Supplemental Digital Content is available in the text
Keywords: elder age onset rheumatoid arthritis, genotype, HLA-DRB1, younger age onset rheumatoid arthritis
Abstract
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by joint destructions and human leukocyte antigen (HLA)-DRB1 is an important genetic risk factor for RA and influences the phenotype of RA. The clinical features of elder age onset RA (EORA) were known to be different from those of younger age onset RA (YORA). Previous studies reported the different association pattern of DRB1 alleles with YORA or EORA. The associations of DRB1 genotype with these RA subsets remained almost unknown. We investigated the genotype association of DRB1 with YORA or EORA in Japanese populations.
HLA genotyping was performed in Japanese RA patients and the association of allele or genotype carrier frequencies were analyzed.
The genotype frequency of DRB1∗04:05/DRB1∗04:06 (P = .0204, OR 7.69, 95%CI 1.39–42.72), DRB1∗04:05/DRB1∗12:01 (P = .0050, OR 5.53, 95%CI 1.71–17.88), and DRB1∗04:05/DRB1∗15:01 (P = .0124, OR 3.34, 95%CI 1.39–8.02) in YORA was higher than EORA. However, the frequencies of DRB1∗01:01/DRB1∗04:05 in YORA was tended to be lower than EORA (P = .0784, OR 0.14, 95%CI 0.01–2.42). The gene dosage effect of the shared epitope alleles was detected in EORA, but not in YORA. Trans-complementing DQ heterodimer molecules, formed by DQA1 and DQB1 of the haplotypes with and without shared epitope alleles, might explain the higher genotype frequencies of “shared epitope /not shared epitope”. Linear regression analyses showed the primary role of DQB1∗04:01 allele for the age at onset of RA.
This is the first report for the associations of DRB1 genotype with YORA or EORA in the Japanese population and the differential distribution of the genotypes was noted between these RA subsets. The involvement of DQ molecules for the age at onset of RA was suggested.
1. Introduction
Rheumatoid arthritis (RA) is an inflammatory disease characterized by structural destruction of cartilage and bone. RA patients produced specific auto-antibodies including anti-citrullinated peptide antibodies and rheumatoid factor. The specificity of anti-citrullinated peptide antibodies with RA is higher than that of rheumatoid factor. It was reported that the age at onset of RA had been increased in Japanese populations[1] and the clinical features of elder age onset RA (EORA) were different from those of younger age onset RA (YORA) on the gender distribution, the frequency of acute onset of RA, the involvement of large joints and extra-articular manifestations, the positivity of anti-citrullinated peptide antibody and rheumatoid factor.[2–7]
The etiology of RA is believed to be influenced by genetic and environmental factors. In particular, human leukocyte antigen (HLA)-DRB1 is one of the most important genetic risk factors for RA. Diverse DRB1 alleles are associated with RA in different ethnic populations. In Japanese populations, DRB1∗04:05 is associated with RA[8] and DRB1∗04:01 in European populations.[9] In the RA-associated DRB1 alleles, amino acid sequences at position 70 to 74 in the DRβ chain were conserved. These alleles were defined as the shared epitope (SE) alleles.[9] The homozygosity for the SE alleles conferred higher risk for the susceptibility of RA than the heterozygosity for them. This gene dosage effect was a distinctive feature of the SE alleles. Previous studies reported the different association pattern of DRB1 alleles with YORA or EORA. The gene dosage effect of the SE alleles was not confirmed in stratified analyses with the age at onset of RA.[10,11]DRB1∗04 was strongly associated with YORA,[12,13] but the frequency of DRB1∗04 was lower in EORA compared with YORA.[14–16] Moreover, DRB1∗01 was associated with EORA.[13,16] However, the sample sizes of these studies were modest, the resolution of the genotyping methods used in these studies was lower and DRB1 genotype was not analyzed. Thus, larger scale studies on the association of DRB1 genotype with YORA or EORA should be conducted to validate these results. In the present study, we investigated the association of DRB1 genotype with Japanese YORA and EORA, using the genotyping methods with higher resolution.
2. Materials and methods
2.1. Materials
In this study, RA patients were recruited in Hyogo College of Medicine, Miyakonojo Medical Center, Nagasaki Medical center, Nagoya Medical Center, Sagamihara National Hospital, and Tochigi Rheumatology Clinic. The healthy individuals (n = 1026) were recruited in Kanazawa University, Sagamihara Hospital, Teikyo University, University of Tokyo or by Pharma SNP Consortium (Tokyo, Japan).[17,18] The patients and the healthy individuals were native Japanese living in Japan. All the patents fulfilled American College of Rheumatology criteria for RA[19] or Rheumatoid Arthritis Classification Criteria.[20] Anti-citrullinated peptide antibody was detected by Mesacup-2 test CCP (Medical & Biological Laboratories, Nagoya, Japan). Rheumatoid factor was measured by N-latex RF kit (Siemens Healthcare Diagnostics, München, Germany). RA patients with age at onset lower than 30 years old and equal or higher than 16 were defined as YORA[21] to eliminate juvenile idiopathic arthritis. RA patients with age at onset lower than 60 years old and equal or higher than 30 years old were defined as moderate age onset RA (MORA) based on the distribution of age at onset in Japanese RA patients.[1] RA patients with age at onset equal or higher than 60 years old were defined as EORA.[16] Steinbrocker stage and class were evaluated to measure the disease progression and the activities of daily living of RA patients.[22]
This study was reviewed and approved by Hyogo College of Medicine Research Ethics Committee (178, 2012), Miyakonojo Medical Center Research Ethics Committee (approval number was not provided, 2009), Nagasaki Medical Center Research Ethics Committee (22081, 2010), Nagoya Medical Center Research Ethics Committee (2012–526, 2012), Sagamihara National Hospital Research Ethics Committee (2009061621, 2009), University of Tsukuba Research Ethics Committee (250, 2015), and Tokyo National Hospital Research Ethics Committee (190010, 2019). Written informed consent was obtained from all the participants. This study was conducted in accordance with the principles expressed in the Declaration of Helsinki.
2.2. Genotyping
HLA genotyping was performed by the polymerase chain reaction with sequence-specific oligonucleotide probes (WAKFlow HLA typing kits, Wakunaga, Akitakata, Japan), using Bio-Plex system (Bio-Rad, Hercules, CA). DRB1∗01:01, DRB1∗04:01, DRB1∗04:04, DRB1∗04:05, DRB1∗04:10, DRB1∗10:01, DRB1∗14:02, and DRB1∗14:06 were included in the SE alleles.[9] Genotyping results for some of the RA patients and the healthy controls were reported in previous studies.[8,23]
2.3. Statistical analysis
Differences of demographic features of RA patients were analyzed by Fisher exact test using 2X2 contingency tables. Associations of HLA-DRB1 allele carrier frequencies, genotype frequencies, or amino acid residue carrier frequencies were tested by Fisher exact test using 2X2 contingency tables. Simple linear regression analyses under the additive model were conducted to analyze whether each allele contributes to the age at onset of RA. The deviation from zero was tested for partial regression coefficient (PRC) and P values were calculated. Alleles with PRC more than 0 contribute to increase the age at onset of RA and alleles with PRC less than 0 decrease. Multiple linear regression analyses of an allele were performed to identify the independent association of the allele from another allele. The deviation from zero was tested for PRC of an allele and P values were calculated, when conditioned on another allele. Multiple comparisons were corrected by Bonferroni method; the corrected P (Pc) values were obtained by multiplying the P values by the number of alleles or amino acid residues tested.
3. Results
3.1. Demographic features of YORA and EORA
The characteristics of the RA patients were shown in Table 1. Eighty-nine patients were defined as YORA, 714 as MORA, and 329 as EORA. The ratio of male was higher in EORA than YORA. Steinbrocker stage and class were higher in YORA than EORA.
Table 1.
Characteristics of RA patients.

3.2. HLA-DRB1 allele carrier frequency in YORA and EORA
HLA genotyping was performed to compare allele carrier frequency (Table 2, Supplementary Table S1). DRB1∗04:01 was associated with the susceptibility of EORA (P = .0004, Pc = .0119, odds ratio [OR] 3.11, 95% confidence interval [CI] 1.69–5.73). DRB1∗04:05 was associated with the susceptibility of YORA (P = 8.33 × 10–14, Pc = 2.42 × 10–12, OR 5.47, 95% CI 3.47–8.61), MORA (P = 1.02 × 10–31, Pc = 3.15 × 10–30, OR 3.35, 95% CI 2.73–4.12), and EORA (P = 2.45 × 10–12, Pc = 7.11 × 10–11, OR 2.57, 95%CI 1.98–3.34), compared with the controls. Furthermore, the allele carrier frequency of DRB1∗01:01 in YORA was tended to be lower than EORA and that of DRB1∗04:05 in YORA was tended to be higher than EORA. The association pattern of DRB1 allele carrier frequency in MORA was similar to that of RA per se.[8] Thus, the association pattern of YORA with DRB1 alleles was different from that of EORA.
Table 2.
HLA-DRB1 allele carrier frequency in RA patients and the controls.
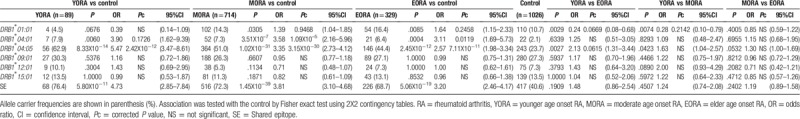
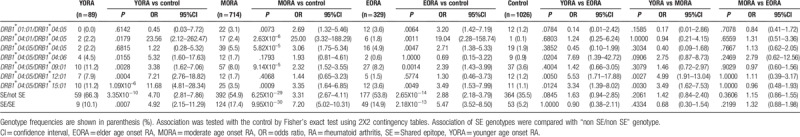
3.3. HLA-DRB1 genotype frequency in YORA and EORA
The DRB1 genotype frequencies in the RA patients were analyzed (Table 3, Supplementary Table S2). The genotype frequencies of DRB1∗04:01/DRB1∗04:05 (YORA: P = .0179, OR 23.56, 95%CI 2.12–262.47, EORA: P = .0011, OR 19.04, 95%CI 2.28–158.74) and DRB1∗04:05/DRB1∗09:01 (YORA: P = .0028, OR 3.38, 95%CI 1.62–7.06, EORA: P = .0014, OR 2.39, 95%CI 1.43–3.99) were increased in both YORA and EORA, compared with the controls. The genotype frequency of DRB1∗04:05/DRB1∗12:01 (P = .0004, OR 7.21, 95%CI 2.76–18.82) was increased in YORA, compared with the controls, but not in EORA. The frequencies of DRB1∗01:01/DRB1∗04:05 (P = .0064, OR 3.20, 95%CI 1.42–7.19) and DRB1∗04:05/DRB1∗04:05 (P = .0047, OR 2.71, 95%CI 1.38–5.33) were increased in EORA, compared with the controls, but not in YORA. Furthermore, the frequency of DRB1∗04:05/DRB1∗04:06 (P = .0204, OR 7.69, 95%CI 1.39–42.72), DRB1∗04:05/DRB1∗12:01 (P = .0050, OR 5.53, 95%CI 1.71–17.88), and DRB1∗04:05/DRB1∗15:01 (P = .0124, OR 3.34, 95%CI 1.39–8.02) in YORA was higher than EORA. However, the frequencies of DRB1∗01:01/DRB1∗04:05 in YORA was tended to be lower than EORA (P = .0784, OR 0.14, 95%CI 0.01–2.42). The homozygosity for the SE alleles conferred higher risk for EORA than the heterozygosity (SE/not SE: P = 2.65X10–14, OR 2.88, 95%CI 2.18–3.79, SE/SE: P = 2.18X10–13, OR 5.47, 95%CI 3.52–8.50), though this gene dosage effect was not observed in YORA (SE/not SE: P = 3.35X10–10, OR 4.70, 95%CI 2.81–7.86, SE/SE: P = .0007, OR 4.92, 95%CI 2.15–11.29). The association pattern of DRB1 genotype frequency in MORA was similar to those of YORA or EORA. Thus, the association pattern of YORA with DRB1 genotypes was different from that of EORA.
Table 3.
HLA-DRB1 genotype frequency in RA patients and the controls.
3.4. Associations of amino acid residues in the HLA-DRβ chain with YORA and EORA
The associations of amino acid residues in the HLA-DRβ chain with RA were shown in Figure 1. Tyrosine at position 37 (37Y, P = 8.10X10–8, OR = 4.07, Pc = 2.75X10–6, 95% CI 2.31–7.19) in the DRβ chain was associated with YORA (Fig. 1A), but not with EORA (Fig. 1C). On the other hand, leucine at position 67 (67L) and aspartic acid at position 70 (70D) were not associated with YORA (Fig. 1A), though these amino acid residues were associated with EORA (Fig. 1C, 67L: P = 3.20X10–8, OR = 2.19, Pc = 1.10X10–6, 95% CI 1.65–2.92, 70D: P = 1.50X10–8, OR = 0.48, Pc = 5.12X10–7, 95% CI 0.37–0.62). The association pattern of MORA (Fig. 1B) was similar to that of RA per se.[8] Thus, the association pattern of YORA with amino acid residues in the DRβ chain was different from that of EORA.
Figure 1.

Associations of amino acid residues in the DRβ chain with YORA (A), MORA (B), and EORA (C). Associations were established by Fisher exact test using 2X2 contingency tables. Corrected P (Pc) values were calculated by multiplying the P value by the number of amino acid residues tested. Positive associations are indicated by filled circles and negative associations by open circles. RA = rheumatoid arthritis, YORA = younger age onset RA, MORA = moderate age onset RA, EORA = elder age onset RA.
3.5. Linear regression analysis of HLA alleles for the age at onset of RA
Simple linear regression analyses of HLA alleles were conducted to reveal the effects of alleles for the age at onset of RA (Table 4, Supplementary Table S3). DQB1∗04:01 was associated with the age at onset of RA (P = .0005, Pc = 0.0079, PRC –2.36), though some other alleles were also tended to be associated (Table 4). The haplotype including B∗44:03, DRB1∗13:02, DQB1∗06:04, and DPB1∗04:01 was known[24]; DRB1∗01:01 and DQB1∗05:01 were in strong linkage disequilibrium in Japanese populations.[25]DRB1∗04:05, DQB1∗04:01, and DPB1∗02:01 were also included in the other haplotype.[25] Multiple linear regression analyses of HLA alleles were performed to identify the independent association of these alleles for the age at onset of RA (Supplementary Table S4). The significant association of B∗44:03, DRB1∗13:02, DQB1∗06:04, and DPB1∗04:01 was disappeared, when conditioned on each allele. These findings supported the strong linkage disequilibrium in the haplotype and the primary allele could not be detected in this analysis. The association of DQB1∗04:01 still remained significant (P = .0081, PRC –4.89), when conditioned on DRB1∗04:05, suggesting the independent association of DQB1∗04:01 from DRB1∗04:05. However, the association of DRB1∗04:05 was not detected (P = .1403, PRC 2.83), when conditioned on DQB1∗04:01, suggesting the dependent effects of DRB1∗04:05 on DQB1∗04:01. These data suggested the primary role of DQB1∗04:01 for the age at onset of RA. The association of DRB1∗04:05 (P = .0044, PRC –2.02) and DQB1∗04:01 (P = .0003, PRC –2.44) still remained significant, when conditioned on DPB1∗02:01. The association of DPB1∗02:01 still remained significant, when conditioned on DRB1∗04:05 (P = .0051, PRC –1.86) or DQB1∗04:01 (P = .0052, PRC –1.85). These data suggested the independent association of DPB1∗02:01 form DQB1∗04:01. The significant association of DRB1∗01:01 and DQB1∗05:01 was disappeared, when conditioned on each allele. These also suggested the strong linkage disequilibrium between DRB1∗01:01 and DQB1∗05:01 and the primary allele could not be detected in the analysis. Thus, some HLA haplotypes or alleles were detected to be responsible for the age at onset of RA.
Table 4.
Linear regression analysis of HLA alleles for the age at onset of RA.
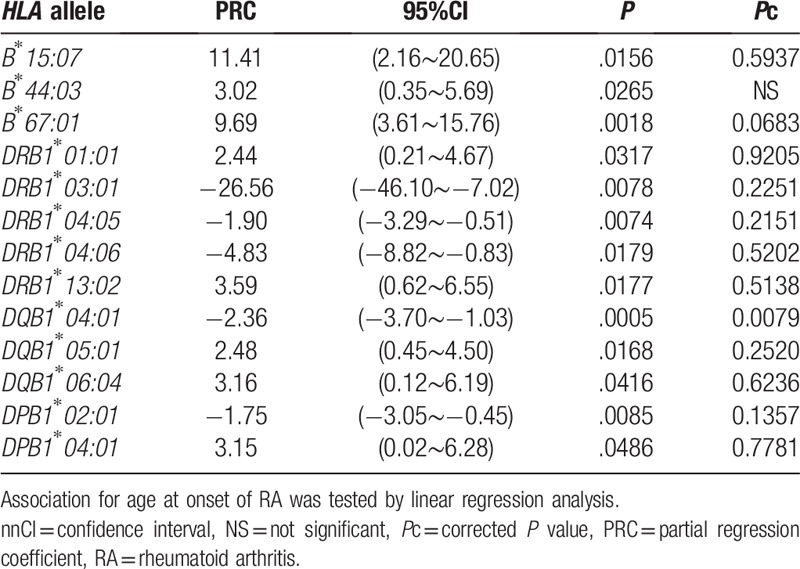
4. Discussion
Many reports on the associations of DRB1 with RA were published, so far. However, a few studies on the role of DRB1 on YORA or EORA were reported. The associations of DRB1 genotype with these RA subsets remained almost unknown. It was reported that the frequencies of DRB1∗04 in EORA were lower than those in YORA[14–16] and that DRB1∗01 was associated with EORA.[13,16] Similar tendencies on DRB1 allele carrier frequencies were observed in this study (Table 2), confirming the results of the previous studies. These results suggested the differential roles of DRB1∗04:05 and DRB1∗01:01 in the pathogenesis of YORA and EORA, respectively. The differential diagnosis is difficult in some patients with EORA from polymyalgia rheumatica and DRB1∗01 and DRB1∗04 were reported to be associated with polymyalgia rheumatica.[13,26–28] The susceptible DRB1 alleles were shared between EORA and polymyalgia rheumatica, suggesting the common etiological bases between them. Thus, this association study on DRB1 alleles with YORA or EORA sheds light on the pathogenesis of the subsets of RA.
DRB1∗04:05, one of the SE alleles, is considered to be the most important risk factor for RA in Japanese.[8] In the present study, the frequencies of some genotypes including DRB1∗04:05 were differently distributed between YORA and EORA, though the frequencies of some genotypes were reported to be increased in overall RA.[29–33] The genotype frequencies of DRB1∗04:01/DRB1∗04:05, DRB1∗04:05/DRB1∗09:01, DRB1∗04:05/DRB1∗12:01, and DRB1∗04:05/DRB1∗15:01 were increased in YORA (Table 3). In these genotypes increased in YORA, all the genotypes except DRB1∗04:01/DRB1∗04:05 were “SE/not SE”. These results suggested the important roles of the heterozygous genotypes of “SE/not SE” in the susceptibility of YORA and explained that the gene dosage effect of the SE alleles was not found in YORA. The heterozygous genotypes of “SE/not SE” might increase the variety of the self-antigens presented by DR molecules; more than two types of self-antigens, antigens presented by the SE alleles and those by the non-SE alleles, would increase the susceptibility risk of YORA. Alternatively, trans-complementing DQ heterodimer molecules, formed by DQA1 and DQB1 of the haplotypes with and without SE, might explain the higher genotype frequencies of “SE/not SE”, as proposed in the studies on other diseases.[23,34] The primary effect of DQB1∗04:01 for the age at onset of RA detected in the multiple linear regression analyses also support the role of DQ molecules. The frequencies of DRB1∗01:01/DRB1∗04:05, DRB1∗04:01/DRB1∗04:05, DRB1∗04:05/DRB1∗04:05, DRB1∗04:05/DRB1∗09:01, and DRB1∗04:05/DRB1∗15:01 were increased in EORA (Table 3). In these genotypes increased in EORA, all the genotypes except DRB1∗04:05/DRB1∗09:01 and DRB1∗04:05/DRB1∗15:01 were “SE/SE”. These results suggested the important roles of the homozygous genotypes of “SE/SE” in the susceptibility of EORA and interpreted the gene dosage effect of the SE alleles in EORA. Thus, the differential distribution of the genotypes including DRB1∗04:05 were noted between YORA and EORA, suggesting the different pathogenesis in YORA from EORA.
The amino acid residues of 11V, 13H, 33H, 57S, and 96Y in the DRβ chain were associated with both of YORA and EORA (Fig. 1A and 1C). The predominant roles of DRB1∗04:05 on the susceptibility of YORA and EORA could account for the results. However, leucine at position 67 (67L) was associated with EORA, but not with YORA, suggesting the effects of DRB1∗01:01 allele on EORA. Similarly, DRB1∗12:01might explain that aspartic acid at position 70 (70D) was not associated with YORA. Additionally, tyrosine at position 37 (37Y) was associated with YORA, but not with EORA, supported by the higher frequency of DRB1∗04:05 in YORA. Some HLA alleles were predicted to be selected in the pathogen-driven manner.[24,35] Analogically, SE alleles would be selected in the similar manner, increasing the susceptibility of YORA.
Linear regression analyses revealed the role of some HLA haplotypes or alleles for the age at onset of RA. Because of the strong linkage disequilibrium of HLA region, it is difficult to identify the primary role of the responsible allele for the age at onset of RA. DQB1∗04:01 was the sole significantly associated allele with the age at onset of RA in simple linear regression analyses. In multiple linear regression analyses, the association of DQB1∗04:01 still remained significant, when conditioned on DRB1∗04:05. However, the association of DRB1∗04:05 was not detected, when conditioned on DQB1∗04:01. Thus, the results of the multiple linear regression analyses showed the primary role of DQB1∗04:01 for the age at onset of RA, suggesting the involvement of DQ molecules on the age at onset. It is possible that other HLA loci than DRB1 might contribute to the age at onset of RA. However, this primary association of DQB1∗04:01 should be confirmed in future replication studies, as DQA1 is a risk factor for RA in Chinese populations.[36] The association of DRB1 genotypes with YORA and EORA should also be validated in larger scale studies, because of the limited sample size of this study. Since the distribution pattern of DRB1 is different in other ethnic populations, DRB1 genotypes of YORA and EORA should be investigated in other populations.
In the present study, it was revealed that the association pattern of DRB1 genotype was widely different between YORA and EORA. The genotype frequencies of DRB1∗04:05/DRB1∗12:01 and DRB1∗04:05/DRB1∗15:01 were increased in YORA. On the other hand, the frequencies of DRB1∗01:01/DRB1∗04:05 and DRB1∗04:05/DRB1∗04:05 were increased in EORA. The gene dosage effect of the SE alleles was detected in EORA, but not observed in YORA.
5. Conclusion
To the best of our knowledge, this is the first report for these associations of DRB1 genotype with these RA subsets in the Japanese population and the differentially associated DRB1 genotypes were detected between these subsets. Although accumulated genetic risk factors were thought to cause autoimmune diseases in younger age, our results suggested that different genetic risk factors contribute to the pathogenesis of YORA from EORA. The involvement of DQ molecules on the age at onset of RA was suggested.
Acknowledgments
We thank Ms. Mayumi Yokoyama and Ms. Satomi Hanawa (Clinical Research Center for Allergy and Rheumatology, Sagamihara National Hospital) for secretarial assistance.
Author contributions
Conceptualization: Hiroshi Furukawa, Shigeto Tohma.
Data curation: Shomi Oka, Hiroshi Furukawa.
Formal analysis: Shomi Oka, Hiroshi Furukawa.
Funding acquisition: Hiroshi Furukawa, Atsushi Hashimoto, Shigeto Tohma.
Investigation: Shomi Oka, Hiroshi Furukawa.
Methodology: Shomi Oka.
Resources: Kota Shimada, Atsushi Hashimoto, Akiko Komiya, Shinichiro Tsunoda, Koichiro Saisho, Naoyuki Tsuchiya, Masao Katayama, Satoshi Shinohara, Toshihiro Matsui, Naoshi Fukui, Hajime Sano, Kiyoshi Migita, Shigeto Tohma.
Supervision: Kiyoshi Migita, Shigeto Tohma.
Validation: Shomi Oka, Hiroshi Furukawa.
Visualization: Shomi Oka, Hiroshi Furukawa.
Writing – original draft: Shomi Oka.
Writing – review & editing: Hiroshi Furukawa, Shigeto Tohma.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: CI = confidence interval, EORA = elder age onset RA, HLA = human leukocyte antigen, MORA = moderate age onset RA, OR = odds ratio, Pc = corrected P, PRC = partial regression coefficient, RA = rheumatoid arthritis, SE = shared epitope, YORA = younger age onset RA.
How to cite this article: Oka S, Furukawa H, Shimada K, Hashimoto A, Komiya A, Tsunoda S, Saisho K, Tsuchiya N, Katayama M, Shinohara S, Matsui T, Fukui N, Sano H, Migita K, Tohma S. Association of HLA-DRB1 genotype with younger age onset and elder age onset rheumatoid arthritis in Japanese populations. Medicine. 2019;98:48(e18218).
The work was supported by Grants-in-Aid for Scientific Research (B, C) (26293123, 22591090, 15K09543, 18K08402) and for Young Scientists (B) (24791018) from the Japan Society for the Promotion of Science, Health and Labour Science Research Grants from the Ministry of Health, Labour, and Welfare of Japan, Grants-in-Aid of the Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) from Japan Agency for Medical Research and Development, Grants-in-Aid for Clinical Research from National Hospital Organization, Research Grants from Daiwa Securities Health Foundation, Research Grants from Japan Research Foundation for Clinical Pharmacology, Research Grants from The Nakatomi Foundation, Research Grants from Takeda Science Foundation, Research Grants from Mitsui Sumitomo Insurance Welfare Foundation, Bristol-Myers K.K. RA Clinical Investigation Grant from Bristol-Myers Squibb Co., and research grants from the following pharmaceutical companies: Abbott Japan Co., Ltd., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Mitsuibishi Tanabe Pharma Corporation, Merck Sharp and Dohme Inc., Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, Teijin Pharma Limited. The funders had no role in study design, data collection and analysis, decision to publish, or preparing the manuscript.
Conflict of interest statement: HF has the following conflicts, and the following funders are supported wholly or in part by the indicated pharmaceutical companies. The Japan Research Foundation for Clinical Pharmacology is run by Daiichi Sankyo, the Takeda Science Foundation is supported by an endowment from Takeda Pharmaceutical Company and the Nakatomi Foundation was established by Hisamitsu Pharmaceutical Co., Inc. The Daiwa Securities Health Foundation was established by Daiwa Securities Group Inc. and Mitsui Sumitomo Insurance Welfare Foundation was established by Mitsui Sumitomo Insurance Co., Ltd. HF was supported by research grants from Bristol-Myers Squibb Co. HF received honoraria from Ajinomoto Co., Inc., Daiichi Sankyo Co., Ltd., Dainippon Sumitomo Pharma Co., Ltd., Pfizer Japan Inc., and Takeda Pharmaceutical Company, Luminex Japan Corporation Ltd., and Ayumi Pharmaceutical Corporation. ST was supported by research grants from 9 pharmaceutical companies: Abbott Japan Co., Ltd., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Merck Sharp and Dohme Inc., Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, Teijin Pharma Limited. ST received honoraria from Asahi Kasei Pharma Corporation, Astellas Pharma Inc., AbbVie GK., Chugai Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Pfizer Japan Inc.
The other authors have no conflicts of interests to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Imanaka T, Shichikawa K, Inoue K, et al. Increase in age at onset of rheumatoid arthritis in Japan over a 30 year period. Ann Rheum Dis 1997;56:313–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Terkeltaub R, Esdaile J, Decary F, et al. A clinical study of older age rheumatoid arthritis with comparison to a younger onset group. J Rheumatol 1983;10:418–24. [PubMed] [Google Scholar]
- [3].Deal CL, Meenan RF, Goldenberg DL, et al. The clinical features of elderly-onset rheumatoid arthritis. A comparison with younger-onset disease of similar duration. Arthritis Rheum 1985;28:987–94. [DOI] [PubMed] [Google Scholar]
- [4].Mavragani CP, Moutsopoulos HM. Rheumatoid arthritis in the elderly. Exp Gerontol 1999;34:463–71. [DOI] [PubMed] [Google Scholar]
- [5].Lopez-Hoyos M, Ruiz de Alegria C, Blanco R, et al. Clinical utility of anti-CCP antibodies in the differential diagnosis of elderly-onset rheumatoid arthritis and polymyalgia rheumatica. Rheumatology (Oxford) 2004;43:655–7. [DOI] [PubMed] [Google Scholar]
- [6].Turkcapar N, Demir O, Atli T, et al. Late onset rheumatoid arthritis: clinical and laboratory comparisons with younger onset patients. Arch Gerontol Geriatr 2006;42:225–31. [DOI] [PubMed] [Google Scholar]
- [7].Arnold MB, Bykerk VP, Boire G, et al. Are there differences between young- and older-onset early inflammatory arthritis and do these impact outcomes? An analysis from the CATCH cohort. Rheumatology (Oxford) 2014;53:1075–86. [DOI] [PubMed] [Google Scholar]
- [8].Oka S, Furukawa H, Kawasaki A, et al. Protective effect of the HLA-DRB1∗13:02 allele in Japanese rheumatoid arthritis patients. PLoS One 2014;9:e99453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Reveille JD. The genetic contribution to the pathogenesis of rheumatoid arthritis. Curr Opin Rheumatol 1998;10:187–200. [DOI] [PubMed] [Google Scholar]
- [10].Stavropoulos C, Spyropoulou M, Koumantaki Y, et al. HLA-DRB1 alleles in Greek rheumatoid arthritis patients and their association with clinical characteristics. Eur J Immunogenet 1997;24:265–74. [DOI] [PubMed] [Google Scholar]
- [11].Del Rincon I, Battafarano DF, Arroyo RA, et al. Ethnic variation in the clinical manifestations of rheumatoid arthritis: role of HLA-DRB1 alleles. Arthritis Rheum 2003;49:200–8. [DOI] [PubMed] [Google Scholar]
- [12].Wu H, Khanna D, Park G, et al. Interaction between RANKL and HLA-DRB1 genotypes may contribute to younger age at onset of seropositive rheumatoid arthritis in an inception cohort. Arthritis Rheum 2004;50:3093–103. [DOI] [PubMed] [Google Scholar]
- [13].Gonzalez-Gay MA, Hajeer AH, Dababneh A, et al. Seronegative rheumatoid arthritis in elderly and polymyalgia rheumatica have similar patterns of HLA association. J Rheumatol 2001;28:122–5. [PubMed] [Google Scholar]
- [14].Inoue K, Shichikawa K, Nishioka J, et al. Older age onset rheumatoid arthritis with or without osteoarthritis. Ann Rheum Dis 1987;46:908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Terkeltaub R, Decary F, Esdaile J. An immunogenetic study of older age onset rheumatoid arthritis. J Rheumatol 1984;11:147–9. [PubMed] [Google Scholar]
- [16].Yukioka M, Wakitani S, Murata N, et al. Elderly-onset rheumatoid arthritis and its association with HLA-DRB1 alleles in Japanese. Br J Rheumatol 1998;37:98–101. [DOI] [PubMed] [Google Scholar]
- [17].Kamatani N, Kawamoto M, Kitamura Y, et al. Establishment of B-cell lines derived from 996 Japanese individuals. Tissue Culture Res Commun 2004;23:71–80. [Google Scholar]
- [18].Kamitsuji S, Matsuda T, Nishimura K, et al. Japan PGx data science consortium database: SNPs and HLA genotype data from 2994 Japanese healthy individuals for pharmacogenomics studies. J Hum Genet 2015;60:319–26. [DOI] [PubMed] [Google Scholar]
- [19].Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. [DOI] [PubMed] [Google Scholar]
- [20].Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 2010;62:2569–81. [DOI] [PubMed] [Google Scholar]
- [21].Petty RE, Southwood TR, Manners P, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol 2004;31:390–2. [PubMed] [Google Scholar]
- [22].Steinbrocker O, Traeger CH, Batterman RC. Therapeutic criteria in rheumatoid arthritis. J Am Med Assoc 1949;140:659–62. [DOI] [PubMed] [Google Scholar]
- [23].Oka S, Furukawa H, Yasunami M, et al. HLA-DRB1 and DQB1 alleles in Japanese type 1 autoimmune hepatitis: The predisposing role of the DR4/DR8 heterozygous genotype. PLoS One 2017;12:e0187325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kawashima M, Ohashi J, Nishida N, et al. Evolutionary analysis of classical HLA class I and II genes suggests that recent positive selection acted on DPB1∗04:01 in Japanese population. PLoS One 2012;7:e46806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nakajima F, Nakamura J, Yokota T. Analysis of HLA haplotypes in Japanese, using high resolution allele typing. MHC 2001;8:1–32. [Google Scholar]
- [26].Sakkas LI, Loqueman N, Panayi GS, et al. Immunogenetics of polymyalgia rheumatica. Br J Rheumatol 1990;29:331–4. [DOI] [PubMed] [Google Scholar]
- [27].Guerne PA, Salvi M, Seitz M, et al. Molecular analysis of HLA-DR polymorphism in polymyalgia rheumatica. Swiss Group for Research on HLA in Polymyalgia Rheumatica. J Rheumatol 1997;24:671–6. [PubMed] [Google Scholar]
- [28].Dababneh A, Gonzalez-Gay MA, Garcia-Porrua C, et al. Giant cell arteritis and polymyalgia rheumatica can be differentiated by distinct patterns of HLA class II association. J Rheumatol 1998;25:2140–5. [PubMed] [Google Scholar]
- [29].Lee HS, Lee KW, Song GG, et al. Increased susceptibility to rheumatoid arthritis in Koreans heterozygous for HLA-DRB1∗0405 and ∗0901. Arthritis Rheum 2004;50:3468–75. [DOI] [PubMed] [Google Scholar]
- [30].Liu SC, Chang TY, Lee YJ, et al. Influence of HLA-DRB1 genes and the shared epitope on genetic susceptibility to rheumatoid arthritis in Taiwanese. J Rheumatol 2007;34:674–80. [PubMed] [Google Scholar]
- [31].Balandraud N, Picard C, Reviron D, et al. HLA-DRB1 genotypes and the risk of developing anti citrullinated protein antibody (ACPA) positive rheumatoid arthritis. PLoS One 2013;8:e64108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lenz TL, Deutsch AJ, Han B, et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet 2015;47:1085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Terao C, Okada Y, Ikari K, et al. Genetic landscape of interactive effects of HLA-DRB1 alleles on susceptibility to ACPA(+) rheumatoid arthritis and ACPA levels in Japanese population. J Med Genet 2017;54:853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sun H, Yang Z, Lin K, et al. The adaptive change of HLA-DRB1 allele frequencies caused by natural selection in a Mongolian population that migrated to the South of China. PLoS One 2015;10:e0134334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Guo J, Zhang T, Cao H, et al. Sequencing of the MHC region defines HLA-DQA1 as the major genetic risk for seropositive rheumatoid arthritis in Han Chinese population. Ann Rheum Dis 2019;78:773–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


