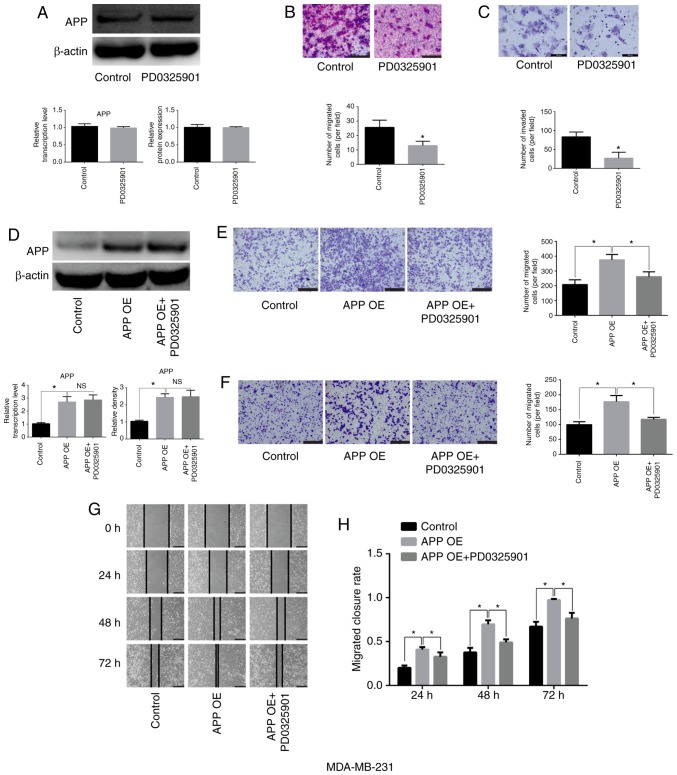
MEK inhibitor PD0325901 does not affect the expression of APP but impairs the migratory and invasive ability of breast cancer cells
MDA-MB-231 cells were cultured and seeded into 6-well plates at a density of 5×105 cells/well. They were then treated with the MEK inhibitor PD0325901 (1 nM) for 48 h. Cells not treated with the inhibitor were used as the control group. After 48 h, total protein was collected to detect APP protein expression. In addition, Transwell and Matrigel assays were performed. PD0325901 treatment alone did not affect the expression levels of APP (Fig. 5A), but significantly inhibited the migratory and invasive abilities of breast cancer cells compared with the control group (Fig. 5B and C; P<0.05). Moreover, following APP overexpression, MDA-MB-231 cells (Fig. 5D) were treated with MEK inhibitor PD0325901, and it was found that PD0325901 could significantly reduce the migratory and invasive ability increased following APP overexpression, as shown by migration, invasion and gap closure assays (Fig. 5E-H; P<0.05).
Figure 5.
Effects of the MEK inhibitor PD0325901 on the expression level of APP and the migratory and invasive abilities of MDA-MB-231 breast cancer cells. (A) MEK inhibitor exerted no effect on the mRNA and protein expression levels of APP, as indicated by RT-qPCR and western blot analysis. (B) Representative staining images showing the effect of PD0325901 on the migratory ability of breast cancer cells and statistical analysis of the number of migrated cells. Magnification, ×200. Scale bar, 100 µm. (C) Representative staining images showing the effect of PD0325901 on the invasive ability of breast cancer cells and statistical analysis of the number of invaded cells. Magnification, ×400. Scale bar, 50 µm. (D) PD0325901 exerted no effect on APP-overexpressing MDA-MB-231 breast cancer cells. Following APP overexpression and treatment with the MEK inhibitor PD0325901, PD0325901 significantly reduced the migratory and invasive ability of breast cancer cells, as indicated by the (E) migration and (F) invasion assays. Scale bar, 200 µm. Magnification, ×100. (G) Representative gap closure assay images and (H) statistical analysis. Magnification, ×40. Scale bar, 250 µm. In total, five representative fields were randomly imaged and quantified for each well in the cell migration and invasion assays. All the experiments were performed in triplicate. Data are presented as the mean ± SD. *P<0.05. APP, amyloid precursor protein; MEK, mitogen-activated protein kinase kinase; OE, overexpression; RT-qPCR, reverse transcription-quantitative PCR; NS, not significant.
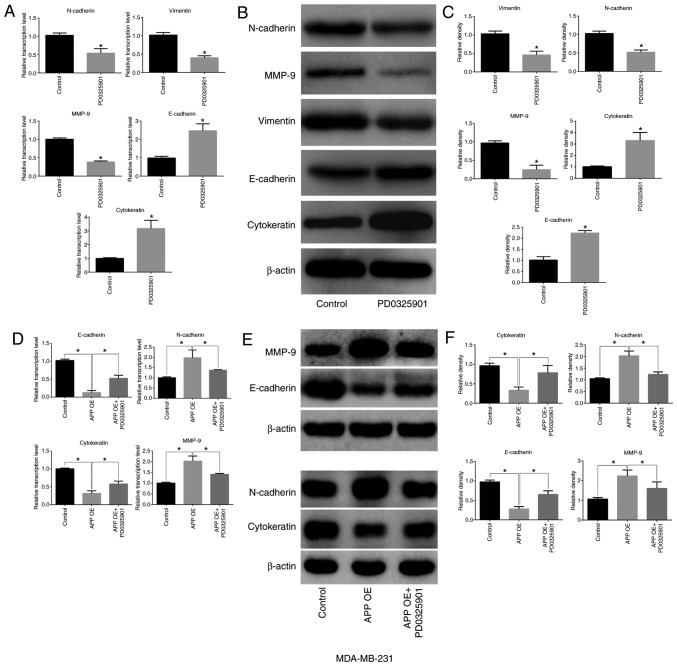
MEK inhibitor PD0325901 regulates the expression of EMT-related factors in breast cancer cells
PD0325901 treatment alone significantly inhibited the expression levels of MMP-9, N-cadherin and vimentin in MDA-MB-231 breast cancer cells (P<0.05), but significantly increased the expression levels of E-cadherin and cytokeratin (Fig. 6A-C; P<0.05). A decreased expression of epithelial-associated markers (E-cadherin and Cytokeratin) and increased expression of mesenchymal markers (MMP-9 and N-cadherin) was identified in APP overexpressing breast cancer cells compared with the control group (Fig. 6D-F; P<0.05). Conversely, PD0325901 treatment significantly reversed the expression levels of EMT-related genes caused by APP overexpression (Fig. 6D-F; P<0.05).
Figure 6.
Effect of the MEK inhibitor PD0325901 on the expression levels of EMT-related markers in APP-overexpressing MDA-MB-231 breast cancer cells. (A) Effect of PD0325901 on the mRNA expression levels of EMT-related genes in breast cancer cells by RT-qPCR. (B) Effect of PD0325901 on the protein expression levels of EMT-related factors in breast cancer cells by western blot analysis. (C) Statistical analysis of the protein expression levels of EMT-associated factors. (D) Effect of PD0325901 treatment on the transcription level of EMT-related genes in breast cancer cells following APP overexpres-sion by RT-qPCR. (E) Representative images of the effect of PD0325901 on the expression of EMT-related molecules in breast cancer cells following APP overexpression by western blot analysis, and (F) statistical analysis of western blot bands. All the experiments were performed in triplicate. Data are presented as the mean ± SD. *P<0.05. MEK, mitogen-activated protein kinase kinase; EMT, epithelial-to-mesenchymal transition; APP, amyloid precursor protein; RT-qPCR, reverse transcription-quantitative PCR; MMP, matrix metalloproteinase; OE, overexpression.
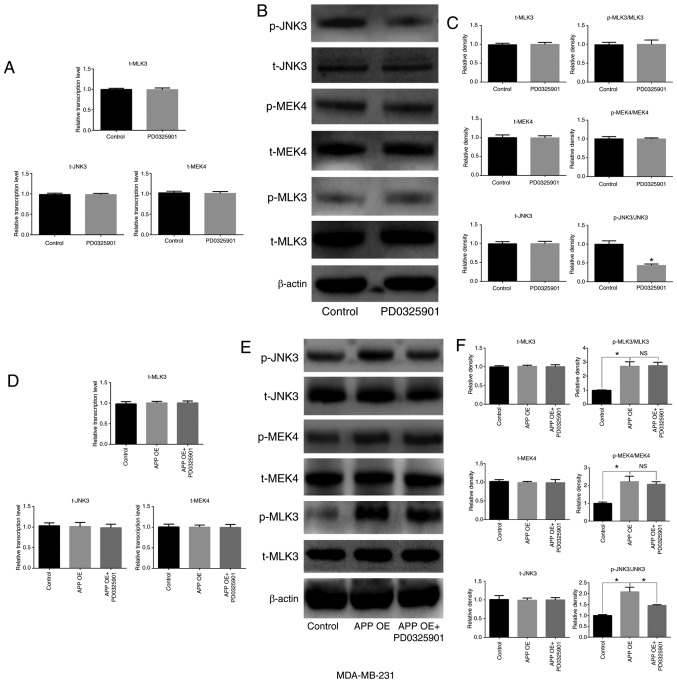
MEK inhibitor PD0325901 is involved in the expression and phosphorylation of MAPK signaling pathway-related factors in breast cancer cells
The present results suggested that PD0325901 treatment significantly inhibited the activation of JNK3, which is downstream of MEK (Fig. 7A-C; P<0.05), but did not affect the expression of total JNK3 compared with untreated cells. In addition, no significant differences were observed in the expression and activation levels of MEK4 and its upstream factor, MLK3. The overexpression of APP increased the phosphorylation level of MEK, MLK3 and JNK3. In addition, in MDA-MB-231 cells transfected with APP overexpressing plasmid and treated with the MEK inhibitor PD0325901 (1 nM), the phosphorylation levels of MEK4 and MLK3 were not significantly changed compared with the overexpression group (Fig. 7F). By contrast, PD0325901 treatment markedly reversed the phosphorylation of JNK3 caused by APP overexpression (P<0.05). Moreover, cell transfection and drug treatment had no effect on the protein expression levels of the unphosphorylated forms of MEK4, JNK3 and MLK3 in MDA-MB-231 cells (Fig. 7D-F).
Figure 7.
Effects of the MEK inhibitor PD0325901 on the expression and phosphorylation of MAPK signaling pathway-related molecules in MDA-MB-231 breast cancer cells. (A) Effect of PD0325901 on the mRNA expression levels of MAPK signaling pathway-related genes in breast cancer cells by RT-qPCR. (B) Effect of PD0325901 on the protein expression levels of MAPK signaling pathway-related molecules in breast cancer cells by western blot analysis, and (C) statistical analysis of the western blot bands. APP overexpression in breast cancer cells treated with PD0325901. (D) mRNA expression level of MAPK signaling pathway-related genes in breast cancer cells was detected by RT-qPCR. (E) Protein expression levels of MAPK signaling pathway-related molecules in breast cancer cells were determined by western blot analysis. (F) Statistical analysis of the relevant western blot bands. All the experiments were performed in triplicate. Data are presented as the mean ± SD. *P<0.05. MEK, mitogen-activated protein kinase kinase; MAPK, mitogen-activated protein kinase; APP, amyloid precursor protein; RT-qPCR, reverse transcription-quantitative PCR; MEK4, mitogen-activated protein kinase kinase 4; OE, overexpression; p-, phosphorylated; t-, total; NS, not significant.
Discussion
Breast cancer metastasis is associated with a variety of factors (38,39). Previous studies have suggested that cancer metastasis mainly occurs at the later stages, when the primary tumor reaches a certain size and tumor cells escape from its periphery (9). However, it has been reported that tumor metastasis also occurs at the early stages, as explained by the parallel metastasis theory (9). It was previously demonstrated that overexpression of APP can promote hematogenous metastasis in melanoma and lung cancer cells (30). When tumor cells penetrate the vascular endothelial barrier, they bind to death receptor 6 expressed on the endothelial cell through the APP extracellular domain fragment on the cell surface, inducing programmed necrosis of endothelial cells and allowing tumor cells to enter the bloodstream and metastasize to distant sites (30). However, whether APP affects the migratory and invasive abilities of tumor cells remains unclear.
APP has been shown to be upregulated in a variety of cancer types, including breast, lung and cervical cancer (40-44), but the effect of APP overexpression on the biological features of tumor cells has not been clearly determined. In the present study, following APP overexpression, the migration and invasion of breast cancer cells were significantly increased. Conversely, APP silencing reduced the migratory and invasive ability of breast cancer cells. It has been previously reported that APP promotes cancer cell migration and invasion, and a similar phenotype has been observed in prostate cancer (44). By silencing APP in LNCaP and DU145 prostate cancer cells, cell proliferation was impaired, and cell migration and invasion were notably attenuated (44). Moreover, APP silencing in prostate cancer cells significantly inhibited the expression of EMT-related genes, in particular of MMP family members (44). However, it is unclear how the expression of EMT-related genes is regulated by APP silencing.
EMT is a necessary step in the process of tumor metastasis and affects the prognosis of patients with tumors (23). When tumor cells undergo EMT, their migratory and invasive abilities are significantly increased, promoting cancer metastasis (22). The present study investigated the effect of APP on EMT-related gene expression in breast cancer cells. Both RT-qPCR and western blot analysis results indicated that the overexpression of APP in MDA-MB-231 cells increased the expression levels of the mesenchymal markers MMP-9, MMP-2, MMP-3, N-cadherin and vimentin, whereas the expression levels of the epidermal-associated markers N-cadherin and cytokeratin were significantly reduced. Therefore, the present results suggested that APP affected the migration and invasion of breast cancer cells by regulating the expression of EMT-related proteins. Various studies have reported that MCF-7 cells, as a metastatic cell line, have migratory and invasive properties (45,46). The present results suggested that MCF-7, MDA-MB-231 and BT474 cells presented similar responses to altered APP expression.
The present results suggested that APP affected the expression of EMT-related genes in breast cancer cells; however, the specific mechanism of action remains unclear. A previous study demonstrated that the intracellular domain fragment of the APP protein activates the MAPK signaling pathway (47), which is associated with EMT activation (48). When MAPK is activated, its downstream transcription factors enter the nucleus and promote the expression of mesenchymal-related genes, thereby promoting the progression of EMT (22,49). The present study indicated that the overexpression of APP activated MAPK signaling pathway components, including MLK3, MEK4 and JNK3, whose phosphorylation levels were found to be significantly upregulated following APP overexpression. However, APP overexpression exerted no effect on the total expression levels of MLK3, MEK4 and JNK3. Additionally, PD0325901 (MEK inhibitor) treatment significantly reversed the expression levels of EMT-related genes and the phosphorylation of the MAPK signaling pathway-related factor JNK3 induced by APP overexpression. Collectively, the regulation of the MAPK signaling pathway may be a possible mechanism through which APP regulates EMT in breast cancer cells, affecting their migratory and invasive abilities. In conclusion, the present results suggested that APP could regulate the expression of EMT-related genes in breast cancer cells by activating MAPK signaling pathway-related proteins. The activation of these factors could promote the EMT of breast cancer cells, thus increasing the migration and invasion of breast cancer cells. However, the present study presents certain limitations, including the limited number of human tissues and the lack of animal models, which are required to further investigate whether APP may be used as a therapeutic target for metastatic breast cancer.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CL conceived and designed the study. XW and SC performed the experiments. CL provided experimental technical guidance. All authors performed the data analysis and drafted the manuscript. All authors have read and approved the final version of the manuscript for publication.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Xiamen University (approval no. 2016-318S). Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Antoni MH, Dhabhar FS. The impact of psychosocial stress and stress management on immune responses in patients with cancer. Cancer. 2019;125:1417–1431. doi: 10.1002/cncr.31943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cedolini C, Bertozzi S, Londero AP, Bernardi S, Seriau L, Concina S, Cattin F, Risaliti A. Type of breast cancer diagnosis, screening, and survival. Clin Breast Cancer. 2014;14:235–240. doi: 10.1016/j.clbc.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Criscitiello C, Viale G, Curigliano G. Peptide vaccines in early breast cancer. Breast. 2019;44:128–134. doi: 10.1016/j.breast.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Feng C, Zhang H, Chen J, Wang S, Xin Y, Qu Y, Zhang Q, Ji W, Yamashita F, Rui M, Xu X. Ratiometric co-encapsulation and co-delivery of doxorubicin and paclitaxel by tumor-targeted lipo-disks for combination therapy of breast cancer. Int J Pharm. 2019;560:191–204. doi: 10.1016/j.ijpharm.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Balazy KE, Benitez CM, Gutkin PM, Jacobson CE, von Eyben R, Horst KC. Association between primary language, a lack of mammographic screening, and later stage breast cancer presentation. Cancer. 2019;125:2057–2065. doi: 10.1002/cncr.32027. [DOI] [PubMed] [Google Scholar]
- 7.Peart O. Breast intervention and breast cancer treatment options. Radiol Technol. 2015;86:535M–558M. quiz 559-562. [PubMed] [Google Scholar]
- 8.Castaneda SA, Strasser J. Updates in the treatment of breast cancer with radiotherapy. Surg Oncol Clin N Am. 2017;26:371–382. doi: 10.1016/j.soc.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 10.Yousefi M, Nosrati R, Salmaninejad A, Dehghani S, Shahryari A, Saberi A. Organ-specific metastasis of breast cancer: Molecular and cellular mechanisms underlying lung metastasis. Cell Oncol (Dordr) 2018;41:123–140. doi: 10.1007/s13402-018-0376-6. [DOI] [PubMed] [Google Scholar]
- 11.Sharma R, Sharma R, Khaket TP, Dutta C, Chakraborty B, Mukherjee TK. Breast cancer metastasis: Putative therapeutic role of vascular cell adhesion molecule-1. Cell Oncol (Dordr) 2017;40:199–208. doi: 10.1007/s13402-017-0324-x. [DOI] [PubMed] [Google Scholar]
- 12.Uemura MI, French JT, Hess KR, Liu D, Raghav K, Hortobagyi GN, Arun BK, Valero V, Ueno NT, Alvarez RH, et al. Development of CNS metastases and survival in patients with inflammatory breast cancer. Cancer. 2018;124:2299–2305. doi: 10.1002/cncr.31336. [DOI] [PubMed] [Google Scholar]
- 13.Szarszewska M, Markowska A, Jach R, Marszałek A, Filas V, Bednarek W, Olejek A, Tomczak P, Sajdak S, Nowak-Markwitz E, et al. Significance of BRCA1 expression in breast and ovarian cancer patients with brain metastasis-A multicentre study. Adv Med Sci. 2019;64:235–240. doi: 10.1016/j.advms.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Fu B, Zhang A, Li M, Pan L, Tang W, An M, Liu W, Zhang J. Circular RNA profile of breast cancer brain metastasis: Identification of potential biomarkers and therapeutic targets. Epigenomics. 2018;10:1619–1630. doi: 10.2217/epi-2018-0090. [DOI] [PubMed] [Google Scholar]
- 15.Rice SL, Friedman KP. Clinical PET-MR imaging in breast cancer and lung cancer. PET Clin. 2016;11:387–402. doi: 10.1016/j.cpet.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Li Z, Deng M, Liu Q, Zhang T, Guo W, Li P, Qiao W. Prognostic and clinicopathological value of CXCL12/SDF1 expression in breast cancer: A meta-analysis. Clin Chim Acta. 2018;484:72–80. doi: 10.1016/j.cca.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 17.BacalbaȘa N, Balescu I, Dima S, Popescu I. Long-term survivors after liver resection for breast cancer liver metastases. Anticancer Res. 2015;35:6913–6917. [PubMed] [Google Scholar]
- 18.Ma R, Feng Y, Lin S, Chen J, Lin H, Liang X, Zheng H, Cai X. Mechanisms involved in breast cancer liver metastasis. J Transl Med. 2015;13:64. doi: 10.1186/s12967-015-0425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holen I, Lefley DV, Francis SE, Rennicks S, Bradbury S, Coleman RE, Ottewell P. IL-1 drives breast cancer growth and bone metastasis in vivo. Oncotarget. 2016;7:75571–75584. doi: 10.18632/oncotarget.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM, Shaw CA, Wu MF, Hilsenbeck SG, White LD, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8:15773. doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, Wang S, Wang Y, Wu H, Frank JA, Zhang Z, Luo J. Role of p38γ MAPK in regulation of EMT and cancer stem cells. Biochim Biophys Acta Mol Basis Dis. 2018;1864:3605–3617. doi: 10.1016/j.bbadis.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian M, Schiemann WP. TGF-β stimulation of EMT programs elicits non-genomic ER-α activity and anti-estrogen resistance in breast cancer cells. J Cancer Metastasis Treat. 2017;3:150–160. doi: 10.20517/2394-4722.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutlu M, Saatci Ö, Ansari SA, Yurdusev E, Shehwana H, Konu Ö, Raza U, Şahin Ö. miR-564 acts as a dual inhibitor of PI3K and MAPK signaling networks and inhibits proliferation and invasion in breast cancer. Sci Rep. 2016;6:32541. doi: 10.1038/srep32541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhou X, Li G, Zhang Y, Wu Y, Song W. Modifications and trafficking of APP in the pathogenesis of Alzheimer's disease. Front Mol Neurosci. 2017;10:294. doi: 10.3389/fnmol.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hefter D, Draguhn A. APP as a protective factor in acute neuronal insults. Front Mol Neurosci. 2017;10:22. doi: 10.3389/fnmol.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatami A, Monjazeb S, Milton S, Glabe CG. Familial Alzheimer's disease mutations within the amyloid precursor protein alter the aggregation and conformation of the amyloid-β peptide. J Biol Chem. 2017;292:3172–3185. doi: 10.1074/jbc.M116.755264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkins HM, Swerdlow RH. Amyloid precursor protein processing and bioenergetics. Brain Res Bull. 2017;133:71–79. doi: 10.1016/j.brainresbull.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habib A, Sawmiller D, Tan J. Restoring soluble amyloid precursor protein α functions as a potential treatment for Alzheimer's disease. J Neurosci Res. 2017;95:973–991. doi: 10.1002/jnr.23823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strilic B, Yang L, Albarrán-Juárez J, Wachsmuth L, Han K, Müller UC, Pasparakis M, Offermanns S. Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature. 2016;536:215–218. doi: 10.1038/nature19076. [DOI] [PubMed] [Google Scholar]
- 31.Lei H, Zhang Y, Huang L, Xu S, Li J, Yang L, Wang L, Xing C, Wang X, Peng Y. L-3-n-Butylphthalide regulates proliferation, migration, and differentiation of neural stem cell in vitro and promotes neurogenesis in APP/PS1 mouse model by regulating BDNF/TrkB/CREB/Akt pathway. Neurotox Res. 2018;34:477–488. doi: 10.1007/s12640-018-9905-3. [DOI] [PubMed] [Google Scholar]
- 32.Harder H, Holroyd P, Burkinshaw L, Watten P, Zammit C, Harris PR, Good A, Jenkins V. A user-centred approach to developing bWell, a mobile app for arm and shoulder exercises after breast cancer treatment. J Cancer Surviv. 2017;11:732–742. doi: 10.1007/s11764-017-0630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Stadio CS, Altieri F, Minopoli G, Miselli G, Rippa E, Arcari P. Role of human GKN1 on APP processing in gastric cancer. Biochimie. 2017;135:149–153. doi: 10.1016/j.biochi.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Tsang JYS, Lee MA, Ni YB, Chan SK, Cheung SY, Chan WW, Lau KF, Tse GMK. Amyloid precursor protein is associated with aggressive behavior in nonluminal breast cancers. Oncologist. 2018;23:1273–1281. doi: 10.1634/theoncologist.2018-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takagi K, Ito S, Miyazaki T, Miki Y, Shibahara Y, Ishida T, Watanabe M, Inoue S, Sasano H, Suzuki T. Amyloid precursor protein in human breast cancer: An androgen-induced gene associated with cell proliferation. Cancer Sci. 2013;104:1532–1538. doi: 10.1111/cas.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Y, Zhang Y, Fan S, Chen L, Tang L, Chen X, Lyu J. GALNT6 promotes tumorigenicity and metastasis of breast cancer cell via β-catenin/MUC1-C signaling pathway. Int J Biol Sci. 2019;15:169–182. doi: 10.7150/ijbs.29048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz RS, Erban JK. Timing of metastasis in breast cancer. N Engl J Med. 2017;376:2486–2488. doi: 10.1056/NEJMcibr1701388. [DOI] [PubMed] [Google Scholar]
- 39.Peart O. Metastatic breast cancer. Radiol Technol. 2017;88:519M–539M. [PubMed] [Google Scholar]
- 40.Pandey P, Sliker B, Peters HL, Tuli A, Herskovitz J, Smits K, Purohit A, Singh RK, Dong J, Batra SK, et al. Amyloid precursor protein and amyloid precursor-like protein 2 in cancer. Oncotarget. 2016;7:19430–19444. doi: 10.18632/oncotarget.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golde TE, Cai XD, Shoji M, Younkin SG. Production of amyloid beta protein from normal amyloid beta-protein precursor (beta APP) and the mutated beta APPS linked to familial Alzheimer's disease. Ann N Y Acad Sci. 1993;695:103–108. doi: 10.1111/j.1749-6632.1993.tb23036.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L, He D, Jiao M, Kong L, Shao C, Chen J, Fang Z, Ma X, Chen H, Li L, et al. Overexpression of histone deacetylase and amyloid precursor protein in hepatocellular carcinoma. Technol Cancer Res Treat. 2017;16:586–594. doi: 10.1177/1533034616661664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobol A, Galluzzo P, Liang S, Rambo B, Skucha S, Weber MJ, Alani S, Bocchetta M. Amyloid precursor protein (APP) affects global protein synthesis in dividing human cells. J Cell Physiol. 2015;230:1064–1074. doi: 10.1002/jcp.24835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazaki T, Ikeda K, Horie-Inoue K, Inoue S. Amyloid precursor protein regulates migration and metalloproteinase gene expression in prostate cancer cells. Biochem Biophys Res Commun. 2014;452:828–833. doi: 10.1016/j.bbrc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y, Lv F, Liang D, Yang Q, Zhang B, Lin H, Wang X, Qian G, Xu J, You W. HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed Pharmacother. 2017;90:555–561. doi: 10.1016/j.biopha.2017.03.054. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J, Zhang WW, Peng F, Sun JY, He ZY, Wu SG. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544. doi: 10.2147/CMAR.S155923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi C, Zhu X, Wang J, Long D. Estrogen receptor α promotes non-amyloidogenic processing of platelet amyloid precursor protein via the MAPK/ERK pathway. J Steroid Biochem Mol Biol. 2014;144(Pt B):280–285. doi: 10.1016/j.jsbmb.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Zhao XW, Zhou JP, Bi YL, Wang JY, Yu R, Deng C, Wang WK, Li XZ, Huang R, Zhang J, Tao DT. The role of MAPK signaling pathway in formation of EMT in oral squamous carcinoma cells induced by TNF-α. Mol Biol Rep. 2019;46:3149–3156. doi: 10.1007/s11033-019-04772-0. [DOI] [PubMed] [Google Scholar]
- 49.Kumar K, Chow CR, Ebine K, Arslan AD, Kwok B, Bentrem DJ, Eckerdt FD, Platanias LC, Munshi HG. Differential regulation of ZEB1 and EMT by MAPK-interacting protein kinases (MNK) and eIF4E in pancreatic cancer. Mol Cancer Res. 2016;14:216–227. doi: 10.1158/1541-7786.MCR-15-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.