In 670 children treated with PPIs, children with normal CYP2C19 metabolic function had more infection events than did children with increased CYP2C19 function.
Abstract
OBJECTIVES:
Proton pump inhibitors (PPIs) are often used in pediatrics to treat common gastrointestinal disorders, and there are growing concerns for infectious adverse events. Because CYP2C19 inactivates PPIs, genetic variants that increase CYP2C19 function may decrease PPI exposure and infections. We tested the hypothesis that CYP2C19 metabolizer phenotypes are associated with infection event rates in children exposed to PPIs.
METHODS:
This retrospective biorepository cohort study included individuals aged 0 to 36 months at the time of PPI exposure. Respiratory tract and gastrointestinal tract infection events were identified by using International Classification of Diseases codes in the year after the first PPI mention. Variants defining CYP2C19 *2, *3, *4, *8, *9, and *17 were genotyped, and all individuals were classified as CYP2C19 poor or intermediate, normal metabolizers (NMs), or rapid or ultrarapid metabolizers (RM/UMs). Infection rates were compared by using univariate and multivariate analyses.
RESULTS:
In all, 670 individuals were included (median age 7 months; 44% girls). CYP2C19 NMs (n = 267; 40%) had a higher infection rate than RM/UMs (n = 220; 33%; median 2 vs 1 infections per person per year; P = .03). There was no difference between poor or intermediate (n = 183; 27%) and NMs. In multivariable analysis of NMs and RM/UMs adjusting for age, sex, PPI dose, and comorbidities, CYP2C19 metabolizer status remained a significant risk factor for infection events (odds ratio 0.70 [95% confidence interval 0.50–0.97] for RM/UMs versus NMs).
CONCLUSIONS:
PPI therapy is associated with higher infection rates in children with normal CYP2C19 function than in those with increased CYP2C19 function, highlighting this adverse effect of PPI therapy and the relevance of CYP2C19 genotypes to PPI therapeutic decision-making.
What’s Known on This Subject:
Proton pump inhibitors are commonly used in children and may increase infection events. Differences in the CYP2C19 enzyme affect medication exposure, but the clinical impact has not been assessed in unselected pediatric cohorts.
What This Study Adds:
In a retrospective cohort of 670 children treated with proton pump inhibitors, children with normal CYP2C19 function had more infection events than did children with increased CYP2C19 function. Risk in infection during proton pump therapy is modified by CYP2C19 functional status.
Proton pump inhibitors (PPIs) are among the most prescribed medications in the United States.1 Within the pediatric population, and in particular among infants, PPI use continues to rise.2–4 Gastroesophageal reflux disease is perhaps the most common indication for PPIs in children; however, PPIs are used in a variety of inflammatory conditions of the upper intestinal tract.5 In their activated form, PPIs bind to and inactivate proton pumps in the stomach, suppressing acid release and increasing gastric pH.6 Reduced gastric acidity permits mucosal healing and is the primary therapeutic benefit of this drug class. However, PPI efficacy is dependent on the drug’s plasma concentrations and is therefore directly related to its pharmacokinetics.7 PPIs are primarily inactivated in the liver by microsomal enzyme CYP2C19, and genetic variation in the CYP2C19 gene determines enzyme activity.6 Common genetic variants give rise to several metabolizer phenotypes, ranging from slow to normal to rapid drug inactivation.8,9 Individuals with no or decreased-function variants are termed poor metabolizers (PMs) or intermediate metabolizers, resulting in higher drug exposure compared with normal metabolizers (NMs) given an equivalent dose.10–12 Conversely, individuals with increased-function alleles are rapid or ultrarapid metabolizers (RM/UMs) and have reduced exposure to the active drug for a given dose of PPI.13–17
Despite the wide therapeutic index of PPIs, differences in CYP2C19 activity may have clinical significance. Several studies have demonstrated an association between CYP2C19 metabolizer status and PPI treatment outcomes for a variety of conditions, including gastroesophageal reflux disease, Helicobacter pylori gastritis, and esophageal eosinophilia.18–23 Conversely, adverse outcomes, including vitamin and mineral deficiencies, bone fractures, development of allergic diseases in childhood, and respiratory tract infections (RTIs) and gastrointestinal tract infections (GTIs), may also be impacted by differential CYP2C19 activity.24–29 Infectious outcomes related to PPI use are hypothesized to be secondary to reduced gastric acidity and resultant dysbiosis of the gastric microflora, permitting colonization of pathogenic microbes.30,31 The potentially infectious gastric contents may reflux into the esophagus and oropharynx, and microaspiration within the respiratory tract or within the distal gastrointestinal tract may occur, leading to RTIs and GTIs, respectively.27,28,32–34 On the basis of data from children with asthma, CYP2C19 PMs may experience higher rates of infections compared with NMs at equivalent doses.35,36 CYP2C19 genotype-based PPI dosing guidelines are in development, but CYP2C19 gene-based therapeutic decision-making is not routinely performed.37,38 Given the relative paucity of pediatric data to support CYP2C19-based PPI dosing and management guidelines, we sought to further investigate the role of CYP2C19 metabolizer phenotypes on rates of RTIs and GTIs in children on PPI therapy.
Methods
Study Population
The retrospective study was performed by using BioVU, the Vanderbilt University Medical Center DNA biorepository linked to deidentified electronic health record (EHR) data.39,40 This study was reviewed by the Vanderbilt Institutional Review Board and determined to be nonhuman subjects research. Inclusion criteria for the study were as follows: (1) PPI exposure, defined as any mention of the generic or trade name of any of the PPIs available in the United States with at least 3 mentions on 3 separate dates within 1 year; (2) age 0 to 36 months at the time of first PPI exposure; and (3) available DNA in BioVU. There were no exclusion criteria. There was no requirement that individuals receive primary care or all medical care within Vanderbilt University Medical Center.
Outcomes and Covariates
The primary outcome for analysis was total infection events in the year after PPI start. To define infection events, all International Classification of Diseases (ICD) codes for RTIs and GTIs listed in Supplemental Table 3 were identified for each individual for the time period beginning 1 week after PPI start and ending 12 months after PPI start. Infection events within this window were counted, requiring a minimum of 14 days between events to avoid duplicate entries for a single infection (Supplemental Fig 2). Separately, the total RTIs and GTIs were each measured as secondary outcomes. Demographic covariates (sex, race, ethnicity, and age at PPI start) were extracted from the deidentified EHR. Congenital heart disease, chronic lung disease, prematurity, gastrointestinal motility disorders, structural gastrointestinal disorders, chronic diarrheal disorders, and prematurity were identified as comorbid conditions on the basis of ICD codes (Supplemental Table 4). Outcome and covariate assessments were performed blinded to CYP2C19 genotype or phenotype.
PPI Dose
PPI dose was determined by extracting lines of text surrounding every mention of PPI from inpatient and outpatient clinical notes, electronic prescriptions, inpatient orders, and problem lists. Text strings were discarded if they did not contain the PPI drug name or if they had no numeric data indicating dosing information. When multiple entries were available for the same date, a single entry per date was identified by prioritizing electronic prescriptions, inpatient orders, and clinical notes (both inpatient and outpatient). The total daily dose was then determined via manual review by using a crowdsourcing strategy implemented in VBOSSA, an institutionally derived version of PYBOSSA, an open-source data collection technology.41,42 In brief, text strings were displayed together with a dosing calculator on a Web-based platform. Five workers separately reviewed each text entry, 20% of which were reviewed by multiple workers to ensure efficacy. Of the overlapped tasks, workers were in congruence 75% of the time. Additionally, a select set of known dosages were provided, for which the workers responded accurately 81% of the time. Discordant entries were manually reviewed to determine accurate information. For each dose, the nearest recorded weight (kg) was then used to calculate the daily dose by weight (mg/kg per day). Each individual’s annual weighted average was calculated on the basis of the duration of time on each dose.
Genotyping
Genotyping was performed by the Vanderbilt Technologies for Advanced Genomics core laboratory using the Sequenom MassArray platform (Agena Bioscience, San Diego, CA). Six CYP2C19 single-nucleotide variants were genotyped to identify CYP2C19 haplotypes: *2 (rs4244285), *3 (rs4986893), *4 (rs28399504), *8 (rs41291556), *9 (rs17884712), and *17 (rs12248560). Metabolizer status was assigned on the basis of current CYP2C19 diplotype-to-phenotype tables from the Clinical Pharmacogenetics Implementation Consortium by using the currently suggested consensus nomenclature.43,44 Individuals were classified as NMs if they carried 2 functional *1 alleles. RMs are those with 1 functional allele and 1 increased-function allele (CYP2C19*17), and ultrarapid metabolizers are those with 2 increased-function alleles. These were analyzed together as RM/UMs. Individuals were classified as poor or intermediate metabolizers (PM/IMs) if they carried 1 or more alleles with no function or decreased function (*2, *3, *4, *8, and *9) even if the other allele was increased function (eg, *2/*17; Supplemental Table 5).
Data Analysis
The association of CYP2C19 metabolizer phenotype and each of the covariates to the outcome of total infection events was tested via univariate analysis by using 2-sided χ2 tests for categorical variables and the Kruskal-Wallis test for continuous variables. Multivariate analysis was performed by using ordinal logistic regression to test for association between CYP2C19 metabolizer status and infection events, adjusting for age at the time of PPI start, sex, PPI dose (average mg/kg per day), and comorbidities (including a dichotomous variable for the presence or absence of comorbidities as well as dichotomous variables for the presence or absence of each of the following: congenital heart disease, chronic lung disease, gastrointestinal motility disorder, gastrointestinal structural disorder, chronic diarrhea disorder, and prematurity). Data were analyzed by using Stata version 15.1 (Stata Corp, College Station, TX). P <.05 was considered statistically significant.
Results
We identified 670 individuals who met inclusion criteria. The median age of the cohort was 7 months (interquartile range [IQR] 3–13), and the majority were boys (Table 1). Most (n = 561; 84%) of the individuals had at least 1 of the assessed comorbidities. In the year after starting PPI therapy, individuals had a median of 1 infection event (IQR 0–3).
TABLE 1.
Demographics of the Study Cohort and Subsets by CYP2C19 Metabolizer Phenotype
| All (N = 670) | PM/IMs (N = 183) | NMs (N = 267) | RM/UMs (N = 220) | Pa | |
|---|---|---|---|---|---|
| Age, mo, median (IQR) | 7 (3–13) | 7 (4–15) | 7 (4–13) | 8 (4–14) | .18 |
| Female sex, n (%) | 292 (44) | 71 (39) | 119 (45) | 102 (46) | .29 |
| Race, n (%) | .50 | ||||
| White | 553 (83) | 147 (80) | 219 (82) | 187 (85) | |
| African American | 76 (11) | 24 (13) | 27 (10) | 25 (11) | |
| Asian American and/or Pacific Islander | 11 (2) | 5 (3) | 5 (2) | 1 (0.5) | |
| Otherb or unknown | 30 (5) | 7 (4) | 16 (6) | 7 (3) | |
| Ethnicity, n (%) | .10 | ||||
| Hispanic | 34 (5) | 9 (5) | 19 (7) | 6 (3) | |
| Non-Hispanic | 617 (92) | 168 (92) | 238 (89) | 211 (96) | |
| Unknown | 19 (3) | 6 (3) | 10 (4) | 3 (1) | |
| Comorbidities, n (%) | |||||
| Any | 561 (84) | 157 (86) | 223 (84) | 181 (82) | .63 |
| Congenital heart disease | 307 (46) | 84 (46) | 134 (50) | 89 (40) | .10 |
| Chronic lung disease | 330 (49) | 95 (52) | 131 (49) | 104 (47) | .65 |
| Gastrointestinal motility disorder | 75 (11) | 20 (11) | 31 (12) | 24 (11) | .96 |
| Gastrointestinal structural disorder | 303 (45) | 91 (50) | 127 (48) | 85 (39) | .05 |
| Chronic diarrhea disorder | 101 (15) | 24 (13) | 39 (15) | 38 (17) | .50 |
| Prematurity | 150 (22) | 42 (23) | 60 (23) | 48 (22) | .96 |
P from Kruskal-Wallis test for continuous variable (age) and χ2 test for categorical variables.
Includes American Indian and Alaskan native.
In all, 183 (27%) patients in the cohort were CYP2C19 PM/IMs, 267 (40%) were NMs, and 220 (33%) were RM/UMs. CYP2C19 allele frequencies were consistent with previously published data (Supplemental Table 6).45 CYP2C19 metabolizer phenotype groups were similar in age, sex, race, ethnicity, and the assessed comorbidities (Table 1).
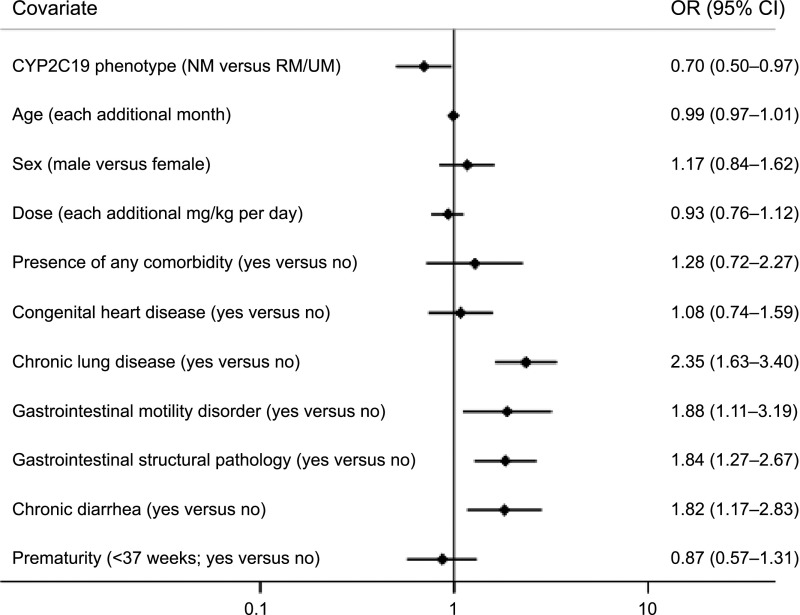
A total of 1419 infection events were identified (1087 RTIs and 332 GTIs). When infection event rates were compared across CYP2C19 metabolizer groups, NMs had more total infection events than did RM/UMs (Table 2). When RTIs and GTIs were evaluated separately, NMs experienced more of each infection type than did RM/UMs, but the difference was not statistically significant. There were also no significant differences between PM/IMs and NMs for total infection events, RTIs, or GTIs. In a multivariable analysis of NMs and RM/UMs adjusting for age, sex, PPI dose, and comorbidities, CYP2C19 metabolizer status remained a significant risk factor for total infection events (odds ratio [OR] 0.70 [95% confidence interval (CI) 0.50–0.97] for RM/UM versus NM; Fig 1). The comorbidities of chronic lung disease, gastrointestinal motility disorder, gastrointestinal structural pathology, and chronic diarrhea were also associated with increased total infection events (Fig 1). A similar multivariable analysis for RTIs and GTIs demonstrated no significant difference between CYP2C19 RM/UMs and NMs (Supplemental Fig 3) but did reveal that chronic lung disease and gastrointestinal structural pathology were associated with increased RTIs, and comorbid gastrointestinal disease (motility disorder, structural pathology, and chronic diarrhea) was associated with increased GTIs.
TABLE 2.
Infection Outcomes by CYP2C19 Metabolizer Status
| PM/IM (N = 183) | NM (N = 267) | RM/UM (N = 220) | PM/IM Versus NM, P | NM Versus RM/UM, P | |
|---|---|---|---|---|---|
| Total infection events | |||||
| Mean (±SD) | 2.05 (2.49) | 2.38 (2.60) | 1.85 (2.24) | — | — |
| Median (IQR) | 1 (0–3) | 2 (0–3) | 1 (0–3) | 0.10 | 0.03 |
| Respiratory infection events | |||||
| Mean (±SD) | 1.56 (2.01) | 1.82 (2.16) | 1.44 (1.87) | — | — |
| Median (IQR) | 1 (0–2) | 1 (0–3) | 1 (0–2) | 0.17 | 0.07 |
| Gastrointestinal infection events | |||||
| Mean (±SD) | 0.49 (0.89) | 0.56 (1.01) | 0.42 (0.83) | — | — |
| Median (IQR) | 0 (0–1) | 0 (0–1) | 0 (0–1) | 0.4 | 0.1 |
P from Kruskal-Wallis test. —, not applicable.
FIGURE 1.
Multivariable analysis of total infection events in CYP2C19 NMs versus RM/UMs. Shown are the ORs (diamonds) and 95% CIs (horizontal lines) for each of the variables included in the ordinal regression model for association to infection events in the 670 children treated with PPIs. ORs are for CYP2C19 phenotype (NM versus RM/UM), age (each additional month), sex (male versus female), and dose (each additional mg/kg per day) and 7 additional dichotomous variables (presence of comorbidity, congenital heart disease, chronic lung disease, gastrointestinal motility disorder, gastrointestinal structural pathology, chronic diarrhea, and prematurity, all yes versus no). Point estimates for the ORs and 95% CIs are listed to the right of each plot.
Discussion
Because PPIs are metabolized by the polymorphic CYP2C19 enzyme, we hypothesized that individuals capable of rapid metabolism of these drugs would be protected from the increased infection events seen with PPI exposure. Consistent with this hypothesis, in this cohort of 670 children, we found that CYP2C19 RM/UMs had fewer total infection events than NMs (1.85 ± 2.24 vs 2.38 ± 2.6 events per child in the first year of PPI use). This finding is consistent in both univariate analysis of infection events and multivariate analysis, adjusting for factors that may contribute to the susceptibility to RTIs and GTIs. These findings demonstrate that the differences in metabolism of PPIs due to CYP2C19 variation have clinical consequences. The difference of ∼0.5 infection events per child per year may be negligible for individual, otherwise healthy children; however, across the entire population of PPI-exposed children (1.6% of all newborns and infants seen in the outpatient setting)4 and for children with tenuous health due to chronic diseases, this increased risk is clinically meaningful and highlights the importance of judicious PPI use. The multivariate analyses also indicated several associations of comorbid conditions to RTIs and GTIs while on PPI therapy, identifying children at increased risk for infectious adverse outcomes with acid suppression.
Our data add to the growing evidence for the impact of CYP2C19 function on PPI exposure and on outcomes of PPI therapy. An early pharmacokinetic study of 24 children observed that NMs had lower exposure to pantoprazole than PM/IMs.11 Additional data have demonstrated lower exposure for RM/UMs versus NMs for pantoprazole in 40 children13 and for RM/UMs versus NMs versus PMs for lansoprazole in a study of 244 children.46 A recent study of 41 children with obesity also observed lower exposure in NMs versus PM/IMs.47 The effect of these differences in drug exposure on adverse and therapeutic clinical outcomes has also been observed. In children with asthma, there was a higher frequency of RTIs in the 136 children treated with lansoprazole versus the 135 who received a placebo; furthermore, RTIs were most frequent among the 45 PM/IMs in the lansoprazole arm and less frequent in the 91 NMs.35 In another study of children with asthma, CYP2C19 PMs treated with lansoprazole had worsened asthma control compared with NMs.36 A study of therapeutic response to PPI in children showed that among 74 children with pH testing while on PPI therapy, CYP2C19 NMs had more complete acid suppression than RM/UMs.23 There is also an overrepresentation of CYP2C19 RM/UMs among children who fail PPI therapy and proceed to antireflux surgery,22 and CYP2C19 RM phenotype is an independent risk factor for loss of response to PPI therapy in PPI-responsive esophageal eosinophilia.18 Taken together with our findings, it is apparent that differential exposure to PPIs due to variability in CYP2C19 functional status has clinical consequences: individuals whose metabolic phenotypes allow greater exposure to the drug have greater therapeutic benefits but are at greater risk for adverse clinical events.
The clinical implications of these findings depend on the clinical context of PPI use. These data demonstrate that PPI exposure has the potential for adverse effects. When evaluating the need for these medications, the increased risk of infection events should be considered, particularly for those at risk for life-threatening infection events (eg, those with chronic lung disease or congenital heart disease). In instances in which the potential benefit of PPIs outweighs the risk, genotype-guided therapy may be helpful to achieve therapeutic goals. CYP2C19 PM/IMs, representing ∼1 in 4 patients, may achieve acid suppression at the lowest recommended dose. For these patients, higher doses are unlikely to provide additional benefit. In contrast, the one-third of children who are CYP2C19 RM/UMs likely require doses at least at the high end of the recommended range to achieve adequate exposure for therapeutic effect. There are published recommendations for genotype-guided PPI dosing. For CYP2C19 RM/UMs, the Dutch Pharmacogenetics Working Group recommends increasing the pantoprazole dose by 400%, lansoprazole by 200%, omeprazole by 100% to 200%, and esomeprazole by 50% to 100%; of note, these guidelines are not specifically for pediatric patients.38 For children, we have previously suggested dose increases of 50% for RMs and 100% for ultrarapid metabolizers, regardless of which PPI is prescribed, and reducing the dose by 60% for PM/IMs.9
In our data, we did not find a difference between PM/IMs and NMs. This may have been due to inadequate sample size to detect a difference in infection rates between these subgroups. There may also be unmeasured differences between metabolizer groups, such as compliance with the PPI regimen. Given the same PPI dose, NMs have lower exposure than PM/IMs; thus, NMs may have higher compliance because symptoms after missed doses serve as a reminder to take the medicine. PM/IMs may not have this reinforcement because they experience sustained benefit even after missing doses. We can only speculate on differences in compliance across metabolizer groups because we have no measures of compliance in this retrospective cohort, but this concept illustrates the potential impact of unmeasured confounders. It is also possible that the pharmacokinetic difference between PM/IMs and NMs is within the therapeutic window of these PPI drugs, and thus, there is no clinical difference between the groups. However, as discussed above, previous studies in prospective cohorts have demonstrated differences between PM/IMs and NMs.35,36 A large prospective trial with protocol-driven dosing and uniform assessment of compliance and outcome measures may definitively determine the clinical impact of CYP2C19 PM, intermediate metabolizer, and NM status.
To determine if there was a specific infection type (GTI or RTI) that was driving the difference between CYP2C19 NMs and RMs, we investigated RTI and GTI events separately. We did not find a significant difference for either RTIs or GTIs, although this analysis was limited by the low number of events for either infection type. We did find expected differences in the risk factor profile for RTIs and GTIs. Namely, chronic lung disease and structural gastrointestinal diseases were associated with increased RTIs, and comorbid gastrointestinal diseases were associated with increased GTIs. These findings highlight the need for vigilance in these high-risk patient populations.
In our multivariable analysis, CYP2C19 NMs had more infection events than RM/UMs even after adjusting for PPI dose. If there were an easily measured biomarker for PPI efficacy, we would expect PPI dose titration-to-effect (higher dosing with increased CYP2C19 function) and attenuation of the effect of genotype. However, there is no such biomarker for PPI effect. pH probe monitoring is an invasive, expensive, and uncomfortable test for a small child and is not part of routine care after starting a PPI. In contrast to adults, infants and small children treated with PPIs are not able to provide subjective information about improvement in their symptoms. Because of this lack of information, it is not surprising that the PPI dose is not informative for the outcome. Preprescription genotyping can be particularly helpful in this situation because it may identify CYP2C19 RM/UMs who require a higher dose for PPI efficacy.
Our study has several limitations. This retrospective study ascertained exposures, outcomes, and covariates from EHR data. It is likely that our observed infection events underrepresent the total number of infection events; events would not be recorded in the EHR if the parent and/or family did not seek medical care or sought care outside of our health care system. We also may have incomplete ascertainment of some covariate data and did not include adjustment for inhaled corticosteroids, which may increase infection risk. We expect that these factors are independent of CYP2C19 genotype. Our adjustment for comorbid conditions, such as congenital heart disease and gastrointestinal disorders, may not fully capture the impact of these conditions, which are slightly more common among CYP2C19 NMs, although the difference does not achieve statistical significance. Additionally, some infection event ICD codes are nonspecific (eg, codes for “cough” and “diarrhea”) but often used by providers when a causative pathogen is unknown. These codes may represent symptom exacerbation for individuals with chronic conditions rather than an infection event, which is a limitation in our study. We performed genotyping of the most common CYP2C19 variants leading to decreased or increased enzyme function, but it is possible that additional rare genetic variants are present in some individuals in our cohort. These would also not be ascertained by most clinical pharmacogenetic tests, which focus on commonly known variants. Our data come from a single tertiary-care children’s hospital and may not be generalizable across all practice settings and populations.
Conclusions
In this retrospective cohort of 670 infants and children treated with PPIs, CYP2C19 NMs had more frequent infections in the year after starting therapy than did CYP2C19 RM/UMs. The previously observed differences in drug disposition and drug exposure due to CYP2C19 genetic variation translates into clinically observable differences in adverse event rates in pediatric patients. The potential risk for an increased number of infections should be considered before the start of PPI therapy, particularly in the high-risk groups identified by this study. In patients who require PPI treatment, preprescription pharmacogenetic testing may assist in achieving an effective dosing regimen.
Glossary
- CI
confidence interval
- EHR
electronic health record
- GTI
gastrointestinal tract infection
- ICD
International Classification of Diseases
- IQR
interquartile range
- NM
normal metabolizer
- OR
odds ratio
- PM
poor metabolizer
- PM/IM
poor or intermediate metabolizer
- PPI
proton pump inhibitor
- RM/UM
rapid or ultrarapid metabolizer
- RTI
respiratory tract infection
Footnotes
Dr Bernal conceptualized and designed the study and data collection instruments, collected data, conducted the initial analyses, drafted the initial manuscript, and reviewed and revised the manuscript; Ms Aka, Dr Carroll, and Mr Coco designed the data collection instruments, collected data, conducted the initial analyses, and reviewed and revised the manuscript; Drs Lima, Acra, and Roden designed the study, supervised data collection, and critically reviewed the manuscript for important intellectual content; Dr Van Driest conceptualized and designed the study, drafted the initial manuscript, and reviewed and revised the manuscript; and all authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
FINANCIAL DISCLOSURE: Dr Roden has received licensing fees for the use of phenome-wide association study technology; the other authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Supported by the Burroughs Wellcome Fund (Innovation in Regulatory Science Award 1015006), the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1 TR000445), BioVU, and institutional funding, private agencies, and federal grants (S10RR025141, UL1TR002243, UL1TR000445, and UL1RR024975). Dr Bernal was funded by the National Institute of General Medical Sciences of the National Institutes of Health (T32 GM007569). This study used Vanderbilt University Medical Center’s Research Electronic Data Capture. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
COMPANION PAPER: A companion to this article can be found online at www.pediatrics.org/cgi/doi/10.1542/peds.2019-2544.
References
- 1.Agency for Healthcare Research and Quality Prescribed drugs: number of people with purchase in thousands by therapeutic class, United States, 1996–2016. 2019. Available at: https://meps.ahrq.gov/mepstrends/hc_pmed/#plot-tab. Accessed February 18, 2019
- 2.Barron JJ, Tan H, Spalding J, Bakst AW, Singer J. Proton pump inhibitor utilization patterns in infants. J Pediatr Gastroenterol Nutr. 2007;45(4):421–427 [DOI] [PubMed] [Google Scholar]
- 3.Blank ML, Parkin L. National study of off-label proton pump inhibitor use among New Zealand infants in the first year of life (2005–2012). J Pediatr Gastroenterol Nutr. 2017;65(2):179–184 [DOI] [PubMed] [Google Scholar]
- 4.Illueca M, Alemayehu B, Shoetan N, Yang H. Proton pump inhibitor prescribing patterns in newborns and infants. J Pediatr Pharmacol Ther. 2014;19(4):283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbons TE, Gold BD. The use of proton pump inhibitors in children: a comprehensive review. Paediatr Drugs. 2003;5(1):25–40 [DOI] [PubMed] [Google Scholar]
- 6.Ward RM, Kearns GL. Proton pump inhibitors in pediatrics : mechanism of action, pharmacokinetics, pharmacogenetics, and pharmacodynamics. Paediatr Drugs. 2013;15(2):119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yacyshyn BR, Thomson ABR. The clinical importance of proton pump inhibitor pharmacokinetics. Digestion. 2002;66(2):67–78 [DOI] [PubMed] [Google Scholar]
- 8.Wedlund PJ. The CYP2C19 enzyme polymorphism. Pharmacology. 2000;61(3):174–183 [DOI] [PubMed] [Google Scholar]
- 9.Lima JJ, Franciosi JP. Pharmacogenomic testing: the case for CYP2C19 proton pump inhibitor gene-drug pairs. Pharmacogenomics. 2014;15(11):1405–1416 [DOI] [PubMed] [Google Scholar]
- 10.Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006;58(3):521–590 [DOI] [PubMed] [Google Scholar]
- 11.Kearns GL, Blumer J, Schexnayder S, et al. Single-dose pharmacokinetics of oral and intravenous pantoprazole in children and adolescents. J Clin Pharmacol. 2008;48(11):1356–1365 [DOI] [PubMed] [Google Scholar]
- 12.Knebel W, Tammara B, Udata C, et al. Population pharmacokinetic modeling of pantoprazole in pediatric patients from birth to 16 years. J Clin Pharmacol. 2011;51(3):333–345 [DOI] [PubMed] [Google Scholar]
- 13.Kearns GL, Leeder JS, Gaedigk A. Impact of the CYP2C19*17 allele on the pharmacokinetics of omeprazole and pantoprazole in children: evidence for a differential effect. Drug Metab Dispos. 2010;38(6):894–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunfeld NG, Mathot RA, Touw DJ, et al. Effect of CYP2C19*2 and *17 mutations on pharmacodynamics and kinetics of proton pump inhibitors in Caucasians. Br J Clin Pharmacol. 2008;65(5):752–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshpande N, Vuddagiri S, Ravi Kanth VV, et al. Rapid and ultra-rapid metabolizers with CYP2C19*17 polymorphism do not respond to standard therapy with proton pump inhibitors. Meta Gene. 2016;9:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gawrońska-Szklarz B, Adamiak-Giera U, Wyska E, et al. CYP2C19 polymorphism affects single-dose pharmacokinetics of oral pantoprazole in healthy volunteers. Eur J Clin Pharmacol. 2012;68(9):1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin RM, Ohlsson S, Pedersen RS, et al. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol. 2008;65(5):767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina-Infante J, Rodriguez-Sanchez J, Martinek J, et al. Long-term loss of response in proton pump inhibitor-responsive esophageal eosinophilia is uncommon and influenced by CYP2C19 genotype and rhinoconjunctivitis. Am J Gastroenterol. 2015;110(11):1567–1575 [DOI] [PubMed] [Google Scholar]
- 19.Settin A, Abdalla AF, Al-Hussaini AS, El-Baz R, Galal A. Cure rate of Helicobacter pylori infection in Egyptian children related to CYP2C19 gene polymorphism. Indian J Gastroenterol. 2014;33(4):330–335 [DOI] [PubMed] [Google Scholar]
- 20.Furuta T, Shirai N, Sugimoto M, et al. Influence of CYP2C19 pharmacogenetic polymorphism on proton pump inhibitor-based therapies. Drug Metab Pharmacokinet. 2005;20(3):153–167 [DOI] [PubMed] [Google Scholar]
- 21.Shirai N, Furuta T, Moriyama Y, et al. Effects of CYP2C19 genotypic differences in the metabolism of omeprazole and rabeprazole on intragastric pH. Aliment Pharmacol Ther. 2001;15(12):1929–1937 [DOI] [PubMed] [Google Scholar]
- 22.Franciosi JP, Mougey EB, Williams A, et al. Association between CYP2C19 extensive metabolizer phenotype and childhood anti-reflux surgery following failed proton pump inhibitor medication treatment. Eur J Pediatr. 2018;177(1):69–77 [DOI] [PubMed] [Google Scholar]
- 23.Franciosi JP, Mougey EB, Williams A, et al. Association between CYP2C19*17 alleles and pH probe testing outcomes in children with symptomatic gastroesophageal reflux. J Clin Pharmacol. 2018;58(1):89–96 [DOI] [PubMed] [Google Scholar]
- 24.Bavishi C, Dupont HL. Systematic review: the use of proton pump inhibitors and increased susceptibility to enteric infection. Aliment Pharmacol Ther. 2011;34(11–12):1269–1281 [DOI] [PubMed] [Google Scholar]
- 25.Canani RB, Cirillo P, Roggero P, et al. ; Working Group on Intestinal Infections of the Italian Society of Pediatric Gastroenterology, Hepatology and Nutrition (SIGENP) . Therapy with gastric acidity inhibitors increases the risk of acute gastroenteritis and community-acquired pneumonia in children. Pediatrics. 2006;117(5). Available at: www.pediatrics.org/cgi/content/full/117/5/e817 [DOI] [PubMed] [Google Scholar]
- 26.Mitre E, Susi A, Kropp LE, et al. Association between use of acid-suppressive medications and antibiotics during infancy and allergic diseases in early childhood. JAMA Pediatr. 2018;172(6):e180315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark CM, Nylund CM. Side effects and complications of proton pump inhibitors: a pediatric perspective. J Pediatr. 2016;168:16–22 [DOI] [PubMed] [Google Scholar]
- 28.Heidelbaugh JJ, Goldberg KL, Inadomi JM. Adverse risks associated with proton pump inhibitors. Gastroenterol Hepatol (NY). 2009;5(10):725–734 [PMC free article] [PubMed] [Google Scholar]
- 29.De Bruyne P, Ito S. Toxicity of long-term use of proton pump inhibitors in children. Arch Dis Child. 2018;103(1):78–82 [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Lin HJ, Perng CL, et al. The effect of H2-receptor antagonist and proton pump inhibitor on microbial proliferation in the stomach. Hepatogastroenterology. 2004;51(59):1540–1543 [PubMed] [Google Scholar]
- 31.Freedberg DE, Lebwohl B, Abrams JA. The impact of proton pump inhibitors on the human gastrointestinal microbiome. Clin Lab Med. 2014;34(4):771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen R, Hu L, Amirault J, et al. 16S community profiling identifies proton pump inhibitor related differences in gastric, lung, and oropharyngeal microflora. J Pediatr. 2015;166(4):917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dial S, Delaney JA, Barkun AN, Suissa S. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA. 2005;294(23):2989–2995 [DOI] [PubMed] [Google Scholar]
- 34.Turco R, Martinelli M, Miele E, et al. Proton pump inhibitors as a risk factor for paediatric Clostridium difficile infection. Aliment Pharmacol Ther. 2010;31(7):754–759 [DOI] [PubMed] [Google Scholar]
- 35.Lima JJ, Lang JE, Mougey EB, et al. Association of CYP2C19 polymorphisms and lansoprazole-associated respiratory adverse effects in children. J Pediatr. 2013;163(3):686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang JE, Holbrook JT, Mougey EB, et al. ; American Lung Association-Airways Clinical Research Centers . Lansoprazole is associated with worsening asthma control in children with the CYP2C19 poor metabolizer phenotype. Ann Am Thorac Soc. 2015;12(6):878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cicali EJ, Blake K, Gong Y, et al. Novel implementation of genotype-guided proton pump inhibitor medication therapy in children: a pilot, randomized, multisite pragmatic trial. Clin Transl Sci. 2019;12(2):172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673 [DOI] [PubMed] [Google Scholar]
- 39.McGregor TL, Van Driest SL, Brothers KB, et al. Inclusion of pediatric samples in an opt-out biorepository linking DNA to de-identified medical records: pediatric BioVU. Clin Pharmacol Ther. 2013;93(2):204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010;3(1):42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye C, Coco J, Epishova A, et al. A crowdsourcing framework for medical data sets. AMIA Jt Summits Transl Sci Proc. 2018;2017:273–280 [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez D, Marvin R, Keegan M, et al. Scifabric/Pybossa. Geneva, Switzerland: Zenodo; 2017 [Google Scholar]
- 43.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19(2):215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.CPIC CPIC guideline for Clopidogrel and CYP2C19. Available at: https://cpicpgx.org/guidelines/guideline-for-clopidogrel-and-cyp2c19/. 2018. Accessed May 8, 2018
- 45.PharmGKB. Gene-specific information tables for CYP2C19. Available at: https://www.pharmgkb.org/page/cyp2c19RefMaterials. Accessed February 26, 2019
- 46.Gumus E, Karaca O, Babaoglu MO, et al. Evaluation of lansoprazole as a probe for assessing cytochrome P450 2C19 activity and genotype-phenotype correlation in childhood. Eur J Clin Pharmacol. 2012;68(5):629–636 [DOI] [PubMed] [Google Scholar]
- 47.Shakhnovich V, Smith PB, Guptill JT, et al. ; Best Pharmaceuticals for Children Act: Pediatric Trials Network . Obese children require lower doses of pantoprazole than nonobese peers to achieve equal systemic drug exposures. J Pediatr. 2018;193:102–108.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]