Abstract
The digestive system cancers are aggressive cancers with the highest mortality worldwide. In this study, we undertook a systematic investigation of the tumor immune microenvironment to identify diagnostic and prognostic biomarkers. The fraction of 22 immune cell types of patients were estimated using CIBERSORT. The least absolute shrinkage and selection operator (LASSO) analysis was carried out to identify important immune predictors. By comparing immune cell compositions in 801 tumor samples and 46 normal samples, we constructed the diagnostic immune score (DIS), showing high specificity and sensitivity in the training (area under the receiver operating characteristic curve [AUC] = 0.929), validation (AUC = 0.935), and different cancer type cohorts (AUC > 0.70 for all). We also established the prognostic immune score (PIS), which was an effective prognostic factor for relapse‐free survival in training, validation, and entire cohorts (P < .05). In addition, PIS provided a higher net benefit than TNM stage. A composite nomogram was built based on PIS and patients' clinical information with well‐fitted calibration curves (c‐index = 0.84). We further used other cohorts from Gene Expression Omnibus databases and obtained similar results, confirming the reliability and validity of the DIS and PIS. In addition, the unsupervised clustering analysis using immune cell proportions revealed 6 immune subtypes, suggesting that the immune types defined as having relatively high levels of M0 or/and M1 macrophages were the high‐risk subtypes of relapse. In conclusion, this study comprehensively analyzed the tumor immune microenvironment and identified DIS and PIS for digestive system cancers.
Keywords: diagnosis, digestive system cancer, immune cell, prognosis, TCGA
Our study comprehensively analyzed the utility of consideration of immune cells in the diagnosis and prognosis of digestive system cancers. The diagnostic immune score and prognostic immune score signature might serve as biomarkers for early diagnosis and predicting survival.

Abbreviations
- CIBERSORT
cell type identification by estimating relative subsets of RNA transcripts
- c‐index
Harrell's concordance index
- COAD
colon adenocarcinoma
- DIS
diagnostic immune score
- DSC
digestive system cancer
- ESCA
esophageal squamous cell carcinoma
- GEO
Gene Expression Omnibus
- LASSO
least absolute shrinkage and selection operator
- LIHC
liver hepatocellular carcinoma
- PAAD
pancreatic adenocarcinoma
- PIS
prognostic immune score
- READ
rectum adenocarcinoma
- RFS
relapse‐free survival
- ROC
receiver operating characteristic
- STAD
stomach adenocarcinoma
- TCGA
The Cancer Genome Atlas
1. INTRODUCTION
The digestive system consists of digestive tract and digestive accessory organs. The DSCs, a worldwide health problem, share many characteristics, suggesting common etiological pathways or mechanisms.1 The DSCs are the leading cause of cancer‐related death worldwide. Among all the cancer types, colon cancer, stomach cancer, liver cancer, rectum cancer, and esophageal cancer are the top 10 most commonly diagnosed cancers and the top 10 most lethal types2 because of difficulty of early stage diagnosis and limitations of current therapeutic methods. Therefore, it is critical to find effective biomarkers for prompt diagnosis and prognosis.
Increasing studies suggest the importance of the immune microenvironment on DSC development,3, 4, 5, 6 which could provide potential biomarkers to improve the reliability and precision of diagnosis and prognosis. However, because of the complex and dynamic nature of the immune microenvironment, our understanding of its role remains incomplete. Tumor‐infiltrating immune cells are a part of the complex microenvironment. They play a key role in inhibiting or supporting the growth and development of tumors, which can be effectively targeted by drugs and are related to the survival time of patients.7, 8 Although the immune microenvironment has been recently analyzed in pan‐cancer or specific cancers,8, 9 no studies have been carried out to provide the comprehensive immune profile specifically for DSCs.
A new algorithm for enumeration of immune cell subsets,10 CIBERSORT, provides the possibility to identify immune biomarkers for diagnosis and prognosis. CIBERSORT contains 547 genes and is highly sensitive and specific to 22 phenotypes of human immune cells, which can properly determine the diversity and landscape of tumor‐infiltrating immune cells. Because of the superior performance of CIBERSORT, increasing attention has been aroused in many studies on the tumor microenvironment.11, 12
In the present study, we used CIBERSORT to estimate the proportions of 22 immune cells based on the gene expression profiling of DSC patients. Then we applied LASSO analysis to identify important immune predictors and build diagnostic and prognostic immune signature, which can provide a powerful means of early detection and predicting survival.
2. METHODS AND MATERIALS
2.1. Patients and datasets
This study used data from the public domain. The cohort of DSCs for identifying the immune signature consists of 1874 patients in TCGA. Six major cancer types were included: ESCA, STAD, LIHC, PAAD, COAD, and READ. Level 3 data of gene expression profiles of patients were downloaded from TCGA database (https://tcga-data.nci.nih.gov/tcga/), a large‐scale public data platform portal, measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform (October 13, 2017). At the same time, patients' clinical characteristics were also obtained from TCGA data portal, including age, gender, survival time, outcome, and TNM stage. The data were extracted from TCGA database, strictly following the publication guidelines approved by TCGA. Therefore, there was no requirement for ethics committee approval.
2.2. Estimation of immune cell type fractions
To quantify the proportions of immune cells, the CIBERSORT algorithm was used. The normalized gene expression data were uploaded to the CIBERSORT web portal, running with the LM22 signature and 1000 permutations.10 LM22, 22 infiltrating immune cells, includes B cells, T cells, natural killer cells, macrophages, dendritic cells, and myeloid subsets. CIBERSORT derives a P value for the deconvolution of each sample using Monte Carlo sampling, providing a measure of confidence in the results.11 The CIBERSORT results of samples with P < .05 indicate that the inferred fractions of immune cell populations produced by CIBERSORT are accurate. Therefore, only patients with CIBERSORT P < .05 were considered eligible for further analysis. For each sample, the final CIBERSORT output estimates were normalized and immune cell type fractions summed up to one.
2.3. Diagnostic analysis
According to the inclusion criteria: (i) patients' CIBERSORT results were P < .05; and (ii) samples were primary or normal tissues, eligible samples were selected for further diagnostic analysis. Subsequently, the patients were randomly separated into training and validation cohorts (3:1) using the “Sample function” in R software. Then LASSO analysis, as a statistical method, was carried out to identify the most important immune cells, whose predictive accuracy could be improved significantly. The DIS was built based on the coefficients of LASSO‐logistic analysis in the training cohort, which could differentiate tumor and normal tissues. In addition, the sensitivity and specificity of the diagnostic models were analyzed by ROC curves.
2.4. Prognostic analysis
The primary endpoint for prognostic analysis in the study was RFS, defined as the time from the date of the first diagnosis to the date of the earliest recurrence. According to the inclusion criteria: (i) patients' CIBERSORT results were P < .05; (ii) samples were primary tissues; and (iii) patients' RFS information was complete. Subsequently, the eligible patients were randomly separated into training and validation cohorts (3:1). In addition, the optimal cut‐off values of each immune cell type were evaluated based on the patients' RFS and cell fraction using the “survminer” package. The LASSO‐Cox analysis was then used to obtain the most useful predictive features from the training cohort. Similarly, the PIS was built based on the corresponding coefficients. To evaluate the distinguishing ability of patients' outcome, the Kaplan‐Meier survival curves were used, and time‐dependent ROC (survival ROC) curves were applied to assess the PIS prognostic power. Moreover, the clinical usefulness of PIS was determined by quantifying the net benefits under different threshold probabilities using decision curve analysis.13 Finally, to provide a quantitative tool for predicting the individual probability of patients' prognosis, we built a prognostic nomogram on the basis of PIS and patients' clinical information. Calibration curves were plotted to compare the expected and observed survival probabilities. The discrimination of the TNM stage, PIS, and prognostic nomogram was measured and compared by the c‐index.
2.5. Validation of DIS and PIS using GEO datasets
To confirm the reliability and validity of the DIS/PIS, we used other cohorts from GEO databases according to the following inclusion criteria: (i) diagnosis of patients with DSCs; (ii) mRNA expression level in tissue samples; (iii) for DIS validation, the dataset provided tumor and normal samples; and (iv) for PIS validation, the dataset provided patients' survival information. The exclusion criteria were: (i) datasets with small sample size (n < 50); and (ii) datasets that used cell line or animal samples. Therefore, we selected http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23400, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13195, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22058, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62452, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90627, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53625, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26253, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76427, and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38832 to validate the results from TCGA database.
2.6. Discovery of immune subtypes
To investigate possible associations between distinct classes of immune cell infiltration and patient prognosis, we carried out unsupervised clustering of the infiltrating immune cells, dividing patients into different immune subtypes. According to patients' clinical information, the survival analysis of immune subtypes was also carried out. Combining PIS distribution and survival analysis, high‐risk immune subtypes were identified.
2.7. Statistical analysis
All data were expressed as mean ± SD. Group comparisons were undertaken for continuous variables using Student's t test or one‐way ANOVA. The LASSO analysis was undertaken using the “glmnet” package. Survival ROC was plotted using the “survivalROC” package. Decision curve analysis was carried out with the “rmda” package. Nomograms and calibration plots were undertaken with the “rms” package. Survival analyses and c‐index calculations were carried out using the “survival” package. Unsupervised clustering analysis was carried out using the “cluster” package. The above analyses were undertaken using R software 3.5 and SPSS software 23.0, and all statistical tests were 2‐sided. P < .05 was considered statistically significant.
3. RESULTS
3.1. Patient characteristics
According to the inclusion criteria, 801 patients diagnosed with DSC from TCGA datasets (99 ESCA samples, 225 STAD samples, 57 LIHC samples, 122 PAAD samples, 229 COAD samples, and 69 READ samples) were analyzed in this study. Patients were 64.65 ± 12.86 years old, with 481 men (60.05%) and 320 (39.95%) women. Detailed patient characteristics are given in Table S1. The patient selection scheme and analytic flowcharts of the study are shown in Figure S1.
3.2. Composition of immune cells in DSCs
Figure S2 provides a summary of the immune cell composition within and across clinical subgroups of DSCs. In general, the 5 most common immune cell fractions in DSCs were M0 macrophages, M2 macrophages, CD4 memory resting T cells, CD8 T cells, and M1 macrophages, whose sum of mean proportions was more than 55% in all clinical subgroups.
However, the distribution of immune cell fractions in normal tissues was different from that in tumor tissues, showing that plasma cells, CD4 T memory resting cells, M2 macrophages, CD8 T cells, and resting mast cells were the five most common cells. In addition, the fractions of M0 and M1 macrophages, activated mast cells, CD4 memory activated T cells, and resting natural killer cells were significantly higher in the tumor tissues than those of the normal tissues, and the fractions of plasma cells, resting mast cells, CD4 memory resting T cells, monocytes, naive and memory B cells, and resting dendritic cells were significantly lower in the tumor tissues than those of normal tissues (Figure 1A).
Figure 1.
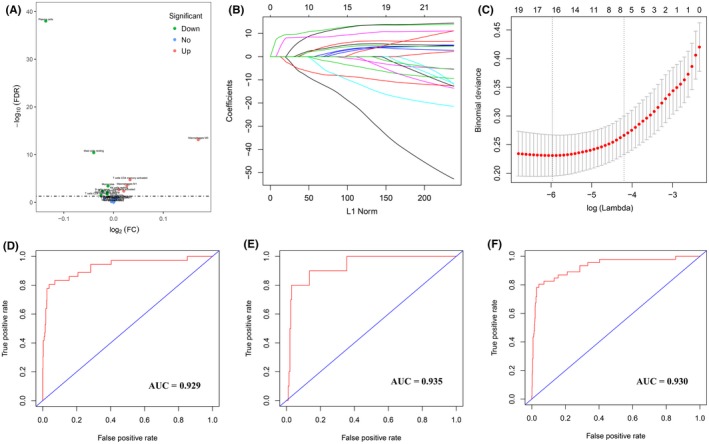
Construction and validation of the diagnostic immune score (DIS) in patients with digestive system cancer. A, Volcano plot visualizing the differentially infiltrated immune cells between tumor tissues and normal tissues. Red and green plots represent differentially statistical significance (P < .05). FC, Fold Change; FDR, False Discovery Rate. B, Least absolute shrinkage and selection operator (LASSO) coefficient profiles of the fractions of 22 immune cell types. C, Tenfold cross‐validation for tuning parameter selection in the LASSO model. D‐F, Receiver operating characteristic (ROC) curves of DIS in the training (D), validation (E), and entire (F) cohorts. AUC, area under ROC curve
3.3. Diagnostic signature building
We separated all samples (801 tumor samples and 46 normal samples) into training and validation cohorts. Six potential predictors in the training cohort were identified and were features with nonzero coefficients in the LASSO‐logistic regression model (Figure 1B,1, Table S2). We calculated DIS as the diagnostic signature as the following: DIS = (fraction level of naive B cells × 0.520) + (fraction level of plasma cells × 9.710) + (fraction level of CD4 memory activated T cells × −5.601) + (fraction level of monocytes × 5.640) + (fraction level of M0 macrophages × −4.857) + (fraction level of resting mast cells × 8.246). Distribution of the DIS in tumor and normal tissues is given in Figure S3. Moreover, we evaluated the ability of DIS in differentiating between tumor and normal tissues, suggesting that DIS had a high accuracy of prediction (AUC = 0.929, 0.935, and 0.930 in training, validation, and entire cohorts, respectively; Figure 1D‐F). Similarly, DIS showed high sensitivity and specificity for differentiating tumors from normal tissues in all cancer subtypes (Figure S4).
3.4. Prognostic signature building
According to the inclusion criteria for prognostic analysis, 644 patients were included in the present study. These patients were randomly divided into training and validation cohorts, and optimal cut‐off values for the fraction of each immune cell were generated in the training cohort (Table S3). Then LASSO‐Cox analysis was undertaken to identify key prognostic markers, and PIS was built in the training cohort (Figure 2A,2). The formula for the model was based on the coefficients of immune cells (Table S4): PIS = (naive B cells × −0.207) + (CD8 T cells × −0.008) + (CD4 naive T cells × 0.195) + (CD4 memory activated T cells × −0.221) + (follicular helper T cells × −0.083) + (regulatory T cells × 0.055) + (M2 macrophages × 0.272) + (resting mast cells × 0.045) + (eosinophils × −0.101). In this formula, the immune cell fraction level was valued as 0 or 1. A value of 1 was assigned when the fraction of one type of cell was over the corresponding cut‐off value and a value of 0 otherwise.
Figure 2.
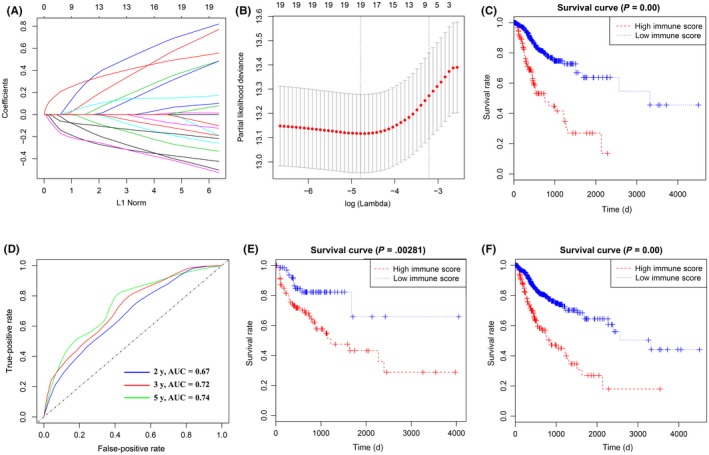
Construction of prognostic immune score (PIS) in patients with digestive system cancer. A, Least absolute shrinkage and selection operator (LASSO) coefficient profiles of the fractions of 22 immune cell types. B, Tenfold cross‐validation for tuning parameter selection in the LASSO model. C, Kaplan‐Meier curves for relapse‐free survival by PIS group in the training cohorts. D, PIS measured by survival receiver operating characteristic (ROC) curves in the training cohort. The area under the ROC curve (AUC) was 0.67, 0.72, and 0.74 at 2, 3, and 5 y, respectively. E, F, Kaplan‐Meier curves for relapse‐free survival by PIS group in the validation (E) and entire cohorts (F)
Distributions of the PIS in training and validation cohorts were given in Figure S5. In addition, patients in the training cohort were divided into high‐PIS and low‐PIS groups using the cut‐off value (0.059). The Kaplan‐Meier curves were plotted to confirm that the patients with high PIS had a significantly higher risk of relapse in the training cohort (P < .001, Figure 2C). Furthermore, the PIS showed the strong predictive power of 2‐, 3‐, and 5‐year survival in the training cohort (AUC = 0.67, 0.72, and 0.74, respectively; Figure 2D).
3.5. Validation of PIS in validation and entire cohorts
To confirm that the PIS has similar prognostic value in different populations, the same formula was applied in the validation and entire cohorts. The patients were also assigned to high‐PIS or low‐PIS groups using the cut‐off value obtained from the corresponding cohort (validation cohort, 0.019; entire cohort, 0.047). Consistent with the results in the training cohort, patients with high PIS had a significantly lower RFS time than those with low PIS in both the validation cohort (P = .003, Figure 2E) and the entire cohort (P < .001, Figure 2F). Then survival ROC curves were applied to confirm the strong predictive power of PIS (AUC of validation cohort = 0.61, 0.65, and 0.65, respectively; AUC of entire cohort = 0.66, 0.69, and 0.73, respectively; Figure S6), suggesting that the PIS had good sensitivity and specificity in predicting patients' 2‐, 3‐, and 5‐year RFS. In addition, the Kaplan‐Meier curves suggested that patients with high PIS had higher risk in different cancer types (Figure S7).
3.6. Clinical use of prognostic signature
To examine the association of the PIS with clinical features, the distribution of PIS in clinical characteristics and cancer subtypes were assessed (Table S5). Moreover, univariate and multivariate Cox regression analyses were undertaken in the training, validation, and entire cohorts. The multivariate Cox regression model showed that PIS could become a potential independent prognostic indicator in the training (P = .005), validation (P = .03), and entire cohorts (P = .003) (Table 1). The results of the univariate Cox analysis are shown in Table S6.
Table 1.
Multivariable Cox regression analysis of prognosis immune score and patients' characteristics in different cohorts of patients with digestive system cancer
| Training cohort | Validation cohort | Entire cohort | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | P value | Hazard ratio | P value | Hazard ratio | P value | |
| Age | 0.99 (0.97‐1.01) | .230 | 1.01 (0.98‐1.03) | .620 | 0.99 (0.98‐1.01) | .611 |
| Gender | ||||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| Male | 1.27 (0.82‐1.96) | .290 | 1.72 (0.83‐3.54) | .140 | 1.28 (0.89‐1.84) | .180 |
| Tumor stage | ||||||
| I | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| II | 1.30 (0.67‐2.56) | .440 | 1.89 (0.60‐5.97) | .280 | 1.44 (0.82‐2.53) | .210 |
| III | 1.20 (0.57‐2.53) | .630 | 2.52 (0.72‐8.83) | .150 | 1.48 (0.79‐2.75) | .220 |
| IV | 1.38 (0.58‐3.27) | .470 | 0.66 (0.13‐3.34) | .610 | 1.13 (0.53‐2.41) | .750 |
| Cancer status | ||||||
| Tumor‐free | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| With tumor | 12.42 (7.35‐21.01) | <.001 | 20.64 (8.42‐50.58) | <.001 | 13.57 (8.71‐21.14) | <.001 |
| Residual tumor | ||||||
| R0 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| R1 + R2 | 0.68 (0.39‐1.17) | .160 | 0.72 (0.25‐2.12) | .550 | 0.70 (0.43‐1.128) | .140 |
| PIS | 3.89 (1.50‐10.08) | .005 | 2.35 (1.05‐5.26) | .030 | 3.39 (1.51‐7.59) | .003 |
The decision curve analysis for PIS and TNM stage is presented in Figure 3. The decision curve showed that using the PIS to predict patients' RFS added more benefit than TNM stages. In addition, the value of PIS c‐index was higher than that of the TNM stage, indicating that PIS was a better predictor for patients (Table 2). A similar tendency was also obtained in both the validation and entire cohorts (Table 2). All the above results suggested that the PIS has good predictive ability in clinical use.
Figure 3.
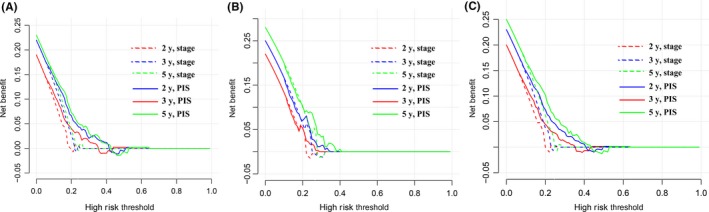
Decision curve analyses of the prognostic immune score (PIS) and TNM stage for 2‐, 3‐, and 5‐y risk in the training (A), validation (B), and entire (C) cohorts of patients with digestive system cancer
Table 2.
Harrell's concordance indexes of prognosis immune score (PIS), stage, and nomogram in different cohorts of patients with digestive system cancer
| Cohort | PIS | TNM stage | Nomogram |
|---|---|---|---|
| Training | 0.69 (0.64‐0.74) | 0.53 (0.48‐0.58) | 0.84 (0.81‐0.86) |
| Validation | 0.61 (0.57‐0.70) | 0.54 (0.50‐0.58) | 0.83 (0.79‐0.86) |
| Entire | 0.67 (0.63‐0.71) | 0.53 (0.51‐0.55) | 0.82 (0.79‐0.86) |
3.7. Nomogram construction
To provide a quantitative tool to predict the individual probability of relapse, we constructed the prognostic nomogram on the basis of PIS and clinical information using the training cohort (Figure 4A). The calibration curve of the prognostic nomogram showed good agreement between prediction and observation in the training cohort (Figure 4B). Good calibration was also observed for the probability of relapse in the validation and entire cohorts (Figure 4C,4). The c‐index for the prognostic nomogram was 0.84, 0.83, and 0.82 in the training, validation, and entire cohorts, which improved the prognostic accuracy compared with that of the TNM stage and PIS (Table 2).
Figure 4.
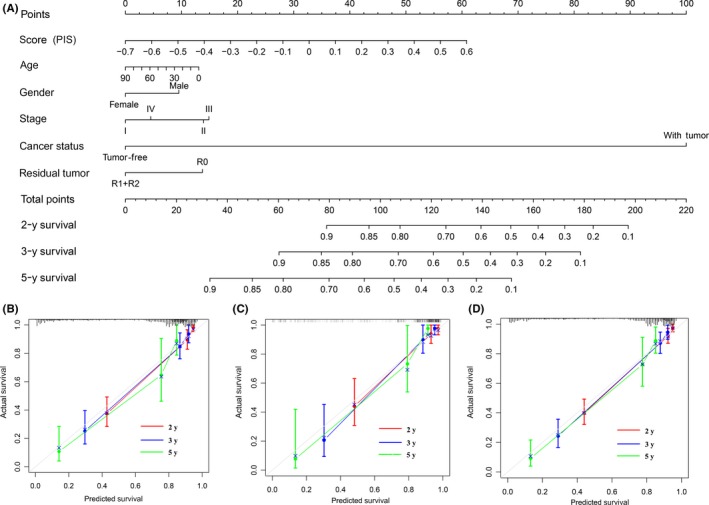
Construction and validation of nomogram in patients with digestive system cancer. A, Nomogram for predicting 2‐, 3‐, and 5‐y relapse‐free survival for patients in the training cohort. PIS, prognostic immune score. B‐D, Calibration curves of nomograms in terms of agreement between predicted and observed 2‐, 3‐, and 5‐y outcomes in the training (B), validation (C), and entire (D) cohorts. Dashed line at 45° represents perfect prediction, and the actual performances of our nomogram are red, blue, and green lines
3.8. Validation of DIS and PIS using GEO datasets
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23400 (ESCA), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13195 (STAD), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22058 (LIHC), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62452 (PAAD), and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90627 (COAD and READ) were obtained from the GEO database to validate the DIS. We evaluated the ability of DIS to differentiate between tumor and normal tissues using these datasets, showing a high accuracy for diagnosis (AUC = 0.871, 0.867, 0.812, 0.751, and 0.978, respectively; Table S7).
Additionally, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53625 (ESCA), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26253 (STAD), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76427 (LIHC), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62452 (PAAD), and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38832 (COAD and READ) were used to validate the PIS. Consistent with the results in TCGA database, DSC patients with high PIS had poor survival compared to those with low PIS (hazard ratio = 1.52, P < .001; Table S8).
3.9. Immune subtypes
To discern distinct patterns of immune infiltration, we undertook unsupervised clustering using 644 patients based on the immune cell proportions. The optimal number of clusters was 6 (Figure S8) and 6 immune subtypes were identified. The relationship between cancer types and the immune subtype re shown in Table S9. The ESCA patients were evenly distributed among the 6 immune subtypes, most STAD patients were in immune subtypes 1, and LIHC patients were mainly immune subtypes 1 and 6. More than half of PAAD patients were immune subtype 6. Most COAD patients were immune subtype 3 and a few COAD patients were immune subtype 2, which was the same as the distribution of READ patients. The cell proportions of each immune subtype are shown in Figure 5A. Among them, immune subtype 1 was defined by high levels of CD8 T cells, immune subtype 2 was defined by high levels of naive B cells, and immune subtype 3 was defined by moderate levels of M0 macrophages. Immune subtype 4 was defined by moderate levels of CD4 memory resting T cells and M0 macrophages, immune subtype 5 was defined by more than 50% M0 macrophages, and immune subtype 6 was defined by a relatively high level of M2 macrophages. In addition, immune subtypes were associated with distinct patterns of survival (2 = 15.55, P = .008, Figure 5B). Immune subtypes 5 and 6 were both associated with poor RFS. Moreover, the distribution of PIS in different immune subtypes was assessed, showing that subtypes 5 and 6 had higher PIS than others (Figure 5C, Table S5). Combining the PIS distribution and survival analyses, immune subtypes 5 and 6 were the high‐risk subtypes of relapse.
Figure 5.
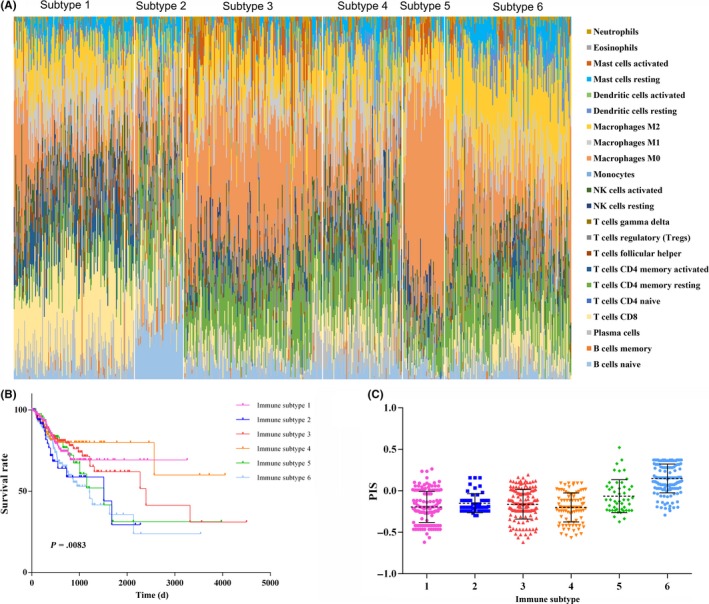
Immune subtypes in patients with digestive system cancer. A, Unsupervised clustering of all samples based on immune cell proportions. Stacked bar charts of samples ordered by cluster assignment. NK, natural killer. B, Survival analysis of patients within different immune subtypes. C, Prognostic immune score (PIS) in different immune subtypes
4. DISCUSSION
Digestive system cancers are the most prevalent cancers with the highest mortality worldwide. The current therapeutic methods, including surgery, radiotherapy, and immunotherapy, are constantly improving. However, because of hidden early symptoms, quick development, and invasiveness, the average survival time of patients with advanced DSC remains very low. Therefore, researchers are committed to discoveries of new signatures for diagnosis or prognosis. Huang et al14 used clinicopathologic risk factors to build the radiomics signature and radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer, which can conveniently improve the preoperative individualized prediction. Zhang et al15 used high‐throughput microRNA data in TCGA database to obtain a 5‐microRNA signature for predicting survival time of gastric cancer patients. A 6‐gene signature was identified by using univariate Cox regression analysis and the LASSO‐Cox regression model to predict overall survival for hepatocellular carcinoma.16 In recent years, studies on the tumor immune microenvironment have been in the leading position in cancer research.17, 18 In the present study, we explored the tumor immune microenvironment to find a more effective and precise signature for DSCs.
First, we applied the newly developed algorithm CIBERSORT to estimate the fractions of immune cells. Because the distribution of immune cell fractions in normal tissues was significantly different from that in tumor tissues, we established the DIS. The high AUC value indicated that DIS was an effective predictor of diagnosis, indicating that the immune system participated in tumor carcinogenesis. Similarly, Zhou et al19 established a diagnostic immune risk score for the diagnosis of colon cancer. This method opens the door to new diagnostic strategies from the perspective of immune inflammation.
Recurrence after primary resection is one of the important factors affecting the overall survival rate of patients, suggesting that it is essential to assess the relapse risk accurately for improving prognosis. Several immune score models based on immunohistochemistry have been built to quantify the immune structure and to provide a strong parameter for prognosis in patients with various tumors, including DSCs.20, 21 However, immunohistochemistry was limited by small sample size and few cell types. In the present study, candidate immune cells used to construct PIS were estimated based on high‐throughput gene expression generated by CIBERSORT. Then, we used the LASSO‐Cox analysis to select key immune biomarkers for constructing the PIS. The PIS mainly included naive B cells, T cells, M2 macrophages, resting mast cells, and eosinophils. In the present studies, CD4 naive T cells, regulatory T cells, M2 macrophages, and resting mast cells were associated with poor prognosis. In contrast, naive B cells, CD8 T cells, CD4 memory activated T cells, follicular helper T cells, and eosinophils were the protective factors of patients. T cells are very complex and heterogeneous with constant renewal in vivo, and can exist at different developmental stages or functional subgroups at the same time. In the immune response, different subsets of T cells play different roles, such as releasing lymphokines, killing target cells, assisting immune response, and memory‐specific antigen stimulation.22 Naive B cells represent one of the differentiation stages of B cells, which is one of the mechanisms of autoimmune tolerance. Similar to our results, Zhang et al23 also found that naive B cells were associated with superior survival of LIHC patients. Eosinophils are components of the immune microenvironment that modulate tumor initiation and progression,24 playing an antitumorigenic role in gastric, colorectal, and many other cancers.25, 26, 27 The results showed the relapse risk of patients with high PIS was higher than that of patients with low PIS. In addition, the survival ROC and c‐index analyses confirmed the powerful prediction ability of PIS for prognosis. To determine its clinical usefulness, we further assessed whether the PIS would improve patient outcomes. The decision curve analysis showed that PIS could add more benefit than TNM stages.28
To improve the accuracy of predicting prognosis, we recommended that the nomogram integrate PIS, age, gender, TNM stage, cancer status, and residual tumor, with satisfactory discrimination achieved (c‐index > 0.8). The nomogram took into account markers from different aspects, including the immune microenvironment, basic characteristics, and clinical information, which could be a promising approach to change clinical management.29 Similarly, Toiyama et al30 combined serum biomarkers with clinical risk factors in predictive models, improving the predictive accuracy of colon cancer. In addition, the calibration curve showed that the prognostic‐immune‐score could predict the clinical consequences of patients.
The effect of the tumor immune microenvironment on survival rate has been well reported in many cancer types.31, 32 In our study, we identified 6 immune subtypes by unsupervised clustering based on the abundance of immune cells, and understood the distribution of patients in different immune subtypes. The distribution of the patients with upper digestive tract tumors (ESCA and STAD), digestive gland tumors (PAAD and LIHC), and lower digestive tract tumors (COAD and READ) was similar in different immune subtypes. Different parts of the digestive system perform different basic physiological functions (ingesting, transporting and digesting food, absorbing nutrients, and excreting waste), which could influence the immune cell proportions. The results of analysis were consistent with the actual clinical situation. We also found that immune subtypes with high levels of M0 macrophages or M2 macrophages were the high‐risk subtypes of relapse. Macrophages, as important cells of the immune system, show different genotypes and functions in different microenvironments.33 Pereira et al12 revealed the landscape of glioblastoma, showing that patients can be clustered in different subtypes with M0 and M2 macrophages representing major contributors to the tumor microenvironment. Xiong et al34 found that M2 macrophages were the independent prognostic factors in colorectal cancer by univariate and multivariable Cox regression analysis. Ali's team divided patients with breast cancer into 8 clusters. The cluster defined by high levels of M0 and M2 macrophages and the cluster defined by a high level of M2 macrophages were both associated with poor prognosis.11
Several studies about the immune microenvironment have been reported. The present study differed from recent reports in several important aspects and had its own advantages. First, no studies provided comprehensive immune profiles specifically for DSCs. In a pan‐cancer analysis, Chen et al35 discovered 4 immune types across 14 solid cancer types, including COAD, LIHC, STAD, and other system cancers. In addition, studies of the tumor immune microenvironment on colorectal cancer,36 esophageal cancer,37 pancreatic cancer,38 and gastric cancer39 have been reported. We focused on DSCs that have many similar characteristics and risk factors, systematically analyzing the immune environment and providing predictors of DSCs. Second, we not only comprehensively analyzed the tumor environment, as other studies have done,34, 40 but also used tumor‐infiltrating immune cells to establish diagnosis and prognosis signatures. Third, CIBERSORT discriminates immune cell phenotypes with highly sensitive and specific discrimination, which was the most accurate method. The LASSO‐Cox analysis was further used as a statistical method for screening cell variables to establish DIS and PIS, which could significantly improve the accuracy of prediction.41, 42 Finally, we assessed the reproducibility of DIS and PIS in the validation and entire TCGA cohorts and in other cohorts from GEO databases, which are more rigorous and reliable.
Nevertheless, the study retains a few limitations. First, it was based on TCGA database, and lacked some patients' information. Therefore, some patients with acute infection or immune system disorders were included in the study, which might affect analysis results. Second, some risk factors, including living environment, smoking, drinking, Helicobacter pylori infection, family history, and other factors, were incomplete, which might be more meaningful for diagnosis and prognosis. Finally, the potential bias relating to unbalanced clinicopathological features with treatment heterogeneity could not be ignored, because all samples were from the retrospective collection. Further prospective studies are required to validate the results. In conclusion, our study comprehensively analyzed the utility of consideration of immune cells in the diagnosis and prognosis of DSCs. The DIS and PIS signatures could serve as biomarkers for early diagnosis and predicting survival.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest to report.
ETHICAL APPROVAL
The data were extracted from TCGA database, strictly following the publication guidelines approved by TCGA. Therefore, there was no requirement for ethics committee approval.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study were derived from the following resource: TCGA database, https://www.cancer.gov/tcga.
Supporting information
ACKNOWLEDGEMENTS
The present study was supported by The National Natural Science Foundation of China (Grant Nos. 81673132 and 81972998).
Yang S, Liu T, Cheng Y, Bai Y, Liang G. Immune cell infiltration as a biomarker for the diagnosis and prognosis of digestive system cancer. Cancer Sci. 2019;110:3639–3649. 10.1111/cas.14216
REFERENCES
- 1. Xie WQ, Wang XF. MiR‐146a rs2910164 polymorphism increases the risk of digestive system cancer: a meta‐analysis. Clin Res Hepatol Gastroenterol. 2017;41:93‐102. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Zeng D, Li M, Zhou R, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7:737‐750. [DOI] [PubMed] [Google Scholar]
- 4. Masugi Y, Abe T, Ueno A, et al. Characterization of spatial distribution of tumor‐infiltrating CD8+ T cells refines their prognostic utility for pancreatic cancer survival. Mod Pathol. 2019;32(10):1495‐1507. [DOI] [PubMed] [Google Scholar]
- 5. Xue J, Yu X, Xue L, Ge X, Zhao W, Peng W. Intrinsic beta‐catenin signaling suppresses CD8(+) T‐cell infiltration in colorectal cancer. Biomed Pharmacother. 2019;115:108921. [DOI] [PubMed] [Google Scholar]
- 6. Ahtiainen M, Wirta EV, Kuopio T, et al. Combined prognostic value of CD274 (PD‐L1)/PDCDI (PD‐1) expression and immune cell infiltration in colorectal cancer as per mismatch repair status. Mod Pathol. 2019;32(6):866‐883. [DOI] [PubMed] [Google Scholar]
- 7. Domingues P, Gonzalez‐Tablas M, Otero A, et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun. 2016;53:1‐15. [DOI] [PubMed] [Google Scholar]
- 8. Tamborero D, Rubio‐Perez C, Muinos F, et al. A Pan‐cancer Landscape of Interactions between Solid Tumors and Infiltrating Immune Cell Populations. Clin Cancer Res. 2018;24:3717‐3728. [DOI] [PubMed] [Google Scholar]
- 9. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48:812‐830 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: a gene‐expression‐based retrospective study. PLoS Medicine. 2016;13:e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pereira MB, Barros L, Bracco PA, et al. Transcriptional characterization of immunological infiltrates and their relation with glioblastoma patients overall survival. Oncoimmunology. 2018;7:e1431083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol. 2016;34:2157‐2164. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Dong Y, Hua J, et al. A five‐miRNA signature predicts survival in gastric cancer using bioinformatics analysis. Gene. 2019;699:125‐134. [DOI] [PubMed] [Google Scholar]
- 16. Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of a six‐gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int. 2019;19:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fridman WH, Pages F, Sautes‐Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298‐306. [DOI] [PubMed] [Google Scholar]
- 18. Bhatia S, Oweida A, Lennon S, et al. Inhibition of EphB4‐Ephrin‐B2 signaling reprograms the tumor immune microenvironment in head and neck cancers. Can Res. 2019;79:2722‐2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou R, Zhang J, Zeng D, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I‐III colon cancer. Cancer Immunol Immunother. 2019;68:433‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Lin HC, Huang MY, et al. The Immunoscore system predicts prognosis after liver metastasectomy in colorectal cancer liver metastases. Cancer Immunol Immunother. 2018;67:435‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang Y, Zhang Q, Hu Y, et al. ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg. 2018;267:504‐513. [DOI] [PubMed] [Google Scholar]
- 22. Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16:599‐611. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Z, Ma L, Goswami S, et al. Landscape of infiltrating B cells and their clinical significance in human hepatocellular carcinoma. Oncoimmunology. 2019;8:e1571388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varricchi G, Galdiero MR, Loffredo S, et al. Eosinophils: The unsung heroes in cancer? Oncoimmunology. 2018;7:e1393134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain M, Kasetty S, Sudheendra US, Tijare M, Khan S, Desai A. Assessment of tissue eosinophilia as a prognosticator in oral epithelial dysplasia and oral squamous cell carcinoma‐an image analysis study. Pathol Res Int. 2014;2014:507512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harbaum L, Pollheimer MJ, Kornprat P, Lindtner RA, Bokemeyer C, Langner C. Peritumoral eosinophils predict recurrence in colorectal cancer. Mod Pathol. 2015;28(3):403‐413. [DOI] [PubMed] [Google Scholar]
- 27. Cuschieri A, Talbot IC, Weeden S, Influence of pathological tumour variables on long‐term survival in resectable gastric cancer. Br J Cancer. 2002;86:674‐679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thirunavukarasu P, Talati C, Munjal S, Attwood K, Edge SB, Francescutti V. Effect of incorporation of pretreatment serum carcinoembryonic antigen levels into AJCC staging for colon cancer on 5‐year survival. JAMA Surgery. 2015;150:747‐755. [DOI] [PubMed] [Google Scholar]
- 29. Birkhahn M, Mitra AP, Cote RJ. Molecular markers for bladder cancer: the road to a multimarker approach. Expert Rev Anticancer Ther. 2007;7:1717‐1727. [DOI] [PubMed] [Google Scholar]
- 30. Toiyama Y, Inoue Y, Shimura T, et al. Serum angiopoietin‐like protein 2 improves preoperative detection of lymph node metastasis in colorectal cancer. Anticancer Res. 2015;35:2849‐2856. [PubMed] [Google Scholar]
- 31. Li B, Cui Y, Diehn M, Li R. Development and validation of an individualized immune prognostic signature in early‐stage nonsquamous non‐small cell lung cancer. JAMA Oncology. 2017;3:1529‐1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anichini A. Progress in understanding complexity and determinants of immune‐related prognostic subsets in primary melanoma. Can Res. 2019;79:2457‐2459. [DOI] [PubMed] [Google Scholar]
- 33. Xu Y, Zhang M, Wang Q, Li Z. In situ detecting changes in membrane lipid phenotypes of macrophages cultured in different cancer microenvironments using mass spectrometry. Anal Chim Acta. 2018;1026:101‐108. [DOI] [PubMed] [Google Scholar]
- 34. Xiong Y, Wang K, Zhou H, Peng L, You W, Fu Z. Profiles of immune infiltration in colorectal cancer and their clinical significant: A gene expression‐based study. Cancer Med. 2018;7:4496‐4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen YP, Zhang Y, Lv JW, et al. Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: immunotherapeutic implications. Theranostics. 2017;7:3585‐3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roelands J, Kuppen P, Vermeulen L, et al. Immunogenomic classification of colorectal cancer and therapeutic implications. Int J Mol Sci. 2017;18(10):2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin EW, Karakasheva TA, Hicks PD, Bass AJ, Rustgi AK. The tumor microenvironment in esophageal cancer. Oncogene. 2016;35:5337‐5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wei C, Liang Q, Li X, et al. Bioinformatics profiling utilized a nine immune‐related long noncoding RNA signature as a prognostic target for pancreatic cancer. J Cell Biochem. 2019;120(9):14916‐14927. [DOI] [PubMed] [Google Scholar]
- 39. Lazar DC, Avram MF, Romosan I, Cornianu M, Taban S, Goldis A. Prognostic significance of tumor immune microenvironment and immunotherapy: Novel insights and future perspectives in gastric cancer. World J Gastroenterol. 2018;24:3583‐3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao JJ, Zhou ZQ, Wang P, et al. Orchestration of immune checkpoints in tumor immune contexture and their prognostic significance in esophageal squamous cell carcinoma. Cancer Manag Res. 2018;10:6457‐6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Agesen TH, Sveen A, Merok MA, et al. ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560‐1567. [DOI] [PubMed] [Google Scholar]
- 42. Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385‐395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study were derived from the following resource: TCGA database, https://www.cancer.gov/tcga.


