Abstract
Circular RNAs (circRNAs) have a covalently closed circular conformation and are structurally stable. Those circRNAs with tumor‐suppressive properties play an important role in tumorigenesis and metastasis and thus may be used as therapeutic targets of cancers. Herein, we review the current understanding of the classification of circRNAs and summarize the functions and mechanisms of circRNAs that have tumor‐suppressive roles in various cancers, including liver cancer (circARSP91, circADAMTS13, circADAMTS14, circMTO1, hsa_circ_0079299, and circC3P1), bladder cancer (circFNDC3B, circITCH, circHIPK3, circRNA‐3, cdrlas, and circLPAR1), gastric cancer (circLARP4, circYAP1, hsa_cric_0000096, hsa_circ_0000993, and circPSMC3), breast cancer (circ_000911, hsa_circ_0072309, and circASS1), lung cancer (hsa_circ_0000977, circPTK2, circ_0001649, hsa_circ_100395, and circ_0006916), glioma (circ_0001946, circSHPRH, and circFBXW7), and colorectal cancer (circITGA7 and hsa_circ_0014717). Thanks to their structural stability, these tumor‐suppressive circRNAs may be used as potential and potent therapeutic targets. Moreover, we propose a new method for the classification of circRNAs. Based on whether they can be translated, circRNAs can be divided into noncoding circRNAs and coding circRNAs.
Keywords: biogenesis, cancer, circular RNA, mechanism, therapeutic target
Tumor‐suppressive circRNAs may be used as potential and potent therapeutic targets. Moreover, we can propose a new method for the classification of circRNAs: based on whether they can be translated, circRNAs can be divided into noncoding circRNAs and coding circRNAs.
Abbreviations
- Ago2
Argonaute 2
- Alu
Arthrobacter luteus
- AR
androgen receptor
- ASS1
argininosuccinate synthetase 1
- BCa
bladder cancer
- BMI1
B lymphoma Mo‐MLV insertion region 1 homolog
- ceRNAs
competitive endogenous RNAs
- circRNAs
circular RNAs
- ciRNAs
circular intronic RNAs
- CRC
colorectal cancer
- DHX9
DExH‐box helicase 9
- ecircRNAs
exonic circRNAs
- EIciRNAs
exon‐intronic circRNAs
- FAT1
FAT atypical cadherin 1
- GBM
glioblastoma
- GC
gastric cancer
- HCC
hepatocellular carcinoma
- HN
human protein
- IRES
internal ribosome entry site
- LARP4
La‐related protein 4
- LATS1
large tumor suppressor kinase 1
- lncRNAs
long noncoding RNAs
- MIBC
muscle invasive bladder cancer
- miRNA
microRNA
- ncRNAs
noncoding RNAs
- NF1
neurofibromin 1
- Notch1
notch homolog 1
- NSCLC
non‐small cell lung cancer
- OS
overall survival
- PCK1
phosphoenolpyruvate carboxykinase 1
- PDAC
pancreatic ductal adenocarcinoma
- RBP
RNA‐binding protein
- RCAN1
regulation of Down syndrome critical region gene 1
- sncRNAs
short noncoding RNAs
- TIMP3
metalloproteinase inhibitor 3
- USP28
ubiquitin‐specific peptidase 28
- YBX1
Y box binding protein 1
1. INTRODUCTION
Those RNAs that do not encode proteins are called ncRNAs.1 Regulatory ncRNAs are divided into lncRNAs and sncRNAs. LncRNAs can be linear or circular.2
Circular RNAs were first discovered in 1976.3 Since then, a variety of circRNAs have been discovered.4, 5 CircRNAs are more resistant to exonuclease and are more stable than other lncRNAs.3, 6 They exist in exosomes and plasma.6, 7 Li et al identified more than 1000 circRNAs in human serum exosomes.6 Li et al found 343 differentially expressed circRNAs in the plasma.2 Their expression profiles are specific in different cell types and developmental stages.3, 4
In the present review, we first briefly summarize the characteristics of circRNAs, highlight the relationships between tumor‐suppressive circRNAs and cancers, and finally illustrate the mechanisms underlying circRNA‐mediated inhibition of cancer occurrence and development.
2. CLASSIFICATION AND BIOGENESIS OF circRNAs
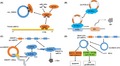
According to their composition, circRNAs can be divided into three categories (Figure 1A): ecircRNAs,8 ciRNAs,9 and EIciRNAs.10 According to their location, circRNAs can be divided into five categories (Figure 1B): ecircRNAs, ciRNAs, antisense circRNAs, sense‐overlapping circRNAs, and intergenic circRNAs.11
Figure 1.
Categories of circular RNAs (circRNAs). A, Classification of circRNAs based on their compositions: Exonic circRNAs (ecircRNAs), circular intronic RNAs (ciRNAs), and exon‐intronic circRNAs (EIciRNAs). B, Classification of circRNAs based on their positions and their adjacent mRNAs: ecircRNAs, ciRNAs, antisense circRNAs, sense overlapping circRNAs, and intergenic circRNAs. C, Coding circRNAs may have at least one internal ribosome entry site (IRES) and have an open reading frame (ORF)
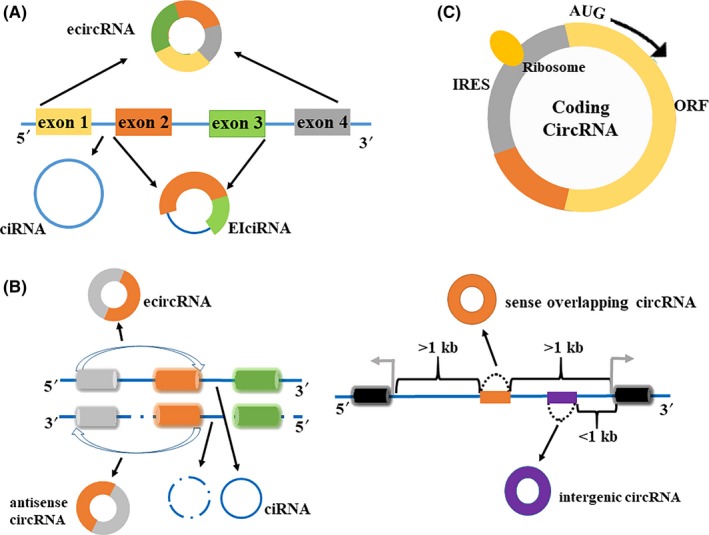
Simultaneously, recent studies have shown the potential of circRNAs in protein translation.12, 13, 14 CircMbl3 can be translated in a splicing‐dependent but cap‐independent way in fly head extract.12 A single N6‐methyladenosine residue in circRNA may be sufficient to drive translation.13 Here, we propose a new method for the classification of circRNAs: based on whether they can be translated, circRNAs may be divided into noncoding circRNAs and coding circRNAs (Figure 1C). Specifically, coding circRNAs may have at least one IRES or have specific m6A site, which allows the ribosome to initiate translation directly in the mRNA sequence. In addition, coding circRNAs have an ORF. Merely meeting these two points is sufficient for classification as coding circRNAs. Most circRNAs known today are produced by reverse splicing of pre‐mRNA (Figure 2A).15, 16 There are two other biogenesis methods, intron‐pairing circulation and RBP‐induced circulation (Figure 2B,C).17, 18, 19 For some circRNAs, the reverse complementary Alu can be used to boost circularization.20, 21, 22
Figure 2.
Biogenesis of circular RNAs (circRNAs). A, Produced by reverse splicing of pre‐mRNA. B, Intron pairing cycle produces circRNA. C, RNA‐binding protein (RBP) induces circulation
3. BIOLOGICAL FUNCTIONS OF circRNAs
Functions of circRNAs include the following: (i) regulation of transcription; (ii) competition as endogenous RNA or miRNA sponges; (iii) translation of proteins; and (iv) interaction with RBP.23 In addition, some ecircRNAs may affect alternative splicing.
As the formation of some circRNAs and linear RNAs share some common exons, they may compete with each other.24 For example, circZKSCAN1 (hsa_circ_0001727) from the zinc finger protein with KRAB and SCAN domains 1 (ZKSCAN1) gene may retain endogenous RNA as a competitive inhibitor and regulate tumor cell proliferation and metastasis‐related gene expression. Both ZKSCAN1 and its related circRNA (circZKSCAN1) inhibit HCC growth, migration, and invasion but through different signaling pathways.25
CircRNAs may function as ceRNAs to retain endogenous RNA and regulate the expression of related genes.26, 27 For example, circLARP4 from the LARP4 pre‐mRNA inhibits the occurrence and progression of GC by affecting the expression of cavernous miR‐424 and increasing the expression of LATS1.28
Increasing numbers of studies have shown that circRNAs have a potential role in translation.12, 29, 30 CircRNAs that can be translated to peptides or proteins mainly have the following characteristics: (i) they have an ORF with a longer length; (ii) they have an ORF that crosses reverse junctions; and (iii) they have some essential regulatory elements upstream of the ORF at the start of translation, such as N6‐methyladenosine (m6A) near the initiation codon.13 For example, circSHPRH from exons 26 and 29 of the SNF2 histone linker PHD RING helicase (SHPRH) pre‐mRNA uses an overlapping genetic code to encode a 17‐kDa protein SHPRH‐146aa. This novel protein functions as a tumor suppressor by protecting its associated full‐length SHPRH.31
Other circRNAs may also interact with different proteins, regulate the transcription of parental genes, and promote protein‐protein interactions.20 For example, circFAT1(e2) (hsa_circ_0001461) from exon 2 of FAT atypical cadherin 1 (FAT1) can directly bind to YBX1 and then inhibit the progression of GC.32
More importantly, a related primary consideration for circRNA function is their localization. EcircRNAs have been shown to be predominantly cytoplasmic,33 but ciRNAs and EIciRNAs are localized to the nucleus, cytoplasm, or both.
4. CHARACTERISTICS OF TUMOR‐SUPPRESSIVE circRNAs
Many circRNAs have been found to exert tumor‐suppressive effects in several types of cancer (Table 1). A few of them, such as circFAT1(e2), are produced by a single exon.32 However, most circRNAs are produced by multiple exons. For example, cSMARCA5 (hsa_circ_0001445) is from exons 15 and 16 of the SWI/SNF‐related matrix‐associated actin‐dependent regulator of the chromatin subfamily A member 5 (SMARCA5) gene,34 and circASS1 (hsa_circ_0089105) is derived from exons 9, 10, and 11 of the ASS1 gene.35 Regarding localization, tumor‐suppressive circRNAs are mainly located in the cytoplasm.
Table 1.
Summary of circRNAs as a tumor suppressor in various tumors
| Cancer type | CircRNA | Function | Mechanism | Reference |
|---|---|---|---|---|
| Hepatocellular carcinoma | CSMARCA5 | MiRNA sponge | As sponges of miR‐17‐3p and miR‐181b‐5p to regulate miR‐17‐3p/miR‐181b‐5p‐TIMP3 axes | 34 |
| CircZKSCAN1 | Regulating the transcription of linear RNA | Acting as a competitive inhibitor to retain endogenous RNA | 25 | |
| CircADAMTS14 | MiRNA sponge | Acting as a sponge of miR‐572 to regulate expression of RCAN1 | 37 | |
| Hsa_circ_0079299 | Interaction with protein | Inhibiting cell proliferation through PI3K/AKT/mTOR signaling pathway | 40 | |
| CircMTO1 | MiRNA sponge | Binding with miR‐9 and regulating P21 expression | 39 | |
| CircARSP91 | Interaction with protein | Suppressed by androgen receptor | 41 | |
| CircC3P1 | MiRNA sponge | Acting as a sponge of miR‐4641 to promote PCK1 expression | 42 | |
| CircADAMTS13 | MiRNA sponge | Acting as a sponge of miR‐484 | 38 | |
| Bladder cancer | CircFNDC3B | MiRNA sponge | Inhibiting G3BP2 expression and SRC/FAK phosphorylation by binding with miR‐1178‐3p | 43 |
| CircITCH | MiRNA sponge | Acting as sponges of miR‐17 and miR‐224 to upregulate the expression of P21 and PTEN | 44 | |
| CircLPAR1 | MiRNA sponge | Acting as a sponge of miR‐762 | 48 | |
| CircHIPK3 | MiRNA sponge | Acting as a sponge of miR‐558 and inhibiting heparanase expression, thereby inhibiting invasion and metastasis | 45 | |
| CircRNA‐3 (BCRC‐3) | MiRNA sponge | Interacting with miR‐182‐5p and subsequently promoting P27 activity | 46 | |
| Cdr1as | MiRNA sponge | Binding to miR‐135a | 47 | |
| Gastric cancer | CircFAT1 (e2) | MiRNA sponge and interacting with RBP | Acting as a sponge of miR‐548g to regulate the expression of RUNX1; interacting with YBX1 | 32 |
| CircYAP1 | MiRNA sponge | Acting as a sponge of miR‐367‐5p to upregulate the expression of P27 | 49 | |
| CircLARP4 | MiRNA sponge | Acting as a sponge of miR‐424 to regulate the expression of LATS1 | 28 | |
| Hsa_circ_0000993 | MiRNA sponge | Inhibit metastasis by chelation of miR‐214‐5p | 51 | |
| Hsa_circ_0000096 | Interaction with protein | Regulating the expression of cyclin D1, CDK6, MMP‐2, and MMP‐9 | 50 | |
| CircPSMC3 | MiRNA sponge | Acting as a sponge of miR‐296‐5p to regulate the expression of PTEN | 52 | |
| Breast cancer | CircASS1 | MiRNA sponge | Inhibiting the expression of miR‐4443 by sponge activity, and upregulating the expression of ASS1 | 35 |
| Hsa_ circ_0072309 | MiRNA sponge | Acting as a sponge of miR‐492 | 54 | |
| Circ000911 | MiRNA sponge | Promoting Notch1 expression by acting as a sponge of miR‐449a | 53 | |
| Lung cancer | CircNOL10 | Interaction with protein | Promoting the expression of human protein | 55 |
| Circ0006916 | MiRNA sponge | Combining to miR‐522‐3p to upregulate the expression of PHLPP1, thereby inhibiting cell cycle progression and inhibiting cancer progression | 59 | |
| Hsa_circ_100395 | MiRNA sponge | Regulating miR‐1228/TCF21 axis | 58 | |
| Non‐small cell lung cancer | CircPTK2 | MiRNA sponge | Acting as sponges of miR‐429/miR‐200b‐3p, targeting TIF1γ to inhibit TGFβ‐induced EMT | 56 |
| Circ_0001649 | MiRNA sponge | Directly ejecting miR‐331‐3p and miR‐338‐5p | 57 | |
| Glioblastoma multiforme | Circ_0001946 | MiRNA sponge | Inhibiting miR‐6715p to promote the expression of CDR1 | 60 |
| CircFBXW7 | Translation | Encoding FBXW7‐185aa and then reducing the half‐life of c‐Myc | 61 | |
| CircSHPRH | Translation | Encoding a 146‐aa protein, which protects its associated full‐length SHPRH | 31 | |
| Colorectal cancer | CircIGA7 | MiRNA sponge | Competitively binding miR‐370‐3p to upregulate NF1 translation and then inhibit Ras signaling pathway; upregulating the transcription of its host gene ITGA7 by inhibiting RREB1 by Ras | 62 |
| Hsa_circ_0014717 | Unknown | Acting as a potential tumor suppressor | 63 | |
| Oral squamous cell carcinoma | Hsa_circ_0008309 | MiRNA sponge | Regulating miR‐382‐5P/ATXN1 axis | 64 |
| Tube cancer | Circ0043898 | Unknown | May serve as a target of histone H3 and BMI18 | 65 |
circRNA, circular RNA; EMT, epithelial‐mesenchymal transition; LATS1, large tumor suppressor kinase 1; miRNA, microRNA; PTEN, phosphatase and tensin homolog; TGF, transforming growth factor.
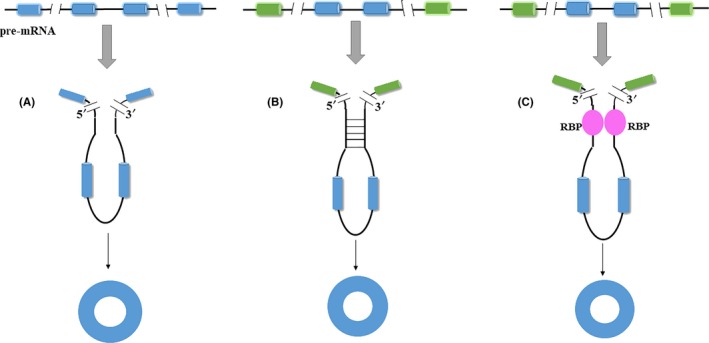
From a functional point of view, tumor‐suppressive circRNAs mainly have the following functions (Figure 3): (a) acting as a sponge of miRNAs, (b) interacting with proteins, (c) translating proteins, and (d) regulating the transcription of linear RNAs. In addition, some circRNAs may have several roles. For example, acting as sponges of miRNA and protein, cSMARCA5 inhibits HCC progression by modulating DHX9 and sponges miR‐17‐3p and miR‐181b‐5p, which target TIMP3.34
Figure 3.
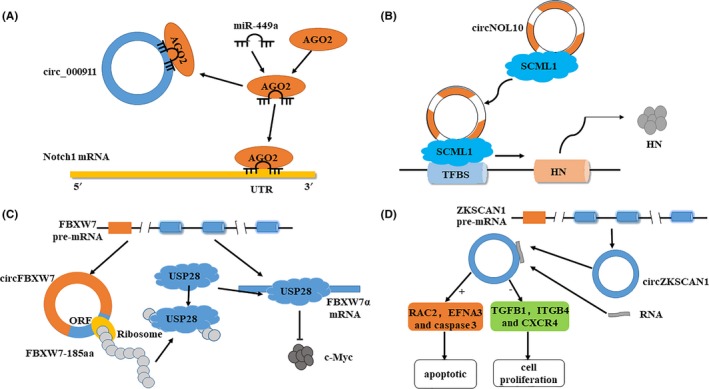
Biological functions of cancer‐suppressive circular RNAs (circRNAs). A, Acting as a sponge of microRNA (miRNA). For example, circ_000911 competitively binds with miR‐449a, which binds with Argonaute 2 (Ago2) and targets notch homolog 1 (Notch 1) mRNA. B, Interacting with proteins. For example, circNOL10 first binds with sex comb on midleg‐like 1 (SCML1), then moves transcription factor binding sites (TFBS), and finally promotes the expression of human protein (HN) in lung cancer. C, Translating proteins. For example, the pre‐mRNA of F‐box and WD repeat domain containing 7 (FBXW7) may produce a circRNA called circFBXW7 and FBXW7α mRNA. The open reading frame (ORF) in circFBXW7, which is driven by the internal ribosome entry site, encodes a protein, FBXW7‐185aa. FBXW7‐185aa can bind with ubiquitin‐specific peptidase 28 (USP28) to prevent USP28 binding with FBXW7α mRNA, thereby reducing the half‐life of c‐Myc and reducing the stability of c‐Myc. D, Regulating the transcription of linear RNA. For example, the pre‐mRNA of zinc finger protein with KRAB and SCAN domains 1 (ZKSCAN1) may produce a circRNA called circZKSCAN1 that acts as a competitive inhibitor to retain endogenous RNA and then regulates the expression of cell proliferation and apoptosis‐related genes, including apoptotic genes RAS‐associated C3 botulinum toxin substrate 2 (RAC2), ephrin‐A3 (EFNA3), and caspase 3, transforming growth factor beta 1 (TGFB1), integrin beta 4 (ITGB4), and CXC motif chemokine receptor 4 (CXCR4)
5. TUMOR‐SUPPRESSIVE circRNAs AND CANCERS
5.1. Liver cancer
Liver cancer is the sixth most common cancer in the world and the fourth‐leading cause of cancer death. The most common types of liver cancer are HCC and intrahepatic cholangiocarcinoma; there are also other rare types.36
CircADAMTS14, which is downregulated in the HCC cell line, acts as a sponge of miR‐572 to regulate the expression of RCAN1 and thereby inhibit HCC progression.37 It has also been found that circADAMTS13 acts as a tumor suppressor during HCC progression through a functional pathway as a miR‐484 sponge.38 Another study found that circMTO1 acts as a sponge of miR‐9 and then upregulates the expression of P21 and inhibits the progression of HCC.39 Compared with adjacent normal tissues, hsa_circ_0079299 expression is downregulated in HCC tissues, and it inhibits cell proliferation and blocks cell cycle progression in the G2/M phase through the PI3K/AKT/mTOR pathway.40 CircARSP91 is one of the circRNAs downregulated by AR in a double‐stranded RNA‐specific adenosine deaminase 1 (ADAR1)‐dependent method. In fact, AR is thought to play an important role in prostate cancer. Interestingly, circARSP91 inhibits HCC tumor growth via the AR/ADAR1/circARSP91 axis.41 As a tumor suppressor, circC3P1 acts as a sponge of miR‐4641, promotes PCK1 expression, and inhibits migration and invasion of HCC cells in vitro and in vivo.42
5.2. Bladder cancer
Bladder cancer is the tenth most common cancer in the world and the ninth largest cause of cancer death.36
A study found that circFNDC3B is significantly downregulated in BCa tissues and is associated with pathological T staging, grading, lymphatic invasion, and overall patient survival.43 Mechanistically, circFNDC3B binds directly to miR‐1178‐3p, which targets oncogene G3BP stress granule assembly factor 2 (G3BP2) mRNA.43 Another BCa‐associated circRNA, circITCH was found to be reduced in BCa tissues and cell lines, and expression of P21 and phosphatase and tensin homolog (PTEN) was upregulated by sponging of miR‐17 and miR‐224.44 CircITCH inhibits migration and invasion in vitro and tumorigenesis in vivo.44 Li et al found that circHIPK3 inhibited heparanase expression by targeting miR‐558, thereby inhibiting the invasion and metastasis of BCa cells.45 In addition, circRNA‐3 (BCRC‐3) acts as a tumor suppressor to inhibit BCa proliferation via the miR‐182‐5p/P27 axis.46 As the first identified circRNA that has a miRNA sponge role, Cdr1as is significantly downregulated in BCa tissue compared with adjacent normal tissues.47 In vitro and in vivo experiments further found that Cdr1as inhibits proliferation, invasion, and migration of BCa by binding to miR‐135a.47 In another study, circLPAR1 was found to be downregulated in MIBC tissues.48 Patients with low expression levels of circLPAR1 had shorter disease‐specific survival than patients with high expression levels.48 At the same time, it was found that circLPAR1 affected MIBC invasion and metastasis by sponging miR‐762.48
5.3. Gastric cancer
Gastric cancer is the fifth‐leading cause of cancer death.36 Tumor‐suppressive circLARP4 inhibits the development and progression of GC through a regulatory network of the circLARP4/miR‐424/LATS1 axis.28 The expression level of circYAP1 in GC tissues was significantly lower than that in adjacent normal tissues, and the survival time of patients with low expression of circYAP1 was shorter than that of patients with high expression of circYAP1.49 Moreover, circYAP1 acts as a sponge of miR‐367‐5p to inhibit P27Kip 1 expression and inhibits GC cell growth and invasion.49 Our group found that hsa_cric_0000096 inhibits the growth and migration of GC cells through regulating the expression of cyclin D1, cyclin‐dependent kinase 6 (CDK6), MMP‐2, and MMP‐9.50 Acting as a sponge of miR‐214‐5p, hsa_circ_0000993 may be used as a target for the treatment of GC.51 It has also been found that circPSMC3 acted as a sponge of miR‐296‐5p to regulate PTEN expression in GC.52
5.4. Breast cancer
Breast cancer is the second most common cancer and is the most common cancer in women.36 By acting as a sponge of miR‐449a, circ_000911 promotes the expression of notch homolog 1 (Notch1), thereby inhibiting the progression of breast cancer (Figure 3A).53 Another study found that overexpression of hsa_circ_0072309 inhibits miR‐492 activity and thus inhibits breast cancer progression.54 In addition, circASS1 inhibits breast tumorigenesis and progression through the miR‐4443/ASS1 axis.35
5.5. Lung cancer
Lung cancer is the most common cancer in the world. NSCLC accounts for 80% to 85% of all lung cancer cases.36
CircNOL10 (hsa_circ_0000977) affects mitochondrial function by promoting the expression of the HN polypeptide family in lung cancer (Figure 3B).55 Alterations in mitochondrial function trigger a variety of signaling pathways and ultimately inhibit cell proliferation and cell cycle progression and promote apoptosis in lung cancer cells, thereby significantly inhibiting lung cancer progression.55 It was found that circPTK2 inhibited transforming growth factor β‐induced mesenchymal‐epithelial transition (EMT) and cell invasion via the miR‐429/miR‐200b‐3p/TIF1γ axis in NSCLC.56 CircPTK2 overexpression also decreases Snail expression and inhibits the progression of NSCLC.56 Circ_0001649 has a tumor‐suppressive effect by sponging miR‐331‐3p and miR‐338‐5p.57 This means that the circ_0001649/miR‐331‐3p and circ_0001649/miR‐338‐5p regulatory axis may contribute to tumorigenesis and progression of NSCLC. Hsa_circ_100395 is downregulated in lung cancer tissues and cells and acts as a sponge of miR‐1228 to regulate transcription factor 21 (TCF21) expression, thus inhibiting the proliferation activity, migration, and invasion of lung cancer cells.58 Another study found that circ_0006916 bound miR‐522‐3p and then directly targeted the PH domain and leucine‐rich repeat protein phosphatases (PHLPP1).59 Therefore, circ0006916 may be used as a possible therapeutic target for lung cancer.
5.6. Glioma
Glioma is one of the deadliest tumors, and two‐thirds of patients with glioma specifically have GBM.36 The mortality rate of glioma is very high.
Circ_0001946 promotes the expression of CDR1 by inhibiting miR‐671‐5p and inhibits the malignant proliferation of GBM cells.60 CircSHPRH, which translates to SHPRH146aa using an overlapping genetic code, is used as a protective “bait” for SHPRH to prolong the half‐life of the relevant full‐length SHPRH, thereby reducing the malignant proliferation and phenotype of glioma.31 In another similar study, cross‐linked ORF in circFBXW7, driven by the IRES, was found to encode a new 21‐kDa protein called FBXW7‐185aa.61 FBXW7‐185aa can bind with USP28 to prevent USP28 binding with FBXW7α mRNA, thereby reducing the half‐life of c‐Myc and inhibiting proliferation and cell cycle progression, significantly inhibiting glioma progression (Figure 3C).61
5.7. Other cancers
Many studies have found that circRNAs play an important role in the development of other tumors. CircITGA7 plays a role in the regulation of NF1 translation by competitive binding to miR‐370‐3p.62 CircITGA7 inhibits the Ras signalling pathway, thus affecting the progression of CRC.62 In addition, hsa_circ_0014717 inhibits CRC growth, possibly by upregulating P16 expression.63 By regulating miR‐136‐5p/ATXN1 and miR‐382‐5p/ATXN1 networks, hsa_circ_0008309 regulates cell proliferation and EMT in various cancers.64 In addition, circ004389 may be a target of histone H3 and BMI1 proto‐oncogene in esophageal cancer. It inhibits cell proliferation, migration, and invasion and induces cell death.65
6. TUMOR‐SUPPRESSIVE CIRCRNAS MAY BE USED AS TUMOR BIOMARKERS
The following characteristics of circRNAs indicate that they may be used as potential tumor biomarkers. (i) Stability: circRNAs are resistant to RNase R.10 (ii) Specificity: circRNAs are expressed in a tissue‐specific and developmental stage‐specific way. In particular, many studies have shown that circRNAs are distinctively expressed between cancerous and noncancerous tissues.26 (iii) Universality: circRNAs are considered to be the most widely distributed molecules in human cells. (iv) Conservatism: circRNAs are evolutionally conserved in different species.
In GC, our group found that there were differences in the expression of hsa_circ_002059 between GC tissues and nontumor tissues and pairs of plasma samples before and after surgery.66 In another study, it was found that, compared with non‐tumor tissue, hsa_circ_0001649 was significantly downregulated in GC tissues, and its levels in serum samples of postoperative GC patients were significantly higher than those from preoperative patients.67 These results suggest that hsa_circ_002059 and hsa_circ_0001649 may be used as new biomarkers for GC.
Another study found that the expression of circFBXW7 was positively correlated with OS of patients with GBM.61 OS of the group with higher expression of circFBXW7 was approximately 12.5 months longer than that of the group with low circFBXW7 expression.61 Thus, circ‐FBXW7 may be a potential prognostic biomarker of GBM.
Expression levels of circITCH were positively correlated with histological grade in BCa but were not related to age, tumor lymph node metastasis, or tumor size.44 Thus, circITCH may be used as a valuable biomarker for the detecting prognosis of BCa and ovarian cancer.
7. CONCLUSION AND FURTHER PERSPECTIVES
Circular RNAs play a crucial role in the development of various tumors. One of the mechanisms underlying tumor‐suppressive circRNAs in cancer is that they act as sponges of miRNAs through the circRNA‐miRNA‐mRNA regulatory network. Tumor‐suppressive circRNAs can also interact with proteins to affect their biological functions. In addition, tumor‐suppressive circRNAs also function to regulate the transcription of linear RNAs.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article. The funders had no role in study design, data collection, data analysis, interpretation, or writing of the report.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (no. 81772279), the Scientific Innovation Team Project of Ningbo (no. 2017C110019), and the KC Wong Magna Fund in Ningbo University.
Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630–3638. 10.1111/cas.14211
REFERENCES
- 1. Mo X, Wu Y, Chen L, et al. Global expression profiling of metabolic pathway‐related lncRNAs in human gastric cancer and the identification of RP11‐555H23.1 as a new diagnostic biomarker. J Clin Lab Anal. 2019;33:e22692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT‐PCR detection. J Mol Med (Berl). 2018;96:85‐96. [DOI] [PubMed] [Google Scholar]
- 3. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci USA. 1976;73(11):3852‐3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11‐20. [DOI] [PubMed] [Google Scholar]
- 5. Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32:e22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32:e22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427:2414‐2417. [DOI] [PubMed] [Google Scholar]
- 9. Aucamp J, Bronkhorst AJ, Pretorious A. A historical and evolutionary perspective on circulating nucleic acids and extracellular vesicles: Circulating nucleic acids as homeostatic genetic entities. Adv Exp Med Biol. 2016;924:91‐95. [DOI] [PubMed] [Google Scholar]
- 10. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256‐264. [DOI] [PubMed] [Google Scholar]
- 11. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(9–21):e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)‐methyladenosine. Cell Res. 2017;27:626‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Heesch S, Witte F, Schneider‐Lunitz V, et al. The translational landscape of the human heart. Cell. 2019;178(242–260):e29. [DOI] [PubMed] [Google Scholar]
- 15. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205‐211. [DOI] [PubMed] [Google Scholar]
- 16. Chen YG, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67(228–238):e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dong R, Ma XK, Chen LL, Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu T, Cui L, Zhou Y, et al. Transcriptome‐wide investigation of circular RNAs in rice. RNA. 2015;21:2076‐2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X, Liu C‐X, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67(2):214‐227.e7. [DOI] [PubMed] [Google Scholar]
- 20. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. CircRNA biogenesis competes with pre‐mRNA splicing. Mol Cell. 2014;56:55‐66. [DOI] [PubMed] [Google Scholar]
- 21. Zhang X‐O, Wang H‐B, et al. Complementary sequence‐mediated exon circularization. Cell. 2014;159:134‐147. [DOI] [PubMed] [Google Scholar]
- 22. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428‐442. [DOI] [PubMed] [Google Scholar]
- 24. Hou LD, Zhang J. Circular RNAs: An emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 2018;7:3101‐3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR‐424‐5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Legnini I, Di Timoteo G, Rossi F, et al. Circ‐ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22‐37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805‐1814. [DOI] [PubMed] [Google Scholar]
- 32. Fang J, Hong H, Xue X, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR‐548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222‐232. [DOI] [PubMed] [Google Scholar]
- 33. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214‐1227. [DOI] [PubMed] [Google Scholar]
- 35. Hou JC, Xu Z, Zhong SL, et al. Circular RNA circASS1 is downregulated in breast cancer cells MDA‐MB‐231 and suppressed invasion and migration. Epigenomics. 2019;11:199‐213. [DOI] [PubMed] [Google Scholar]
- 36. Bray F, Ferla y J, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 37. Song C, Li D, Liu H, et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA‐572/regulator of calcineurin 1. J Cell Physiol. 2019;234:2460‐2470. [DOI] [PubMed] [Google Scholar]
- 38. Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR‐484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13:441‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Han D, Li J, Wang H, et al. Circular RNA MTO1 acts as the sponge of miR‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151. [DOI] [PubMed] [Google Scholar]
- 40. Zheng H, Chen T, Li C, et al. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag Res. 2019;11:443‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi L, Yan P, Liang Y, et al. Circular RNA expression is suppressed by androgen receptor (AR)‐regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR‐4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499:1044‐1049. [DOI] [PubMed] [Google Scholar]
- 43. Liu H, Bi J, Dong W, et al. Invasion‐related circular RNA circFNDC3B inhibits bladder cancer progression through the miR‐1178‐3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17(1):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang C, Yuan W, Yang X, et al. Circular RNA circ‐ITCH inhibits bladder cancer progression by sponging miR‐17/miR‐224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR‐558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646‐1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie F, Li Y, Wang M, et al. Circular RNA BCRC‐3 suppresses bladder cancer proliferation through miR‐182‐5p/p27 axis. Mol Cancer. 2018;17:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li P, Yang X, Yuan W, et al. CircRNA‐Cdr1as exerts anti‐oncogenic functions in bladder cancer by sponging microRNA‐135a. Cell Physiol Biochem. 2018;46:1606‐1616. [DOI] [PubMed] [Google Scholar]
- 48. Lin G, Sheng H, Xie H, et al. CircLPAR1 is a novel biomarker of prognosis for muscle‐invasive bladder cancer with invasion and metastasis by miR‐762. Oncol Lett. 2019;17:3537‐3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu H, Liu Y, Bian Z, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR‐367‐5p/p27 (Kip1) axis. Mol Cancer. 2018;17:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhong S, Wang J, Hou J, et al. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics. 2018;10:1301‐1313. [DOI] [PubMed] [Google Scholar]
- 52. Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR‐296‐5p. Mol Cancer. 2019;18:25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA‐000911/miR‐449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR‐492. Cancer Manag Res. 2019;11:1033‐1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nan A, Chen L, Zhang N, et al. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1‐mediated transcriptional regulation of the humanin polypeptide family. Adv Sci (Weinh). 2019;6:1800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF‐beta‐induced epithelial‐mesenchymal transition and metastasis by controlling TIF1gamma in non‐small cell lung cancer. Mol Cancer. 2018;17:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR‐331‐3p and miR‐338‐5p. Biochem Biophys Res Commun. 2018;503(3):1503‐1509. [DOI] [PubMed] [Google Scholar]
- 58. Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR‐1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17:2080‐2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dai X, Zhang N, Cheng Y, et al. RNA‐binding protein trinucleotide repeat‐containing 6A regulates the formation of circular RNA 0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39:981‐992. [DOI] [PubMed] [Google Scholar]
- 60. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR‐671‐5p and CDR1. J Cell Physiol. 2019;234:13807‐13819. [DOI] [PubMed] [Google Scholar]
- 61. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI‐J Nat Cancer Inst. 2018;110:304‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246:166‐179. [DOI] [PubMed] [Google Scholar]
- 63. Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775‐782. [DOI] [PubMed] [Google Scholar]
- 64. Li B, Wang F, Li X, et al. Hsa_circ_0008309 may be a potential biomarker for oral squamous cell carcinoma. Dis Markers. 2018;2018:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang W, Ma J, Lu J, et al. Circ0043898 acts as a tumor inhibitor and performs regulatory effect on the inhibition of esophageal carcinoma. Cancer Biol Ther. 2018;19(12):1117‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132‐136. [DOI] [PubMed] [Google Scholar]
- 67. Li WH, Song YC, Zhang H, et al. Decreased expression of hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. [DOI] [PMC free article] [PubMed] [Google Scholar]


