Abstract
Somatic mutations together with immunoediting drive extensive heterogeneity within non-small-cell lung cancer (NSCLC). Herein we examine heterogeneity of the T cell antigen receptor (TCR) repertoire. The number of TCR sequences selectively expanded in tumors varies within and between tumors and correlates with the number of nonsynonymous mutations. Expanded TCRs can be subdivided into TCRs found in all tumor regions (ubiquitous) and those present in a subset of regions (regional). The number of ubiquitous and regional TCRs correlates with the number of ubiquitous and regional nonsynonymous mutations, respectively. Expanded TCRs form part of clusters of TCRs of similar sequence, suggestive of a spatially constrained antigen-driven process. CD8+ tumor-infiltrating lymphocytes harboring ubiquitous TCRs display a dysfunctional tissue-resident phenotype. Ubiquitous TCRs are preferentially detected in the blood at the time of tumor resection as compared to routine follow-up. These findings highlight a noninvasive method to identify and track relevant tumor-reactive TCRs for use in adoptive T cell immunotherapy.
NSCLC is characterized by the progressive emergence of mutations, a number of which provide a source of neoantigens that, together with tumor-associated antigens, stimulate an antitumoral immune response. The genomic and immunological heterogeneity of tumors is shaped by a variety of mechanisms through which tumors escape this immune response. This immunological editing includes selection for DNA copy number loss of clonal neoantigens, transcriptional repression of neoantigens, loss or mutation of human leukocyte antigen (HLA) and antigen-presentation components, and mutations in interferon (IFN) and interleukin (IL)-2 signaling components[1, 2]. Tumor heterogeneity can influence disease progression; for example, a high burden of clonal nonsynonymous mutations is associated with reduced disease recurrence and improved response to checkpoint blockade[2, 3, 4].
Antigen-specific T cell responses are a central feature of the antitumoral response and are fundamental in understanding the intricate relationship between the tumor and host. Although the majority of targeted tumor antigens remain unknown, the TCR repertoire provides a way to assess the breadth and strength of the T cell immune response. We therefore set out to examine the NSCLC intratumoral TCR repertoire, to document the spatial heterogeneity of individual TCRs within tumors and to examine how this heterogeneity relates to genomic heterogeneity.
The lung TRACERx (Tracking Cancer Evolution Through Therapy) study provides a rare setting in which to systematically study genomic and immunological intratumoral heterogeneity in the context of NSCLC. Lung TRACERx is a large (>700 patients recruited to date) prospective multi-institutional study[5] with a primary goal of mapping the genetic evolution of NSCLC from early- to late-stage disease and assessing the impact of tumor genomic and immune heterogeneity on disease progression through the analysis of multiregion tumor specimens. In a parallel paper[2], we used transcriptional profiling to characterize the intratumoral immune infiltrate, linking this to the mutational landscape, and highlighted diverse mechanisms of tumor immunoediting. Here we extend this analysis to the intratumoral TCR repertoire.
Several studies have used TCR sequencing to examine the intratumoral T cell response in solid cancers[6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25], including in NSCLC[10, 26, 27]. TCR repertoire analysis has also been used as a biomarker, in the context of checkpoint blockade[28, 29, 30, 31, 32, 33, 34, 35]. We have recently published an experimental and computational pipeline for TCR repertoire sequencing[36, 37]. In contrast to the standard commercially available TCR sequencing protocols, our method incorporates unique molecular identifiers (UMIs) into each cDNA molecule, enabling accurate computational correction for sequencing and PCR errors and bias[36, 38]. The pipeline therefore achieves a high level of quantitative precision. We used this pipeline to carry out a quantitative comparison of the TCR repertoires in multiregion tumor samples and in matched nontumor lung and blood from a cohort of 72 TRACERx patients with early-stage untreated NSCLC.
Results
NSCLC tumors contain expanded TCRs that are differentially expressed in tumors as compared to nontumor lung and whose numbers correlate with tumor mutational burden
We sequenced the α-chain and β-chain TCR repertoires from 220 tumor regions, 64 matched nontumor lung tissue samples and 56 peripheral blood mononuclear cell (PBMC) samples taken at the time of primary NSCLC surgery, collected from 72 patients within the lung TRACERx cohort[39] (Extended Data Figs. 1 and 2). The median number of unique α-chain and β-chain transcripts per tumor sample was 2,339 and 3,711, respectively (Extended Data Fig. 2), reflecting a highly diverse polyclonal population of intratumoral T cells.
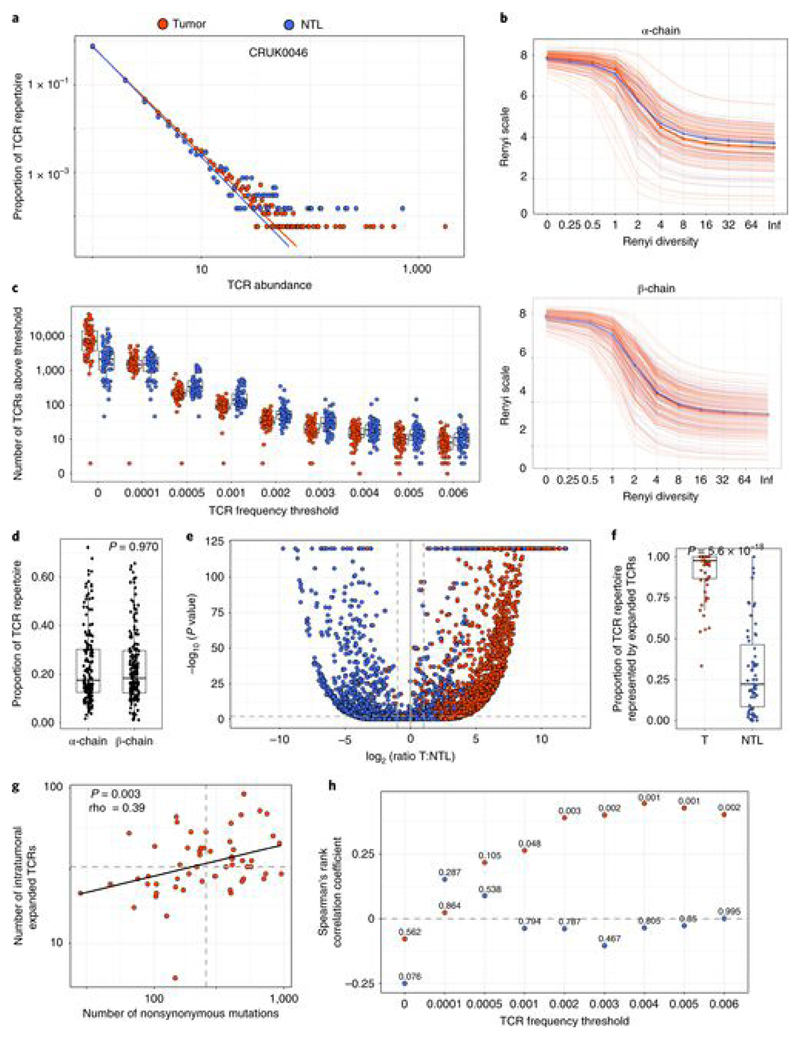
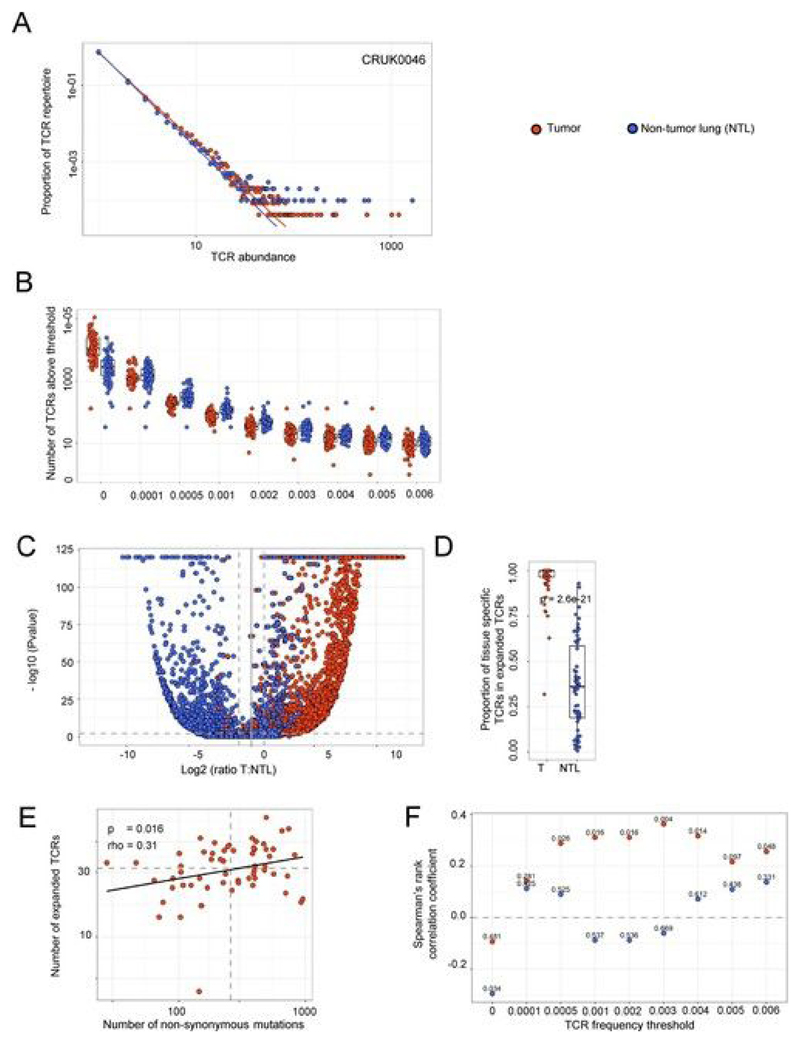
Recent publications have highlighted the presence of a large number of ‘bystander’ T cells within tumors[25, 40] that may reflect the continuous migration of effector memory T cells through the tumor tissue, driven by local inflammation. In line with this, the abundance distribution of different TCRs in tumor followed an approximate discrete power law distribution similar to that described previously for circulating effector memory T cells in blood[36] (Fig. 1a and Extended Data Fig. 3a; the power law parameter, the negative of which corresponds to the slope on a log–log plot, was 2.43 ± 0.05 for α-chains and 2.47 ± 0.05 for β-chains, mean + s.e.m.; TCR repertoires from n = 71 patients). The distribution was similar to that of the TCR repertoire in nontumor lung (Fig. 1a and Extended Data Fig. 3a; 2.59 ± 0.05 for α-chains and 2.59 ± 0.06 for β-chains, mean + s.e.m.; n = 66). The diversities of the tumor and nontumor lung TCR repertoires, as captured by different Rényi entropies, were also not significantly different (P > 0.1 at all Rényi orders) (Fig. 1b).
Fig. 1. NSCLC tumors contain expanded TCRs that are differentially expressed in tumor as compared to nontumor lung and whose numbers correlate with tumor mutational burden.
a, The frequency distribution of TCR α-chain abundance (TCR β-chains are shown in Extended Data Fig. 3a) was fitted to a discrete power law (f(k) = Ck−α) by maximum likelihood. The Figure shows a representative plot (from patient CRUK0046) for TCR α-chain sequences in tumor (red circles) and matched nontumor lung (NTL; blue circles). The average power law parameter α was 2.43 ± 0.05 for tumor and 2.59 ± 0.05 for nontumor TCR α-chains. The parameters for the patient shown were 2.43 for tumor and 2.53 for nontumor lung. The distribution parameters fitted to tumor and to nontumor samples did not differ significantly: P = 0.16, two-sided Mann–Whitney test. The x axis represents TCR abundance (size of clone), and the y axis represents the proportion of the repertoire. b, The Rényi entropies of order 0 to infinity for the α-chain (top) and β-chain (bottom) TCR repertoires of tumor regions (multiple tumor regions were pooled from an individual patient; red) and nontumor lung samples (blue). Each repertoire was subsampled to 5,000 TCRs 100 times before calculating the Rényi entropy (see Methods). The Rényi index of order 1 corresponds to the Shannon diversity index. c, The number of α-chain sequences detected above a given frequency threshold is shown for tumor (n = 72 patients, with multiple tumor regions pooled from an individual patient; red circles) and matched nontumor lung (n = 64 patients; blue circles). The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. d, The proportion of the total intratumoral TCR repertoire (total number of TCRs, with pooled tumor regions from each patient) accounted for by the expanded (frequency ≥ 2/1,000) α-chain and β-chain TCRs. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The proportions for α-chain and β-chain sequences were not significantly different (two-sided Mann–Whitney test); n = 204 tumor regions from 49 patients for α-chains and 214 tumor regions from 52 patients for β-chains. e, A volcano plot showing the likelihood (–log10 (P value)) of a TCR being sampled from two populations of equal mean in tumor and nontumor lung, plotted against differential expression in tumor versus nontumor lung. If the log likelihood was >120, it was given a value of 120 for plotting purposes. Blue circles represent α-chain sequences expanded (>0.002) in nontumor lung (n = 3,197 TCRs from 59 patients) and red circles represent α-chain sequences expanded in tumor (n = 2,193 TCRs from 59 patients); the equivalent plot for TCR β-chain sequences is shown in Extended Data Fig. 3c. The horizontal dashed line corresponds to P = 0.01; the vertical dashed lines indicate a twofold differential in expression between the tissues. f, The proportion of expanded tumor or nontumor lung α-chain sequences that are specific to their respective tissue, defined on the volcano plot as TCRs that had P < 0.01 and a differential abundance of at least two between the tissues. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The two proportions were significantly different, with the two-sided Mann–Whitney test P value shown; n = 59 patients (tumor) and n = 59 patients (nontumor lung). The equivalent plot for TCR β-chain sequences is shown in Extended Data Fig. 3d. g, The correlation between the number of nonsynonymous mutations and the number of unique intratumoral expanded α-chain sequences (frequency ≥ 2/1,000) for each patient. The Spearman’s rank correlation coefficient and P value are shown (n = 59 patients). The equivalent plot for β-chain sequences is shown in Extended Data Fig. 3e. h, The Spearman’s rank correlation coefficient and P value (shown above each point; n = 59 patients) for the relationship between the number of nonsynonymous mutations and the number of unique intratumoral (red circles) or nontumor lung (blue circles) expanded α-chain sequences at different frequencies (ranging from all TCRs (threshold of zero) up to those found at a frequency of ≥ 8/1,000). The equivalent plot for β-chain sequences is shown in Extended Data Fig. 3f.
We hypothesized that tumor-specific T cells would be enriched within the set of expanded T cells in the tumor, owing to clonal expansion and tissue retention of these cells. We selected the most expanded TCRs, with a threshold corresponding to TCRs within the top 1% of the empirical tumor frequency distribution (TCR frequency cutoff value of 2/l,000 (0.002); hereafter referred to as ‘expanded TCRs’), for further in-depth analysis (Fig. 1C and Extended Data Fig. 3b). Reflecting the key ‘heavy tail’ of power law distributions, the expanded TCRs comprised on average only 1% of the unique TCR sequences observed, but accounted for almost 20% (one in every five TCRs) of the total observed TCRs (Fig. 1d; median for α-chains = 18% and median for β-chains = 17%).
Nontumor lung also contained expanded TCRs defined as above (Fig. 1c). We therefore investigated whether the expanded intratumoral TCRs were enriched in tumor samples as compared to matched nontumor lung. We calculated the likelihood that each expanded intratumoral TCR observed R times in tumor and N times in nontumor lung was in fact derived from the same population by random sampling (assuming a random Poisson sampling distribution with sampling from a distribution with the mean (R + N)/2)) and plotted log likelihood versus log relative abundance for expanded TCRs from both tumor and nontumor lung (Fig. 1e,f and Extended Data Fig. 3c,d). The majority of expanded intratumoral TCRs were differentially expressed in tumor samples (median proportion of differentially expressed α-chains = 0.97 ± 0.02 and β-chains = 1 ± 0.02, mean ± s.e.m.), while a smaller proportion (median proportion of differentially expressed α-chains = 0.21 ± 0.03 and β-chains = 0.36 ± 0.08) of the TCRs expanded in nontumor lung were preferentially expressed in nontumor lung as compared to tumor samples. The differences between tumor and nontumor lung were significant for both α-chain and β-chain genes (P < 2 × 10−16, Mann–Whitney test; n as above).
We tested whether there was a relationship between the number of expanded TCRs in tumor samples and the number of nonsynonymous mutations, as determined by whole-exome sequencing of tumor tissue. The number of distinct expanded intratumoral α-chain and β-chain sequences was correlated with the number of mutations (Fig. 1g (α-chains: rho = 0.39, P = 0.003, tumors from n = 59 patients) and Extended Data Fig. 3e (β-chains: rho = 0.31, P = 0.016, tumors from n = 61 patients)). The correlations were robust over a range of TCR expansion frequency thresholds (Fig. 1h and Extended Data Fig. 3f). As expected, we observed no significant correlation between the number of expanded TCRs in nontumor lung and the number of nonsynonymous mutations (Fig. 1h and Extended Data Fig. 3f).
NSCLC tumors contain expanded ubiquitous and regional TCRs, which reflect the tumor mutational landscape
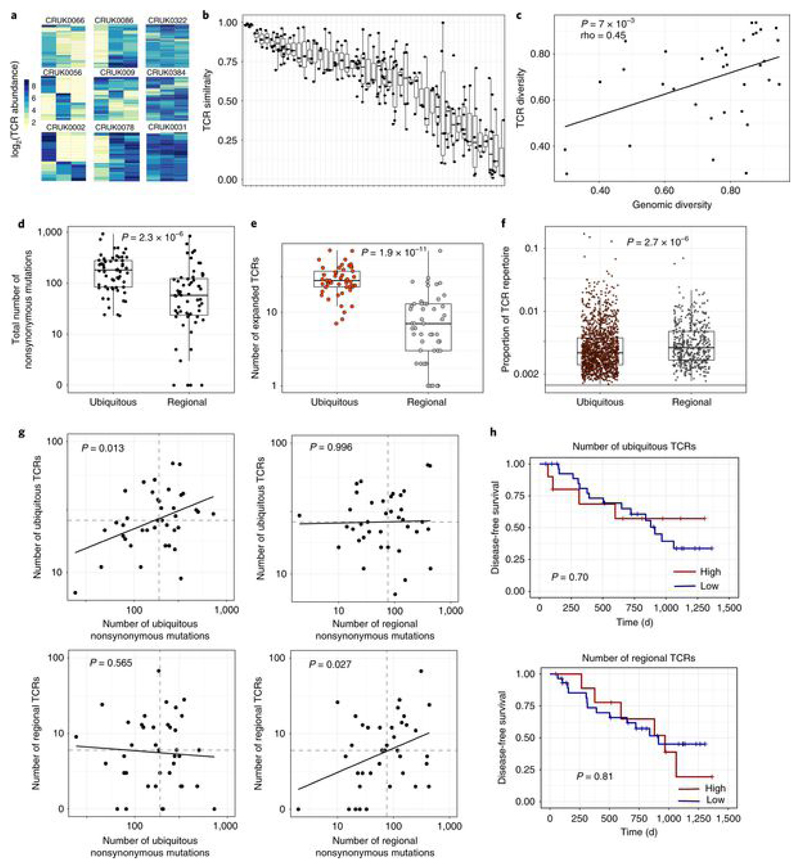
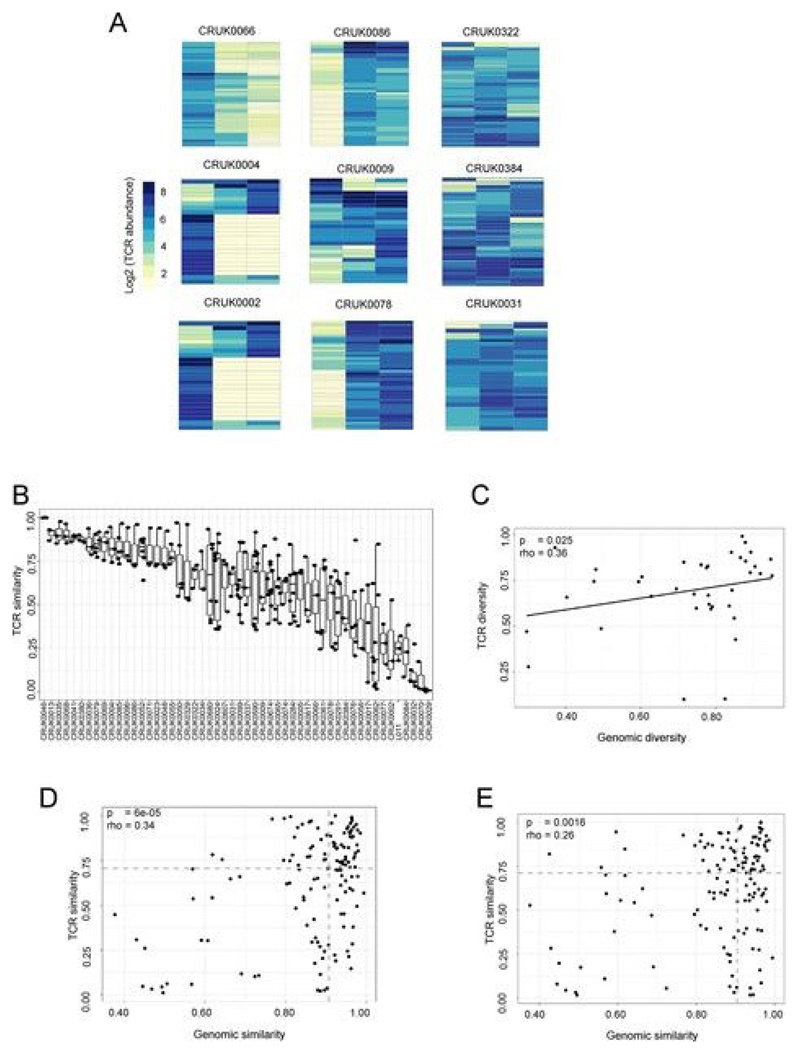
The abundance of the expanded TCRs in different tumor regions of tumors from several patients is shown in Fig. 2a and Extended Data Fig. 4a. The heterogeneity of the expanded TCR profile differed markedly between patients, with tumors from some patients showing profound differences between individual regions of the tumor and those from other patients showing a more homogeneous TCR expression pattern. We quantified TCR intratumoral heterogeneity in two ways. We defined an inter-region similarity index for pairs of tumor regions (the cosine distance; see Methods), which varied widely both within and between tumors (Fig. 2b and Extended Data Fig. 4b). We also measured the average normalized Shannon diversity for all TCRs across all tumor regions (Fig. 2c and Extended Data Fig. 4c; patients with three or more regions). Similarly, we defined pairwise genomic similarity or average normalized Shannon genomic diversity metrics on the basis of the prevalence of nonsynonymous mutations in different tumor regions (Extended Data Fig. 5; see Methods). Both TCR similarity (Extended Data Fig. 4d,e; α-chains: rho = 0.34, P = 6 × 10−5; β-chains: rho = 0.26, P = 0.0016) and TCR spatial diversity (Fig. 2c and Extended Data Fig. 4c; α-chains: rho = 0.45, P = 7 × 10−3; β-chains: rho = 0.36, P = 0.025) were significantly correlated with their equivalent genomic metrics, providing support for the hypothesis that TCR spatial heterogeneity reflects genomic intratumoral heterogeneity.
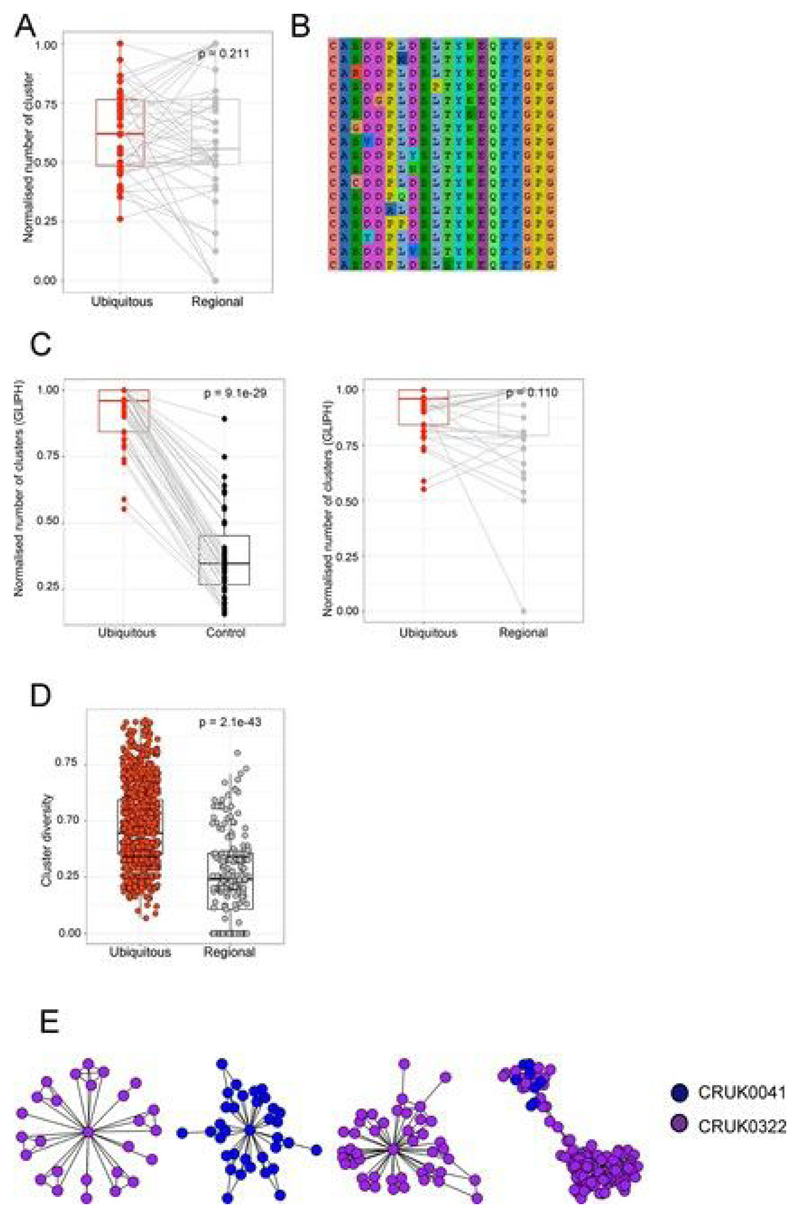
Fig. 2. NSCLC tumors contain expanded ubiquitous and regional TCRs, which reflect the tumor mutational landscape.
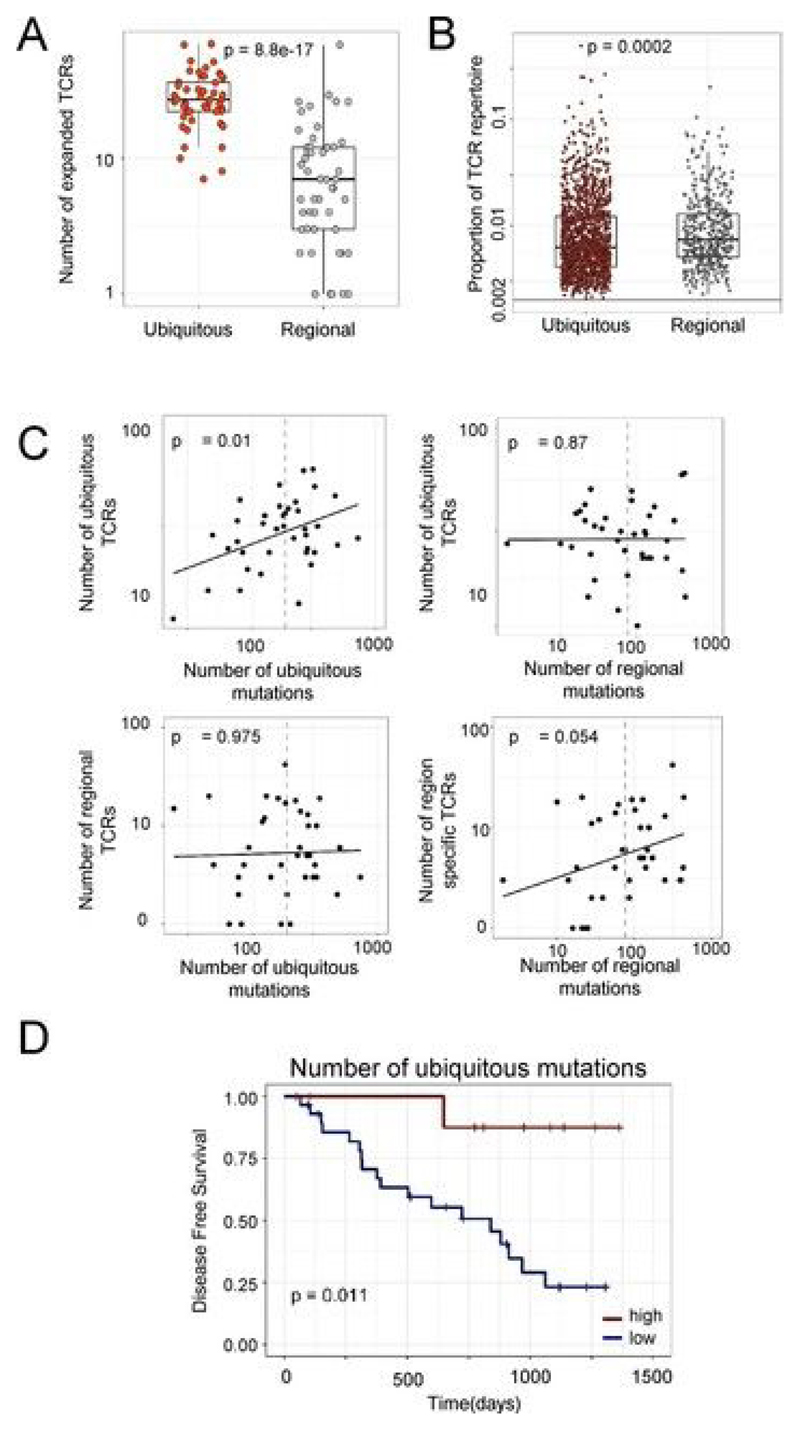
a, Heat maps showing the abundance (log2 of the number of times each TCR was found) of expanded intratumoral α-chain sequences (frequency ≥ 2/1,000) in different tumor regions for several patients. Patient ID is shown above each heat map. Each row represents one unique sequence, and each column represents one tumor region. Equivalent plots for β-chain sequences are shown in Extended Data Fig. 4a. b, The TCR repertoires of multiple regions from a patient’s tumor were sequenced and a pairwise comparison of the repertoires of different regions from the same tumor was performed by using the cosine similarity (see Methods). The pairwise intratumoral TCR repertoire similarity (α-chain sequences; the equivalent plot for β-chain sequences is in Extended Data Fig. 4b) is shown for each patient. Each circle represents a comparison between two regions from the same patient’s tumor (n = 226 total comparisons from 49 patients). For all box plots, the minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. Patients are ordered along the x axis by descending mean intratumoral TCR similarity. c, TCR repertoire (α-chain sequences) diversity plotted against genomic diversity for each patient. The diversity measurement was calculated as the normalized Shannon entropy (see Methods). The Spearman’s rank correlation coefficient and P value are shown; n = 38 patients. The equivalent plot for β-chain sequences is shown in Extended Data Fig. 4c. d, The numbers of ubiquitous and regional nonsynonymous mutations for each tumor region. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The number of ubiquitous mutations is greater than the number of regional mutations, with the two-sided Mann–Whitney test P value shown; n = 60 patients. e, The numbers of expanded (frequency ≥ 2/1,000) ubiquitous (red circles) and regional (gray circles) α-chain sequences for each tumor region. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The number of ubiquitous TCRs is greater than the number of regional TCRs, with the two-sided Mann–Whitney test P value shown; n = 49 patients. The equivalent plot for β-chain sequences is shown in Extended Data Fig. 6a. f, The frequency distribution of the intratumoral expanded α-chain ubiquitous (red circles; n = 1,379 individual TCRs combined from 49 patients) and regional (gray circles; n = 446 individual TCRs from 44 patients) TCRs. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. Expanded regional TCRs are expressed at a higher abundance than ubiquitous TCRs when compared by two-sided Mann–Whitney test (P = 2.7 × 10−6). The equivalent plot for β-chain sequences is shown in Extended Data Fig. 6b. g, The number of expanded ubiquitous (top) or regional (bottom) α-chain sequences plotted against the number of ubiquitous (left) or regional (right) nonsynonymous mutations for each tumor region. The Spearman’s rank correlation and associated P value are shown; dashed lines correspond to median values. n = 39 patients. The equivalent plot for β-chain sequences is shown in Extended Data Fig. 6c. h, Patients were stratified according to the number of expanded intratumoral ubiquitous α-chain sequences (top) or the number of expanded intratumoral regional α-chain sequences (bottom). n = 39 patients. The red line indicates a ratio above the median, and the blue line indicates a ratio below the median. Kaplan–Meier P values are shown.
We classified expanded intratumoral TCRs into ubiquitous and regional TCRs by using a statistical model to define regional TCRs. For expanded intratumoral TCRs that were absent from one or more regions of a tumor (we analyzed tumors with TCR data for at least three regions), we defined a probability p that the absence of a TCR in that region was due to sampling (the null hypothesis). We defined regional TCRs as TCRs for which p < 0.05 (see Methods).The numbers of ubiquitous and regional nonsynonymous mutations (Fig. 2d; see Methods and Extended Data Fig. 5) and ubiquitous and regional expanded TCRs (Fig.2e and Extended Data Fig. 6a) were determined for each patient. Expanded regional TCRs were expressed at a higher abundance than ubiquitous TCRs (Fig.2f and Extended Data Fig. 6b; Mann–Whitney test, P =2.7 × 10−6 for α-chains and P = 0.0002 for β-chains).
We reexamined the relationship between expanded TCRs and nonsynonymous mutational burden, in the context of classification as ubiquitous or regional. The number of expanded intratumoral ubiquitous TCRs was correlated with the number of ubiquitous mutations but showed no correlation with the number of regional mutations (compare Fig. 2g and Extended Data Fig. 6c, upper left and right; in multivariate regression, ubiquitous TCRs were significantly associated with ubiquitous mutations (P = 0.013 for α-chains and P = 0.01 for β-chains), but not with regional mutations (P = 0.99 for α-chains and P = 0.87 for β-chains); 39 degrees of freedom).
In contrast, the number of expanded intratumoral regional TCRs was correlated with the number of regional mutations, but showed no correlation with the number of ubiquitous mutations (compare Fig. 2g and Extended Data Fig. 6c, lower left and right; in multivariate regression, regional TCRs were significantly associated with regional mutations for α-chains (P = 0.027) and showed a trend toward correlation for β-chains (P = 0.054), but were not correlated with ubiquitous mutations (P = 0.565 for α-chains and P = 0.975 for β-chains); 39 degrees of freedom).
Finally, we examined the relationship between tumor mutational load, the number of expanded intratumoral TCRs and clinical outcome, measured as time to first recurrence. As expected from previous studies[3], improved clinical outcome was found for patients with a high number of total or ubiquitous nonsynonymous mutations (Extended Data Fig. 6d; multivariate Cox regression, P = 0.011; n = 39 patients). However, no difference in outcome was observed in patients with high numbers of expanded ubiquitous or regional intratumoral TCRs (Fig. 2h; multivariate Cox regression, expanded ubiquitous or regional intratumoral TCRs, P > 0.5; n = 39 patients).
Expanded intratumoral TCR CDR3 sequences identify clusters of related TCRs and show enhanced convergent recombination
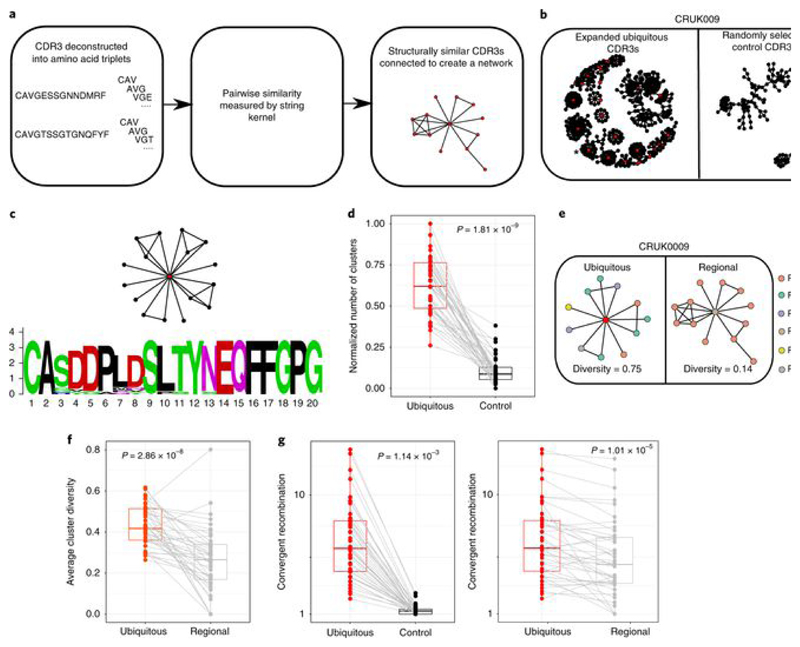
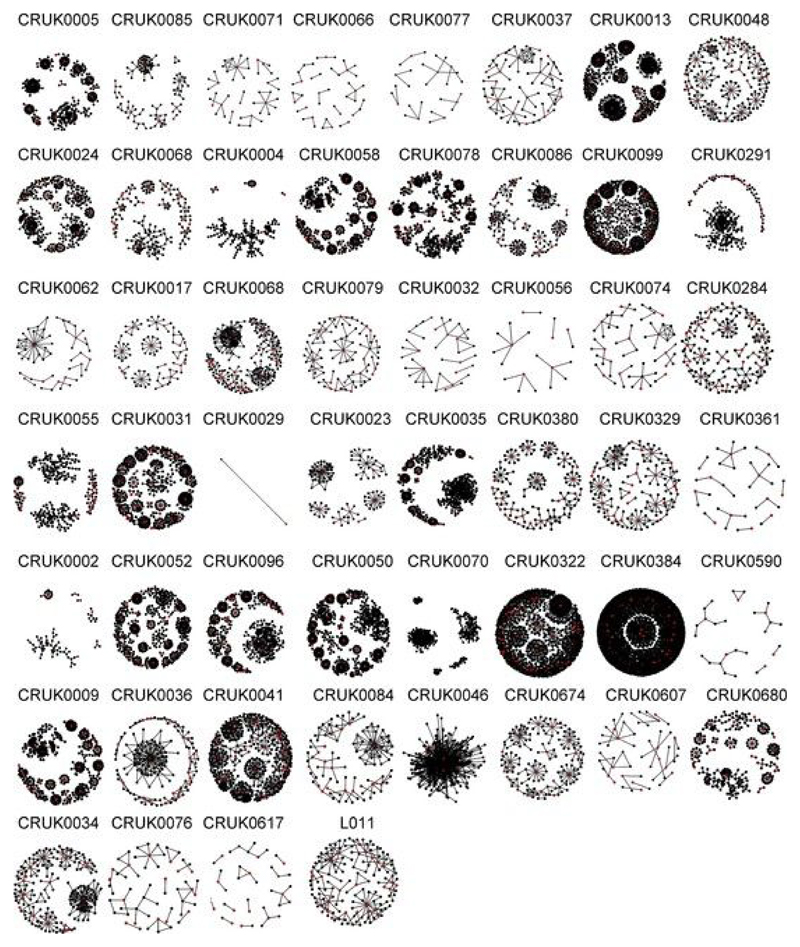
Previous work from our lab[41, 42] and those of others[43, 44] has highlighted the importance of short protein motifs in defining the antigen specificity of TCRs. We therefore searched for clusters of related TCR sequences within the tumor repertoire, on the basis of sharing of amino acid triplets (Fig. 3a; CDR3 cluster definition described in the Methods). Many of the expanded intratumoral ubiquitous TCR sequences were observed to form part of a cluster of highly related TCR sequences (representative example for β-chain sequences from patient CRUK0009 shown in Fig. 3b, left; cluster networks for all patients in Extended Data Fig. 7). In contrast, much less clustering was observed with a random sample of CDR3s from the same tumor repertoire (Fig. 3b, right, and Fig. 3d). A representative example of an alignment of CDR3 sequences from a single cluster is shown in Fig. 3c (full alignment in Extended Data Fig. 8b), illustrating a highly related set of TCR sequences. GLIPH (grouping of lymphocyte interactions by paratope hotspots)[44], an alternative published clustering algorithm for TCRs, also detected much more clustering when using the expanded ubiquitous TCRs in comparison to randomly sampled sets of CDR3s from the same repertoire (Extended Data Fig. 8c, left).
Fig. 3. Expanded intratumoral TCR CDR3 sequences identify clusters of related TCRs and show enhanced convergent recombination.
a, Diagram illustrating the CDR3 similarity network construction process. Individual CDR3s are deconstructed into overlapping series of contiguous amino acid triplets, and the pairwise similarity between two CDR3s is calculated as the normalized string (triplet) kernel. CDR3s that have a pairwise similarity of >0.82 are connected by an edge. b, Representative network diagrams of intratumoral CDR3 β-chain sequences for patient CRUK0009. Both panels show the network of TCR CDR3 β-chain sequences that are connected to at least one other TCR within the tumor. Left, clustering around expanded intratumoral ubiquitous TCRs (red circles). An asterisk indicates a cluster whose CDR3 sequences are analyzed in c. Right, clustering around a random sample of TCRs from the same repertoire (same numbers as for the expanded ubiquitous TCRs). c, A representative example of alignment of CDR3 sequences from a single cluster (full alignment in Extended Data Fig. 8b). The alignment ts shown as a sequence logo (https://weblogo.berkeley.edu/logo.cgi). d, The clustering algorithm was run on all patients, and the number of clusters for the networks containing expanded ubiquitous and control randomly selected β-chain sequences is shown. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The expanded TCRs exhibit greater clustering, with the one-sided Mann–Whitney test P value shown; n = 46 patients. e, Representative clustering around a ubiquitous or regional expanded TCR from CRUK0009, with nodes colored according to the regions in which each TCR was found. f, The average cluster Shannon diversity (see Methods) for clusters containing ubiquitous or regional expanded TCRs. The one-sided Mann–Whitney test P value is shown; n = 42 patients. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. g, The amount of convergent recombination was calculated as the average number of distinct DNA TCR sequences that contributed to each observed TCR CDR3 amino acid sequence for each expanded ubiquitous TCR, as compared to a randomly selected set of TCRs (left) or regional TCRs (right) from each patient intratumoral repertoire. One-sided Mann–Whitney P values are shown; n = 43 patients.The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. All panels in this figure refer to TCR β-chain sequences, as these showed more diversity and a lower background rate of clustering.
Expanded intratumoral regional TCRs also showed clustering, and the numbers of clusters formed by regional and ubiquitous TCRs were not significantly different (Extended Data Fig. 8a), once normalized for the number of expanded TCRs. However, the regional composition of the clusters containing expanded ubiquitous TCRs was different from that of clusters containing expanded regional TCRs. Clusters with regional TCRs contained TCRs from fewer regions of the tumor than those with ubiquitous TCRs (Fig. 3e,f and Extended Data Fig. 8d). This observation supports the hypothesis that regional TCRs are responding to regional antigens and provides additional independent evidence that ubiquitous and regional TCRs are responding to antigens with distinct spatial distributions.
The average number of different DNA sequences encoding each expanded ubiquitous TCR was significantly higher than the average number encoding either expanded regional TCRs or a random selection of TCRs from the same repertoire (Fig. 3g). This phenomenon (‘convergent recombination’) is a further characteristic of antigen-driven responses.
The presence of clusters of CDR3 sequences related to the expanded intratumoral TCRs and the demonstration of increased convergent recombination for these sequences are both consistent with the hypothesis that this subset of TCRs identifies a core set of antigen-specific T cells.
Expanded intratumoral ubiquitous TCRs are associated with a TH1 and CD8+ T cell transcriptional signature in the tumor and have a phenotype consistent with tissue-resident tumor-antigen-reactive T cells
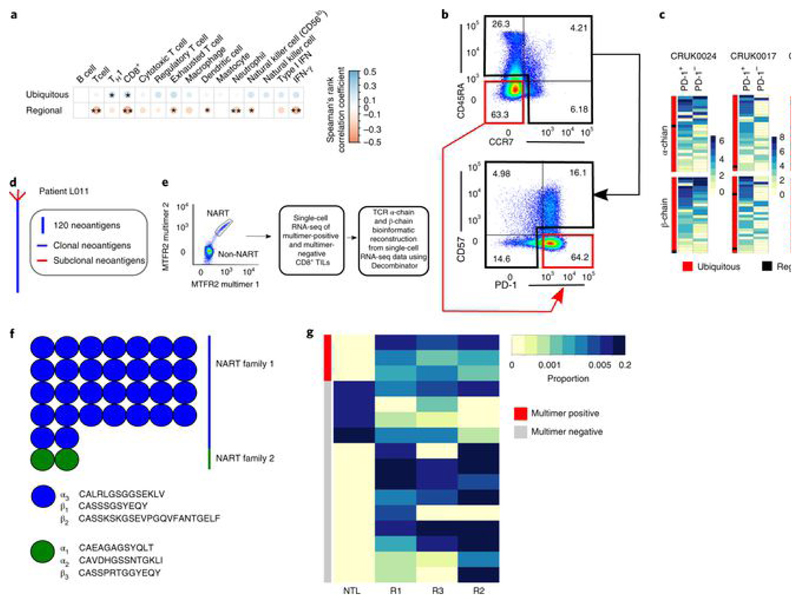
We examined the functional properties of intratumoral T cells displaying the expanded TCRs by several indirect approaches. We first looked at the tumor transcriptional landscape, calculating a transcriptional score for sets of genes defining specific cell types and functional states (see Methods) for each region of the tumor. The number of expanded intratumoral ubiquitous TCRs was significantly correlated with the transcriptional score of type 1 T helper (TH1) and CD8+ gene sets (Fig. 4a and Extended Data Fig. 9a). There was a nonsignificant trend toward correlation with the transcriptional signatures of exhausted T cells, natural killer (NK) cells (reaching significance for β-chains) and an IFN-γ response signature. In contrast, and unexpectedly, the number of expanded regional TCRs was negatively correlated with the transcriptional score of T cells, CD8+ T cells, exhausted T cells, NK cells, dendritic cells, neutrophils and the IFN-γ gene module. We conclude that tumor regions with a strong CD8+ T cell response are enriched for expanded ubiquitous T cells but are depleted of expanded regional T cells.
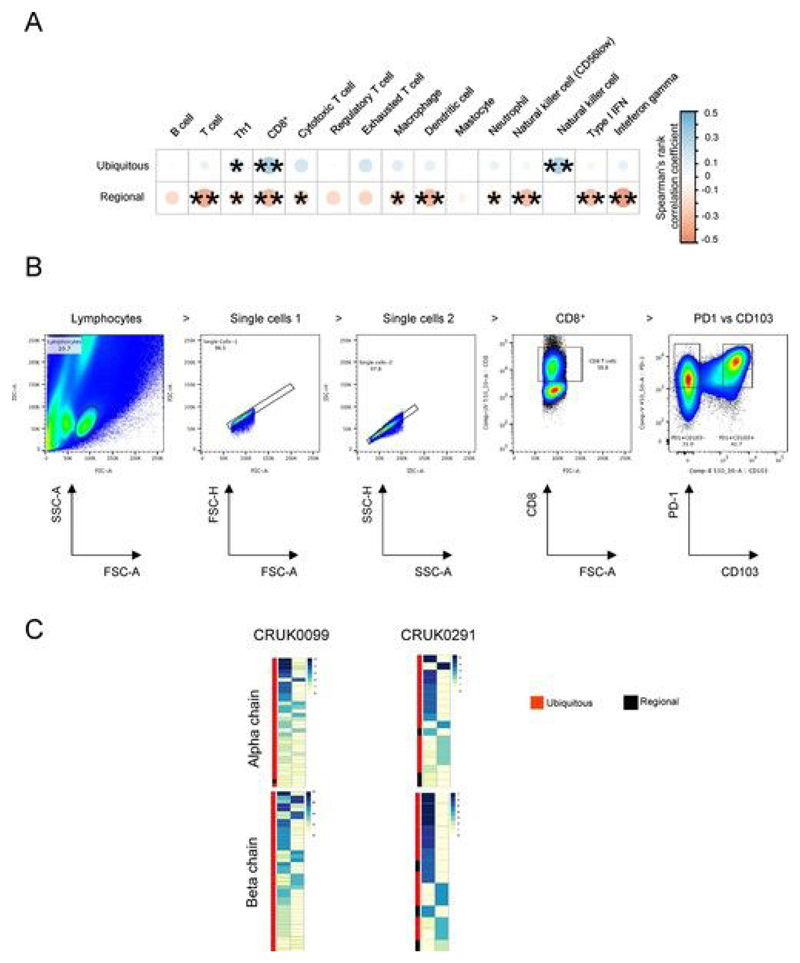
Fig. 4. Expanded intratumoral ubiquitous TCRs are associated with a TH1 and CD8+ T cell transcriptional signature in the tumor and have a phenotype consistent with tumor antigen reactivity.
a, Correlation between the numbers of expanded intratumoral ubiquitous and regional α-chain TCR sequences and the transcriptional expression score (geometric mean) for various immune-related gene sets, characterizing cell types or functional states (names appear above the heat map). Details of how the transcriptional scores are calculated are in the Methods. The area and color of the circles correspond to the magnitude of the Spearman’s rank correlation coefficient. *P < 0.05; **P < 0.01; after Bonferroni correction. n = 78 tumor regions. b, CD8+ TILs from CRUK0024, CRUK0017 and CRUK0069 were sorted into two populations: CD45RA−CCR7−PD-1+CD57− and all other CD8+ TILs (referred to as PD-1+ and PD-1− subpopulations, respectively). The flow cytometry gating strategy for a representative patient is shown (pre-gated on live > singlets > CD3+ > CD8+ T cells). c, RNA from sorted populations was extracted and sequenced, and the RNA-seq data were mined for the presence of expanded ubiquitous and regional α-chain and β-chain sequences. The heat maps show the number of times each expanded ubiquitous or regional TCR CDR3 sequence was found in the RNA-seq data from PD-1+ or PD-1− cells, as a proportion of the number of times a constant region sequence of the same length was detected. The color key gives the proportions scaled for each row, where each row represents a distinct expanded TCR sequence. d, CD8+ T cells reactive to a clonal neoantigen were isolated from a patient with a high clonal nonsynonymous mutational load (patient L011). Phylogenetic tree of tumor adapted from ref. [3]. e, Representative dot plot of the TIL peptide:multimer sorting strategy (left) and the workflow for single-cell RNA-seq (right). CD8+ TILs were screened for neoantigen-reactive T cells (NARTs) reactive to a peptide arising from the mutated MTFR2 gene and sorted for single-cell RNA-seq (pre-gated on lymphocyte > single cell > viable > CD3+CD8+ T cells). f, TCR α-chain and β-chain sequences were bioinformatically reconstructed from the single cells with Decombinator (see Methods). The neoantigen-reactive T cells comprised two families of cells with distinct CDR3 sequences and abundances. g, Heat map showing the abundance of antigen-specific β-chain sequences in tumor regions (Rl–R3) and nontumor lung from multimer-positive (red) and multimer-negative (gray) cells within the bulk TCR sequencing data. Each row represents a unique TCR found in the multimer-positive or multimer-negative single cells.
Expression of high levels of PD-1 and absence of CD57 are characteristic of tumor-specific dysfunctional CD8+ T ce1ls[40, 45, 46, 47]. We therefore sorted CD8+ tumor-infiltrating lymphocytes (TILs) by flow cytometry (Fig. 4b) from the tumors of three patients, CRUK0024, CRUK0017 and CRUK0069, into PD-1+CD45RA−CCR7−CD57− cells (referred to as PD-1+) and all other CD8+ T cells (referred to as PD-1−) and performed RNA-seq. These transcriptomes were mined for expanded intratumoral ubiquitous and regional TCRs. We used TCR constant region sequences to normalize the samples for the total number of TCRs detected in the RNA-seq data (see Methods). Interestingly, expanded ubiquitous TCRs were frequently detected in both the PD-1+ and PD-1− compartments, suggesting that the same clonotype differentiates into both phenotypes (Fig. 4c). 33 ± 6% of the TCR CDR3 RNA reads detected could be attributed to expanded intratumoral ubiquitous TCRs in the PD-1+ population, whereas 16 ± 3% could be attributed to expanded intratumoral ubiquitous TCRs in the PD-1− population (significantly different by t-test, P = 0.02; n = 3 patients). The expanded regional TCRs were detected at low abundance in the RNA-seq data in tumors from the three patients studied, perhaps reflecting a low average abundance of these TCRs in the RNA-seq samples.
We were able to generate CD8+ TIL RNA-seq data for two further patient samples, CRUK0099 and CRUK0291. CD8+ TILs from these patients’ tumors were sorted by flow cytometry into PD-1+CD103+ (where CD103 is a marker of tissue-resident cells) and PD-1+CD103− populations (Extended Data Fig. 9b). The expanded intratumoral ubiquitous TCRs could be detected in both cell populations, suggesting that these expanded TCRs are present as a mixture of tissue-resident and migratory T cells (Extended Data Fig. 9c). Of the total TCRs detected in the RNA-seq data from both T cell populations, 50 ± 0.5% could be attributed to the expanded ubiquitous TCRs in the PD-1+CD103+ population, whereas 20 ± 5% could be attributed to expanded ubiquitous TCRs in the PD-1−CD103− population. The expanded regional TCRs were again detected at low abundance (less than 2% in any population) in the TIL RNA from both patients.
Previously published work from our group has demonstrated the presence of neoantigen-reactive CD8+ T cells in multiregion tumor specimens sampled from patient L011 (ref. [3]). This patient’s tumor DNA encoded 400 putative neoantigens, 90% of which were clonal (Fig. 4d). CD8+ TILs were sorted by fluorescent MHC multimers bound to a peptide encoded by a ubiquitous mutation in the MTFR2 gene (Fig. 4e), and the sorted cells were then subjected to single-cell RNA-seq. Analysis of the TeR sequences from the single-cell RNA-seq data identified two families of TCRs, on the basis of the sharing of TCR α-chain and/or β-chain CDR3 sequences (Fig. 4f). Comparison of the single-cell β-chain sequences to the bulk TCR sequencing data from the same patient showed that both families of TCRs were expanded and enriched in the tumor, and were ubiquitously expressed throughout the tumor (Fig. 4g). This functional observation supports the hypothesis that expanded intratumoral ubiquitous TCRs may recognize ubiquitously expressed neoantigens.
Expanded intratumoral TCR sequences can be identified in matched blood samples at the time of primary tumor resection and can persist in the blood long term
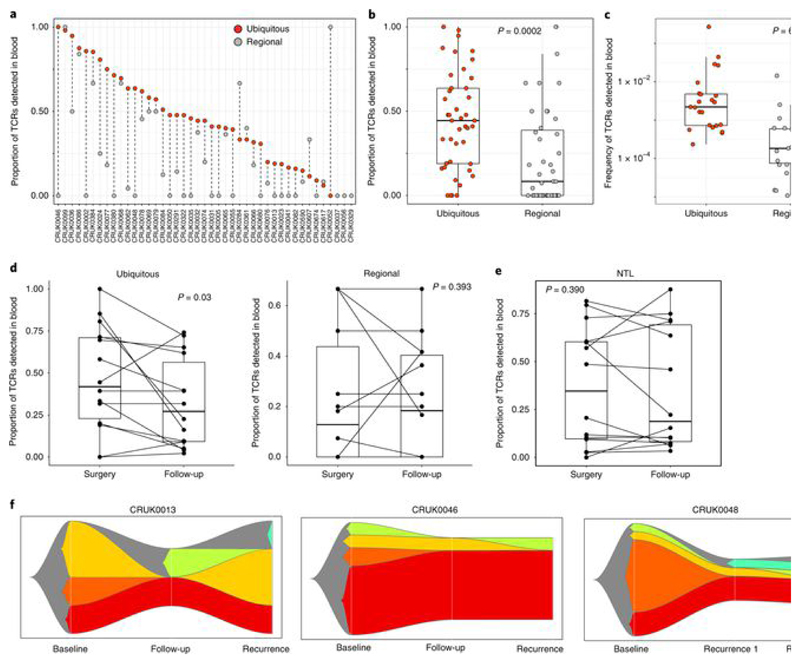
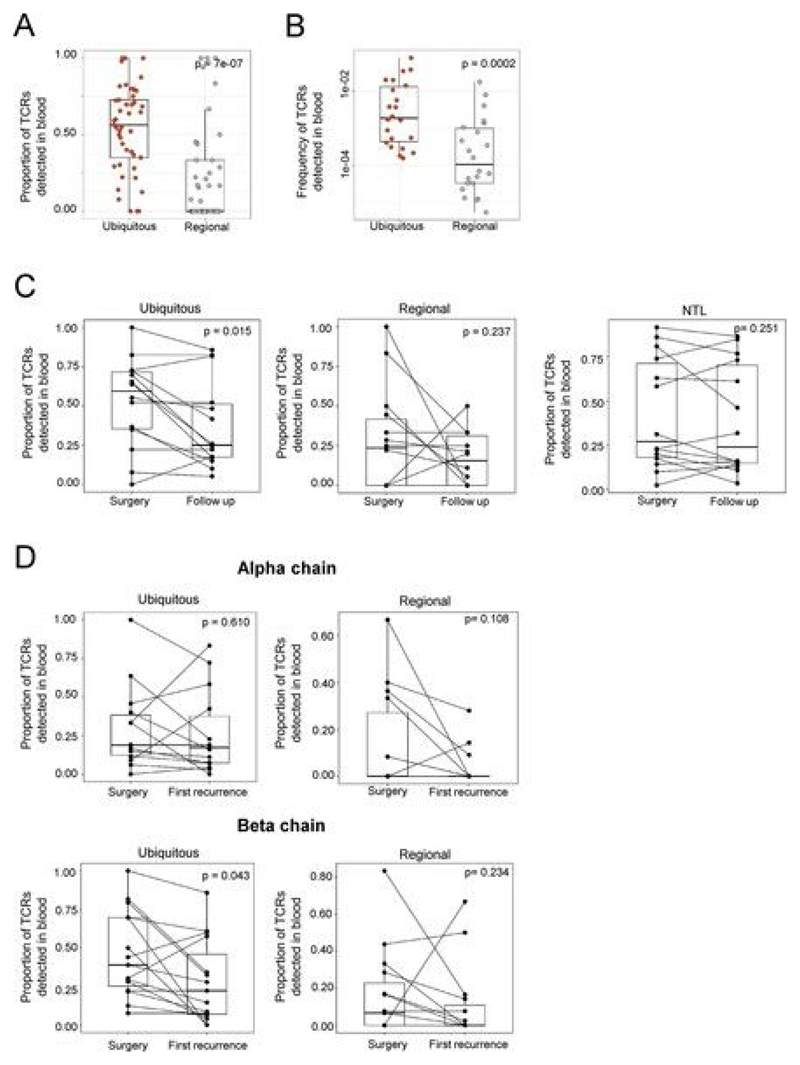
We next searched for the set of expanded intratumoral TCRs in matched blood samples. Remarkably, many of the expanded intratumoral TCRs were detected in blood samples taken at tumor resection (Fig. 5a). Both the proportion (Fig. 5b and Extended Data Fig. 10a; α-chains: P = 0.0002, n = 43; β-chains: P = 7 × 10−7, n = 45) and frequency (Fig. 5c and Extended Data Fig. 10b; α-chains: P = 6.8 × 10−7, n = 23; β-chains: P = 0.0002, n = 22) of expanded intratumoral ubiquitous TCRs detected in the blood were higher than for regional TCRs.
Fig. 5. Expanded intratumoral TCR sequences can be identified in matched blood samples at the time of primary tumor resection and can persist in the blood long term.
a, The proportion of expanded intratumoral ubiquitous and regional TCRs (α-chain) detected in peripheral blood from the same patient at the time of primary NSCLC surgery, with patients ordered by descending proportion. The proportions were obtained by dividing the number of TCRs found in the blood by the total number of TCRs for each category. We thus ensured that the difference observed was not due to the size of the input. b, Summary of the proportion of expanded intratumoral ubiquitous (red circles) and expanded intratumoral regional (gray circles) TCRs (α-chain) detected within the blood for all patients (the one-sided Mann–Whitney test P value is shown; n = 43 individual patients). The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The equivalent plot for β-chain sequences is shown in Extended Data Fig. 10a. c, The frequency (number of TCR sequences detected, as a proportion of the total number of TCRs) of expanded intratumoral ubiquitous (red circles) and regional (gray circles) TCRs (α-chain) in the peripheral blood at the time of NSCLC surgery (the one-sided Mann–Whitney test P value is shown; n = 23 individual patients). Only TCRs actually detected in blood were used for this analysis. The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The equivalent plot for β-chains is shown in Extended Data Fig. 10b. d, The proportion of expanded intratumoral ubiquitous (left) and regional (right) TCRs (α-chain) that were detected in the blood at the time of primary NSCLC surgery and at routine follow-up (median time to follow-up was just under 2 years; one-sided Mann–Whitney test P values are shown; n = 14 individual patients for ubiquitous and regional TCRs). The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The equivalent plot for β-chains is shown in Extended Data Fig. 10c. e, The proportion of expanded nontumor lung TCRs (α-chain) that were detected in the blood at the time of primary NSCLC surgery and at routine follow-up (the one-sided Mann–Whitney test P value is shown; n = 14 individual patients). The minimum and maximum are indicated by the extreme points of the box plot; the median is indicated by the thick horizontal line; and the first and third quartiles are indicated by box edges. The equivalent plot for β-chain sequences is shown in Extended Data Fig. 10c. f, The relative proportion of expanded intratumoral ubiquitous TCRs (α-chain) with different patterns of occurrence in peripheral blood taken at three longitudinal time points for patients CRUK0013 (left), CRUK0046 (middle) and CRUK0048 (right). Expanded intratumoral ubiquitous TCRs found at all three time points are shown in red. TCRs found in baseline blood only are shown in orange.
We next investigated the presence of the expanded TCRs in non-recurrence follow-up blood samples. The proportion of expanded intratumoral ubiquitous TCRs in the blood was significantly lower at routine follow-up after tumor resection as compared to baseline (Fig. 5d, left, and Extended Data Fig. 10c, left; α-chains: P = 0.03, n = 14; β-chains: P = 0.01, n = 14), perhaps reflecting tumor debulking and a consequent drop in antigen load. No significant differences were detected in the proportion of expanded intratumoral regional TCRs (Fig. 5d, right, and Extended Data Fig. 10c, middle) or the number of expanded nontumor lung TCRs (Fig. 5e and Extended Data Fig. 10c, right) between baseline and follow-up blood. Although the population of expanded intratumoral ubiquitous TCRs detected in blood decreased after surgery, many intratumoral TCRs (both ubiquitous and regional) were detected in the blood many months later (median time to follow-up was greater than 2 years), suggestive of an established and stable memory response. Interestingly, the changes in the repertoire at disease recurrence (Extended Data Fig. 10d) showed varied patterns, with some expanded TCRs increasing in frequency while others decreased in frequency, perhaps reflecting the dynamic nature of the tumor antigenic landscape.
The dynamic nature of the tumor immune response was further illustrated in patients for whom we had three longitudinal blood samples (Fig. 5f). For patients CRUK0013 and CRUK0046, we observed the disappearance of a number of expanded intratumoral ubiquitous TCRs in follow-up blood and their reappearance in blood taken at disease recurrence. In patient CRUK0048, for whom blood samples were available at two time points during disease recurrence, we observed sequential disappearance and reappearance of expanded intratumoral ubiquitous TCRs after disease recurrence. In all three cases, a number of expanded intratumoral ubiquitous TCRs were observed at all three time points studied.
Discussion
We have used a robust, economical and quantitative TCR gene sequencing pipeline[36] to characterize the TCR repertoire in multiregion primary tumor samples, nontumor lung and PBMCs from patients with early-stage, treatment-naive NSCLC.
We uncover a rich repertoire in both tumor and nontumor lung samples, comprising thousands of different TCR sequences, many present at low frequency. This repertoire may contain many irrelevant bystander T cells[40], attracted by a strong intratumoral inflammatory response. We hypothesized that T cells that recognize tumor antigens would be enriched within the tumor microenvironment, owing to antigen-driven expansion and antigen-dependent T cell arrest[40, 46, 48]. We therefore focused on TCR sequences that were found at the highest frequency within the tumor. Although this expanded population contains only a very small subset of the sequences, it is a dominant feature of the intratumoral immune microenvironment, accounting for almost 20% of the total TCRs that we observe, and is therefore very stable to sampling effects.
The vast majority of the expanded intratumoral TCRs were differentially expressed in the tumor as compared to adjacent nontumor lung. Furthermore, the number of expanded intratumoral TCRs significantly correlated with the number of nonsynonymous mutations, suggesting that this population may contain neoantigen-specific TCRs. We therefore focused on the spatial heterogeneity of this population, as a proxy for the tumor-specific immune response. Spatial heterogeneity in this expanded intratumoral TCR repertoire was correlated with spatial mutational heterogeneity, suggesting that this expanded TCR repertoire is driven by the intratumoral neoantigen landscape, sculpted by focal HLA loss or antigen processing defects.
We therefore further classified expanded intratumoral TCRs into ubiquitous (present in all regions) and regional (present in a subset of regions) TCRs. A rigorous statistical framework was used to ensure that this classification did not simply reflect incomplete sampling. This TCR classification bore close parallels to the distribution pattern of mutation prevalence, where we could similarly define ubiquitous and regional mutations. Strikingly, the number of expanded intratumoral ubiquitous TCRs correlated with the number of ubiquitous nonsynonymous mutations, but not with the number of regional nonsynonymous mutations. Conversely, the number of expanded intratumoral regional TCRs correlated with the number of regional nonsynonymous mutations, but not with the number of ubiquitous nonsynonymous mutations. We therefore hypothesize that a proportion of ubiquitously expressed expanded intratumoral TCRs may recognize mutations that are ubiquitously found throughout the tumor and document a functional example by single-cell RNA-seq of tetramer-sorted neoantigen-reactive CD8+ T cells. Similarly, we hypothesize that some regional TCRs may recognize regional mutations. Additional single-cell paired TCR α-chain and β-chain sequencing will be required to confirm this hypothesis at a functional level.
The hypothesis that expanded intratumoral TCRs are expressed on tumor-reactive T cells was further supported by the observation that these TCRs formed clusters of sequence-related TCRs within the tumor and by evidence for convergent recombination in this population. Both TCR clustering and convergent recombination are characteristic of antigen-specific T cell immune responses[43, 44]. Furthermore, expanded intratumoral ubiquitous TCRs constituted a predominant proportion of the PD-1+CD57− and PD-1+CD103+ effector tissue-resident CD8+ T cells, in line with ongoing local antigen-driven T cell activation, tumor specificity and T cell dysfunction[40, 45].
This study highlights the dynamic complexity of the antitumoral immune response in NSCLC, across space and time, with important clinical implications. The use of neoantigen-reactive T cells or tumor-specific TCRs for adoptive cellular immunotherapy is being actively explored in many laboratories. However, as we demonstrate, the intratumoral T cell repertoire is complex, comprising T cells that may respond to both ubiquitous and regional neoantigens. Furthermore, while a high number of clonal neoantigens is associated with better clinical outcome, we found no evidence for a similar protective effect of a larger number of expanded intratumoral TCRs, whether total, ubiquitous or regional. The dysfunctional phenotype of the clonally expanded intratumoral T cells may suggest that this T cell population fails over time and, in the absence of therapeutic intervention, is unable to control tumor growth. Harnessing intratumoral T cells for therapeutic purposes will therefore depend on identifying ubiquitous TCRs and being able to successfully correct their dysfunctional state. Encouragingly, many of the ubiquitously expressed expanded intratumoral TCRs were frequently detected in the blood at the time of tumor resection. The ability to track and isolate these cells from the blood rather than tumor tissue may help improve clinical outcomes for patients with NSCLC by providing a noninvasive method for monitoring and accessing neoantigen-specific T cells for personalized immunotherapeutic strategies.
Methods
Patient cohort
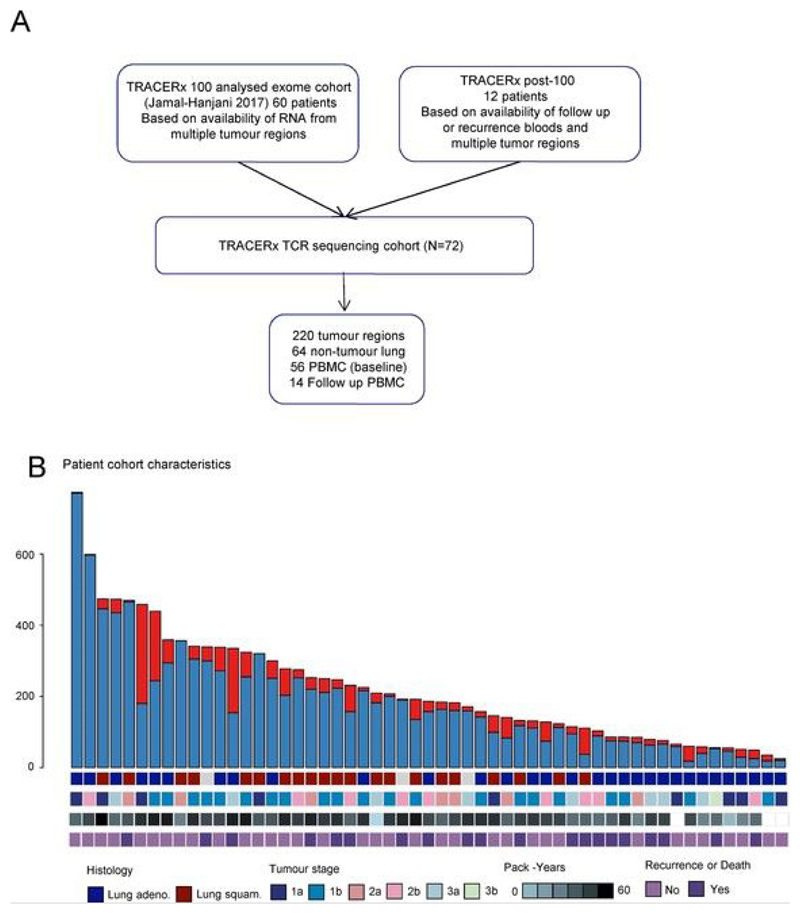
All patients within the TCR sequencing study were recruited to the lung TRACERx study (Research Ethics Committee no. 13/L0/1546). Patients with sufficient RNA from at least two tumor regions were selected for the TCR sequencing study. Samples from adjacent nontumor lung and PBMCs taken at the time of resection, as well as a number of follow-up PBMC samples, were sequenced whenever these were available. All tissue specimens were reviewed by a lung pathologist before being selected, as previously described[39]. The clinical characteristics of the patients as on 15 April 2018 are provided in Extended Data Fig. 1b, and this was used as a censor date in all the analysis.
TCR sequencing
TCR α-chain and β-chain sequencing was performed by utilizing whole RNA extracted from NSCLC tumor samples and nontumor lung tissue or from cryopreserved PBMC samples, by using a quantitative experimental and computational TCR sequencing pipeline described previously[36, 41]. An important feature of this protocol is the incorporation of a UMI attached to each cDNA TCR molecule that enables correction for PCR and sequencing errors[36, 38]. The suite of tools used for TCR identification, error correction and CDR3 extraction is freely available at https://github.com/innate2adaptive/Decombinator. The raw DNA fastq files and the processed TCR sequences will be available on the NCBI Short Read Archive and Github, respectively, following publication.
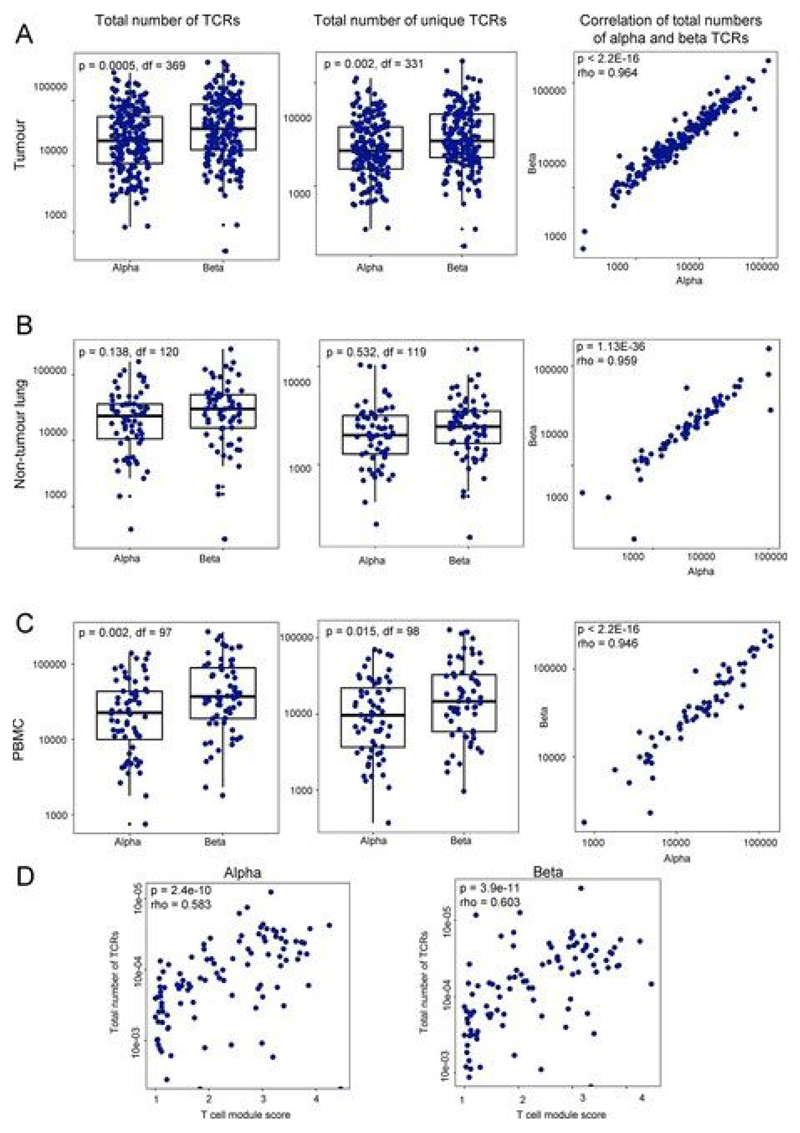
The numbers of TCR α-chain and β-chain transcripts sequenced from multiregion tumor specimens, nontumor lung and PBMCs were highly correlated (Extended Data Fig. 2a–c, right). We consistently detected more β-chains than α-chains, most likely owing to the higher number of β-chain transcripts[36]. To validate the sequencing efficiency, we correlated the number of α-chain and β-chain transcripts with matched bulk RNA-seq data for the tumor regions studied, quantifying T cell infiltration with a previously validated T cell transcriptional module[49]. The TCR transcripts were highly correlated with the T cell module score (Extended Data Fig. 2d). We note that, on average, each unique TCR:UMI combination was seen more than ten times in the raw uncorrected data, making it unlikely that these singletons arose from sequencing errors.
We used a previously described qPCR method to measure the total number of TCR α-chain or β-chain transcripts in the tumor samples, as a proxy for the total number of TCRs expressed. By standardizing these measurements against TILs, where the absolute number of T cells was calculated from flow cytometry (CD3) and cell counting, we could estimate the number of T cells present in the tumor samples. We could then compare the total number of T cells, as estimated by the qPCR method, with the total number of TCRs we obtained by the TCR-seq protocol outlined above. We obtained an estimated coverage of 7 ± 2% for α-chains and 13 ± 3% for β-chains (n = 8). The higher efficiency of the β-chain may reflect the higher number of transcripts per cell, as mentioned above.
Rényi entropy
The Rényi entropy is a generalized measure of diversity given by
where α is a scale of values, ranging from zero to infinity. As α approaches zero, more weight is given to the rare items in the sample (in this case, rare TCRs); the closer α gets to infinity, the more weight is given to the more common items (more abundant TCRs). α = 0 corresponds to the ecological diversity measure of ‘richness’, the proportion of all individual items that only occur once. α = 1 corresponds to the widely used Shannon entropy, while α = 2 corresponds to the Simpson diversity. We calculated the Rényi diversity at a range of α values by using the renyi function from the vegan package in R (https://cran.r-project.org/web/packages/vegan/index.html). Rényi values are sensitive to sample size, so all repertoires were repeatedly (100 times) subsampled to the same number of TCRs (5,000) before calculating the Rényi. The plots show the average over subsamples, for each tumor region or nontumor lung.
TCR frequency distribution
The frequency distribution of TCR abundance (the number of times we observed TCRs (once, twice, etc.)) fell on an approximate straight line on a log–log plot of frequency versus abundance (see example in Fig. 1). We fitted a discrete power law distribution (of the form f(k) = Ck−α where ζ denotes the Reimann zeta function and f(k) is the frequency of TCRs detected k times), by maximum-likelihood estimation as described in ref. [50].
Classification of expanded TCRs in tumor and nontumor lung
We counted the number of TCRs detected with frequencies above a range of frequency thresholds in the tumors or nontumor lung samples (Fig. 1c). To focus on the most expanded TCRs, we examined those present above a threshold frequency of 2/1,000 (corresponding to the top 1% of the empirical TCR frequency distribution) in at least one region of the tumor. However, the results we obtained for correlation with the number of mutations held true over a broad range of cutoff values, above a minimum frequency of around 1–2 in 1,000. In Fig. 1e and Extended Data Fig. 3c, we calculate the relative abundance of the TCR in the tumor (averaged over all regions) versus the abundance in paired nontumor lung from the same patient. The P value for the difference in abundance between tumor and nontumor lung was calculated with the poisson.test function in R, as the data were counts.
Repertoire intratumoral similarity and diversity measures
The similarity between two TCR repertoires was assessed with the normalized dot product (also known as the cosine similarity) between the vectors of TCR abundance. This measure is a well-established metric widely used in machine learning to compare numerical vectors and gives a value between 0 (no similarity, that is, orthogonal vectors) and 1 (complete similarity, from vectors with an identical direction in the feature space). Each pair of repertoires is represented as two vectors of equal length, indexed by the union of TCRs found in both repertoires and containing the number of times each TCR is detected in each of the two repertoires (each position contains an integer ≥0). The similarity between the two vectors is given as
where TCRl and TCR2 are the abundance vectors, • represents the vector product and paired vertical bars represent the Euclidean norm of the vector.
A similar index was used to calculate the genomic mutational similarity. In this case, the two vectors contained the corrected mutation prevalence in the tumor region.
The normalized Shannon diversity was estimated for each expanded TCR by using the command entropy.empirical from the entropy R package, on the basis of the observed frequency of the TCR across all regions (Hnorm = −(∑pi × log(pi))/ln(N), where H is the diversity, pi is the probability of being observed in the ith region and N is the number of regions). pi is obtained by dividing the observed frequency in region i by the sum of the frequencies across regions such that ∑pi = 1. Hnorm lies between 0 (TCR found in one region only) and 1 (TCR evenly found across all regions). To derive a metric for each patient, we computed the average of the diversity scores obtained for all expanded TCRs.
A similar index was used to calculate the genomic mutational diversity. In this case, Shannon diversity was obtained for each nonsynonymous mutation with the same formula, where pi was obtained by dividing the observed corrected prevalence of the mutation in the ith region by the sum of the corrected prevalence across regions such that ∑pi = 1.
Ubiquitous and regional TCR definitions
Expanded intratumoral TCRs were subsequently classified as ubiquitous or regional. We first determined the probability that a TCR was absent from a region owing to sampling. For each TCR, we compared the likelihood of the data given two alternative models. In model 1 (the null model), the TCR counts are drawn from a single Poisson distribution with the mean equal to the mean of all regions. In model 2, the TCR counts are drawn from a mixed distribution, where the one or more regions has no TCR with probability of 1, and the remaining regions are drawn from a Poisson distribution. We then calculate the log-likelihood ratio between the two models. Finally, for each TCR, we ran both models 1,000 times, drawing independent deviates from a Poisson distribution with the mean equal to the mean of all regions. We calculated the proportion (p) of simulations in which a log-likelihood ratio was observed that was greater or equal to the one observed with the real data. This procedure gave us a nonparametric estimation of the P value, correcting for the increased complexity of model 2. The algorithm was implemented in R and was run on all TCRs in each tumor. A TCR was deemed absent if the P value (corrected for multiple testing) was less than 0.05 (TCRs where the null model was significantly less likely to explain the data than the alternative model 2).
Expanded TCRs were then classified as regional if they were absent from at least one region of the tumor and as ubiquitous if otherwise. Thus, ubiquitous TCRs can therefore be absent from the data for specific regions, but this is attributed to sampling rather than true spatial heterogeneity.
CDR3 clustering and convergent recombination
The CDR3 protein sequences of expanded ubiquitous TCRs were identified with the package CDR3translator (https://github.com/innate2adaptive/Decombinator). The pairwise similarity between pairs of TCRs was measured on the basis of amino acid triplet sharing[42]. Sharing was quantified using, the normalized string kernel function stringdot (with parameters stringdot (type = ‘spectrum’, length = 3, normalized = TRUE) from the Kernlab package[51]. The kernel is calculated as the number of amino acid triplets (sets of three consecutive amino acids) shared by two CDR3s, normalized by the number of triplets in each CDR3 being compared. The TCR similarity matrix was converted into a network diagram by using the iGraph package in R[52]. Two TCRs were considered connected if the similarity index was >0.82. We explored a range of thresholds and chose the lowest threshold that consistently gave few large (>3) clusters with random samples of TCRs from the study.
As an additional control in the TCR clustering analysis, we took expanded ubiquitous TCRs from two patients and mixed them in silico, and we then looked to see whether the resulting clusters were primarily composed of TCRs from individual patients. We analyzed three pairs of patients in whom we observed prominent clustering in this way. The proportion of clusters that were ‘specific’ (that is, no mixing of TCRs from different patients) was 84%, 89% and 80%. Thus, most clusters segregated between repertoires. In the clusters in which we observed mixing, there was usually an overwhelming majority of TCRs from one or the other patient (Extended Data Fig. 8e).
Calculation of cluster diversity
We wanted to capture whether each cluster was composed predominantly of TCRs found in the same region or contained TCRs that were present in many different regions. For each CDR3 cluster, we therefore calculated the Shannon diversity, which captured the contribution of each possible combination of regions to the cluster. If n is the number of regions for a given patient, there are N = 2n – 1 possible combinations of regions of any given size. The Shannon diversity is given by Hnorm = −(∑pi × log(pi))/ln(N), where H is the diversity, pi represents the relative contribution of the ith combination to the cluster and N is the number of combinations. pi is obtained by dividing the count of CDR3s belonging to the ith combination by the total number of CDR3s in the cluster such that ∑pi = 1. Hnorm lies between 0 (a cluster composed of CDR3s belonging to one combination only) and 1 (a cluster evenly composed of all possible combinations). To derive a metric for each patient, we computed the average of diversity scores obtained for all clusters.
GLIPH
Using the TCR similarity matrix mentioned above, for each patient, we ran GLIPH (https://github.com/immunoengineer/gliph) on the 3,000 CDR3 β-chain sequences most similar to any of the expanded ubiquitous sequences (3,000 was picked because of the consistency of the resulting clustering). We ran the gliph-group-discovery Perl script with parameters that demonstrated efficient and quick clustering results (--simdepth = 100 --kmer_mindepth = 30 --local = 0). Results presented were derived from the convergence-groups.txt output files, obtained individually for each patient. From this output, we asked how many ubiquitous CDR3s fell into one cluster and counted the number of distinct clusters containing such a sequence. As a control, we repeated the process a 100 times, replacing the true ubiquitous TCRs with a same-sized random set of TCRs from the same patient repertoire, and then counted how many clusters contain a ‘false’ ubiquitous CDR3.
Convergent recombination
To measure convergent recombination, we counted the average number of TCR DNA sequences (as determined by Decombinator) that gave rise to each expanded intratumoral ubiquitous CDR3 sequence. As a control, we used either expanded regional intratumoral CDR3 sequences or a set (the same number) of randomly selected CDR3 sequences from the same patient intratumoral repertoire. For each patient, we plotted the average number of DNA sequences per CDR3 sequence.
Multiregion whole-exome sequencing analysis
Whole-exome sequencing of multiregion tumor specimens and matched germline samples derived from whole blood was performed as previously described[39]. The calling of mutations was described in detail in that paper, but is briefly summarized again here.
Raw paired-end reads (100 bp) in fastq format generated by the Illumina pipeline were aligned to the full hg19 genomic assembly (including unknown contigs) obtained from GATK bundle 2.88, by using bwa mem (bwa-0.7.7). Picard tools v1.107 was used to clean, sort and merge files from the same patient region and to remove duplicate reads (http://broadinstitute.github.io/picard). A combination of Picard tools (1.107), GATK (2.8.1) and FastQC (0.10.1) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to produce quality-control metrics. SAMtools mpileup (0.1.19) was used to locate non-reference positions in tumor and germline samples. Bases with a Phred score of <20 or reads with mapping quality <20 were omitted. BAQ computation was disabled, and the coefficient for downgrading mapping quality was set to 50. VarScan2 somatic (v2.3.6) utilized output from SAMtools mpileup to identify somatic variants between tumor and matched germline samples. With the exception of minimum coverage for the germline sample that was set to 10, the minimum variant frequency that was changed to 0.01 and tumor purity that was set to 0.5, default parameters were used. VarScan2 processSomatic was used to extract the somatic variants. The resulting single-nucleotide variant (SNV) calls were filtered for false positives with VarScan2’s associated fpfilter.pl script, initially with default settings and then with min-var-frac = 0.02, having first run the data through bam-readcount (0.5.1) (https://github.com/genome/bam-readcount). MuTect (1.1.4) was also used to detect SNVs with annotation files contained in GATK bundle 2.8. Following completion, variants called by MuTect were filtered according to the filter parameter ‘PASS’.
To further reduce false-positive variant calls, additional filtering was performed. An SNV was considered a true positive if the variant allele frequency (VAF) was greater than 2% and the mutation was called by both VarScan2, with somatic P value ≤ 0.01, and MuTect. Alternatively, a frequency of 5% was required if the variant was only called in VarScan2, again with somatic P value ≤ 0.01. Additionally, sequencing depth in each region was required to be ≥30 and ≥5 sequence reads had to support the variant call. In contrast, the number of reads supporting the variant in the germline data had to be <5 and the VAF was required to be ≤1%. In addition to these sample-specific measures, we also used the cohort to reduce SNP contamination through two independent means. First, all variants designated as ‘germline’ by VarScan2, from all regions, were combined so that every germline variant detected in the cohort had an associated TRACERx population frequency. SNVs were filtered out if they were found to have >1% frequency in the TRACERx cohort. In an effort to reduce the impact of direct sample-to-sample contamination, the SNVs from each patient were compared against the germline SNPs in every other patient independently. If >5% of SNVs were identified as SNPs in another patient, the sample was flagged as contaminated and any such variant that matched a SNP was removed from further analysis. Finally, a blacklist filter, relating to the genomic location of the variant, was applied. The blacklisted genomic regions were obtained from UCSC Genome Table Browser[53] and include regions excluded from the ENCODE Project (both DAC and Duke list), simple repeats, segmental duplications and microsatellite regions. Additionally, M-seq provides the opportunity to increase the sensitivity to detect low-frequency mutations. By sharing the independently called mutations across the multiple regions and reassessing the reads at each position for each tumor region, it is possible to call more mutations and reduce the possibility of over-representing the mutational heterogeneity. Where a somatic variant was not called ubiquitously across tumor regions but was called in one or more regions, read information was extracted from the original alignment file with bam-readcount (0.5.1) (https://github.com/genome/bam-readcount). In such cases, VAF restrictions were reduced to VAF ≥ 1%, allowing for the positive identification of low-frequency variants that would otherwise have been missed.
Copy number for each mutation from paired tumor–nontumor lung data was generated with VarScan2 (v2.3.6). VarScan2 copynumber was run using default parameters except min-coverage = 8 and min-segment-size = 50. The data-ratio parameter was calculated on a per-sample basis as described by Koboldt and colleagues[54]. VarScan2 copynumber produced per-region log(R) values. log(R) values were subsequently corrected for G+C content with a method based on wave-pattern G+C correction by Cheng and colleagues[55]. Homozygous and heterozygous SNPs were called in the germline sample by Platypus v0.8.128 with default parameters apart from the genlndels flag set to FALSE. Tumor regions from the same patient were then genotyped on the basis of the variants identified in the germ line. Only SNPs with a minimum coverage of 20× in the germ line and all tumor regions from the same patient were taken forward for copy number analysis. The B-allele frequency (BAF) of each SNP was calculated as the proportion of reads at that position that contained the reference base versus the variant. All SNPs were filtered by a list of previously classified poor quality SNPs constructed from the germline samples. A SNP was classified as poor quality if it was found in at least 20 samples and the number of times a given SNP showed a BAF value in the germ line that was between 0.1–0.32 or 0.68–0.9 was higher than the number of times the same SNP showed a BAF of either <0.1 or >0.9, multiplied by 0.13469. For SNPs that had undergone allelic imbalance, it was possible to track whether the parental origin of the major allele relative to the minor allele swapped between different tumor regions, indicating microsattelite instability events.
log(R) and BAF values for each tumor region were processed by ASCAT v2.329 with default parameters except that ‘gamma’ was set to 1, to provide segmented allele-specific copy number data plus cellularity and ploidy estimates for all samples. Manual verification was performed of the automatically selected models for ploidy and cellularity by using an orthogonal measure of tumor cellularity based on mutation variant allele fraction, as described below. Floating-point copy number values were used for all copy number analysis.
Finally, all mutations were corrected for copy number variation and tumor purity. To calculate the variant frequency of each mutation, local copy number (obtained from ASCAT), tumor purity (also obtained from ASCAT) and VAF were integrated. In brief, for a given mutation, we first calculated the observed mutation copy number, nmut, describing the fraction of tumor cells carrying a given mutation multiplied by the number of chromosomal copies at that locus by using the following formula
where VAF corresponds to the variant allele frequency at the mutated base and p, CNt and CNn are the tumor purity, the tumor-locus-specific copy number and the normal-locus-specific copy number, respectively (CNn was assumed to be 2 for autosomal chromosomes). We then calculated the expected mutation copy number, nchr, by using the VAF and assigning a mutation to one of the possible local copy number states with maximum likelihood. In this case, only the integer copy numbers were considered. The corrected mutant frequencies of a mutation present in every cell of the tumor is therefore predicted to be 1. Further details of mutation calling and validation are given in the cited manuscript.
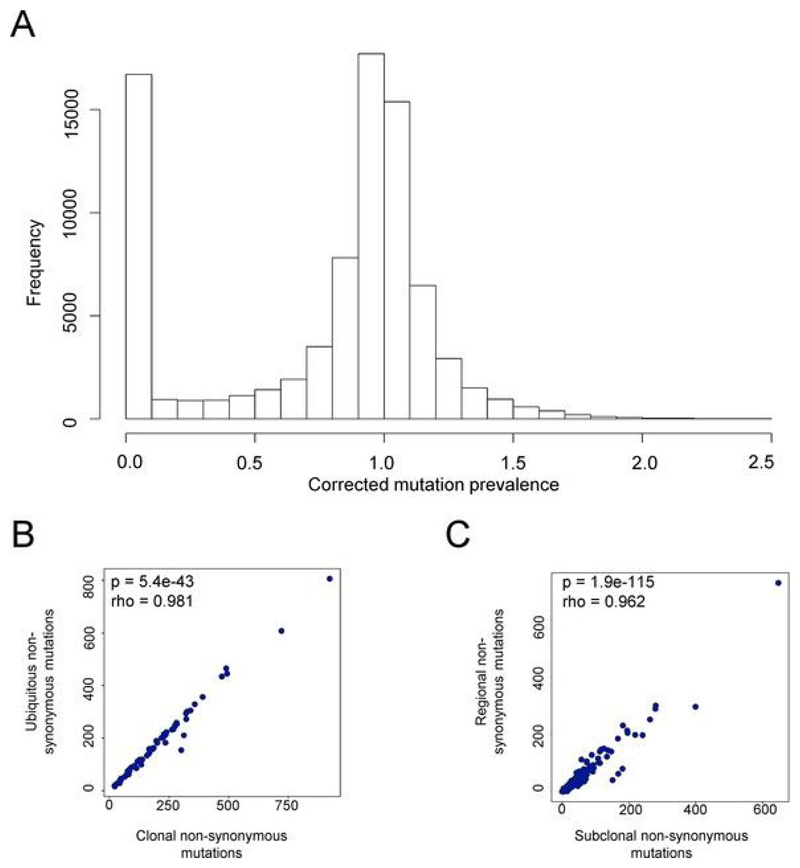
Classification of ubiquitous and regional mutations
We selected for further analysis all mutations classified as nonsynonymous variants, plus all mutations that introduced or removed a stop codon or introduced a frame shift. The distribution of mutation frequencies observed is shown in Extended Data Fig. 5a. The distribution is clearly bimodal, with one peak at very low frequencies and one with a mode of 1. On the basis of this distribution, we therefore defined a hard threshold of 10% and classified all mutations with frequencies of less than 10% as absent and all mutations with frequencies of greater than 10% as present. Because the distribution was sharply biomodal, the actual number of mutations in each category was not much affected by decreasing or increasing the threshold by up to 50% either way. Finally, we classified each mutation as ubiquitous if it was present in all regions of a tumor and regional if it was absent from at least one region. We chose this definition because, in terms of T cell recognition, we felt that the presence or absence of a mutation in a region was a more relevant parameter than the evolutionary history of that mutation (whether it was truncal or subclonal, for example). However, as expected, the number of ubiquitous mutations was highly correlated with the number of clonal mutations (Extended Data Fig. 5b), as defined in Jamal-Hanjani et al.[39], and the number of regional mutations was correlated with the number of subclonal mutations (Extended Data Fig. 5c).
The sequencing data have been deposited in the European Genome–phenome Archive under accession EGAS00001002247.
Multiregion RNA-seq
RNA was extracted from the TRACERx 100 cohort by using a modification of the AllPrep kit (Qiagen) as described in Jamal-Hanjani et al.[39]. RNA integrity (RIN) was assessed by TapeStation (Agilent Technologies). Samples that had an RIN score ≥5 were sent to the Oxford Genomics Centre for whole-RNA (RiboZero-depleted) paired-end sequencing. The ribodepleted fraction was selected from the total RNA provided before conversion to cDNA. Second-strand cDNA synthesis incorporated dUTP. The cDNA was end repaired, A-tailed and adaptor ligated. Before amplification, samples underwent uridine digestion. The prepared libraries were size selected and multiplexed, and underwent quality control before paired-end sequencing. Reads were 75 bp in length. fastq data underwent quality control and were aligned to the hg19 genome with STAR 27. Transcript quantification was performed by using RSEM with default parameters. The RNA-seq data will be deposited in the European Genome–phenome Archive following publication.
Transcriptional modules and RNA-seq
The methods for measuring the transcriptional activity in gene modules are described in detail in Pollara et al.[56]. Briefly, a set of genes that identify either a cell type or a functional state (for example, IFN response) is first defined. In this study, we used gene sets reported and validated in Danaher et al.[49] or in Pollara et al.[56]. The sets of genes for each cell type are listed in Supplementary Table 1. Within each sample, we used the RNA-seq data to calculate the geometric mean of the transcriptional abundance of each gene in the set (in transcripts per million as a normalized measure of transcriptional abundance). This mean defines the transcriptional activity of each module and was then correlated with the number of ubiquitous or regional TCRs detected within that sample. The correlation matrix for ubiquitous and regional TCRs was displayed using the correl function in R, where the area of the circles represents the correlation coefficient and an asterisk indicates the corrected significance P value.
RNA-seq of sorted TIL subsets and identification of specific TCRs
The BD FACSAria II flow cytometer was used to sort CD8+ TILs from NSCLC samples obtained from CRUK0017, CRUK0024, CRUK0069, CRUK0099 and CRUK0291. CD8+ TILs from CRUK0017, CRUK0024 and CRUK0069 tumors were sorted into two populations: (i) CD45RA−CCR7−CD57−PD-l+ cells and (ii) all other CD8+ T cells (‘not gate’). CD8+ TILs from CRUK0099 and CRUK0291 were sorted by flow cytometry into PD-1+CD103+ and PD-1+CD103− populations (Extended Data Fig. 9b). All cells were sorted into TRIzol followed by phenol-chloroform RNA extraction.
Where possible, equivalent amounts of total RNA (100 pg) from all samples were used for first-strand synthesis with the SmartERv3 kit (Takara Clontech) followed by 15–18 cycles of amplification (according to the manufacturer’s instructions). Sequencing libraries were produced from 150 pg input cDNA by using the Illumina Nextera XT library preparation kit. A 1:4 miniaturized version of the protocol was adopted (Fluidigm Single-Cell cDNA Libraries for mRNA sequencing, PN_100-7168_Ll). Libraries were sequenced on the Illumina NextSeq 500 platform with 150-bp paired-end kits according to the manufacturer’s instructions.
To search for tumor-expanded TCRs within the RNA-seq data, we used a tailor-made script in R (this will be made available on GitHub once the paper is published). In brief, for each TCR in the query set (typically the set of expanded TCRs identified in each tumor by TCR-seq), the algorithm selects a sequence of 20 bp that spans the CDR3 region of the TCR and is extended into the V and J region as necessary. This sequence is then searched for in the RNA-seq fastq file, looking at both forward and reverse reads and using both the original 20-bp tag and its reverse complement. The output of the algorithm is the number of hits found for each query TCR. 20-bp tags were found to be the minimum required to give very high specificity. For example, when we searched for expanded TCRs from one patient in the RNA-seq data from a different patient or if we searched for non-expanded TCRs in the RNA-seq data of the same individual, zero hits were recorded. There is therefore no evidence that the tags are mapped to other loci in the human transcriptome or that there is cross-reactivity between TCRs. We also searched for cases where the tags hit two TCRs, with different V or J sequences; no such cases were observed in our dataset. We are confident, therefore, that using 20-bp tags from our known expanded TCRs is the most specific and efficient way of finding these TCRs in the RNA-seq data.
To correct for depth of sequence or differences in RNA extraction and/or amplification, the script also searches for two TCR α-chain constant region and two TCR β-chain constant region tags of the same length. The counts for the two tags (averaged, although in practice they are very similar) give an estimate of the total number of TCRs present in the RNA-seq sample, which can be used to normalize between samples.
The algorithm therefore gives an estimate of the proportion of the TCRs present in a particular sample (for example, the PD-l+CD57− fraction of TILs) that can be attributed to the set of expanded TCRs in the tumor as a whole. Because the number of TCRs detected does not scale linearly with sample size, we cannot obtain a reliable measure of how many of the expanded TCRs are present in a sample, only the proportion of TCRs in that sample that can be assigned to our set of expanded TCRs.
Single-cell RNA-seq and TCR identification of neoantigen-reactive T cells
We have previously identified CD8+ T cells targeted against a clonal neoantigen (arising from the mutated MTFR2 gene) in NSCLC tumor regions derived from patient L011 (ref. [3]). We repeated the staining of neoantigen-reactive T cells based on dual-fluorescent multimer labeling, by using a freshly thawed vial of cryopreserved TILs from the same patient. Multimer-positive and multimer-negative single CD8+ T cells from NSCLC specimens were sorted directly into the Cl Integrated Fluidic Circuit (IFC; Fluidigm). Cell lysis, reverse transcription and cDNA amplification were performed as specified by the manufacturer. The SMARTer v4 Ultra Low RNA kit (Takara Clontech) was used for cDNA synthesis from the single cells. Following cDNA quantification and quality-control checks, Illumina NGS libraries were constructed with the Nextera XT DNA Sample Preparation kit (Illumina), according to the Fluidigm Single-Cell cDNA libraries for mRNA sequencing protocol. Sequencing was performed on the Illumina NextSeq 500 platform with 150-bp paired-end kits.
The bioinformatic reconstruction of TCRs from single-cell RNA-seq data was performed with both TraCeR and a modified version of Decombinator, with the methods showing good agreement in terms of final output. The scripts are available at https://github.com/innate2adaptive/Single-Tag-Decombinator and https://github.com/Teichlab/tracer.
Survival analyses
All survival analyses were performed in R with the survival package (https://cran.r-project.org/web/packages/survival/index.html). We analyzed the data by either Kaplan–Meier plots, splitting the patients according to whether they were above or below the top quartile of mutations or TCRs, or multivariate Cox regression, with the numbers of mutations and/or expanded TCRs as a quantitative explanatory variable.
Statistical analysis
Statistical analysis was performed in R. Correlation was carried out with the Spearman nonparametric rank correlation test. We used multivariate regression when appropriate, with log-transformed data for the number of mutations and the number of TCRs. We used the Mann–Whitney two-tailed paired or non-paired nonparametric tests (as appropriate) to determine whether two independent samples were selected from the same populations. P values were considered significant if less than 0.05, and significance values were corrected for multiple testing by Bonferroni correction when appropriate. Differences in disease-free survival were calculated with the Kaplan–Meier statistic or Cox multivariate regression.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41591-019-0592-2.
Extended Data
Extended Data Fig. 1. Patient selection, mutational burden and clinical characteristics.
a, CONSORT diagram showing the selection of TRACERx patients for TCR sequencing. b, The total number of nonsynonymous mutations (clonal and subclonal) and patient clinical characteristics (histology, stage, smoking status and clinical outcome) for the TCR sequencing cohort are shown.
Extended Data Fig. 2. Tumor and nontumor regions contain a highly diverse polyclonal TCR repertoire.
a–c, Graphs depicting the total number of TCR α-chain and β-chain segments sequenced (left), the number of unique TCR sequences detected (middle) and the correlation between the total number of TCR α-chain and β-chain segments sequenced (right) in multiregion tumors (n = 220) (a), nontumor lung (n = 64) (b) and PBMCs (n = 56) (c). Spearman’s rank correlation P values are shown. d, The relationship between the total number of TCRs in each region (expressed as log2) and the transcriptional score for a set of genes specifically expressed in T cells (see Methods). The Spearman’s rank correlation coefficient and P value are shown; n = 99.
Extended Data Fig. 3. NSCLC tumors contain expanded TCR β-chain sequences that are differentially expressed in tumor as compared to nontumor lung and whose numbers correlate with tumor mutational burden.
a, The frequency distribution of TCR β-chain abundance was fitted to a power law (f=kα) with maximum likelihood. The figure shows a representative plot (patient CRUK0046) for β-chain sequences from pooled tumor regions (red circles) and the matched nontumor lung sample (blue circles). The average power law parameter α, which corresponds to the slope on a log–log plot, was 2.5 ± 0.05 for tumor and 2.6 ± 0.03 for nontumor. The x axis refers to TCR abundance (size of clone), and the y axis refers to the proportion of the repertoire. b, The number of β-chain sequences detected above a given frequency threshold is shown for tumor (n = 72, multiple tumor regions were pooled from an individual patient; red circles) and matched nontumor lung samples (n = 64; blue circles). c, A volcano plot showing the likelihood (−log10 (P value)) of a β-chain sequence being sampled from two populations of equal mean in tumor and nontumor lung, plotted against the differential expression in tumor versus nontumor lung. If the log likelihood was >120, it was given a value of 120 for plotting purposes. Blue circles represent β-chain sequences expanded (>0.002) in nontumor lung; red circles represent β-chain sequences expanded in tumor lung. d, The proportion of expanded tumor α-chain sequences (T) or expanded nontumor lung β-chain sequences (NTL) that are specific to their respective tissue; this is defined on the volcano plot as TCRs that have a P value <0.01 and a differential abundance of at least two between the tissues. The two proportions are significantly different, with the Mann–Whitney P value shown; n(tumor) = 72; n(nontumor lung) = 64. e, The correlation between the number of unique intratumoral expanded β-chain sequences (frequency ≥2/1,000) and the number of nonsynonymous mutations is shown for all patients. The Spearman’s rank correlation and P value are shown (n = 62). f, The Spearman’s rank correlation coefficient and P value (shown above each point; n = 62) are shown for the relationship between the number of unique intratumoral expanded β-chain sequences at different frequencies (ranging from all TCRs (threshold of zero) up to those found at frequency ≥ 8/1,000) and the number of nonsynonymous mutations.
Extended Data Fig. 4. The heterogeneity of TCR repertoires across different regions of tumors differs between patients and correlates with genomic heterogeneity.
a, The heat maps show the abundance (log2 of the number of times each TCR is found) of expanded intratumoral β-chain sequences (frequency ≥ 2/l,000) in different tumor regions for several patients. Patient ID is shown above each heat map. Each row represents one unique sequence. Each column represents one tumor region. b, The TCR repertoire of multiple regions of a patient’s tumor were sequenced and a pairwise comparison of the repertoires of different regions of the same tumor was performed by using the cosine similarity (see Methods). The pairwise intratumoral TCR repertoire similarity (β-chain sequences) is shown for each patient. Each circle represents a comparison between two regions of the same patient’s tumor. Patients are ordered by descending rank of mean intratumoral TCR similarity. c, TCR repertoire (β-chain sequences) diversity plotted against genomic diversity for each patient. The diversity measurement is calculated as the normalized Shannon entropy as described in the Methods. The Spearman’s rank correlation and P value are shown; n = 41. d,e, TCR repertoire for α-chain (d) and β-chain (e) sequence pairwise similarity plotted against genomic similarity for each pair of tumor regions (within patient comparison). The TCR and mutational pairwise similarities are both measured as cosine similarity, as described in the Methods. The Spearman’s rank correlation and P value are shown; n = 226. Dashed lines represent median values.
Extended Data Fig. 5. Mutation prevalence defines ubiquitous and regional mutations in NSCLC.
a, The frequency histogram of corrected mutation prevalence for all mutations in the TRACERx patient cohort analyzed in this paper. Mutation prevalence (number of mutant reads/number of wild-type reads) was corrected for tumor purity and local genomic copy number as described in the Methods. The distribution is bimodal, with peaks at zero (0–10%, very few mutant reads) and 1 (corresponding to every cell in a tumor region carrying the mutation on one chromosome). b, The number of ubiquitous mutations defined as described in the Methods is plotted against the number of clonal mutations, calculated as described in Jamal-Hanjani et al.[39] for all patients analyzed in this study. c, The number of regional mutations defined as described in the Methods is plotted against the number of subclonal mutations, calculated as described in Jamal-Hanjani et al.[39].
Extended Data Fig. 6. The number of ubiquitous and regional TCRs correlates with the number of ubiquitous and regional nonsynonymous mutations, respectively.
a, The numbers of expanded (frequency ≥ 2/l,000) ubiquitous (red circles) and regional TCR (β-chain) sequences (gray circles) is shown for each tumor region. The number of ubiquitous mutations is greater than the number of regional mutations, with the Mann–Whitney P value shown; n = 52. b, The frequency distribution of the intratumoral expanded β-chain ubiquitous (red circles) and regional (gray circles) TCRs is shown. The two distributions were not significantly different when compared by the Kolmogorov–Smirnov test, P = 0.78. c, The number of expanded ubiquitous (top) or regional (bottom) β-chain sequences is plotted against the number of ubiquitous or regional nonsynonymous mutations for each tumor region. The Spearman's rank correlation coefficient and associated P value are shown; the dashed lines indicate median values. n = 42. d, Patients were stratified according to the number of ubiquitous mutations. The red line indicates a ratio above the top quartile and the blue line indicates a ratio below the top quartile. The Kaplan–Meier statistical P value is shown.
Extended Data Fig. 7. Expanded intratumoral ubiquitous TCRs are associated with a TH1 and CD8+ T cell transcriptional signature in the tumor and have a phenotype consistent with tumor antigen reactivity.
a, Correlation between the numbers of expanded intratumoral ubiquitous and regional TCR β-chain sequences and the transcriptional expression score (geometric mean) for various immune-related gene sets, characterizing cell types or functional states (names indicated above heat map). Details of how the transcriptional scores are calculated are in the Methods. The area and color of the circles correspond to the magnitude of the correlation coefficient. The color key indicates Spearman’s rank correlation coefficient. *P < 0.05; **P < 0.0l; after Bonferroni correction. b, CD8+ TILs from CRUK0291 and CRUK0099 were sorted into two populations, PD-1+CD103+ and PD-1+CD103− cells. The flow cytometry gating strategy for a representative patient is shown (pre-gated on live > singlets > CD3+ > CD8+ T cells). RNA was extracted and sequenced from sorted populations as described in the Methods. c, The RNA-seq data were mined for the presence of expanded ubiquitous and regional α-chain and β-chain sequences. The heat maps show the number of times each expanded ubiquitous or regional TCR CDR3 sequence was found in each of the RNA-seq data from PD-1+CD103+ or PD-1+CD103− cells, as a proportion of the number of times a constant region sequence of the same length was detected. These proportions are scaled for each row and color coded. Each row represents a distinct expanded TCR sequence.
Extended Data Fig. 8. Network diagram of clusters of intratumoral CDR3 β-chain sequences shown for all patient CDR3 repertoires.
All panels show the network of TCR CDR3 β-chain sequences that are connected to at least one other expanded intratumoral ubiquitous TCR (shown as red circles). Clusters are defined as networks with at least two nodes. Only those patients with at least one cluster are shown.
Extended Data Fig. 9. Further analysis of TCR clusters.
a, The clustering algorithm was run on all patients, and the number of distinct clusters containing expanded ubiquitous and regional TCRs are shown. The number is normalized for the munber of expanded TCRs of each type. The Mann–Whitney P value is shown; n =46. b, A full alignment of the cluster shown in Fig. 3b,c. c, The GLJPH (https://github.com/immunoengineer/gliph) clustering algorithm was run on all patients. The panels show the number of distinct GLIPH clusters containing expanded ubiquitous, expanded regional and randomly selected CDR3 β-chain sequences. The number is normalized for the number of TCRs of each type. The ubiquitous TCRs show greater clustering than randomly selected TCRs (left), with the Mann–Whitney P value shown; n = 46. There was no significant difference between GLIPH clustering of normalized ubiquitous and regional expanded TCRs (right), with the Mann–Whitney P value shown; n = 46. d, The cluster Shannon diversity (see Methods) for all clusters containing ubiquitous or regional expanded TCRs. The Mann–Whitney P value is shown; n = 46. e, As an additional control in the TCR clustering analysis, we took expanded ubiquitous TCRs from patients CRUK0041 and CRUK0322 and mixed them in silico, and we then looked to see whether the resulting clusters were primarily composed of TCRs from individual patients.We analyzed three pairs of patients in whom we observed prominent clustering in this way. One representative example is shown.
Extended Data Fig. 10. Dynamic occurrence of expanded intratumoral ubiquitous TCRs in blood.
a, The proportion of expanded intratumoral ubiquitous (red circles) and regional (gray circles) TCRs (β-chain) detected within the blood for all patients (the Mann–Whitney P value is shown; n = 45). b, The frequency (number of TCR sequences detected, as a proportion of the total number of TCRs) of expanded intratumoral ubiquitous (red circles) and regional (gray circles) TCRs (β-chain) in the peripheral blood at the time of primary NSCLC surgery (the Mann–Whitney P value is shown; n = 42 for ubiquitous, n = 22 for regional). c, The proportion of expanded intratumoral ubiquitous (left), expanded intratumoral regional (middle) and expanded nontumor lung (right) TCRs (β-chain) that were detected in the blood at the time of primary NSCLC surgery and at routine follow-up (the median time to follow-up was just under 2 years) (the Mann–Whitney P value is shown; n = 14 for ubiquitous, regional and nontumor lung). d, The proportion of expanded intratumoral ubiquitous (left) and regional (right) α-chain (top) and β-chain (bottom) sequences that were detected in the blood at the time of primary NSCLC surgery and at disease recurrence (the median time to first recurrence was 350 d) (the Mann–Whitney P value is shown; n = 14 for α-chains and n = 15 for β-chains).
Editor’s Summary.
A survey of T cell repertoire evolution in the tumors, healthy tissue and blood of patients with early-stage untreated lung cancer offers an opportunity to monitor and identify neoantigen-specific T cells for personalized immunotherapy.
Acknowledgements
This work was undertaken with support from the Cancer Immunotherapy Accelerator Award (CITA-CRUK; C33499/A20265), CRUK’s Lung Cancer Centre of Excellence (C5759/A20465), the National Institute for Health Research UCL Hospitals Biomedical Research Centre (B.C., C.S., S.A.Q., M.N.), a Cancer Research UK (CRUK) Project Grant (B.C.), a CRUK Senior Cancer Research Fellowship (S.A.Q.; C36463/A22246), the Sam Keen Foundation, the Royal Marsden Hospital NHS Foundation Trust and Institute of Cancer Research Biomedical Research Centre, the Royal Marsden Cancer Charity, the UCL Biomedical Research Centre (K.J.), a Cancer Research UK studentship (M.R.D.M.) and an MRC Clinical Infrastructure award (MR/M009033/1). S.A.Q. receives funding from the Rosetrees and Stoneygate Trust (A1388), a CRUK Biotherapeutics Programme grant (C36463/A20764) and a donation from the Khoo Teck Puat UK Foundation via the UCL Cancer Institute Research Trust (539288). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the article. C.S. is Royal Society Napier Research Professor. C.S. is supported by the Francis Crick Institute (FC001169), the Medical Research Council (FC001169) the Wellcome Trust (FC001169), Cancer Research UK (TRACERx and CRUK Cancer Immunotherapy Catalyst Network), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees and Stoneygate Trusts, the Novo Nordisk Foundation (ID 16584), the Breast Cancer Research Foundation (BCRF), the European Research Council Consolidator Grant (FP7-THESEUS-6l7844), European Commission ITN (FP7-PloidyNet-607722), Chromavision (this project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement 665233), the National Institute for Health Research, the UCL Hospitals Biomedical Research Centre and the Cancer Research UK University College London Experimental Cancer Medicine Centre. We thank all the patients who participated in this study and all members of the TRACERx Consortium.
Footnotes
Author contributions
B.C., S.A.Q. and C.S. conceived the project. B.C., S.A.Q., C.S., K.J., M.I. and M.R.D.M. designed the experiments and analysis and wrote the manuscript. B.C., S.A.Q., C.S., T.E., M.N. and K.S.P. contributed to project management and supervision, as well as providing valuable critical discussion. K.J., J.L.R., I.U., A.W., T.O., V.T., A.J.S.F., A.G., Y.N.S.W., A.B.A., M.W.S., S.R.H. and E.H. contributed to the wet lab experiments. R.R., T.P., T.R., N.J.B., G.A.W., J.A.G.-A., J.H., E.G. and N.M. contributed to the bioinformatics analysis. M.J.-H., S.V., C.T.H., C.S., A.H. and the TRACERx Consortium coordinated clinical trials and provided patient samples and patient data.
Data availability
The RNAseq and exome sequence data used during the study is available through the Cancer Research UK & University College London Cancer Trials Centre (ctc.tracerx@ucl.ac.uk) for non-commercial research purposes and access will be granted upon review of a project proposal that will be evaluated by a TRACERx data access committee and entering into an appropriate data access agreement subject to any applicable ethical approvals. The TCRseq Fastq data was deposited at the short read archive (SRA) under accession code UB4501422.
Competing interests C.S. receives grant support from Pfizer, AstraZeneca, BMS and Ventana and has consulted for Boehringer Ingelheim, Eli Lilly, Servier, Novartis, Roche-Genentech, GlaxoSmithKline, Pfizer, BMS, Celgene, AstraZeneca, Illumina and the Sarah Cannon Research Institute. C.S. is a shareholder of Apogen Biotechnologies, Epic Bioscience and GRAIL and has stock options in and is co-founder of Achilles Therapeutics. S.A.Q. is a co-founder of Achilles Therapeutics. R.R., N.M. and G.A.W. have stock options in and have consulted for Achilles Therapeutics. J.L.R. has consulted for Achilles Therapeutics.
Extended data is available for this paper at https://doi.org/10.1038/s4159-019-0592-2.
Peer Review Information Joao Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at http://doi.org/10.1038/s41591-019-0592-2.
Supplementary Table 1 Please add a Supplementary note entitled TRACERx consortium names and affiliations. This file has been uploaded as an additional pdf.
Gene lists for the RNA-seq gene module analysis.
References
- 1.Anagnostou V, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenthal R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao D, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamal-Hanjani M, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol. 2014;12:e1001906. doi: 10.1371/journal.pbio.1001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng L, et al. Heterogeneity of tumor-infiltrating lymphocytes ascribed to local immune status rather than neoantigens by multi-omics analysis of glioblastoma multiforme. Sci Rep. 2017;7 doi: 10.1038/s41598-017-05538-z. 6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T, et al. The different T-cell receptor repertoires in breast cancer tumors, draining lymph nodes, and adjacent tissues. Cancer Immunol Res. 2017;5:148–156. doi: 10.1158/2326-6066.CIR-16-0107. [DOI] [PubMed] [Google Scholar]
- 8.Kuang M, et al. A novel signature for stratifying the molecular heterogeneity of the tissue-infiltrating T-cell receptor repertoire reflects gastric cancer prognosis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-08289-z. 7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin KR, et al. T cell receptor repertoire profiling predicts the prognosis of HBV-associated hepatocellular carcinoma. Cancer Med. 2018;7:3755–3762. doi: 10.1002/cam4.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuben A, et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom Med. 2017;2:10. doi: 10.1038/s41525-017-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, et al. Ultra-deep T cell receptor sequencing reveals the complexity and intratumour heterogeneity of T cell clones in renal cell carcinomas. J Pathol. 2013;231:424–432. doi: 10.1002/path.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62:1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson RO, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J Pathol. 2013;231:433–440. doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiménez-Sánchez A, et al. Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell. 2017;170:927–938. doi: 10.1016/j.cell.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui JH, et al. TCR repertoire as a novel indicator for immune monitoring and prognosis assessment of patients with cervical cancer. Front Immunol. 2018;9:2729. doi: 10.3389/fimmu.2018.02729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai X, et al. Characteristics of tumor infiltrating lymphocyte and circulating lymphocyte repertoires in pancreatic cancer by the sequencing of T cell receptors. Sci Rep. 2015;5 doi: 10.1038/srep13664. 13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balachandran VP, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui C, et al. T cell receptor β-chain repertoire analysis of tumor-infiltrating lymphocytes in pancreatic cancer. Cancer Sci. 2019;110:61–71. doi: 10.1111/cas.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin YB, et al. TCR repertoire profiling of tumors, adjacent normal tissues, and peripheral blood predicts survival in nasopharyngeal carcinoma. Cancer Immunol Immunother. 2018;67:1719–1730. doi: 10.1007/s00262-018-2237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gros A, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasetto A, et al. Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer lmmunol Res. 2016;4:734–743. doi: 10.1158/2326-6066.CIR-16-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavin Y, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thommen DS, et al. A transcriptionally and functionally distinct PD-1+CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmadzadeh M, et al. Tumor-infiltrating human CD4+ regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci Immunol. 2019;4:eaao4310. doi: 10.1126/sciimmunol.aao4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheper W, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 2019;25:89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, et al. TCR repertoire intratumor heterogeneity of CD4+ and CD8+ T cells in centers and margins of localized lung adenocarcinomas. Int J Cancer. 2019;144:818–827. doi: 10.1002/ijc.31760. [DOI] [PubMed] [Google Scholar]
- 27.Reuben A, et al. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 2017;7:1088–1097. doi: 10.1158/2159-8290.CD-17-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert L, et al. CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin Cancer Res. 2014;20:2424–2432. doi: 10.1158/1078-0432.CCR-13-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cha E, et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med. 2014;6:238ra270. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kvistborg P, et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6:254ra128. doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- 31.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder A, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-Ll blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS Med. 2017;14:e1002309. doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yusko E, et al. Association of tumor microenvironment T-cell repertoire and mutational load with clinical outcome after sequential checkpoint blockade in melanoma. Cancer Immunol Res. 2019;7:458–465. doi: 10.1158/2326-6066.CIR-18-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogan SA, et al. Peripheral blood TCR repertoire profiling may facilitate patient stratification for immunotherapy against melanoma. Cancer lmmunol Res. 2019;7:77–85. doi: 10.1158/2326-6066.CIR-18-0136. [DOI] [PubMed] [Google Scholar]
- 35.Hopkins AC, et al. T cell receptor repertoire features associated with survival in immunotherapy-treated pancreatic ductal adenocarcinoma. JCI Insight. 2018;3 doi: 10.1172/jci.insight.122092. 122092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakes T, et al. Quantitative characterization of the T cell receptor repertoire of naïve and memory subsets using an integrated experimental and computational pipeline which is robust, economical, and versatile. Front lmmunol. 2017;8:1267. doi: 10.3389/fimmu.2017.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uddin I, et al. An economical, quantitative, and robust protocol for high-throughput T cell receptor sequencing from tumor or blood. Methods Mol Biol. 2019;1884:15–42. doi: 10.1007/978-1-4939-8885-3_2. [DOI] [PubMed] [Google Scholar]
- 38.Best K, Oakes T, Heather JM, Shawe-Taylor J, Chain B. Computational analysis of stochastic heterogeneity in PCR amplification efficiency revealed by single molecule barcoding. Sci Rep. 2015;5 doi: 10.1038/srep14629. 14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamal-Hanjani M, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 40.Simoni Y, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 41.Thomas N, Heather J, Ndifon W, Shawe-Taylor J, Chain B. Decombinator: a tool for fast, efficient gene assignment in T-cell receptor sequences using a finite state machine. Bioinformatics. 2013;29:542–550. doi: 10.1093/bioinformatics/btt004. [DOI] [PubMed] [Google Scholar]
- 42.Sun Y, et al. Specificity, privacy, and degeneracy in the CD4 T cell receptor repertoire following immunization. Front lmmunol. 2017;8:430. doi: 10.3389/fimmu.2017.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dash P, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glanville J, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bengsch B, et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 2018;48:1029–1045. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bengsch B, et al. Coexpression of PD-1, 2B4, CD160 and KLRGl on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2017;6:e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganesan AP, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18:940–950. doi: 10.1038/ni.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danaher P, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clauset A, Shalizi CR, Newman ME. Power law distibutions in empirical data. J Soc Ind Appl Math. 2009;54:661–703. [Google Scholar]
- 51.Karatzoglou A, Smola A, Hornik K, Achim Z. Kernlab - An S4 Package for Kernal Methods in R. J Stat Software. 2004;11 pages. [Google Scholar]
- 52.Csardi G, Nepusz T. The igraph software complex for network research. Complex Systems. 2006;1695 [Google Scholar]
- 53.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng J, et al. Single-cell copy number variation detection. Genome Biol. 2011;12 doi: 10.1186/gb-2011-12-8-r80. R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollara G, et al. Validation of Immune cell modules in malticellular transcriptomic data. PLoS One. 2017;12:e0169271. doi: 10.1371/journal.pone.0169271. [DOI] [PMC free article] [PubMed] [Google Scholar]