Abstract
Psoriasis is one of the most common immune-mediated inflammatory diseases of the skin. The identification of the pivotal molecular mechanisms responsible for the disease pathogenesis may lead to the development of novel therapeutic options. The present study aimed to identify pivotal differentially expressed genes (DEGs) and methylated DEGs in psoriasis. The raw data from gene microarrays were obtained from the Gene Expression Omnibus database. The data were processed using packages in Bioconductor. In total, 352 upregulated and 137 downregulated DEGs were identified. The upregulated DEGs were primarily enriched in the 'innate immune defense' response and the 'cell cycle'. The down-regulated DEGs were primarily enriched in 'cell adhesion' and 'tight junction pathways'. A total of 95 methylated DEGs were identified, which were significantly enriched in the 'interleukin (IL)-17 signaling pathway' and the 'response to interferon'. Based on a comprehensive evaluation of all algorithms in cytoHubba, the key epigenetic-associated hub genes (S100A9, SELL, FCGR3B, MMP9, S100A7, IL7R, IRF7, CCR7, IFI44, CXCL1 and LCN2) were screened out. In order to further validate these genes, the present study constructed a model of imiquimod (IMQ)-induced psoriasiform dermatitis using mice. The levels of these hub genes were increased in the IMQ group. The knockdown of methylation-regulating enzyme ten-eleven translocation (TET) 2 expression in mice attenuated the expression levels of S100A9, SELL, IL7R, MMP9, CXCL1 and LCN2. Furthermore, the hydroxymethylated level of S100A9 was highly expressed in the IMQ group and was significantly decreased by TET2 deficiency in mice. On the whole, using an integrative system bioinformatics approach, the present study identified a series of characteristic enrichment pathways and key genes that may serve as potential biomarkers in psoriasis.
Keywords: psoriasis, bioinformatics analysis, differentially expressed genes, differentially methylated genes, S100A9
Introduction
Psoriasis is one of the most chronic inflammatory hypertrophic skin diseases, affecting 2-5% of the population worldwide (1,2). Multiple factors may cause psoriasis, including genetics, epigenetic modifications, environmental triggers and immune dysfunction (3,4). Although there are extensive studies available regarding the etiologies and mechanisms underlying psoriasis, the precise molecular mechanisms remain elusive (5). Therefore, crucial molecular markers of psoriasis that are identifiable with more powerful technologies are urgently required.
The methylation and hydroxymethylation of DNA are the complementary processes of epigenetic regulation; these epigenetic alterations are responsible for the modulation of gene expression (6). Ten-eleven translocation (TET)2 is a key methylation-regulating enzyme that can alter the DNA methylation status by converting 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) and can regulate the expression of certain genes (7). It has previously been reported that TET2 plays an important role in biological processes, involving cell differentiation and proliferation (8). In a recent study, TET2 was also demonstrated to be involved in the progression of psoriatic dermatitis in mice by regulating hydroxymethylation (9). Although some methylation microarrays have been performed to identify differentially methylated genes (DMGs) in psoriasis (10), the comprehensive profile network analysis remains largely elusive.
Gene microarray is a high-throughput gene detection technique that has been widely used for the investigation of differentially expressed genes (DEGs), and is an important method for studying the pathogenesis of a variety of diseases (11). In recent years, a number of microarray gene profile studies on psoriasis have been performed. Using microarray data, the gene expression profiles in psoriasis have been disclosed and described (12). Certain studies have integrated analyses of gene expression microarray and methylation microarray by bioinformatics analysis in different types of cancer (13,14). However, to the best of our knowledge, there are currently no systematic studies available focusing on mining expression profiles and methylated DEGs in psoriasis. The integrated bioinformatics analysis will be more reliable and provide valuable new molecular targets to foster the development of accurate diagnosis and novel therapeutic strategies. The present study downloaded original data from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). The integrated gene expression profile and DNA methylation profile microarrays of psoriasis were analyzed using a series of packages in R in order to identify the DEGs. Subsequently, The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/), the Search Tool for the Retrieval of Interacting Genes (STRING https://string-db.org/) and Cytoscape were used to analyze the functional enrichment, protein-protein interaction (PPI) network, cluster module and hub genes. For further verification of the association of DMGs with epigenetic regulators in vivo, the present study created an animal model of imiquimod (IMQ)-induced psoriasiform dermatitis and knocked down TET2 expression. The results of the present study provide additional potential targets for the diagnosis and therapeutic intervention in patients with psoriasis.
Materials and methods
Data collection
Data on gene expression profiles (GSE13355, GSE14905 and GSE78097) and gene methylation profiles (GSE31835) were downloaded from the GEO database of The National Center for Biotechnology Information (NCBI). The 3 selected gene expression profile microarrays were based on the same platform (GPL570), which can decrease batch differences in the integration analysis. GSE31835 was based on the GPL8490 platform. GSE13355 contained the data from 58 skin lesions of patients with psoriasis and 64 normal healthy controls; GSE14905 contained 33 samples from patients with psoriasis and 21 normal control samples; GSE78097 contained 13 samples from patients with psoriasis and 6 normal control samples; 19 samples from lesions of patients with psoriasis and 10 normal skin samples were contained in GSE31835.
Data preprocessing and the identification of DEGs
The new versions of affymetrix annotation files were download from the official affymetrix website. The raw data of the gene expression profile were pre-processed by the Robust Multi-array Average (RMA) algorithm with the use of the affy package in Bioconductor (https://www.bioconductor.org/) for background correction and normalization (15). Linear models for the microarray data and corrections for the P-values were calculated using the limma package in Bioconductor (16). |log2FC|>1 and P<0.05 were set as the cut-off criteria to indicate statistical significance. The methylation profile microarray GSE31835 was analyzed using GEO2R software. The DMGs were identified with a P-value cut-off of <0.05 and |t| >2-fold. Subsequently, a principal component analysis (PCA) and hierarchical cluster analysis were performed with the scatterplot3d package and gplots package in R software.
Functional enrichment analysis
The DAVID provides gene analysis tools that link genomic information to biological annotations in order to help users identify the most valuable and significant annotations and links (17). DAVID was used for the analysis of DEGs of the KEGG pathway enrichment and Gene Ontology (GO) functional analysis, including cellular components, molecular function and biological processes (18). P<0.05 and counts ≥2 were used as the significance threshold.
PPI network construction and module analysis
STRING is an online tool for searching for known proteins and predicting the interaction between proteins (19). DEGs were mapped into STRING, and a PPI network was established by calculating a combined score of >0.4. The PPI network was constructed with Cytoscape (version 3.5.1) software. Significant modules were selected based on MCODE, with scores >4 and the number of nodes >5 in Cytoscape. The hub genes were identified according to the 12 topological analysis methods of cytoHubba (20).
Animal procedures
A total of 72 female BALB/c mice (weighing 18-20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The animals were housed in a controlled environment (23±2°C, 12-h light/dark cycle) with sufficient food and water at The Animal Centre of Tongji Medical College. Experimental mice were 8-10 weeks of age. All experimental protocols with animals were conducted following the ARRIVE guidelines. All mouse care and procedures were approved by the Ethics Committee for experimental animals, Tongji Medical College (no. SYXK HUBEI 2016-0057).
The BALB/c mice were randomly divided into 3 groups as follows and the mice in all groups were shaved on their backs (average area of 2×3 cm): i) The control group (n=8), mice were topically treated with vaseline and received an intradermal injection of lentivirus-delivered scrambled RNAs (shRNA-scr) on days 1 and 4; ii) the IMQ group (n=8), skin inflammation was induced by IMQ treatment, as previously reported (21,22). Briefly, mice in the IMQ group received a daily topical dose of 62.5 mg of 5% IMQ cream on their shaved backs for 7 consecutive days and received injections of the same volume of shRNA-scr; the IMQ + sh-TET2 group (n=8), mice received intradermal injections of lentivirus-delivered shRNA against TET2 (sh-TET2) followed by the application of IMQ cream for 7 days. The lentivirus-delivered shRNAs were designed and obtained from Shanghai Genechem Co., LTD. The sequences of the shRNAs were 5′-TTC TCC GAA CGT GTC ACG T-3′ (sh-sc), 5′-GAG CGT TCC TCA GTA TCA TTT-3′ (sh-TET2). On day 8, following the application of 20% urethane (1,500-2,000 mg/kg) (23), the dorsal skin lesions of the mice were taken for use in further experiments. Anesthetic mice were unresponsive to external stimuli, with the loss of righting and palpebral reflexes, but had a constant heart rate and respiratory rate (24). Subsequently, the mice were euthanized by rapid decapitation. The experiments were performed in triplicate.
Western blot analysis
The samples were lysed with cell lysis buffer (RIPA Lysis Buffer; cat. no. P0013B; Beyotime), and the supernatant was collected following centrifugation at 13,000 × g for 5 min at 4°C. The concentrations of proteins were measured using a BCA protein assay kit (cat. no. G2026; Servicebio). Equal quantities of protein were separated via SDS-PAGE (stacking gel, 5%; separating gel, 8%-TET2, 12%-S100A9) and then transferred onto polyvinylidene difluoride membranes. After blocking with 5% skim milk at room temperature for 1 h, the membranes were incubated with primary antibodies against GAPDH (1:1,000; cat. no. AF7021; Affinity Biosciences), TET2 (1:1,000; cat. no. ab94580; Abcam) or S100A9 (1:1,000; cat. no. NB110-89726SS; Novus Biologicals) at 4°C overnight, followed by the appropriate horseradish peroxidase-conjugated secondary antibodies (anti-rabbit IgG, HRP-linked antibody/anti-mouse IgG, HRP-linked antibody 1:2,000; cat. no. 7074/7076; Cell Signaling Technology) for 1 h at room temperature. Signals were detected using DNR bio-imaging systems according to the manufacturer's protocol. The densitometric quantification analysis of proteins was performed using ImageJ software (Ver. 1.45s; National Institutes of Health).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the whole skin samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized by a reverse-transcriptase reaction with a PrimeScript RT Reagent kit (Takara Biotechnology). The specific cDNA fragments were amplified using a Real-time PCR Detection System and Power SYBR-Green (Takara Biotechnology) with the relevant primers. The data are expressed as the relative expression to the GAPDH gene. The relative expression level of each target gene was analyzed using the 2−ΔΔCq method and normalized to GAPDH (25). The PCR primer sequences are presented in Table SI.
Hydroxymethylated DNA immunoprecipitation (hmeDIP)
The extraction of genomic DNA was performed using the SimpleDIP™ Hydroxymethylated DNA IP kit (Cell Signaling Technology) according to the manufacturer's protocol. Genomic DNA was fragmented to sub 100-500 bp using an Ultrasonic Homogenizer. The 5-hmC mouse monoclonal antibody (1:1; cat. no. 51660, Cell Signaling Technology) was added to the single stranded DNA samples. The samples were incubated overnight at 4°C with rotation. After washing and eluting, the immunoprecipitated DNA was purified using spin columns. RT-qPCR was then utilized for the detection of specific hydroxymethylated DNA levels. Quantitative analysis was performed using the following formula: 10% × 2(C[T] 10% Input Sample-C[T] IP Sample).
Statistical analysis
Comparisons between groups were performed using an unpaired two-tailed Student's t-test. In some analyses, one-way analysis of variance (ANOVA) was used, as indicated. Tukey's post hoc test was used for multiple comparisons among diverse groups. Data are expressed as the means ± standard deviation, and P<0.05 was considered to indicate a statistically significant difference. GraphPad Prism software (version 6.0; GraphPad Software) was used for statistical analysis.
Results
Identification of DEGs in psoriasis
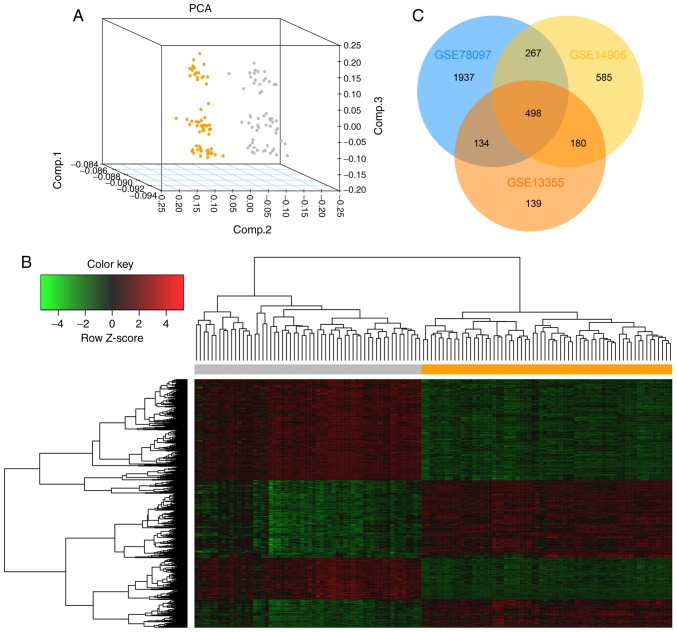
Following quantification and normalization, DEGs were identified based on a threshold of P<0.05 and fold change >2. The present study screened 951, 1,530 and 2,836 DEGs in the psoriasis samples compared with the normal samples in the GSE13355, GSE14905 and GSE78097 datasets, respectively. Using GSE13355 as a representative, the results of the PCA and hierarchical cluster analysis of the DEGs are presented in Fig. 1A and B. The other two PCAs and heatmaps are presented in Fig. S1. In total, 498 common DEGs were screened out, including 352 upregulated, 137 downregulated and 9 non-specific genes (Fig. 1C; Table SII).
Figure 1.
Identification of DEGs in psoriasis. (A) Principal component analysis of the samples in the GSE13355 dataset. The results revealed that psoriasis samples and normal samples could be well distinguished according to these DEGs. Orange dots represent normal samples and gray dots represent psoriasis samples. (B) Hierarchical cluster analysis of DEGs in GSE13355. Gene expression levels were indicated by colors: Red represents high expression level, green represents low expression level. The orange area represents normal samples and the gray area represents psoriasis samples. (C) Venn diagram of overlapping DEGs from 3 microarray datasets (GSE13355, GSE14095 and GSE78097). A |log2FC|>1 and P-value <0.05 were set as the cut-off criteria. DEGs, differentially expressed genes; FC, fold change.
DEGs GO analysis and pathway enrichment analysis
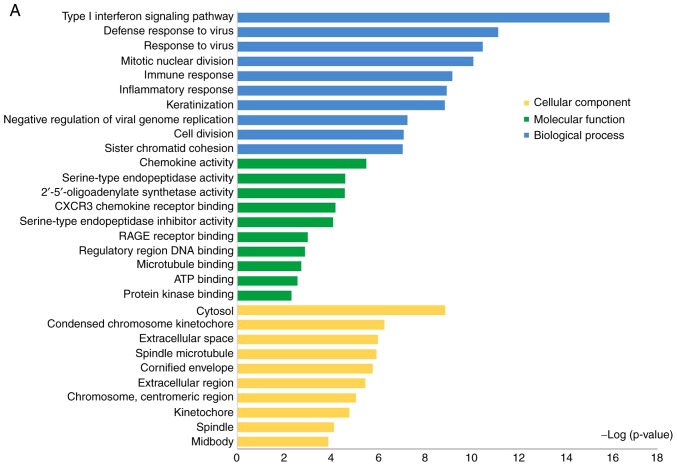
In order to investigate the biological functions of the DEGs, the present study performed a functional pathway enrichment analysis of the DEGs through GO analysis and KEGG pathway enrichment in DAVID. A P<0.05 was considered to indicate a statistically significant difference. As presented in Fig. 2A, in the biological processes group, the upregulated DEGs were primarily involved in 'the type I interferon signaling pathway', 'the defense response to virus', 'mitotic nuclear division', 'the immune response' and 'the inflammatory response'; in the molecular function group, the upregulated DEGs were significantly involved in 'chemokine activity', 'serine-type endopeptidase activity', 'CXC motif chemokine receptor (CXCR) 3 chemokine receptor binding', 'advanced glycosylation end-product specific receptor (RAGE) receptor binding' and 'regulatory region DNA binding'; in the cellular component group, the upregulated DEGs were primarily involved in 'cytosol', 'condensed chromosome kinetochore', 'spindle microtubule' and 'cornified envelope'. The downregulated DEGs were primarily involved in 'cell adhesion', 'positive regulation of protein kinase B signaling', 'RNA polymerase II transcription factor binding', 'transcription factor binding' and 'extracellular matrix' (Fig. 2B).
Figure 2.
Enriched GO terms and KEGG pathways of upregulated and downregulated DEGs identified in psoriasis. (A) Significant enriched GO terms of upregulated genes. Blue, biological processes; green, molecular function; yellow, cellular components. The top 10 significant terms of each category in upregulated DEGs are displayed. (B) Significant enriched GO terms of downregulated genes. Blue, biological processes; green, molecular function; yellow, cellular component. The top 5 significant terms of each category in downregulated DEGs are displayed. (C) Significant enriched KEGG pathways of upregulated and downregulated DEGs associated with psoriasis. P<0.05 was considered to indicate a statistically significant difference. GO, gene ontology; DEGs, differentially expressed genes; KEGG, the Kyoto Encyclopedia of Genes and Genomes.
The significantly enriched KEGG pathways of upregulated DEGs associated with psoriasis were the 'influenza A pathway', 'chemokine signaling pathway', 'cell cycle pathway', 'RIG-I-like receptor signaling pathway' and 'p53 signaling pathway'. The downregulated genes were primarily enriched in 'tight junction pathways' (Fig. 2C).
Module analysis and hub gene selection from the PPI network of DEGs
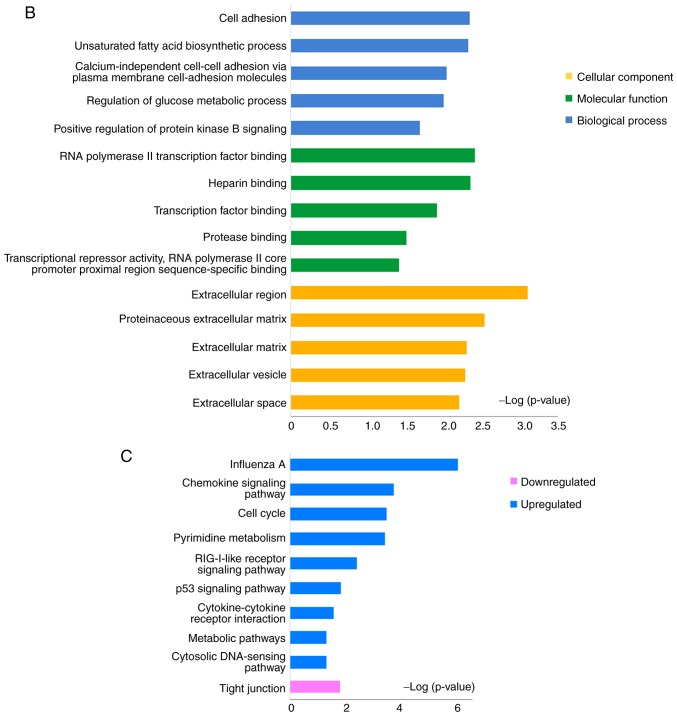
In order to screen out key modules and hub genes, a PPI network of 475 DEGs was established with a combined score of >0.4, including 475 nodes and 2,556 edges (Fig. S2), which was based on the STRING database. A module analysis was performed using MCODE in Cytoscape. MCODE scores >4 and nodes >5 were set as the cut-off criteria. The 4 significant modules are presented in Fig. 3. Subsequently, the present study analyzed the pathways involved in modules 1 and 2 using ClueGO, the plugin of Cytoscape. The 'Cell cycle', 'NOD-like receptor signaling pathway' and 'RIG-I-like receptor signaling pathway' were the significantly enriched pathways (Fig. S3). The hub genes were screened by cytoHubba in Cytoscape (data not shown) and it was revealed that TOP2A, CDK1, IL8, KIAA0101, CCNB1, BIRC5, STAT3, TYMS, UBE2C, ABCA12, TMPRSS11D and S100A9 may all play a pivotal role in the molecular mechanisms underlying psoriasis.
Figure 3.
The top 4 modules from the PPI network. (A) Module 1 contained 48 nodes and 1,106 edges. (B) Module 2 contained 29 nodes and 383 edges. (C) Module 3 contained 13 nodes and 61 edges. (D) Module 4 contained 6 nodes and 12 edges. MCODE scores >4 and nodes >5 were set as the cut-off criteria. PPI, protein-protein interaction; DEGs, differentially expressed genes.
Identification of abnormal methylated-DEGs in psoriasis
The gene methylation microarray GSE31835 was obtained from the public GEO database. Based on the cut-off standards, a total of 2,730 DMGs was primitively identified between the psoriasis samples and healthy controls. By the construction of a Venn diagram (data not shown), the consistent differential genes between DEGs and DMGs were obtained (Table I).
Table I.
Abnormal methylated-differentially expressed genes in psoriasis.
| Term | Gene name |
|---|---|
| Methylated-differentially expressed genes | TNIP3, XDH, HSD17B2, HMGCS2, SELL, FOXM1, C10orf99, IFI27, PPP1R1B, ADRB1, PI15, IRF7, CCND1, ZNF677, PI3, PDK4, CCR7, PDZK1IP1, IL19, ATP11B, CRABP2, GALNT6, CHI3L2, FUT3, CLEC7A, GBP1, NFE2L3, OAS2, KRT16, GZMB, FCGR3B, SMPD3, SLAMF7, LRRC8B, ZDHHC21, STS, ALDH1A3, LRG1, CD47, RORC, S100A7, S100A12, PRELP, SERPINB3, IVL, APOC1, CYP4B1, ARSF, TRIM22, SLC23A2, TIMP3, GJB6, APOL6, SLC2A13, FOXC1, DNASE1L3, KLHDC7B, FADS2, ARNTL2, CCL27, ZBTB16, KYNU, CCL20, CRISPLD1, TMEM40, CHAC1, LYN, BTC, TCN1, ID4, SLC7A5, GDPD3, GJB2, GBP6, WNT2B, ISG20, CXCL1, CDK5R1, TMEM116, LYPD5, IL7R, PTAFR, KLK10, ATP12A, SPRR3, CFB, AIM2, LCN2, KRT6B, UHRF1, MAMDC2, KCNJ15, IFI44, S100A9, MMP9 |
Identification of key candidate genes and pathways with the methylated-DEGs PPI network
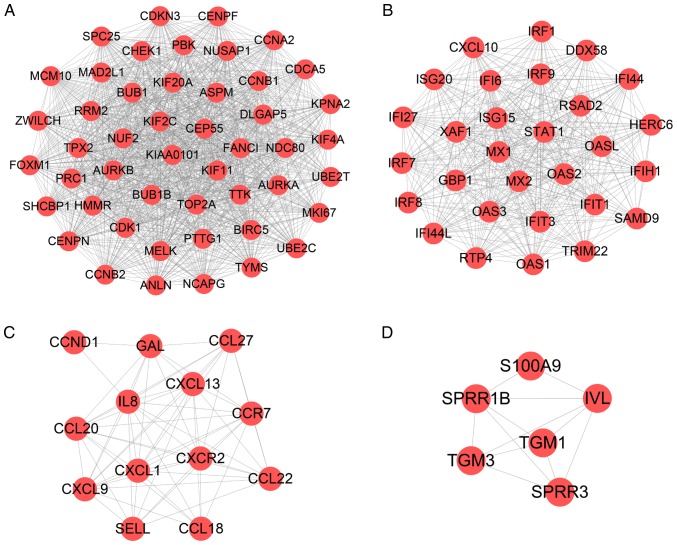
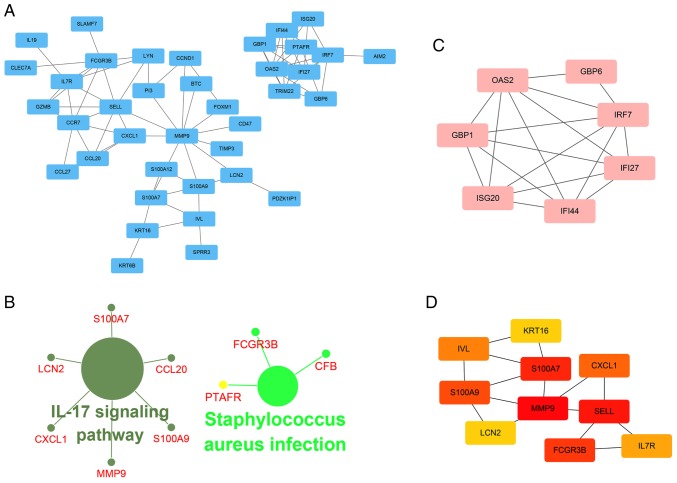
A PPI network of 95 methylated-DEGs was constructed using Cytoscape software (Fig. 4A). According to the ClueGO analysis, these genes were primarily involved in the 'interleukin (IL)-17 signaling pathway', 'Staphylococcus aureus infection', 'response to interferon' and 'granulocyte chemotaxis' (Figs. 4B and S4). GBP1, IRF7, OAS2, IFI44, GBP6, IFI27 and ISG20 were selected as significant module genes using MCODE (Fig. 4C). The top 10 genes, calculated by Betweenness algorithms, are presented in Fig. 4D. S100A9, SELL, FCGR3B, MMP9, S100A7, IL7R, IRF7, CCR7, IFI44, CXCL1 and LCN2 were the top outstanding key genes based on a comprehensive evaluation of 12 algorithms in cytoHubba, which were selected when the genes contained in five or more algorithms.
Figure 4.
The PPI network for methylated-differentially expressed genes. (A) PPI network of 95 methylated-differentially expressed genes was constructed by STRING and reconstructed by Cytoscape. (B) KEGG pathway enrichment analysis of methylated-differentially expressed genes by ClueGO software. P<0.05 was considered to indicate a statistically significant difference. (C) Module analysis of methylated-differentially expressed genes by the MCODE in Cytoscape, including 7 nodes and 17 edges. Scores >4 and nodes >5 were set as the cut off criteria. (D) The top 10 hub genes were calculated by Betweenness algorithms in Cytoscape. The deeper the color, the more important the gene. PPI, protein-protein interaction; KEGG, the Kyoto Encyclopedia of Genes and Genomes; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins.
Evaluation of the hub DEGs in IMQ-induced psoriasiform dermatitis
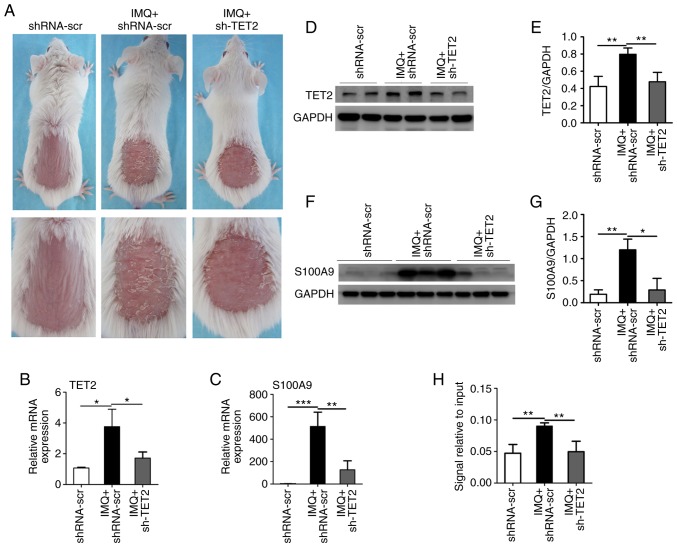
TET2 is a key DNA methylated regulatory enzyme that converts methylation to hydroxymethylation, thereby regulating gene expression. In order to verify the hub DMGs screened in the present study, a mouse model of IMQ-induced psoriasis with or without the knockdown of TET2 expression was established. Compared with the control group, the IMQ group exhibited significant psoriasiform dermatitis of the erythema, thickening and scaling, whereas the skin lesions of lentivirus-delivered shTET2-injected mice were significantly decreased (Fig. 5A). The mRNA levels and protein concentration of TET2 were decreased in the sh-TET2-injected mice compared with the IMQ group (Fig. 5B, D and E). Due to the significantly high expression of S100A9 in the IMQ group, the western blot bands were slightly conjoined, which may affect the accurate quantification of the protein. The expression levels of the hub genes were detected by RT-qPCR. The mRNA expression levels of SELL, FCGR3B, MMP9, S100A7, IL7R, IRF7, CCR7, IFI44, CXCL1 and LCN2 were increased in the IMQ group compared with the control group. The expression levels of SELL, IL7R, MMP9, CXCL1 and LCN2 were suppressed by sh-TET2 treatment (Fig. S5). Furthermore, compared with the control group, the IMQ group exhibited a significantly increased expression of S100A9, which was evidently suppressed by sh-TET2 treatment at both the mRNA and protein levels (Fig. 5C, F and G). When investigating the molecular mechanisms underlying the effects of TET2 on S100A9 expression, it was revealed that the hydroxymethylation level of S100A9 was significantly decreased in the IMQ + sh-TET2 group (Fig. 5H). These data thus indicate that S100A9 may be regulated by epigenetic approaches and may play an important role in the pathogenesis of psoriasis.
Figure 5.
TET2 regulates the expression of S100A9 in a model of psoriasiform dermatitis. (A) Representative lesions of BALB/c mice following continuous treatment for 8 days. (B and C) TET2 and S100A9 mRNA levels in the skin lesions were evaluated by RT-qPCR. The protein expression levels of (D and E) TET2 and (F and G) S100A9 in the skin lesions were measured by western blot analysis. (H) The DNA hydroxymethylation level of S100A9 was analyzed by hMeDIP-qPCR. The results are presented as the means ± standard deviation. All experiments were performed 3 times, and the representative results are presented. *P<0.05, **P<0.01 and ***P<0.001. S100A9, S100 calcium binding protein A9; TET2, ten-eleven translocation 2; IMQ, imiquimod; hMeDIP-qPCR, Hydroxymethylated DNA Immunoprecipitation-quantitative PCR; shRNA-scr, lentivirus-delivered scrambled RNAs; shRNA-scr, control group; IMQ+shRNA-scr, IMQ group; IMQ+sh-TET2, IMQ+sh-TET2 group.
Discussion
The present study analyzed three gene expression profiles and one gene methylation profile. A total of 352 upregulated DEGs and 137 downregulated DEGs were identified. In a comparison between the DEGs and DMGs, 95 overlapping genes were selected. The IL-17 signaling pathway and Staphylococcus aureus infection pathway were the main enriched pathways. The biological processes involved were the response to interferon and granulocyte chemotaxis. S100A9, SELL, FCGR3B, MMP9, S100A7, IL7R, IRF7, CCR7, IFI44, CXCL1 and LCN2 were the top outstanding genes. Furthermore, S100A9 was regulated by epigenetic regulator TET2 in a mouse model of psoriasis.
As the three raw gene expression microarrays had the same normalization and quality control, the present study obtained highly standardized and comparable data. Gene expression datasets combined with comprehensive bioinformatics analysis can provide reliable and new insights into potential crucial diagnostic and therapeutic biomarkers. Researchers have previously focused on key genes and pathways in different diseases (26). According to the analysis of pathways and GO terms in the present study, DEGs were enriched in the 'immune response', 'inflammatory response', 'cell adhesion', 'chemokine signaling pathway' and 'cell cycle', which is consistent with extensive previous studies on psoriasis (5,27,28). Furthermore, all the top 3 significant terms of the biological process analysis and one of the main pathways of the upregulated DEGs were associated with responses to virus, suggesting that the pathogenesis of psoriasis may be closely associated with viral infection. A recent study revealed that activated RIG-I induced the production of IL-23 and triggers psoriasiform dermatitis in mice, highlighting that the dysregulation of antiviral immune responses may initiate the inflammation of psoriasis (29). Extracellular vesicles transport cytosolic proteins, lipids and RNA, mediating intercellular communication between different cells (30). Circulating endothelial- and platelet-derived extracellular vesicles are increased in psoriasis (31) and are decreased following effective anti-TNF-α treatment (32). The present study demonstrated that downregulated DEGs were evidently involved in extracellular vesicles (OGN, APOE, EDIL3 and PRELP) in the GO cell component analysis. Thus, these molecules may be potential biomarkers of psoriasis.
Aberrant DNA methylation can influence the expression of hub genes, and can induce a variety of pathophysiological alterations (33). Therefore, the present study identified 95 abnormally methylated DEGs by overlapping the DEGs and DMGs. These genes were primarily involved in the 'IL-17 signaling pathway', 'response to interferon' and 'granulocyte chemotaxis', which is reasonable, as these biological processes have been demonstrated to increase the risk of developing psoriasis (34). The top 11 hub genes were selected by cyto-Hubba analysis. S100A7 is a calcium-binding protein that participates in a number of biological functions, such as proliferation and inflammation (35). S100A7 has been demonstrated to be upregulated in psoriasis and is also named psoriasin, as it was originally discovered in psoriasis (36). The knockdown of the histone demethylase JMJD3 in keratinocytes has been shown to result in the significant downregulation of the expression of S100A7 (37). MMP9 is an enzyme from the gelatinase family that regulates cell matrix composition and proliferation (38). Elevated concentrations of MMP9 have been described in patients with psoriasis (39). The aberrant methylation of the promoter region of MMP9 has been shown to decrease the levels of RNA and protein secretion in lymphoma cells (40). FCGR3B, IL7R, IRF7, CCR7 and LCN2 have also been shown to be highly expressed in patients with psoriasis (41-45). IRF7 is a key transcriptional regulator of type I interferon (IFN)-dependent immune responses and plays a vital role in the response against viruses (46). TET2 can maintain the hypomethylation of IRF7 to regulate the TLR7/9-elicited IFN response in plasmacytoid dendritic cells (pDCs) (47). Hence, these genes may be potential candidate genes regulated by epigenetics that are involved in the pathogenesis of psoriasis.
S100A9 is a calcium-binding protein of the S100 family, which plays a prominent role during the amplification of the inflammatory response in autoimmunity, including chemotaxis, adhesion and the degranulation of neutrophils (48). Through a comprehensive bioinformatics analysis, S100A9 was one of the identified hub genes of methylated DEGs. Furthermore, the research in the present study demonstrated that the knockdown of epigenetic regulator, TET2, in mice significantly decreased the expression of S100A9, both at the mRNA and protein level. Compared with the IMQ group, the hydroxymethylation level of S100A9 was also significantly decreased in the IMQ + sh-TET2 group. These findings highlight the importance of the regulation of S100A9 by an abnormal methylation status.
To the best of our knowledge, there are limited studies that have used integrated bioinformatics analyses to gather valuable information and provide important clues for future research. Using computational systems biology, Sevimoglu and Arga reported that the JAK/STAT signaling pathway was significant for psoriasis. They screened out several biomarkers and therapeutic candidates by analyzing the raw data of gene expression datasets (49). Certain feature algorithms filtered gene expression profiles in order to acquire classification models of psoriasis (50). Unlike previous studies, the present study comprehensively analyzed DEGs and DMGs to identify active modules and key epigenetically-associated DEGs.
The integrated bioinformatics analysis of both the gene expression profile and the methylation profile is more reliable and provide valuable, novel molecular targets to foster the development of accurate diagnosis and novel therapeutic strategies. However, one limitation of this study was the lack of clinical human specimens. Additional studies with clinical materials and experiment validation are required in order to verify the key candidate genes. Furthermore, the present study analyzed only a single methylation profile microarray; thus, more comparable microarray data are required so that more valuable data can be obtained. The present study selected S100A9 as a representative gene, and preliminarily evaluated the expression and epigenetic regulatory mechanism of S100A9 in psoriasis. Further experiments are required to clarify the exact association between TET2 and S100A9, and the molecular mechanisms of action.
In conclusion, the results from the present study not only identify a series of DEGs and DEMGs, but also analyze the significant modules and key candidate genes. In addition, in order to further verify the bioinformatics analysis data, the present study detected the expression levels of hub genes (S100A9, SELL, FCGR3B, MMP9, S100A7, IL7R, IRF7, CCR7, IFI44, CXCL1 and LCN2) in a mouse model of psoriasiform dermatitis by interfering with the expression of methylation-regulating enzyme TET2. These key genes may serve as potential biomarkers in psoriasis.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81903212, 81673057 and 81502730).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XW, XL, NL and HC conceived and designed the study. XW and NL performed the analysis procedures. XL, NL and HC analyzed the results. XW performed the experiments. XL and HC contributed analysis tools. XW, XL and NL wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All experimental protocols with animals were conducted following the ARRIVE guidelines. All mouse care and procedures were approved by the Ethics Committee for experimental animals, Tongji Medical College (no. SYXK HUBEI 2016-0057).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Raychaudhuri SK, Maverakis E, Raychaudhuri SP. Diagnosis and classification of psoriasis. Autoimmun Rev. 2014;13:490–495. doi: 10.1016/j.autrev.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Duffin KC, Chandran V, Gladman DD, Krueger GG, Elder JT, Rahman P. Genetics of psoriasis and psoriatic arthritis: Update and future direction. J Rheumatol. 2008;35:1449–1453. [PMC free article] [PubMed] [Google Scholar]
- 3.Fogel O, Richard-Miceli C, Tost J. Epigenetic changes in chronic inflammatory diseases. Adv Protein Chem Struct Biol. 2017;106:139–189. doi: 10.1016/bs.apcsb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths CE. The immunological basis of psoriasis. J Eur Acad Dermatol Venereol. 2003;17(Suppl 2):S1–S5. doi: 10.1046/j.1468-3083.17.s2.1.x. [DOI] [PubMed] [Google Scholar]
- 5.Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386:983–994. doi: 10.1016/S0140-6736(14)61909-7. [DOI] [PubMed] [Google Scholar]
- 6.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- 7.Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, et al. The methyl-cytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The ten-eleven translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–496. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Liu X, Duan X, Zhu K, Zhang S, Gan L, Liu N, Jaypaul H, Makamure JT, Ming Z, Chen H. Ten-eleven translocation-2 regulates DNA hydroxymethylation status and psoriasiform dermatitis progression in mice. Acta Derm Venereol. 2018;98:585–593. doi: 10.2340/00015555-2926. [DOI] [PubMed] [Google Scholar]
- 10.Roberson ED, Liu Y, Ryan C, Joyce CE, Duan S, Cao L, Martin A, Liao W, Menter A, Bowcock AM. A subset of methylated CpG sites differentiate psoriatic from normal skin. J Invest Dermatol. 2012;132:583–592. doi: 10.1038/jid.2011.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Sun F, Yang X, Jin Y, Shi M, Wang L, Shi Y, Zhan C, Wang Q. Correlation between RNA-Seq and microarrays results using TCGA data. Gene. 2017;628:200–204. doi: 10.1016/j.gene.2017.07.056. [DOI] [PubMed] [Google Scholar]
- 12.Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Li H, Sun L, Wang Z, Xing C, Yuan Y. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int. 2017;17:75. doi: 10.1186/s12935-017-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huo X, Sun H, Cao D, Yang J, Peng P, Yu M, Shen K. Identification of prognosis markers for endometrial cancer by integrated analysis of DNA methylation and RNA-Seq data. Sci Rep. 2019;9:9924. doi: 10.1038/s41598-019-46195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy-analysis of affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cyto-Hubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, Cornelissen F, Mus AM, Florencia E, Prens EP, Lubberts E. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 22.Schukur L, Geering B, Charpin-El Hamri G, Fussenegger M. Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci Transl Med. 2015;7:318ra201. doi: 10.1126/scitranslmed.aac4964. [DOI] [PubMed] [Google Scholar]
- 23.Taves S, Berta T, Liu DL, Gan S, Chen G, Kim YH, Van de Ven T, Laufer S, Ji RR. Spinal inhibition of p38 MAP kinase reduces inflammatory and neuropathic pain in male but not female mice: Sex-dependent microglial signaling in the spinal cord. Brain Behav Immun. 2016;55:70–81. doi: 10.1016/j.bbi.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gargiulo S, Greco A, Gramanzini M, Esposito S, Affuso A, Brunetti A, Vesce G. Mice anesthesia, analgesia, and care, part I: Anesthetic considerations in preclinical research. ILAR J. 2012;53:E55–E69. doi: 10.1093/ilar.53.1.55. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Dong B, Wang G, Yao J, Yuan P, Kang W, Zhi L, He X. Predicting novel genes and pathways associated with osteosar-coma by using bioinformatics analysis. Gene. 2017;628:32–37. doi: 10.1016/j.gene.2017.06.058. [DOI] [PubMed] [Google Scholar]
- 27.Mabuchi T, Hwang ST. ACKR2: Nature's decoy receptor lures unsuspecting chemokines in psoriasis. J Invest Dermatol. 2017;137:7–11. doi: 10.1016/j.jid.2016.09.035. [DOI] [PubMed] [Google Scholar]
- 28.Reich K. The concept of psoriasis as a systemic inflammation: Implications for disease management. J Eur Acad Dermatol Venereol. 2012;26(Suppl 2):S3–S11. doi: 10.1111/j.1468-3083.2011.04410.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhu H, Lou F, Yin Q, Gao Y, Sun Y, Bai J, Xu Z, Liu Z, Cai W, Ke F, et al. RIG-I antiviral signaling drives interleukin-23 production and psoriasis-like skin disease. EMBO Mol Med. 2017;9:589–604. doi: 10.15252/emmm.201607027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier F, Garnache-Ottou F, Angelot F, Biichlé S, Vidal C, Humbert P, Saas P, Seillès E, Aubin F. Increased levels of circulating endothelial-derived microparticles and small-size platelet-derived microparticles in psoriasis. J Invest Dermatol. 2011;131:1573–1576. doi: 10.1038/jid.2011.57. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier F, Garnache-Ottou F, Biichlé S, Vivot A, Humbert P, Saas P, Seillès E, Aubin F. Effects of anti-TNF-α agents on circulating endothelial-derived and platelet-derived micropar-ticles in psoriasis. Exp Dermatol. 2014;23:924–925. doi: 10.1111/exd.12551. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Chen C, Bi X, Zhou C, Huang T, Ni C, Yang P, Chen S, Ye M, Duan S. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi: 10.1016/j.gene.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 34.Lowes MA, Russell CB, Martin DA, Towne JE, Krueger JG. The IL-23/T17 pathogenic axis in psoriasis is amplified by keratinocyte responses. Trends Immunol. 2013;34:174–181. doi: 10.1016/j.it.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med. 2013;13:24–57. doi: 10.2174/156652413804486214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madsen P, Rasmussen HH, Leffers H, Honoré B, Dejgaard K, Olsen E, Kiil J, Walbum E, Andersen AH, Basse B, et al. Molecular cloning, occurrence, and expression of a novel partially secreted protein 'psoriasin' that is highly up-regulated in psoriatic skin. J Invest Dermatol. 1991;97:701–712. doi: 10.1111/1523-1747.ep12484041. [DOI] [PubMed] [Google Scholar]
- 37.Gschwandtner M, Zhong S, Tschachler A, Mlitz V, Karner S, Elbe-Bürger A, Mildner M. Fetal human keratinocytes produce large amounts of antimicrobial peptides: Involvement of histone-methylation processes. J Invest Dermatol. 2014;134:2192–2201. doi: 10.1038/jid.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monhian N, Jewett BS, Baker SR, Varani J. Matrix metal-loproteinase expression in normal skin associated with basal cell carcinoma and in distal skin from the same patients. Arch Facial Plast Surg. 2005;7:238–243. doi: 10.1001/archfaci.7.4.238. [DOI] [PubMed] [Google Scholar]
- 39.Głażewska EK, Niczyporuk M, Ławicki S, Szmitkowski M, Donejko M, Zajkowska M, Będkowska GE, Przylipiak A. Narrowband ultraviolet B light treatment changes plasma concentrations of MMP-3, MMP-9 and TIMP-3 in psoriatic patients. Ther Clin Risk Manag. 2017;13:575–582. doi: 10.2147/TCRM.S125595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chicoine E, Estève PO, Robledo O, Van Themsche C, Potworowski EF, St-Pierre Y. Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochem Biophys Res Commun. 2002;297:765–772. doi: 10.1016/S0006-291X(02)02283-0. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y, Zhang Z, Tao L, Chen G, Liu F, Wang T, Xue F, Chen Y, He L, Zheng J, Liu Y. A high copy number of FCGR3B is associated with psoriasis vulgaris in Han Chinese. Dermatology. 2014;229:70–75. doi: 10.1159/000360160. [DOI] [PubMed] [Google Scholar]
- 42.Lee SK, Jeon EK, Kim YJ, Seo SH, Kim CD, Lim JS, Lee JH. A global gene expression analysis of the peripheral blood mono-nuclear cells reveals the gene expression signature in psoriasis. Ann Dermatol. 2009;21:237–242. doi: 10.5021/ad.2009.21.3.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raposo RA, Gupta R, Abdel-Mohsen M, Dimon M, Debbaneh M, Jiang W, York VA, Leadabrand KS, Brown G, Malakouti M, et al. Antiviral gene expression in psoriasis. J Eur Acad Dermatol Venereol. 2015;29:1951–1957. doi: 10.1111/jdv.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rittié L, Elder JT. Capturing the finer points of gene expression in psoriasis: Beaming in on the CCL19/CCR7 axis. J Invest Dermatol. 2012;132:1535–1538. doi: 10.1038/jid.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao S, Cao T, Jin L, Li B, Fang H, Zhang J, Zhang Y, Hu J, Wang G. Increased lipocalin-2 contributes to the pathogenesis of psoriasis by modulating neutrophil chemotaxis and cytokine secretion. J Invest Dermatol. 2016;136:1418–1428. doi: 10.1016/j.jid.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 47.Ma S, Wan X, Deng Z, Shi L, Hao C, Zhou Z, Zhou C, Fang Y, Liu J, Yang J, et al. Epigenetic regulator CXXC5 recruits DNA demethylase Tet2 to regulate TLR7/9-elicited IFN response in pDCs. J Exp Med. 2017;214:1471–1491. doi: 10.1084/jem.20161149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 49.Sevimoglu T, Arga KY. Computational systems biology of psoriasis: Are we ready for the age of Omics and systems biomarkers? OMICS. 2015;19:669–687. doi: 10.1089/omi.2015.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo P, Luo Y, Mai G, Zhang M, Wang G, Zhao M, Gao L, Li F, Zhou F. Gene expression profile based classification models of psoriasis. Genomics. 2014;103:48–55. doi: 10.1016/j.ygeno.2013.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.