Abstract
Complex human biomolecular processes are made possible by the diversity of human proteoforms. Constructing proteoform families, groups of proteoforms derived from the same gene, is one way to represent this diversity. Comprehensive, high-confidence identification of human proteoforms remains a central challenge in mass spectrometry-based proteomics. We have previously reported a strategy for proteoform identification using intact-mass measurements, and we have since improved that strategy by mass calibration based on search results, the use of a global post-translational modification discovery database, and the integration of top-down proteomics results with intact-mass analysis. In the present study, we combine these strategies for enhanced proteoform identification in total cell lysate from the Jurkat human T lymphocyte cell line. We collected, processed, and integrated three types of proteomics data (NeuCode-labeled intact-mass, label-free top-down, and multi-protease bottom-up) to maximize the number of confident proteoform identifications. The integrated analysis revealed 5,950 unique experimentally observed proteoforms, which were assembled into 848 proteoform families. Twenty percent of the observed proteoforms were confidently identified at a 3.9% false discovery rate, representing 1,207 unique proteoforms derived from 484 genes.
Keywords: human proteoform, proteoform family, intact-mass, top-down, global PTM discovery, multi-protease, Proteoform Suite, MetaMorpheus, NeuCode, Jurkat
INTRODUCTION
The complex biological processes essential for cell survival, development, and homeostasis require a wide variety of proteins. The human proteome originates from roughly 20,000 protein-coding genes, but the complexity of the proteome is then expanded through genetic variations, alternative splicing, and post-translational modifications (PTMs).1 Capturing and organizing this molecular complexity is aided by the concepts of the “proteoform”, referring to a defined amino acid sequence with a specific set of PTMs, and the “proteoform family”, the set of proteoforms derived from the same gene.2,3 Characterization of proteoforms and elucidation of proteoform families are important emerging areas of proteomic research.
Mass spectrometry (MS)-based analysis of intact protein molecules has developed into a robust and efficient approach to proteoform identification in complex samples.4 Many studies of intact proteoforms have utilized the top-down strategy, where whole proteins are fragmented in the gas phase and analyzed by tandem MS.5 Top-down proteomics is advantageous for proteoform analysis because the molecular context of the PTMs is preserved and fragmentation data can provide sequence evidence for identification.4,6 However, challenges still exist for top-down data acquisition and analysis, as a large fraction of precursor ions are not selected for fragmentation7,8 and limitations in fragmentation can lead to ambiguous proteoform identifications.9,10 Proteoform identification without fragmentation is also possible, by inferring identity from accurate proteoform intact-mass measurements. In such a strategy, identification is achieved by relating the masses of experimentally observed proteoforms to those of theoretical proteoforms in databases.3,7,11 We have previously explored this intact-mass approach to identify proteoforms and proteoform families in both prokaryotic and eukaryotic proteomes3,12–14 and have streamlined the procedure in the Proteoform Suite software (available at https://smith-chem-wisc.github.io/ProteoformSuite).15 Although intact-mass proteomics compensates for some of the limitations of traditional top-down proteomic strategies by making more efficient use of precursor ion data, it is nevertheless challenging to confidently identify proteoforms based on intact-mass measurements alone given the complexity of the proteoform-ome.
We have recently implemented several innovations to improve the number and confidence of proteoform identifications using Proteoform Suite. One of these was NeuCode SILAC (Stable Isotope Labeling by Amino acids in Cell culture),16–18 which was used to count the number of lysine residues in a proteoform as a second piece of information to leverage during identification.3,12,15 Post-acquisition mass calibration based on the software lock-mass concept19 was also introduced to increase the accuracy of intact-mass data, contributing to more proteoform identifications with lower false discovery rates (FDRs).13,14 We also employed bottom-up proteomics to build global PTM discovery (G-PTM-D)20,21 databases for enhanced intact-mass proteoform identification.12 Although bottom-up proteomics by itself does not generally provide sufficient information to identify proteoforms, as intact sequence and PTM context are lost after protease digestion,22 it is an extremely powerful strategy to produce detailed peptide-level data. Tools such as G-PTM-D allow novel PTM sites to be discovered from bottom-up data, information which may then be used to construct richer and more accurate databases of theoretical proteoforms. We have previously shown that the use of a G-PTM-D database generated from tryptic bottom-up data increased the number of Escherichia coli proteoforms that could be confidently identified from intact-mass data12 and have integrated intact-mass and conventional top-down proteomic analyses to increase proteoform identifications in yeast and murine mitochondria.13,14
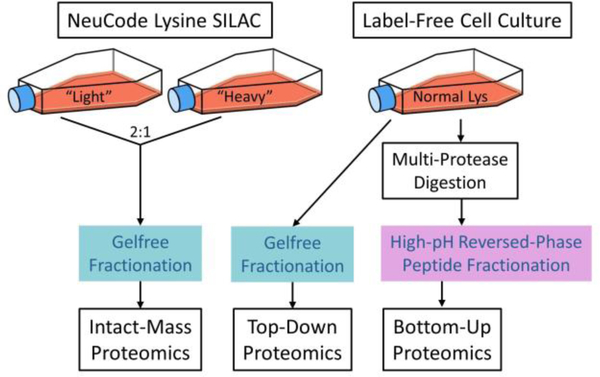
In the present study, we combine these strategies (NeuCode SILAC, post-acquisition intact-mass calibration, G-PTM-D, and incorporation of top-down data) to identify intact proteoforms in human samples using the Jurkat T lymphocyte cell line as a model system.23 We extended the G-PTM-D strategy to use multiple proteases instead of only trypsin digestion to increase proteome coverage.24 Proteoform Suite’s functionality was also expanded to accommodate processing of NeuCode-labeled and label-free data together. Multiple recent studies have explored global human proteoform investigation using conventional top-down proteomics,25–32 and top-down and bottom-up data have been integrated for the purpose of proteoform analysis for more than a decade.29,33,34 Here, we further explored how different types of proteomics schemes (intact-mass, top-down, and bottom-up) can be integrated to yield the most proteoform-level information from the data collected (Figure 1).
Figure 1.
Schematic of sample preparation for intact-mass, top-down, and bottom-up proteomics. In this study, “intact-mass proteomics” refers to MS1-only analysis with no precursor fragmentation, while “top-down proteomics” refers to tandem MS analysis with precursor fragmentation and MS2 analysis.
EXPERIMENTAL PROCEDURES
A detailed account of all materials, including their sources, and experimental procedures employed in this work can be found in the Supporting Information. Brief summaries of these procedures are provided here.
Intact-Mass Proteomics of NeuCode-Labeled Jurkat Cells
NeuCode SILAC Cell Culture.
Jurkat cells were cultured at 37 °C under 5% CO2 in SILAC RPMI-1640 medium supplemented with 10% fetal bovine serum, 1X antibiotic-antimycotic solution, 10 mM HEPES buffer, 1 mM sodium pyruvate, 2 mM GlutaMAX, 1.2 mM L-arginine, and 0.5 mM of either one of two NeuCode lysine isotopologues: “light” (15N213C6) or “heavy” (2H8).18 Cells were grown to a density of ~106 cells/mL at which time they were washed, pelleted, snap-frozen in liquid nitrogen, and stored at −80 °C until use. Cellular incorporation of NeuCode lysine reached ~99% in cells after approximately five doublings, as determined by bottom-up mass spectrometry.
Protein Purification and Fractionation.
Sample preparation was similar to that described in our previous NeuCode proteoform studies of yeast and E. coli.3,12,15 Briefly, light and heavy NeuCode-labeled Jurkat cells were lysed separately and proteins were reduced and alkylated. Proteins were then precipitated with acetone, resuspended, and mixed in a 2:1 light/heavy ratio (Figure 1). The proteins were separated based on molecular weight (MW) using a Gelfree system (Expedeon),35 and 11 fractions were collected. Prior to mass spectrometric analysis, sodium dodecyl sulfate was removed from the fractions via methanol−chloroform precipitation36 and proteins were reconstituted with 5% acetonitrile (ACN) and 0.2% formic acid in water. Three biological replicates of this experiment were performed.
Liquid Chromatography/Mass Spectrometry (LC/MS).
All fractions were analyzed by HPLC-ESI-MS (nanoAcquity, Waters and QE-HF Orbitrap, Thermo Fisher Scientific). Two technical replicate injections of each fraction were performed, yielding a total of 66 raw data files (3 biological replicates × 11 fractions × 2 injections).
Bottom-Up Proteomics of Label-Free Jurkat Cells
Cell Culture and Protein Digestion Using Multiple Proteases in Parallel.
Bottom-up proteomic data had been collected previously for five aliquots of Jurkat cell lysate, each of which had been digested with a different protease (chymotrypsin, GluC, ArgC, AspN, or LysC).37 Briefly, cells were cultured in medium containing normal (i.e., not isotopically labeled) lysine and lysed. Aliquots of lysate were transferred to separate filter units for filter-aided sample preparation (FASP)38 using different proteases. The resultant peptide samples were each separated into 11 fractions via high-pH reversed-phase liquid chromatography. Each fraction was then dried down and reconstituted in 2% ACN and 0.2% formic acid in water. Additionally, as part of a separate work, this process was repeated to collect 10 fractions of peptides from label-free Jurkat cell lysate digested with trypsin.39
LC/MS.
Bottom-up analysis was performed via HPLC-ESI-MS/MS (nanoAcquity, Waters and LTQ Velos Orbitrap, Thermo Fisher Scientific) as described previously.37 The top 10 most intense precursor ions were selected for higher-energy collisional dissociation (HCD) fragmentation via data-dependent acquisition. Dynamic exclusion was enabled. A total of 65 raw data files were collected (10 fractions for trypsin and 11 fractions for each of the other five proteases).
Top-Down Proteomics of Label-Free Jurkat Cells
Cell Culture and Sample Preparation.
Label-free Jurkat cells were cultured as described for the NeuCode-labeled cells, except that normal lysine was substituted for the heavy lysine isotopologues. Cells were lysed and proteins were extracted as described for the NeuCode-labeled samples. After acetone precipitation, proteins were separated via Gelfree and 11 fractions were prepared for mass spectrometry as described for the NeuCode-labeled samples.
LC/MS.
Top-down analysis was performed using HPLC-ESI-MS/MS (nanoAcquity, Waters and QE-HF Orbitrap, Thermo Fisher Scientific). The top three most intense precursor ions were selected for HCD fragmentation via data-dependent acquisition. Dynamic exclusion was enabled. One biological and two technical replicates were performed, generating 22 raw data files.
Processing and Integration of Intact-Mass, Bottom-Up, and Top-Down Data Sets
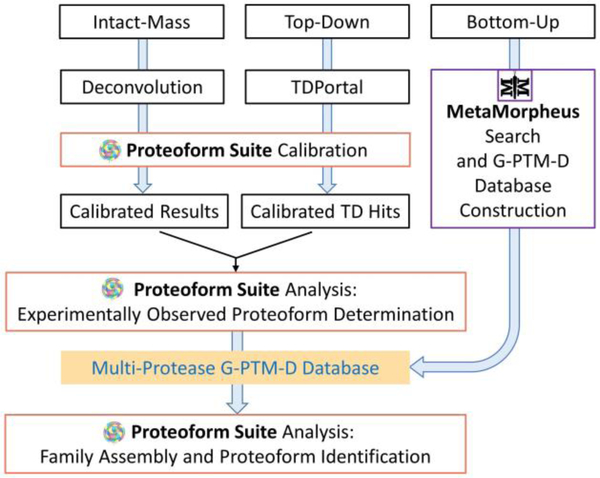
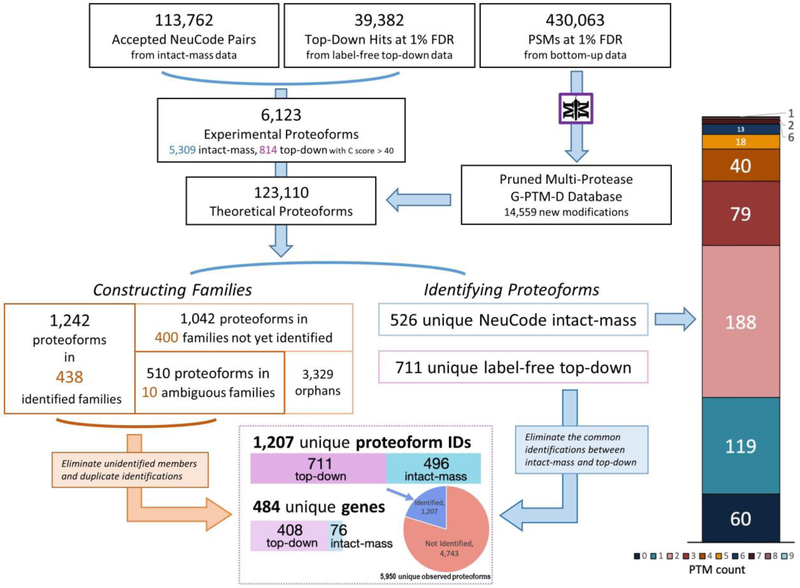
The overall workflow for data processing and proteoform analysis is shown in Figure 2.
Figure 2.
Schematic of data processing and analysis for proteoform identification and family construction using intact-mass, top-down, and bottom-up proteomics data.
Intact-Mass Raw Data Deconvolution.
Intact-mass data files (.raw) were deconvoluted into monoisotopic mass components using Thermo Protein Deconvolution 4.0. The outputs of deconvolution (monoisotopic masses) are referred to herein as “raw mass components”.
Bottom-Up Data Analysis in MetaMorpheus.
The global PTM discovery search workflow20 was performed on the bottom-up raw data files in MetaMorpheus (v0.0.297, available at https://smithchem-wisc.github.io/MetaMorpheus).21 This strategy, which enables discovery of PTMs that are not annotated in UniProt proteome databases, was applied in previous studies to bottom-up data from a single protease (trypsin).12,20 Here, we adapted the MetaMorpheus software to allow raw data files from samples digested with different proteases to be calibrated and searched at the same time. Protease type was specified for each file in the file-specific search parameters. Data were searched against a UniProt human proteome XML database (73,928 entries, downloaded February 2019) and calibrated based on peptide mapping results. The calibrated data were searched again with selected mass errors allowed. These mass errors reflected common biological PTMs, such as phosphorylation, acetylation, and methylation, as well as common artifacts, such as deamidation, sodium adduction, and ammonia loss (Supporting Information, Table S-1). MetaMorpheus added the modification sites revealed by this G-PTM-D search into the database, thereby generating the “multi-protease G-PTM-D database” in XML format. Finally, all of the calibrated files were searched against this new database. Integrating data from multiple proteolytic digestions increases proteome coverage24 and improves protein inference in this final search, thereby decreasing the number of ambiguous protein identifications.39 A “pruned” version of the multi-protease G-PTM-D database was created, limiting the entries to only those proteins that had confidently identified peptides in deep bottom-up data (1% FDR), along with any UniProt-documented modifications and confident G-PTM-D modifications (1% FDR) for those proteins. This pruned version of the multi-protease G-PTM-D database was utilized by Proteoform Suite during the analysis of intact-mass and top-down data, if not otherwise specified.
Top-Down Raw Data Processing.
Top-down raw data files were analyzed using TDPortal (National Resource for Translational and Developmental Proteomics, NRTDP, Northwestern University, Evanston, IL) as previously described.13 Files were searched against the human proteome, and carbamidomethylation of cysteine was set as a fixed modification. A search result table containing all top-down hits (proteoform spectral matches) observed at 1% FDR was generated and used for subsequent data calibration in Proteoform Suite.
Data Calibration with Proteoform Suite.
Deconvoluted NeuCode intact-mass data and top-down hits obtained from TDPortal were calibrated using Proteoform Suite (v0.3.4) to improve mass accuracy for subsequent proteoform family construction (Figure 2). This post-acquisition calibration process utilized a search result-dependent strategy that we initially developed for bottom-up proteomics21 and have since implemented in Proteoform Suite for intact-mass and top-down proteomics. Its previous application for label-free intact-mass data calibration provided an improvement in mass accuracy, which resulted in an increased number of proteoform identifications and a decreased FDR for identifications.13,14 Here, we extend this strategy to calibrate intact-mass data collected from NeuCode-labeled proteoforms. The theoretical light NeuCode-labeled mass was determined for each high-confidence (C-score > 40)40 top-down hit based on the identified sequence’s lysine count. Raw mass components from intact-mass measurements were then selected as calibration points if within 10 ppm and 5 min retention time (RT) of a top-down hit from the same Gelfree fraction number. A random forest machine learning algorithm determined the mass error as a function of m/z, RT, scan total ion current (TIC), and scan injection time to perform a global calibration for each raw file.13 New result tables were generated containing calibrated deconvoluted NeuCode-labeled intact-mass data as well as calibrated label-free top-down hits (Figure 2). The resultant 2,021,232 calibrated raw mass components and 39,382 calibrated top-down hits were used for the subsequent analysis.
Proteoform Family Construction with Proteoform Suite.
Proteoform Suite (v0.3.4) was used to further filter intact-mass and top-down data to generate a list of intact-mass and top-down experimental proteoforms. Additionally, Proteoform Suite was used to make a catalog of theoretical proteoforms from the pruned multi-protease G-PTM-D database generated using bottom-up data.
Deconvoluted and calibrated NeuCode intact-mass data files, calibrated top-down hits, and the pruned multi-protease G-PTM-D database were loaded into Proteoform Suite (Figure 2). Raw mass components from the intact-mass files were first filtered and merged to eliminate errors from missed monoisotopic masses and charge-state harmonics.13,15 Proteoform isotopologue pairs (light and heavy NeuCode pairs) were then identified from the processed mass components. Only those NeuCode pairs with light/heavy intensity ratios between 1.8:1 and 2.5:1 were retained based on the most abundant intensity ratio observed at 2.15:1 (Supporting Information, Figure S-1). The number of lysine residues for each NeuCode pair was calculated using the 36 mDa per lysine residue mass difference. NeuCode pairs were then aggregated to eliminate redundant observations of the same proteoform, allowing mass deviations of up to 10 ppm and RT deviations of up to 5 min. In this way, a list of 5,615 intact-mass experimental proteoforms was created, each with a monoisotopic mass, lysine count, and RT.12–14
Imported top-down hits were filtered by C-score. Those larger than 40 were retained, as they were judged to be confidently identified and extensively characterized.40 The filtered hits were then aggregated using two criteria: (i) the same proteoform record (PFR) number assigned by TDPortal and (ii) an RT tolerance of 5 min,13 generating a list of top-down experimental proteoforms. Each experimental mass was converted to the corresponding light NeuCode-labeled mass based on the number of lysine residues in the identified sequence. This list was combined with the list of intact-mass experimental proteoforms to make a final list of experimental proteoforms.
A catalog of theoretical proteoforms was generated from the pruned multi-protease G-PTM-D protein database, allowing combinations of up to four PTMs on each protein. Note that these theoretical proteoform sequences do not include N-terminal methionine. The strategy of constructing proteoform families has been described previously.3,12–15 Briefly, all experimental proteoforms were compared with the theoretical proteoforms containing the same number of lysines, forming experimental-theoretical (ET) pairs (Supporting Information, Figure S-2). Experimental proteoforms with the same number of lysines and RT differences of less than 2.5 min were also compared to each other, generating experimental-experimental (EE) pairs (Supporting Information, Figure S-3). ET and EE pairs with FDRs no larger than 25% and mass differences corresponding to ~0 Da (exact matches, ET only), known PTMs, PTM combinations, and amino acid residues were accepted. The average FDR of the accepted pairs was determined to be 5% for ET and 8% for EE as previously described.3,12 Proteoforms in accepted pairs were grouped into proteoform families, which were visualized in Cytoscape41,42 (v3.6.0) as networks with nodes representing proteoforms and edges representing mass differences between proteoforms.
This strategy of proteoform family construction is flexible and can accommodate different combinations of input data sets and various protein databases. In this study, we have performed analyses of the NeuCode intact-mass data using a UniProt database, a pruned trypsin-only G-PTM-D database, and a pruned multi-protease G-PTM-D database. The analysis with the trypsin-only G-PTM-D database yielded more proteoform identifications at a fixed FDR than the analysis with the UniProt database, and the number of identifications was further increased when a multi-protease G-PTM-D database was employed. We also integrated NeuCode intact-mass data with label-free top-down data as described to further improve the analysis. Several other types of analyses were performed (i.e., using an unpruned multi-protease G-PTM-D database, using uncalibrated data, and using top-down data only with MS1 spectra as “label-free intact-mass” data) and are presented in the Supporting Information for the interested reader. Proteoform Suite analysis of the data described in this study typically takes ~100 min (Supporting Information, Table S-2)
RESULTS AND DISCUSSION
NeuCode-Labeled Intact-Mass Experimental Proteoforms
Mass spectra from the 66 raw data files obtained from analysis of NeuCode-labeled intact protein samples were deconvoluted and calibrated to provide 2,021,232 mass components. After Proteoform Suite removed the missed monoisotopic and charge-state harmonic errors,13,15 a total of 283,634 NeuCode pairs were revealed. Proteoform Suite accepted 113,762 of these pairs falling within the selected intensity ratio range of 1.8:1 to 2.5:1. This range was a parameter decision seeking to retain the highest number of true NeuCode pairs possible while eliminating likely false NeuCode pairs. The accepted NeuCode pairs were aggregated by mass and RT, yielding 5,615 intact-mass experimental proteoforms (Supporting Information, Table S-3). In each section below, we examine the impact of various analysis strategies on the number of these experimental proteoforms that can be confidently identified.
Multi-Protease G-PTM-D Database Improves Proteoform Identification
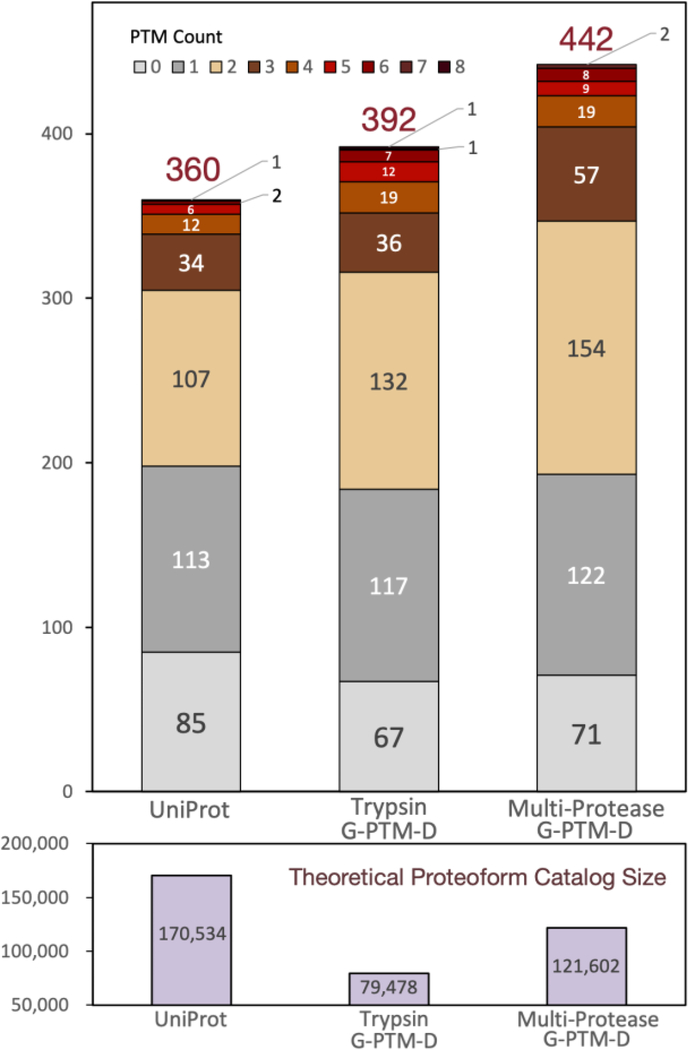
The G-PTM-D strategy was developed and implemented in MetaMorpheus to identify PTMs in bottom-up data and subsequently add newly discovered PTMs to a sample-specific protein database.20,21 We have previously reported that using a G-PTM-D database improves identification of E. coli proteoforms from intact-mass data.12 Here, we demonstrate a further improvement of this strategy by utilizing a pruned multi-protease G-PTM-D database. It is important that the pruned version of the database was used here as the full, unpruned G-PTM-D database contained many proteins from the original UniProt database that were not confidently observed in bottom-up data and therefore were less likely to be observed in intact-mass data. Pruning the database helps to limit the size of the theoretical proteoform catalog, preventing large FDRs in ET comparisons (see the Supporting Information for results from an analysis using an unpruned multi-protease G-PTM-D database). The pruned multi-protease G-PTM-D database contained ~83% fewer proteins than the original UniProt database (12,767 vs 73,928 sequences). The sequences in the pruned database contained 14,559 modified residues that were not documented in the UniProt database (Supporting Information, Table S-4), as these modifications were identified during global PTM discovery. Using the sequences and PTMs from this multi-protease G-PTM-D database (allowing combinations of up to four PTMs on each sequence), a catalog of theoretical proteoforms containing 121,602 entries was built by Proteoform Suite.
Employing the strategy described in the Experimental Procedures, Proteoform Suite constructed 614 proteoform families from accepted ET pairs (6% FDR) and EE pairs (8% FDR) (Supporting Information, Table S-5). A total of 157 families were unambiguously identified, meaning that they were associated with a single gene. These families contained 532 experimental proteoforms. There were also five ambiguous families (associated with multiple genes) assembled, containing 150 experimental proteoforms. Proteoform Suite determined a subset of the proteoforms in these unambiguous and ambiguous families to be “identified experimental proteoforms”, as the program automatically searches for potentially false EE connections (e.g., delta-mass indicating loss of a PTM that is not found in that family) and excludes such proteoforms from the identified list. Proteoform Suite also removes duplicated proteoforms with the same sequence and PTMs but with RT differences larger than 5 min (this RT tolerance is applied in the upstream aggregation step described above), further consolidating this list to 442 “unique proteoform identifications” (Supporting Information, Table S-6). These identifications are depicted in Figure 3 and compared to those obtained from analyses using other databases (discussed below).
Figure 3.
Number of identified NeuCode intact-mass experimental proteoforms from Proteoform Suite analyses using three different protein databases. Identified proteoforms were grouped by PTM count (upper panel). The theoretical proteoform catalog size for each analysis is indicated (lower panel). The overall identification FDR for these three analyses was maintained at ~5%.
The same analysis was repeated using the original UniProt database and a pruned trypsin-only G-PTM-D database (detailed results of these additional Proteoform Suite analyses can be found in the Supporting Information, Table S-7). We found that using the multi-protease database increased the number of unique proteoform identifications by 23% as compared to the original, unmodified UniProt database (442 vs 360), and by 13% as compared to the trypsin-only G-PTM-D database (442 vs 392). In general, the number of PTMs on identified proteoforms also increased (Figure 3), as did the average number of experimental proteoforms in identified families (2.8 in UniProt, 3.0 in trypsin-only G-PTM-D, and 3.4 in multi-protease G-PTM-D). The better performance of the G-PTM-D databases is due to decreased database size, which reduces FDR, as well as to the incorporation of additional PTMs discovered in bottom-up data. The multi-protease G-PTM-D analysis provided more identifications than the trypsin-only G-PTM-D analysis, as the use of multiple proteases provides better proteome coverage, leading to a more comprehensive protein database for proteoform analysis. The superiority of the multi-protease G-PTM-D analysis is also reflected by the highest number of ET proteoform exact matches (~0 Da mass difference) (225 for UniProt, 252 for trypsin-only G-PTM-D, and 317 for multi-protease G-PTM-D).
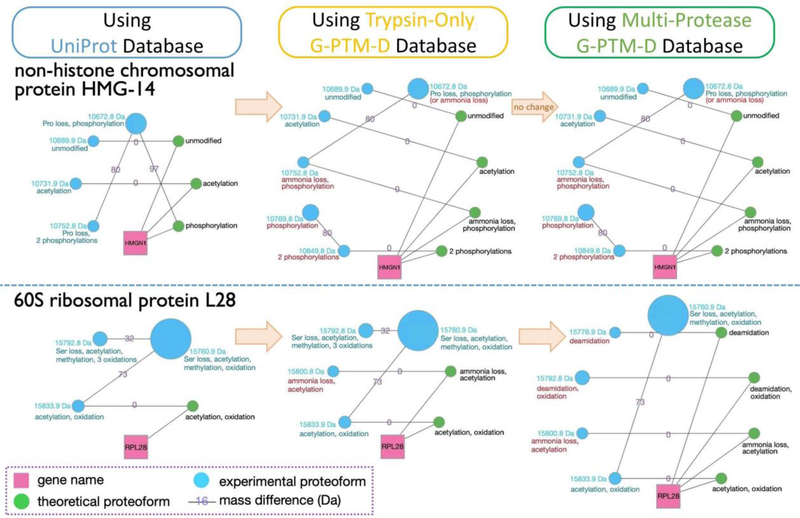
We also examined the results of proteoform family construction from these analyses. In selecting ET pairs, pairs are grouped into “ET peaks”. Each of these peaks has an associated FDR, which reflects the proportion of ET pairs within that peak that are likely false relationships. The ET pairs formed in any given analysis depend on the database used to generate the catalog of theoretical proteoforms. This has a direct impact on the FDR of ET peaks, affecting how many and which of these peaks can be accepted while maintaining the same FDR threshold. This, in turn, determines which theoretical proteoforms are included in proteoform families, and therefore how many experimental proteoforms can be identified. Figure 4 demonstrates how two example families evolved when changing the database utilized in the analysis. The non-histone chromosomal protein HMG-14 family gained three new members when the trypsin-only G-PTM-D database was used (and no further growth was observed in a multi-protease-assisted analysis). The G-PTM-D-assisted analyses updated the identity of the 10,752.8 Da proteoform, as it was not directly connected to a theoretical proteoform in the UniProt analysis but could be connected to a theoretical proteoform in the G-PTM-D analyses (identifications via direct ET connections are used for PTM annotation instead of identifications resulting from daisy-chaining EE connections). The 60S ribosomal protein L28 family gradually increased in size by adding first one and then two identified experimental proteoforms when utilizing the trypsin and multi-protease G-PTM-D databases, respectively. The multi-protease-assisted analysis updated the identity of the 15,792.8 Da proteoform, as it became an exact match to a new theoretical proteoform in the database. These results illustrate how the G-PTM-D strategy improves human proteoform analysis, and how the use of data from multiple proteases further enhances this strategy.
Figure 4.
Two examples of proteoform families constructed using NeuCode intact-mass data. Three separate Proteoform Suite analyses were performed using UniProt, trypsin-only G-PTM-D, and multi-protease G-PTM-D databases. Gene names (pink squares) connect to all theoretical proteoforms (green nodes) in the family. Theoretical proteoforms are labeled “unmodified” or with PTM information and any terminal amino acid losses. Intact-mass experimental proteoforms (blue nodes) are labeled with their masses and PTMs, as deduced by Proteoform Suite. Experimental proteoforms are arranged counterclockwise in ascending order of mass. The size of each node corresponds to the integrated intensity of that proteoform’s spectral peaks. The edges are labeled with the mass difference of the two connected proteoforms (Da). The accepted mass differences are the result of selecting low-FDR ET and EE pairs during the Proteoform Suite analyses. Turquoise annotations are from the UniProt analysis, while red annotations are new findings or PTM corrections gleaned from analyses using G-PTM-D databases.
Top-Down Experimental Proteoforms
The 39,382 calibrated top-down hits obtained from TDPortal analysis of label-free top-down data contained 2,602 unique proteoform record (PFR) numbers. However, this decreased to 1,194 proteoforms (defined by unique PFRs) after filtering for C-scores above 3—the score cutoff that indicates at least partially characterized identifications.40 A total of 711 proteoforms were identified with a C-score above 40, indicating confident characterizations.40 These results were similar to those obtained in a previous top-down study of human proteoforms from a single Gelfree separation.27 In this study, we opted to apply a stringent C-score cutoff of 40 to retain only high-confidence top-down experimental proteoforms for subsequent family construction.
About 100 top-down experimental proteoforms had accession numbers not found in Proteoform Suite’s catalog of theoretical proteoforms. There are a few explanations for this. First, the pruned multi-protease G-PTM-D database used to generate the catalog of theoretical proteoforms only contains proteins that were confidently observed in bottom-up data (i.e., proteins that had a peptide observed at 1% FDR). Thus, proteins that may be present in the sample but did not have a confidently identified peptide would not be included in the pruned database. This explanation accounts for the majority of the 100 accessions that were observed in top-down data but not included in the catalog of theoretical proteoforms. Furthermore, the process of protein inference has a significant influence on which protein sequences are included in the pruned database. When multiple proteins have shared subsequences, those shared peptides are mapped to several possible accessions during the protein inference process. However, based on the principle of Occam’s razor, some of these protein sequences may be excluded from the pruned database if there is stronger support for an alternative protein according to peptide-level evidence (e.g., if a unique peptide was also observed for one of the proteins under consideration). To address this discrepancy between the accessions observed in top-down data and the accessions included in the theoretical proteoform catalog, we made a separate database that contained the sequences and PTMs of the proteoforms that were identified by TDPortal but were not found in the pruned multi-protease G-PTM-D database (see the Supporting Information for further discussion of the entries in this additional database). This “patch database” was imported into Proteoform Suite together with the multi-protease G-PTM-D database. Additionally, top-down identifications whose corresponding theoretical proteoforms were not already present in the catalog were added. The resultant comprehensive catalog contained 123,110 theoretical proteoforms, a modest 1.2% increase in size from the previous catalog, and this catalog was used for the integrated intact-mass/top-down analysis in the next section.
Proteoform Family Construction Using NeuCode Intact-Mass and Top-Down Experimental Proteoforms
Both NeuCode intact-mass and label-free top-down proteomics are useful strategies to identify proteoforms.3,12,15,26–29 However, each of these approaches has its own advantages. NeuCode intact-mass proteomics generates MS1 spectra only, which means that it provides more proteoform observations than top-down proteomics, where instrument time is spent fragmenting precursors and acquiring MS2 spectra. Top-down proteomics, on the other hand, provides better characterized proteoforms because sequence tags can be identified from fragmentation data. Integrating these two types of data for analysis combines the advantages of each strategy. Previously, we were able to expand label-free top-down proteoform identifications by leveraging additional information contained in the MS1 spectra of the top-down data set.13,14 This is in contrast to a typical top-down analysis workflow where a substantial number of peaks in MS1 spectra are ignored because they are never selected for fragmentation. In the current work, we enabled Proteoform Suite to construct families by integrating label-free top-down identifications and NeuCode-labeled intact-mass proteoforms that were obtained from separate MS runs. In this integrated intact-mass/top-down analysis, each identified label-free top-down mass was converted to the corresponding light NeuCode mass using the lysine count of the identified sequence. Proteoform Suite merged the 814 top-down experimental proteoforms with the 5,615 intact-mass experimental proteoforms using a mass tolerance of 10 ppm and an RT tolerance of 5 min. As part of this process, 306 intact-mass experimental proteoforms were replaced by top-down experimental proteoforms of the same mass and RT since the identities of these proteoforms had already been deduced from top-down data. After merging, a final list of 6,123 accepted experimental proteoforms was generated (Supporting Information, Tables S-8 and S-9).
Using this list and the catalog of 123,110 theoretical proteoforms, 848 families were constructed. These included 438 unambiguously identified families, 10 ambiguous families, and 400 unidentified families (Figure 5 and Supporting Information, Table S-10). Overall, we found 526 unique proteoform identifications from the NeuCode intact-mass portion of this integrated analysis, which was an increase from the 442 unique proteoforms identified in the intact-mass-only analysis (Figure 3). The reason for this increase is that when the top-down experimental proteoforms were included in the ET and EE comparisons, they generally increased the number of pairs grouped in delta-mass histogram peaks while providing an overall decrease in the FDR of those peaks. This allowed more peaks to have a low enough FDR to be accepted (Supporting Information, Table S-11), which increased the number of proteoforms that were identified. Numerous proteoforms with multiple PTMs were still present in this analysis (Figure 5, right). The number of identified proteoforms with 2 PTMs or more increased from the intact-mass-only analysis (Figure 3), including one identification with 9 PTMs.
Figure 5.
Stepwise results of the Proteoform Suite integration of intact-mass, top-down, and bottom-up data. Overall, 1,207 unique proteoforms were identified, representing 484 genes. In the bottom box, only the 496 unique intact-mass proteoforms are depicted, so as to eliminate the common identifications between intact-mass and top-down. See the Supporting Information, Table S-12 for more detailed results of this analysis.
Within the 526 unique intact-mass proteoform identifications, 496 were new additions to the list of 711 unique top-down identifications. This represents a 70% increase in identifications as compared to the TDPortal analysis of top-down data alone (C-score cutoff at 40). Thus, a total of 1,207 unique proteoforms representing 484 genes were identified by integrating intact-mass and top-down data (Supporting Information, Table S-13). The overall FDR of the identified proteoforms was 3.9%. After manual removal of the redundant identifications from the original 6,123 experimentally observed proteoforms, 5,950 unique experimental proteoforms remain, 1,207 (20%) of which were confidently identified (Figure 5, bottom box and Supporting Information, Table S-12). These were in a MW range between 1.9 and 30.5 kDa (Supporting Information, Figure S-4). Higher MW proteoforms are not well represented in this study due to limitations of the Orbitrap mass analyzer,43 generally decreased signal-to-noise ratio for high mass proteoforms,44 and the elimination of higher MW species in the Gelfree separation employed.35 The identified proteoforms contained numerous biologically relevant PTMs, including but not limited to methylation, acetylation, and phosphorylation (Supporting Information, Figure S-5). In addition, PTMs that could have either biological or artificial (i.e., sample handling) origin, such as oxidation and deamidation, were also present.
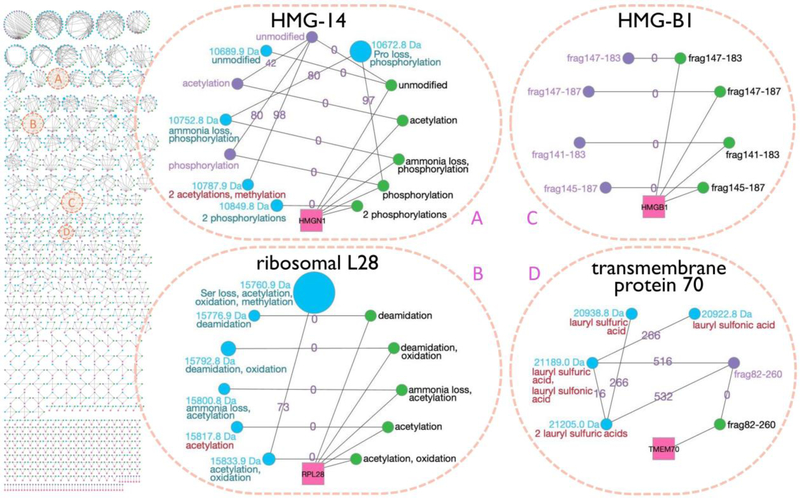
Various functional classes of protein were represented by the proteoforms identified in this integrated intact-mass/top-down analysis, including histones (see the Supporting Information for a discussion of histone proteoforms and Supporting Information, Figure S-6 for a histone H3 family), ribosomal proteins, RNA-/DNA-binding proteins, transcription and translation factors, transmembrane transporters, and ubiquitin-associated proteins (Supporting Information, Tables S-14 and S-15). Among the 438 unambiguously identified families from this integrated analysis (Figure 6), 368 families (84%) contained top-down proteoforms. These included the previously introduced non-histone chromosomal protein HMG-14 family (Figure 4, upper panel) to which three top-down experimental proteoforms were added as part of this analysis (Figure 6A). The acetylated and the singly phosphorylated top-down proteoforms replaced the intact-mass proteoforms with the same mass and RT. The unmodified top-down proteoform did not merge with the unmodified intact-mass proteoform because the two had RTs larger than 5 min apart; nonetheless, these two proteoforms only count as one unique identified proteoform among the 1,207 reported. In addition, the HMG-14 family gained a new intact-mass proteoform: 10,787.9 Da, containing one methylation and two acetylations. This proteoform was identified through the 98 Da EE pair connection with the unmodified top-down proteoform. Figure 6B shows another previously identified family, 60S ribosomal protein L28 (Figure 4, lower panel), which also increased in size as compared to the intact-mass-only analysis. Although no top-down proteoforms were added to the family, the incorporation of top-down data and the changes associated with those data led to an additional intact-mass ET pair that fell within an ET peak with sufficiently low FDR for acceptance. Thus, the 15,817.8 Da intact-mass proteoform was identified via an ET match to the theoretical proteoform with one acetylation.
Figure 6.
Array of the 438 unambiguously identified (i.e., assigned to a single gene) proteoform families that were constructed by the integrated intact-mass/top-down analysis (left) and four example families (right). In addition to the symbols utilized in Figure 4, here we add purple nodes to represent top-down experimental proteoforms, and the blue nodes with red annotations denote new intact-mass identifications arising from the inclusion of top-down data in the Proteoform Suite analysis. Previous versions of families A and B were presented in Figure 4. The versions presented here show new developments in the families upon integrating top-down data. Families C and D are newly identified proteoform families. Note: the families in this figure were modified slightly from the automated output of Proteoform Suite (i.e., some nodes and edges were removed), as described in the Supplementary Experimental Methods.
The integrated intact-mass/top-down analysis also revealed proteoform families not seen in the intact-mass-only analysis, such as high mobility group protein B1 (Figure 6C) and mitochondrial transmembrane protein 70 (Figure 6D). The former contained only experimental proteoforms identified by top-down, which were four protein fragments. The latter family contained one top-down proteoform, which enabled the identification of four intact-mass proteoforms with lauryl sulfuric and lauryl sulfonic acid adducts.
CONCLUSIONS
Analysis of all intact-mass and top-down proteomic data files revealed the presence of 5,950 unique experimental proteoforms. Twelve percent (711) of these were identified and extensively characterized by traditional top-down proteomic analysis. The strategy of constructing intact-mass proteoform families with Proteoform Suite and G-PTM-D further increased the fraction of identified proteoforms to 20% (1,207). Development of new strategies for identification of the remaining 80% of observed experimental proteoforms presents an important challenge to the field of proteomics. These proteoforms, which are manifested in high quality mass spectrometric data, yet remain unidentified, represent the front line in top-down/intact-mass analysis as they have already been observed. Overall, in this study, we identified 1,207 human proteoforms at 3.9% FDR. This work demonstrates that the integration of different types of proteomic data at a high confidence level is an effective strategy to substantially increase the quantity and quality of proteoform identifications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences, NIH grants R01GM114292 and R35GM126914. K.E.B. and R.J.M. were supported in part by the National Human Genome Research Institute grant to the Genomic Science Training Program, 5T32HG002760. L.V.S. was supported by the Biotechnology Training Program, T32GM008349. R.M.M. was supported in part by the NIH Chemistry-Biology Interface Training Grant, T32GM008505.
REFERENCES
- (1).Aebersold R; Agar JN; Amster IJ; Baker MS; Bertozzi CR; Boja ES; Costello CE; Cravatt BF; Fenselau C; Garcia BA; Ge Y; Gunawardena J; Hendrickson RC; Hergenrother PJ; Huber CG; Ivanov AR; Jensen ON; Jewett MC; Kelleher NL; Kiessling LL; Krogan NJ; Larsen MR; Loo JA; Ogorzalek Loo RR; Lundberg E; MacCoss MJ; Mallick P; Mootha VK; Mrksich M; Muir TW; Patrie SM; Pesavento JJ; Pitteri SJ; Rodriguez H; Saghatelian A; Sandoval W; Schlüter H; Sechi S; Slavoff SA; Smith LM; Snyder MP; Thomas PM; Uhlén M; Van Eyk JE; Vidal M; Walt DR; White FM; Williams ER; Wohlschlager T; Wysocki VH; Yates NA; Young NL; Zhang B; How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Smith LM; Kelleher NL; The Consortium for Top Down Proteomics. Proteoform: a single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Shortreed MR; Frey BL; Scalf M; Knoener RA; Cesnik AJ; Smith LM; Elucidating Proteoform Families from Proteoform Intact-Mass and Lysine-Count Measurements. J. Proteome Res. 2016, 15, 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Toby TK; Fornelli L; Kelleher NL; Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Siuti N; Kelleher NL; Decoding protein modifications using top-down mass spectrometry. Nat. Methods 2007, 4, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chen B; Brown KA; Lin Z; Ge Y; Top-Down Proteomics: Ready for Prime Time? Anal. Chem. 2018, 90, 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Durbin KR; Tran JC; Zamdborg L; Sweet SMM; Catherman AD; Lee JE; Li M; Kellie JF; Kelleher NL; Intact mass detection, interpretation, and visualization to automate Top-Down proteomics on a large scale. Proteomics 2010, 10, 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhao Y; Sun L; Zhu G; Dovichi NJ; Coupling Capillary Zone Electrophoresis to a Q Exactive HF Mass Spectrometer for Top-down Proteomics: 580 Proteoform Identifications from Yeast. J. Proteome Res. 2016, 15, 3679–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Park J; Piehowski PD; Wilkins C; Zhou M; Mendoza J; Fujimoto GM; Gibbons BC; Shaw JB; Shen Y; Shukla AK; Moore RJ; Liu T; Petyuk VA; Tolić N; Paša-Tolić L; Smith RD; Payne SH; Kim S; Informed-Proteomics: open-source software package for top-down proteomics. Nat. Methods 2017, 14, 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dang X; Scotcher J; Wu S; Chu RK; Tolić N; Ntai I; Thomas PM; Fellers RT; Early BP; Zheng Y; Durbin KR; Leduc RD; Wolff JJ; Thompson CJ; Pan J; Han J; Shaw JB; Salisbury JP; Easterling M; Borchers CH; Brodbelt JS; Agar JN; Paša-Tolić L; Kelleher NL; Young NL; The first pilot project of the consortium for top-down proteomics: a status report. Proteomics 2014, 14, 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Karabacak NM; Li L; Tiwari A; Hayward LJ; Hong P; Easterling ML; Agar JN; Sensitive and specific identification of wild type and variant proteins from 8 to 669 kDa using top-down mass spectrometry. Mol. Cell. Proteomics 2009, 8, 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Dai Y; Shortreed MR; Scalf M; Frey BL; Cesnik AJ; Solntsev S; Schaffer LV; Smith LM; Elucidating Escherichia coli Proteoform Families Using Intact-Mass Proteomics and a Global PTM Discovery Database. J. Proteome Res. 2017, 16, 4156–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Schaffer LV; Shortreed MR; Cesnik AJ; Frey BL; Solntsev SK; Scalf M; Smith LM; Expanding Proteoform Identifications in Top-Down Proteomic Analyses by Constructing Proteoform Families. Anal. Chem. 2018, 90, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Schaffer LV; Rensvold JW; Shortreed MR; Cesnik AJ; Jochem A; Scalf M; Frey BL; Pagliarini DJ; Smith LM; Identification and Quantification of Murine Mitochondrial Proteoforms Using an Integrated Top-Down and Intact-Mass Strategy. J. Proteome Res. 2018, 17, 3526–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Cesnik AJ; Shortreed MR; Schaffer LV; Knoener RA; Frey BL; Scalf M; Solntsev SK; Dai Y; Gasch AP; Smith LM; Proteoform Suite: Software for Constructing, Quantifying, and Visualizing Proteoform Families. J. Proteome Res. 2018, 17, 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ong SE; Blagoev B; Kratchmarova I; Kristensen DB; Steen H; Pandey A; Mann M; Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- (17).Rhoads TW; Prasad A; Kwiecien NW; Merrill AE; Zawack K; Westphall MS; Schroeder FC; Kimble J; Coon JJ; NeuCode Labeling in Nematodes: Proteomic and Phosphoproteomic Impact of Ascaroside Treatment in Caenorhabditis elegans. Mol. Cell. Proteomics 2015, 14, 2922–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hebert AS; Merrill AE; Bailey DJ; Still AJ; Westphall MS; Strieter ER; Pagliarini DJ; Coon JJ; Neutron-encoded mass signatures for multiplexed proteome quantification. Nat. Methods 2013, 10, 332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cox J; Michalski A; Mann M; Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 2011, 22, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Li Q; Shortreed MR; Wenger CD; Frey BL; Schaffer LV; Scalf M; Smith LM; Global Post-Translational Modification Discovery. J. Proteome Res. 2017, 16, 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Solntsev SK; Shortreed MR; Frey BL; Smith LM; Enhanced Global Post-translational Modification Discovery with MetaMorpheus. J. Proteome Res. 2018, 17, 1844–1851. [DOI] [PubMed] [Google Scholar]
- (22).Chait BT; Chemistry. Mass spectrometry: bottom-up or top-down? Science 2006, 314, 65–66. [DOI] [PubMed] [Google Scholar]
- (23).Schneider U; Schwenk HU; Bornkamm G; Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed nonHodgkin lymphoma. Int. J. Cancer 1977, 19, 621–626. [DOI] [PubMed] [Google Scholar]
- (24).Swaney DL; Wenger CD; Coon JJ; Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 2010, 9, 1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Tran JC; Zamdborg L; Ahlf DR; Lee JE; Catherman AD; Durbin KR; Tipton JD; Vellaichamy A; Kellie JF; Li M; Wu C; Sweet SMM; Early BP; Siuti N; LeDuc RD; Compton PD; Thomas PM; Kelleher NL; Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 2011, 480, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Catherman AD; Durbin KR; Ahlf DR; Early BP; Fellers RT; Tran JC; Thomas PM; Kelleher NL; Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol. Cell. Proteomics 2013, 12, 3465–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Anderson LC; DeHart CJ; Kaiser NK; Fellers RT; Smith DF; Greer JB; LeDuc RD; Blakney GT; Thomas PM; Kelleher NL; Hendrickson CL; Identification and Characterization of Human Proteoforms by Top-Down LC-21 Tesla FT-ICR Mass Spectrometry. J. Proteome Res. 2017, 16, 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Durbin KR; Fornelli L; Fellers RT; Doubleday PF; Narita M; Kelleher NL; Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa. J. Proteome Res. 2016, 15, 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ntai I; LeDuc RD; Fellers RT; Erdmann-Gilmore P; Davies SR; Rumsey J; Early BP; Thomas PM; Li S; Compton PD; Ellis MJC; Ruggles KV; Fenyö D; Boja ES; Rodriguez H; Townsend RR; Kelleher NL; Integrated Bottom-Up and Top-Down Proteomics of Patient-Derived Breast Tumor Xenografts. Mol. Cell. Proteomics 2016, 15, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cai W; Tucholski T; Chen B; Alpert AJ; McIlwain S; Kohmoto T; Jin S; Ge Y; Top-Down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy. Anal. Chem. 2017, 89, 5467–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Toby TK; Fornelli L; Srzentić K; DeHart CJ; Levitsky J; Friedewald J; Kelleher NL; A comprehensive pipeline for translational top-down proteomics from a single blood draw. Nat. Protoc. 2019, 14, 119–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li Z; He B; Kou Q; Wang Z; Wu S; Liu Y; Feng W; Liu X; Evaluation of top-down mass spectral identification with homologous protein sequences. BMC Bioinformatics 2018, 19, 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Millea KM; Krull IS; Cohen SA; Gebler JC; Berger SJ; Integration of multidimensional chromatographic protein separations with a combined “top-down” and “bottom-up” proteomic strategy. J. Proteome Res. 2006, 5, 135–146. [DOI] [PubMed] [Google Scholar]
- (34).Jefferys SR; Giddings MC; Baking a mass-spectrometry data PIE with McMC and simulated annealing: predicting protein post-translational modifications from integrated top-down and bottom-up data. Bioinformatics 2011, 27, 844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tran JC; Doucette AA; Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal. Chem. 2008, 80, 1568–1573. [DOI] [PubMed] [Google Scholar]
- (36).Wessel D; Flügge UI; A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [DOI] [PubMed] [Google Scholar]
- (37).Sheynkman GM; Shortreed MR; Frey BL; Scalf M; Smith LM; Large-scale mass spectrometric detection of variant peptides resulting from nonsynonymous nucleotide differences. J. Proteome Res. 2014, 13, 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wiśniewski JR; Zougman A; Nagaraj N; Mann M; Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- (39).Miller RM; Millikin RJ; Hoffmann CV; Solntsev SK; Sheynkman GM; Shortreed MR; Smith LM; Improved Protein Inference from Multiple Protease Bottom-Up Mass Spectrometry Data. J. Proteome Res. 2019, 18, 3429–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).LeDuc RD; Fellers RT; Early BP; Greer JB; Thomas PM; Kelleher NL; The C-score: a Bayesian framework to sharply improve proteoform scoring in high-throughput top down proteomics. J. Proteome Res. 2014, 13, 3231–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Smoot ME; Ono K; Ruscheinski J; Wang PL; Ideker T; Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T; Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Makarov A; Denisov E; Dynamics of ions of intact proteins in the Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 2009, 20, 1486–1495. [DOI] [PubMed] [Google Scholar]
- (44).Compton PD; Zamdborg L; Thomas PM; Kelleher NL; On the scalability and requirements of whole protein mass spectrometry. Anal. Chem. 2011, 83, 6868–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.