Abstract
Uterine leiomyosarcoma (ULMS) is the major subtype of uterine sarcoma (US) and contributes to uterine cancer deaths. Although preoperative diagnosis of US remains challenging, frequent application of laparoscopic surgery for benign uterine leiomyomas (ULM) requires precise exclusion of US. MicroRNAs are stably present in the bloodstream, and the application of circulating miRNAs as disease biomarkers has been recognized. In the present study, we aimed to identify diagnostic biomarkers for distinguishing US from ULM by focusing on circulating miRNAs. All serum samples were collected preoperatively between 2009 and 2017, and all cases were histopathologically diagnosed. Whole miRNA profiles were obtained using a miRNA microarray. By analyzing expression levels of the miRNAs, candidate miRNAs were selected based on diagnostic performance in discriminating US from ULM, and a diagnostic model was then constructed. A total of 90 serum samples were analyzed, and clustering analyses revealed that the profiles of ULMS were distinct from those of controls. Based on leave‐one‐out cross‐validation, seven miRNAs were selected as biomarker candidates. Based on model construction, the optimal model consisted of two miRNAs (miR‐1246 and miR‐191‐5p), with an area under the receiver operating characteristic curve (AUC) for identifying ULMS of 0.97 (95% confidence interval [CI], 0.91‐1.00). In contrast, serum lactate dehydrogenase had an AUC of only 0.64 (95% CI, 0.34‐0.94). Seven serum miRNAs with high diagnostic performance for preoperative US screening were detected, and a promising diagnostic model for ULMS was generated.
Keywords: circulating microRNA, diagnostic biomarkers, liquid biopsy, uterine leiomyosarcoma, uterine sarcoma
Whole circulating miRNA profiles in uterine sarcoma patient serum are analyzed. The result demonstrates feasibility for application as a novel preoperative biomarker.

1. INTRODUCTION
Uterine sarcomas (US) are rare malignant tumors that account for approximately 5% of all invasive uterine cancers, and uterine leiomyosarcoma (ULMS) is a major subtype that contributes to uterine cancer deaths.1 The 5‐year survival rate for ULMS is estimated as only 15%‐25%, and patient prognosis has not changed in the past several decades.2 Uterine leiomyomas (ULM) are non‐cancerous tumors that arise from the myometrium, and ULM is the most common benign pelvic tumor in women.3 The estimated cumulative incidence of ULM by age 50 is over 70%.3 Even now, it remains challenging to distinguish US precisely from ULM before surgery, as both tumors present in the pelvic cavity as a large mass with similar appearance by imaging. An increasing number of gynecological surgeons are concerned with making this distinction due to frequent application of laparoscopic therapeutic approaches for ULM. When selecting laparoscopic surgery, preoperative prediction of malignant cells is essential because it requires a procedure celled tumor morcellation, which can cause dissemination of cancer cells within the peritoneal cavity.2 Despite recent multimodal examinations, including ultrasound imaging, MRI and blood tests, preoperative diagnosis remains challenging, and improving accurate diagnosis through novel approaches prior to laparoscopic surgery is therapeutically desirable.
MicroRNAs (miRNAs) regulate intracellular expression of various target genes, primarily by manipulating their translation.4 However, recent findings suggest that extracellular miRNAs play a key role in intercellular communication and contribute to many biological mechanisms.5 Among various types of RNA, miRNAs are one of the most promising candidates because they are remarkably stable in human bio‐fluids6; therefore, circulating miRNAs have been the subject of intensive research into their potential as disease biomarkers. Recent evidence indicates that circulating miRNAs can be used as biomarkers because their profiles accurately reflect physiological and pathological status.7, 8 Indeed, our previous work demonstrates the promising potential of serum miRNAs as diagnostic biomarkers.9, 10, 11, 12
Due to the rarity of US, the basic biology of this malignancy has not been thoroughly investigated, and most reports are of retrospective clinical research. Several prior studies identified some putative US‐related miRNAs.13, 14, 15 However, these reports directly analyzed tissue miRNAs to investigate US biology as opposed to diagnosis, and using tissue miRNAs would not be a viable clinical application. In addition, each sample size was limited. In the present study, we aimed to identify preoperative diagnostic biomarkers by focusing on circulating miRNAs, which is non‐invasive, and can be applied in clinical settings. Comprehensive miRNA profiles of 32 serum samples, including 17 benign controls, were obtained by miRNA microarray, allowing for generation of optimal algorithms for predicting US. This is the first report focusing on circulating miRNAs in US patients, and assessing the potential of miRNAs as predictive biomarkers for distinguishing ULMS from ULM preoperatively.
2. MATERIALS AND METHODS
2.1. Clinical samples
All patients with uterine tumors that were referred to the National Cancer Center Hospital (NCCH) from other hospitals due to suspicion of malignant uterine tumors between 2009 and 2017, underwent surgery at the NCCH, and were histopathologically diagnosed as having ULM, ULMS, uterine adenosarcoma, uterine endometrial stromal sarcoma (ESS) or uterine smooth muscle tumor of uncertain malignant potential (STUMP) by pathologists enrolled in the study. Serum samples from patients with benign bone and soft tissue tumors, or with benign ovarian tumors, were also used for analyses. All Serum samples were collected preoperatively, registered in the National Cancer Center (NCC) Biobank, and stored at −20°C until use. The present study was approved by the NCCH Institutional Review Board (2015‐376, 2016‐249). Written informed consent was obtained from each participant.
2.2. MicroRNA expression array in clinical samples
Total RNA was extracted from 300 µL of serum using the 3D‐Gene RNA Extraction Reagent (Toray Industries) according to the manufacturer's instructions. Comprehensive miRNA expression analysis was performed using a 3D‐Gene miRNA Labeling kit and a 3D‐Gene Human miRNA Oligo Chip (Toray Industries), which was designed to detect 2588 miRNAs registered in miRBase release 21 (http://www.mirbase.org/). To ensure quality control of microarray data, a coefficient of variation for the negative control probes >0.15 or a number of flagged probes >10 was considered to indicate a low‐quality result, and samples meeting these criteria were excluded from further analyses. The presence of miRNAs was determined based on a corresponding microarray signal greater than the [mean + 2 × standard deviation] negative control signal from which the top and bottom 5%, ranked according to signal intensity, were removed to exclude false positive candidates. Once a miRNA was considered present, the mean signal of the negative controls of which the top and bottom 5% ranked by signal intensity were removed was subtracted from the miRNA signal. When the signal value was negative (or undetected) after background subtraction, the value was replaced by the lowest signal intensity on the microarray minus 0.1 on a base 2 logarithmic scale. To normalize the signals among microarrays tested, three pre‐selected internal control miRNAs (miR‐149‐3p, miR‐2861 and miR‐4463) were used as described in previous reports.9, 10, 11, 12 All microarray data in this study were in agreement with the Minimum Information About a Microarray Experiment (MIAME) guidelines. The full miRNA expression profiles are stored in the Gene Expression Omnibus (GEO) database (GSE106817, GSE124158 and GSE103708).
2.3. Identification of candidate microRNAs for prediction of uterine leiomyosarcoma
Prior to the analysis, only miRNAs with a signal value >26 in more than 50% of ULMS or other samples were selected in order to identify robust biomarkers, as described in our previous studies.9, 16, 17 A cross‐validation score, which indicates the robustness of discrimination performance between ULMS and other samples, was calculated based on Fisher's linear discriminant analysis for each of the selected miRNAs (Appendix S1). miRNAs with a cross‐validation score of ≥0.75 and |fold change (log2)| ≥0.5 were selected. Subsequently, a cross‐validation score for each two‐miRNA discriminant was calculated. Based on the multiplication of the area under the receiver operating characteristic curve (AUC) and a cross‐validation score, the best two‐miRNA model was selected. The solution of the discriminant (an “index”) ≥0 indicated the presence of ULMS, whereas an index <0 indicated other types of uterine tumors.
2.4. Statistical analysis
Statistical analyses were performed using R version 3.1.2 (R Foundation for Statistical Computing, http://www.R-project.org), compute.es package version 0.2‐4, hash package version 2.2.6, MASS package version 7.3‐45, mutoss package version 0.1‐10, pROC package version 1.8 and IBM SPSS Statistics version 22 (IBM Japan). The limit of statistical significance for all analyses was defined as a two‐sided P‐value of 0.05.
3. RESULTS
3.1. Analyses of whole microRNA profiles in uterine tumor patient serum
A total of 32 serum samples from female patients with ULM, ULMS, adenosarcoma, ESS and STUMP in the uterus were identified, and 3 patients with low‐quality microarray data were excluded. Patient characteristics in the study are shown in Table S1, and representative MRI of ULM and ULMS are shown in Figure 1. In many cases, the appearance of ULM and ULMS in MRI is highly similar, confounding diagnosis of ULMS. To address the potential application of circulating miRNAs as novel biomarkers, serum samples were prepared, and miRNA profiles were generated using a miRNA microarray. According to the signal values of all patients, 385 miRNAs were selected as fulfilling the criteria described in the Methods section, and subjected to further analyses. The results of hierarchical clustering analysis and principal component analysis revealed that the profile of serum from ULMS patients was distinct from that of serum from ULM patients (Figure 2). In contrast, serum from other subtypes, including patients with adenosarcoma, ESS and STUMP, was not classified into unique groups.
Figure 1.
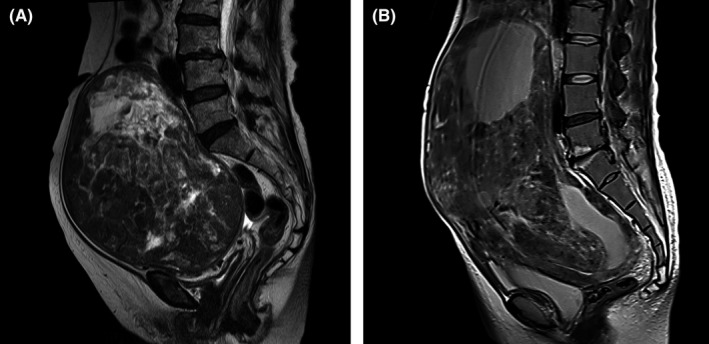
MRI for representative leiomyoma and leiomyosarcoma. Preoperative T2‐weighted MRI for a patient with uterine leiomyosarcoma (A) and leiomyoma (B). Both images were obtained from patients referred to National Cancer Center Hospital
Figure 2.
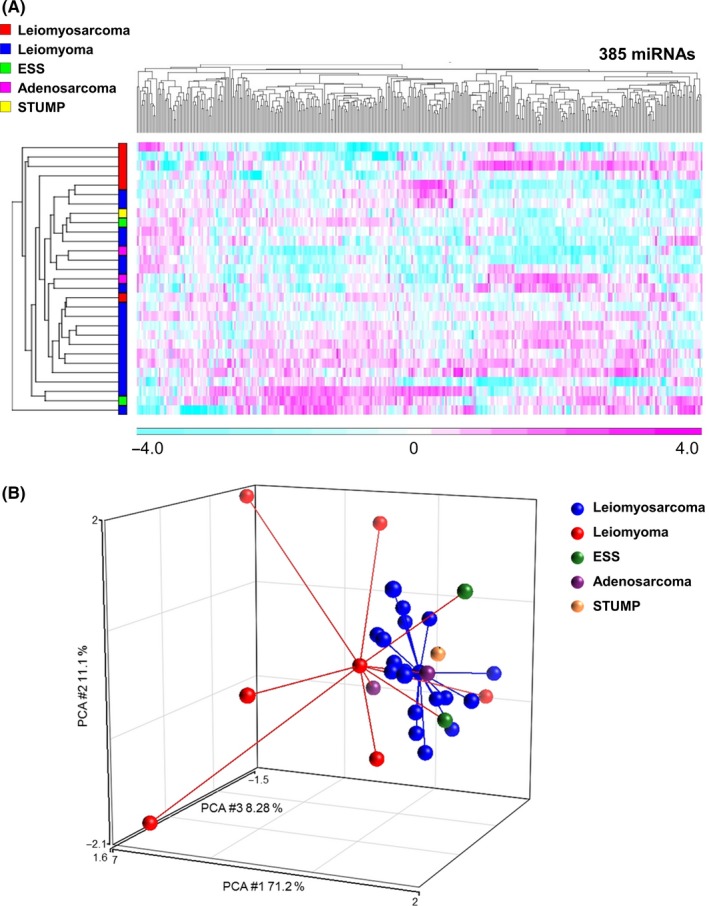
MicroRNA profiles for whole samples. A, Heat map of serum miRNA expression of patients with uterine tumors. For miRNA expression analysis, a one‐way analysis of variance was performed to identify differentially expressed miRNAs (P < .05), resulting in selection of 385 miRNAs. Unsupervised clustering and heat map generation were performed on sorted datasets by Pearson's correlation according to Ward's method using probe sets selected by Partek Genomics Suite 6.6. N = ULMS, 6; ULM, 18; ESS, 2; adenosarcoma, 2; STUMP, 1. B, Principal component analysis mapping for serum miRNA expression of patients with uterine tumors. N = ULMS, 6; ULM, 18; ESS, 2; adenosarcoma, 2; STUMP, 1
3.2. Discovering candidate micrRNAs for uterine sarcoma detection in serum
To identify candidate miRNAs for diagnostic model construction, the performance of each miRNA for US detection was investigated by cross‐validation analysis, as described in the Methods section. The top 20 miRNAs that had the highest cross‐validation score are shown in Table 1. Within those 20 miRNAs, 7 (miR‐4430, miR‐6511b‐5p, miR‐451a, miR‐4485‐5p, miR‐4635, miR‐1246 and miR‐191‐5p) were selected as candidate miRNAs that had a cross‐validation score greater than 0.75 and an absolute value of fold change (log2) >0.5. As shown in the heat map in Figure 3A, the expression level of 7 miRNAs was significantly lower in ULMS, but other subtypes did not have this tendency. To address the specificity of those miRNAs, we also examined expression of age‐matched control samples from patients with benign bone and soft tissue tumors (N = 29), and from patients with benign ovarian tumors (N = 29). As shown in Figure 3B, downregulation of those miRNAs was relatively specific to ULMS. In some cases, however, the difference between ULMS and benign bone and soft tissue tumors was not markedly different. This may be due to the similarity of origin (both tumors arise from stromal cells).
Table 1.
The top 20 microRNAs with the highest cross‐validation score
| miRNA | AUC | CV score | Fold change (log2) |
|---|---|---|---|
| miR‐4430 | 0.71 | 0.82 | −0.64 |
| miR‐6511b‐5p | 0.64 | 0.79 | −1.15 |
| miR‐191‐5p | 0.73 | 0.75 | −2.52 |
| miR‐6737‐5p | 0.72 | 0.75 | −0.31 |
| miR‐451a | 0.71 | 0.75 | −2.88 |
| miR‐4485‐5p | 0.71 | 0.75 | −1.08 |
| miR‐4746‐3p | 0.68 | 0.75 | −0.35 |
| miR‐4466 | 0.64 | 0.75 | 0.11 |
| miR‐4635 | 0.63 | 0.75 | −1.26 |
| miR‐4286 | 0.62 | 0.75 | −0.38 |
| miR‐1246 | 0.61 | 0.75 | −0.91 |
| miR‐6165 | 0.73 | 0.71 | −0.53 |
| miR‐7846‐3p | 0.72 | 0.71 | −1.03 |
| miR‐6822‐5p | 0.72 | 0.71 | −1.40 |
| miR‐24‐3p | 0.70 | 0.71 | −0.72 |
| miR‐6748‐5p | 0.66 | 0.71 | −1.60 |
| miR‐615‐5p | 0.65 | 0.71 | −0.64 |
| miR‐1290 | 0.62 | 0.71 | −1.43 |
| miR‐7845‐5p | 0.61 | 0.71 | −0.39 |
| miR‐7975 | 0.60 | 0.71 | −0.33 |
The selected miRNAs and the columns which met the criteria (a cross‐validation score of ≥0.75 and |fold change (log2)| ≥0.5) are shown in bold.
AUC, area under receiver operating characteristics curve; CV, cross‐validation.
Figure 3.
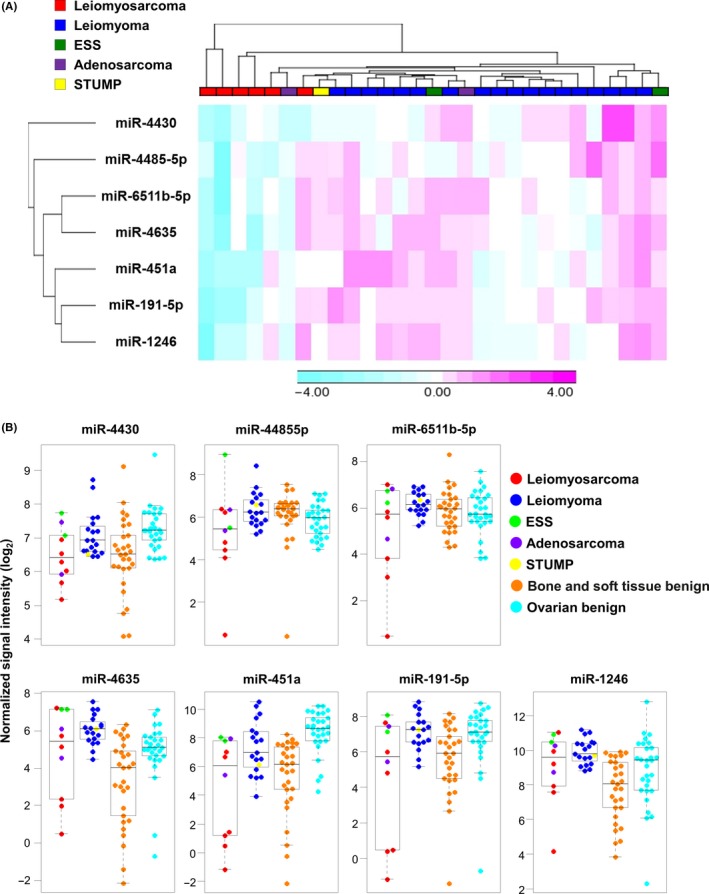
Expression of seven selected microRNAs. A, A heat map for serum miRNA expression of the seven selected miRNAs in patients with uterine tumors. Unsupervised clustering and heat map generation were performed on sorted datasets by Pearson's correlation on Ward's method with selected probe sets by Partek Genomics Suite 6.6. N = ULMS, 6; ULM, 18; ESS, 2; adenosarcoma, 2; STUMP, 1. B, Distribution of seven selected miRNAs across all samples. Serum levels of miRNAs. The dot plots are overlaid with box plots. US included six ULMSs, two ESSs, and two adenosarcomas as one group, and non‐malignant uterine tumors (18 ULMs and one STUMP) as one group. N = 29 benign bone and soft tissue tumors and 29 benign ovarian tumors
3.3. Generating an optimal diagnostic model for uterine sarcoma screening
To identify candidate miRNAs for diagnostic model construction, the performance of each miRNA for US detection was investigated. Using Fisher's linear discriminant analysis, we designed comprehensive discriminants with 1‐2 miRNAs from the 7 selected candidates (Table 2). Based on the multiplied value of the AUC and a cross‐validation score, the best combination was determined, which consisted of two miRNAs (miR‐191‐5p and miR‐1246), and the diagnostic index (DI) of the model was defined as (DI = (−0.479) × miR‐191‐5p + (−0.380) × miR‐1246 + 6.386). When distinguishing all US subtypes from benign ULM controls, the diagnostic performance of the model was higher than that of serum lactate dehydrogenase (LDH), which is a major conventional biomarker for US (model‐AUC: 0.83 [0.65‐1.00], and LDH‐AUC: 0.62 [0.38‐0.85])18 (Figure S1). Notably, when focusing on ULMS screening, the accuracy was much higher and the difference from the LDH level was more significant (model‐AUC: 0.97 [0.91‐1.00], and LDH‐AUC: 0.64 [0.34‐0.94]) (Figure 4A). To consider the combination of miRNAs and LDH values, we generated additional models (Figure 4B). The DI of models comprising (A) LDH, miR‐191‐5p and miR‐1246, (B) LDH and miR‐191‐5p and (C) LDH and miR‐1246 were defined as (DIA = (0.008) × LDH + (−0.747) × miR‐191‐5p + (0.637) × miR‐1246 − 3.330; DIB = (0.011) × LDH + (−0.442) × miR‐191‐5p + 0.943, and DIC = (0.014) × LDH + (−0.653) × miR‐1246 + 2.459), respectively; each ROC curve is shown in Figure 4B. As shown in Figure 4C, the dot plots reveal that all ULMS patients were precisely identified as having a positive DI using the two‐miRNA prediction model. Thus, the identified combination of two miRNAs is a promising biomarker for ULMS screening and distinction from ULM. However, other subtypes of US were not distinguishable using this model. For the DI, statistical analysis for calculating P‐values was not performed due to the small sample number.
Table 2.
Performance of each diagnostic model
| #miRNAs | Diagnostic model | AUC | CV score | AUC × CV score |
|---|---|---|---|---|
| 1 | (−1.39) × miR‐4430 + 8.87 | 0.71 | 0.82 | 0.58 |
| 1 | (−0.77) × miR‐6511b‐5p + 3.86 | 0.64 | 0.79 | 0.5 |
| 1 | (−0.45) × miR‐191‐5p + 2.77 | 0.73 | 0.75 | 0.55 |
| 1 | (−0.38) × miR‐451a + 1.03 | 0.71 | 0.75 | 0.53 |
| 1 | (−0.69) × miR‐4485‐5p + 3.87 | 0.71 | 0.75 | 0.53 |
| 1 | (−0.63) × miR‐4635 + 3.34 | 0.63 | 0.75 | 0.47 |
| 1 | (−0.73) × miR‐1246 + 6.42 | 0.61 | 0.75 | 0.45 |
| 2 | (−0.98) × miR‐4430 + (−0.30) × miR‐4485‐5p + 8.15 | 0.77 | 0.79 | 0.61 |
| 2 | (−0.62) × miR‐4430 + (−0.28) × miR‐451a + 5.34 | 0.76 | 0.79 | 0.60 |
| 2 | (−0.10) × miR‐4485‐5p + (−0.42) × miR‐191‐5p + 3.05 | 0.72 | 0.79 | 0.56 |
| 2 | (−1.09) × miR‐4430 + (−0.27) × miR‐1246 + 9.61 | 0.71 | 0.79 | 0.55 |
| 2 | (−0.67) × miR‐6511b‐5p + (−0.14) × miR‐1246 + 5.22 | 0.65 | 0.79 | 0.51 |
| 2 | (0.82) × miR‐1246 + (−0.84) × miR‐191‐5p − 2.59 | 0.83 | 0.75 | 0.62 |
| 2 | (−0.50) × miR‐4430 + (−0.35) × miR‐191‐5p + 5.45 | 0.76 | 0.75 | 0.57 |
| 2 | (−0.16) × miR‐451a + (−0.29) × miR‐191‐5p + 1.93 | 0.75 | 0.75 | 0.56 |
| 2 | (−0.92) × miR‐4430 + (−0.33) × miR‐4635 + 8.00 | 0.74 | 0.75 | 0.55 |
| 2 | (−0.31) × miR‐451a + (−0.22) × miR‐4485‐5p + 2.98 | 0.74 | 0.75 | 0.55 |
| 2 | (−0.80) × miR‐4430 + (−0.45) × miR‐6511b‐5p + 7.89 | 0.72 | 0.75 | 0.54 |
| 2 | (−0.13) × miR‐6511b‐5p + (−0.40) × miR‐191‐5p + 3.16 | 0.72 | 0.75 | 0.54 |
| 2 | (−0.05) × miR‐4635 + (−0.43) × miR‐191‐5p + 2.89 | 0.72 | 0.75 | 0.54 |
| 2 | (−0.38) × miR‐451a + (0.01) × miR‐4635 + 0.97 | 0.71 | 0.75 | 0.53 |
| 2 | (−0.23) × miR‐6511b‐5p + (−0.30) × miR‐451a + 2.77 | 0.7 | 0.75 | 0.53 |
| 2 | (−0.60) × miR‐6511b‐5p + (−0.20) × miR‐4485‐5p + 4.35 | 0.64 | 0.75 | 0.48 |
| 2 | (−0.32) × miR‐4485‐5p + (−0.40) × miR‐4635 + 3.76 | 0.64 | 0.75 | 0.48 |
| 2 | (−0.51) × miR‐4635 + (−0.19) × miR‐1246 + 4.54 | 0.63 | 0.75 | 0.47 |
| 2 | (−0.66) × miR‐6511b‐5p + (−0.10) × miR‐4635 + 3.86 | 0.62 | 0.75 | 0.47 |
| 2 | (−0.47) × miR‐4485‐5p + (−0.38) × miR‐1246 + 6.02 | 0.72 | 0.71 | 0.51 |
The selected diagnostic model is shown in bold.
AUC, area under receiver operating characteristics curve; CV, cross‐validation.
Figure 4.
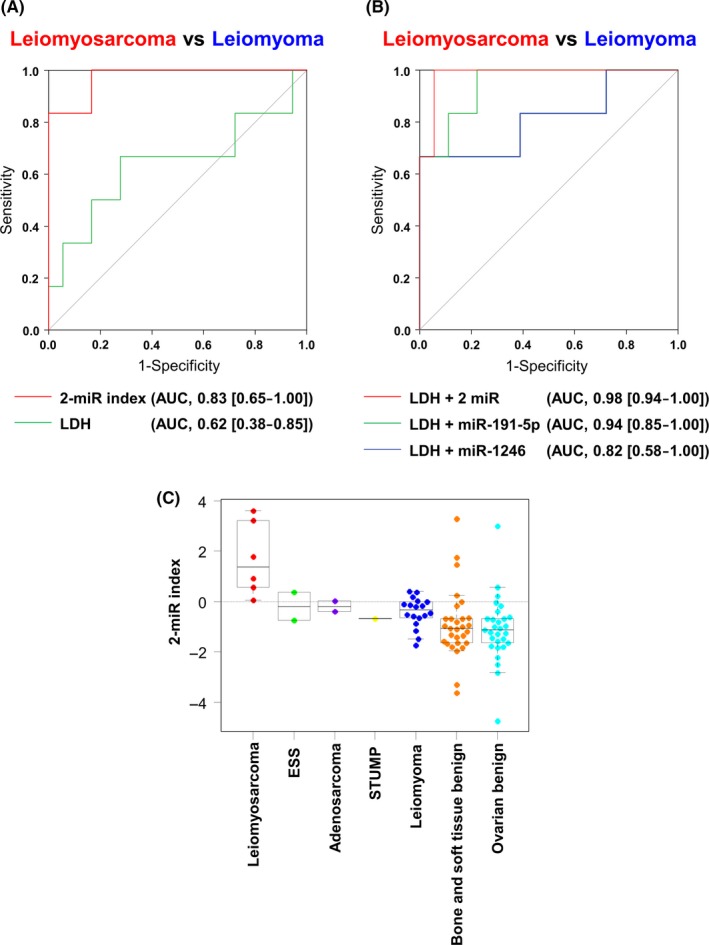
Performance of the diagnostic model for uterine sarcomas. A, Receiver operating characteristics (ROC) curves for distinguishing uterine leiomyosarcoma (ULMS) patients from uterine leiomyoma (ULM) controls using the two‐miRNA strategy that was developed for diagnostic model 1. N = ULMS, 5; ULM, 18. B, ROC curves distinguishing ULMS patients from ULM controls by each diagnostic model based on microRNA and lactate dehydrogenase (LDH) values. N = ULMS, 6; ULM, 18; ESS, 2; adenosarcoma, 2; STUMP, 1. C, Diagnostic index using the 2‐miR diagnostic model. N = ULMS, 6; ULM, 18; ESS, 2; adenosarcoma, 2; STUMP, 1; benign bone and soft tissue tumors, 29; benign ovarian tumors, 29
4. DISCUSSION
Uterine sarcoma is a malignant soft tissue sarcoma, and the characteristics of US subtypes are very diverse. In this study, we initially tried to distinguish all subtypes of US from ULM, and the subtypes of adenosarcoma, ESS and STUMP were also analyzed. However, according to the results of clustering analysis shown in Figures 2 and 3, each subtype may have a different extracellular miRNA profile. Adenosarcoma and ESS are also malignant tumors but were not clearly categorized in ULMS clusters. STUMP in particular is difficult for pathologists to diagnose, and its characteristics, including risk factors and clinical behavior, are poorly understood.19 It was also categorized in non‐cancer clusters. Other US subtypes may have a different profile than that of ULMS, but the number of each sample type was too small to draw any conclusions. In many uterine tumor cases, clinicians may struggle to determine if malignant tumors are present, particularly in distinguishing ULMS from ULM, as shown in Figure 1. For this reason, the model developed in the present study is of considerable therapeutic value for clinical applications.
One of the biggest concerns for surgeons treating ULM is the potential for tumor morcellation (intra‐operative fragmentation) during laparoscopic surgery, causing abdominal spread of malignant cells, which leads to poor prognosis of incidental US.20 Although the laparoscopic approach has many therapeutic advantages, accurate preoperative diagnosis of US is of paramount importance to avoid unnecessary tumor spread. However, accurate preoperative diagnosis remains challenging for this malignancy, and there is no consensus regarding blood‐based biomarkers for US. Furthermore, other imaging approaches, such as ultrasound, MRI and positron emission tomography (PET), are also not sufficient. In addition, the sensitivity of preoperative vaginal endometrial sampling is low (around 35%) because these biopsies originate from the myometrium.21 Serum LDH levels are increased in some US patients, and LDH is a classical biomarker for sarcomas.22, 23 Although the accuracy of LDH elevation is not sufficient, LDH is routinely tested in most cases. In this study, we compared the predictive performance between our diagnostic model and LDH level, and demonstrated that, in this cohort of patients, our newly developed biomarkers had greater predictive accuracy than LDH. Although the number of samples was limited, the substantial performance of the model suggests that it should be validated in a larger patient cohort.
There is no doubt that MR imaging is the best way to assess uterine masses before intervention.24 The accuracy of MR imaging for this purpose is 50%‐100%; however, a new method of MR imaging has been investigated.23, 25 The atypical appearance of ULM caused by degeneration, edema or unusual patterns of growth makes differentiation of ULMS from ULM difficult.26, 27, 28 Although our sample set was small, the detection accuracy of our prediction model is similar to that of MRI. In general, the combination of imaging examinations and blood tests should be a powerful strategy for precise diagnosis; thus, our findings may contribute to development of a liquid biopsy for ULMS.
The miRNA profile of US has been investigated in previous studies, but most analyses have focused on tissue miRNA profiles.15, 29 According to the TCGA database, there are 27 ULMS tissue datasets, and the database data suggest that some miRNAs are significantly related to patient prognosis.30 Very few reports have addressed serum miRNA levels, and only limited targets have been analyzed by quantitative RT‐PCR, thus precluding a comprehensive analysis.31 In addition, no reports have described the diagnostic performances of selected miRNAs. In this study, to accurately model clinical scenarios, we used ULM as a control, and analyzed the predictive performance of each miRNA. As a result, 7 miRNAs that had the highest performances were selected, all of which were decreased in patient serum. Among the 7 selected miRNAs, miR‐191‐5p and miR‐1246 are oncogenes, and others are downregulated in cancer.32, 33 However, the function of most of the identified miRNAs remains unknown or controversial. To understand the functions of the identified miRNAs in US, independent validation experiments will be required and conducted by our group in the near future.
Overall, the current analysis demonstrates the feasibility of serum circulating miRNAs as preoperative biomarkers and is the first report to use miRNA microarray analyses to identify preoperative serum miRNA biomarkers for US. Evidence supporting the utility of circulating miRNAs as biomarkers has been accumulating in the past decade, and many clinical applications are currently being developed. Although future validation and additional optimization of the presently identified model is required, this study could help inform the preoperative diagnosis of US, which would contribute significantly to improved selection of cases appropriate for laparoscopic surgery, thereby improving clinical outcomes.
DISCLOSURE
Satoko Takizawa is an employee of Toray Industries, the provider of the 3D‐Gene system. Yoshiaki Aoki is an employee of Dynacom, the developer of the statistical script for selecting the optimal miRNA combination. All other authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
The authors thank Tomomi Fukuda, Takumi Sonoda, Hiroko Tadokoro, Megumi Miyagi and Tatsuya Suzuki for performing microarray assays, Junpei Kawauchi, Makiko Ichikawa and Satoshi Kondou, for technical support, Noriko Abe and Hiromi Sakamoto for the management of serum samples, Michiko Ohori for the management of personal information, and Hitoshi Fujimiya for developing in‐house analytic tools. Samples used in this study were obtained from the National Cancer Center Biobank, which is supported by the National Cancer Center Research and Development Fund (29‐A‐1). This study was financially supported through a “Development of Diagnostic Technology for Detection of miRNA in Body Fluids” grant from the Japan Agency for Medical Research and Development (to TO, 18ae0101011h0005).
Yokoi A, Matsuzaki J, Yamamoto Y, et al. Serum microRNA profile enables preoperative diagnosis of uterine leiomyosarcoma. Cancer Sci. 2019;110:3718–3726. 10.1111/cas.14215
Akira Yokoi and Juntaro Matsuzaki contributed equally to this work.
REFERENCES
- 1. Brooks SE, Zhan M, Cote T, Baquet CR. Surveillance, epidemiology, and end results analysis of 2677 cases of uterine sarcoma 1989–1999. Gynecol Oncol. 2004;93:204‐208. [DOI] [PubMed] [Google Scholar]
- 2. Ricci S, Stone RL, Fader AN. Uterine leiomyosarcoma: epidemiology, contemporary treatment strategies and the impact of uterine morcellation. Gynecol Oncol. 2017;145:208‐216. [DOI] [PubMed] [Google Scholar]
- 3. Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol. 2003;188:100‐107. [DOI] [PubMed] [Google Scholar]
- 4. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126‐139. [DOI] [PubMed] [Google Scholar]
- 5. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376‐385. [DOI] [PubMed] [Google Scholar]
- 6. Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126:1163‐1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortez MA, Bueso‐Ramos C, Ferdin J, Lopez‐Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yokoi A, Matsuzaki J, Yamamoto Y, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiino S, Matsuzaki J, Shimomura A, et al. Serum miRNA‐based prediction of axillary lymph node metastasis in breast cancer. Clin Cancer Res. 2019;25:1817‐1827. [DOI] [PubMed] [Google Scholar]
- 11. Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110:408‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimomura A, Shiino S, Kawauchi J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kowalewska M, Bakula‐Zalewska E, Chechlinska M, et al. microRNAs in uterine sarcomas and mixed epithelial‐mesenchymal uterine tumors: a preliminary report. Tumour Biol. 2013;34:2153‐2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi G, Perle MA, Mittal K, et al. Let‐7 repression leads to HMGA2 overexpression in uterine leiomyosarcoma. J Cell Mol Med. 2009;13:3898‐3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ravid Y, Formanski M, Smith Y, Reich R, Davidson B. Uterine leiomyosarcoma and endometrial stromal sarcoma have unique miRNA signatures. Gynecol Oncol. 2016;140:512‐517. [DOI] [PubMed] [Google Scholar]
- 16. Sudo K, Kato K, Matsuzaki J, et al. Development and validation of an esophageal squamous cell carcinoma detection model by large‐scale MicroRNA profiling. JAMA Netw Open. 2019;2:e194573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Asano N, Matsuzaki J, Ichikawa M, et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun. 2019;10:1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levy G, Hill MJ, Plowden TC, Catherino WH, Armstrong AY. Biomarkers in uterine leiomyoma. Fertil Steril. 2013;99:1146‐1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guntupalli SR, Ramirez PT, Anderson ML, Milam MR, Bodurka DC, Malpica A. Uterine smooth muscle tumor of uncertain malignant potential: a retrospective analysis. Gynecol Oncol. 2009;113:324‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bretthauer M, Goderstad JM, Løberg M, et al. Uterine morcellation and survival in uterine sarcomas. Eur J Cancer. 2018;101:62‐68. [DOI] [PubMed] [Google Scholar]
- 21. Hinchcliff EM, Esselen KM, Watkins JC, et al. The role of endometrial biopsy in the preoperative detection of uterine leiomyosarcoma. J Minim Invasive Gynecol. 2016;23:567‐572. [DOI] [PubMed] [Google Scholar]
- 22. Seki K, Hoshihara T, Nagata I. Leiomyosarcoma of the uterus: ultrasonography and serum lactate dehydrogenase level. Gynecol Obstet Invest. 1992;33:114‐118. [DOI] [PubMed] [Google Scholar]
- 23. Goto A, Takeuchi S, Sugimura K, Maruo T. Usefulness of Gd‐DTPA contrast‐enhanced dynamic MRI and serum determination of LDH and its isozymes in the differential diagnosis of leiomyosarcoma from degenerated leiomyoma of the uterus. Int J Gynecol Cancer. 2002;12:354‐361. [DOI] [PubMed] [Google Scholar]
- 24. Hricak H, Tscholakoff D, Heinrichs L, et al. Uterine leiomyomas: correlation of MR, histopathologic findings, and symptoms. Radiology. 1986;158:385‐391. [DOI] [PubMed] [Google Scholar]
- 25. Tanaka YO, Nishida M, Tsunoda H, Okamoto Y, Yoshikawa H. Smooth muscle tumors of uncertain malignant potential and leiomyosarcomas of the uterus: MR findings. J Magn Reson Imaging. 2004;20:998‐1007. [DOI] [PubMed] [Google Scholar]
- 26. Ueda H, Togashi K, Konishi I, et al. Unusual appearances of uterine leiomyomas: MR imaging findings and their histopathologic backgrounds. Radiographics. 1999; 19 Spec No:S131‐S145. [DOI] [PubMed] [Google Scholar]
- 27. Tamai K, Koyama T, Saga T, et al. The utility of diffusion‐weighted MR imaging for differentiating uterine sarcomas from benign leiomyomas. Eur Radiol. 2008;18:723‐730. [DOI] [PubMed] [Google Scholar]
- 28. Schwartz LB, Zawin M, Carcangiu ML, Lange R, McCarthy S. Does pelvic magnetic resonance imaging differentiate among the histologic subtypes of uterine leiomyomata? Fertil Steril. 1998;70:580‐587. [DOI] [PubMed] [Google Scholar]
- 29. Gonzalez Dos Anjos L, de Almeida BC, Gomes de Almeida T, et al. Could miRNA signatures be useful for predicting uterine sarcoma and carcinosarcoma prognosis and treatment? Cancers. 2018;10:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research Network .Electronic address edsc , Cancer Genome Atlas Research N . Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017; 171: 950‐965.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tong X, Wang X, Wang C, Li L. Elevated levels of serum MiR‐152 and miR‐24 in uterine sarcoma: potential for inducing autophagy via SIRT1 and deacetylated LC3. Br J Biomed Sci. 2018;75:7‐12. [DOI] [PubMed] [Google Scholar]
- 32. Nagpal N, Kulshreshtha R. miR‐191: an emerging player in disease biology. Front Genet. 2014;5:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Q, Cao LY, Cheng SJ, Zhang AM, Jin XS, Li Y. p53‐induced microRNA‐1246 inhibits the cell growth of human hepatocellular carcinoma cells by targeting NFIB. Oncol Rep. 2015;33:1335‐1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


