Abstract
Hydrolyzable tannins are a class of polyphenolic compounds commonly found in natural products. In this work, we studied the in vitro inhibitory mechanism of six molecules in this class on ALKBH2, an Fe(II)/α-ketogluta-rate-dependent DNA repair enzyme in the AlkB family. We determined the IC50 values of these compounds on the repair of 3-methylcytosine and 1-methyladenine, the prototypical substrates of ALKBH2. A structure—activity relationship was also observed between the strength of inhibition and the number of galloyl moieties in a molecule. In addition, we found that the inhibition by this class of polyphenolic compounds on ALKBH2 is through an iron-chelating mechanism.
Graphical Abstract
INTRODUCTION
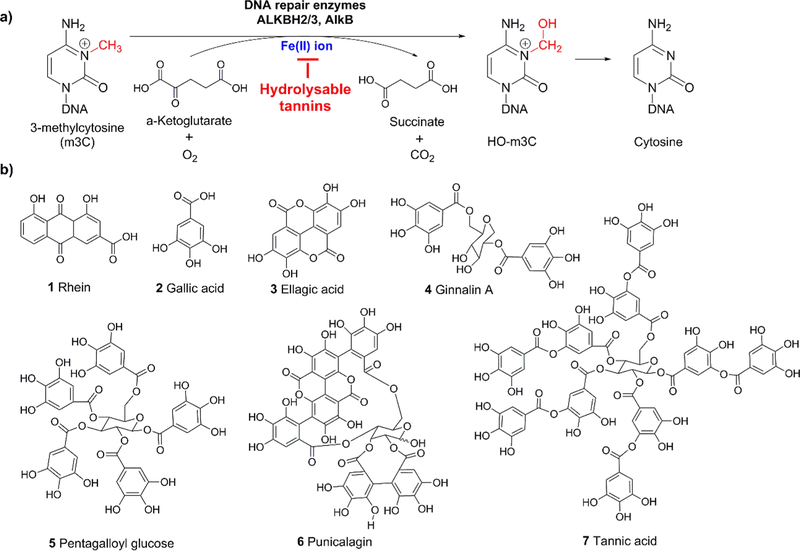
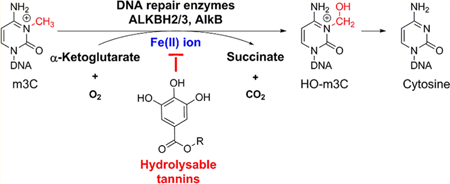
Hydrolyzable tannins including ellagitannins and gallotannins are a class of polyphenolic compounds that are commonly found in a variety of fruits and plants, such as berries and oak barks.1,2 These molecules share a general structural motif of galloyl group(s) and have been shown to exhibit many health benefits including antioxidative, antiproliferative, and neuroprotective effects.3 Recent studies have revealed the involvement of hydrolyzable tannins, as iron chelators, in mitigating iron-overload induced hepatotoxicity and neurological disorders.4 Despite the positive effects, concerns have been raised on the safety of overconsumption of natural products containing hydrolyzable tannins, especially with the lack of evidence in the relationship between cellular uptake of these compounds and activity of many iron-related enzymes. There are about 80 Fe(II)/α-ketoglutarate (αKG)-dependent enzymes in the human body, including TET, JmjC, and AlkB family proteins.5 In this paper, we focus on the DNA repair enzyme ALKBH2 in the AlkB family, which uses an Fe(II)/ αKG-dependent mechanism to repair alkyl nucleic acid lesions induced by endogenous and environmental alkylating agents (Figure 1a).6,7
Figure 1.
Repair mechanism of the AlkB family enzymes and structures of the hydrolyzable tannins and rhein.
The AlkB family proteins include nine human homologues (ALKBH1—8 and FTO). Among them, ALKBH2 has been identified as the major repair enzyme to lesions in both ss- and ds-DNA, and ALKBH3 has activity mainly on lesions in ss- DNA.5,6 A number of alkyl substrates have been reported for the AlkB enzymes including 3-methylcytosine (m3C), 1- methlyadenine (m1A), 1,N-ethenoadenine, 3,N4-ethenocytosine, and other alkyl adducts on the DNA bases. Among these adducts, m3C and m1A have been demonstrated as the best substrates of AlkB.6,8 Considering the important role of the AlkB enzymes in maintaining genome integrity and the previous observations of hydrolyzable tannins as metal chelators, we questioned whether hydrolyzable tannins could affect the activity of the AlkB proteins by chelating the cofactor Fe(II) ion. In this study, we investigated the potential of six hydrolyzable tannins (structures 2 to 7, Figure 1b), as a new class of inhibitor to the AlkB enzymes. We extracted and purified a series of ellagitannins and gallotannins to evaluate their inhibitory effects on the repair efficiency of m3C and m1A by the ALKBH2 protein by removing the Fe(II) ion cofactor in the enzymatic reaction. In addition, we observed a structure—activity relationship between the strength of inhibition and the number of galloyl groups in these hydrolyzable tannins. Our results offer a new perspective into the potential effect of overdosing hydrolyzable tannins that could lead to the inhibition of iron-dependent enzymes, which will be confirmed by in vivo studies with the consideration of bioavailability and metabolism.
EXPERIMENTAL PROCEDURES
Materials.
Rhein (1), gallic acid (2), ellagic acid (3), and tannic acid (7) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Ellagitannin (punicalagin 6) was isolated from a commercially available pomegranate extract (Pomella) provided by Verdure Sciences (Noblesville, IN, USA) using our previously reported methods.9 Gallotannins including ginnalin A (4) and pentagalloyl glucose (5) were isolated from a red maple leaf extract and sumac fruit extract, respectively, as we previously reported.10,11
DNA Synthesis.
Sixteen-mer oligonucleotides with a sequence of 5′-GAAGACCTXGGCGTCC-3′, where X designates m3C or m1A, were synthesized by applying automated solid-phase phosphoramidite chemistry on a Mermade-4 DNA synthesizer.12 The concentration of oligonucleotides were measured by NanoDrop (Thermo Scientific) under UV absorbance of 260 nm. The extinction coefficient (ε) of a lesion is calculated as its unmodified counterpart due to the tiny value difference in a 16mer context. All the oligonucleotides were purified by reverse-phase HPLC with a Phenomenex Luna Semi-Preparative (10 × 250 mm, 5 μm) C18 column. Solvent A was 100 mM 1:1 triethylamine-acetic acid (TEAA), and B was 100% acetonitrile. The oligonucleotides were characterized by LC-ESI-TOF-MS (AB Sciex) under negative ionization mode, the liquid chromatographic separation was conducted using a Phenomenex Luna C18 column (4.6 × 100 mm; 5 μm) at a flow rate of 0.4 mL/min. Solvent A was 10 mM ammonium acetate in water, and solvent B was 100% acetonitrile.
Protein Purification.
The ALKBH2 protein was expressed and purified as previously described.13 Briefly, the ALKBH2 gene was cloned into the pET28a+ vector and then transformed into E. coli BL21 (DE3) pLysS cell for expression. The his-tagged proteins were purified by affinity column chromatography using Fast Protein Liquid Chromatography (GE healthcare). Thrombin was added to digest His-tagged proteins overnight followed by further purification using ion-exchange chromatography. The purified ALKBH2 proteins were stored in an ALKBH buffer containing 300 mM NaCl, 10% glycerol, 50 mM N-[tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid, and 1 mM 2-mercaptoethanol under pH 8.0.
Enzymatic Assay and Inhibition Mechanism Study.
For the enzyme inhibition studies, 5 μM single-stranded m3C/m1A DNA oligonucleotides were incubated with 1.25 μM ALKBH2 proteins in 46.5 mM HEPES buffer (pH8.0) containing 50 μM αKG, 5 μM Fe(NH4)2(SO4)2, and 1.86 mM ascorbic acid in a 20 μL reaction volume at 37 °C for 1 h.13 To test the IC50 values, the concentrations of hydrolyzable tannins used were optimized individually to achieve inhibition percentages that can be accurately measured. For the inhibition mechanism studies, the concentration of αKG was fixed to 93 μM, and different concentrations of Fe(NH4)2(SO4)2 (5, 10, 20, 40, 80 μM) were used for the Fe(II) chelator test. In contrast, varying concentrations of αKG (50, 100, 200, 400, 800 μM) and fixed 10 μM Fe(NH4)2(SO4)2 were used for the α-ketoglutarate competitor test. The enzyme activity was inactivated by adding 10 mM ethylenediaminetetraacetic acid followed by heating up to 95 °C for 5 min. The analyses were performed by HPLC, and all data represent the mean ± standard deviation (SD) of three independent experiments. The inhibition efficiency of a certain compound was obtained by comparing to the solvent control. The 0% inhibition was defined as no difference to the repair efficiency of the solvent control, and the 100% inhibition was defined as 0% repair of a certain substrate. The experimental data of a certain compound were fitted by the nonlinear polynomial fitting from Excel, and the IC50 value was calculated correspondingly.
HPLC Analysis.
m3C/m1A and its repaired product C/A were analyzed by using a DNApac PA-100 anion-exchange column (4 × 250 mm, 13 μm) with solvent A as water and solvent B as 1.5 M ammonium acetate in water. The flow rate was at 1.0 mL/min. A solvent gradient was carried out under the following conditions: 60% of B for 6 min, 60%−80% for 0.5 min, 80% for 1 min, 80%−60% for 0.5 min, and 60% for 4 min.
RESULTS AND DISCUSSION
Our lab has extensively investigated the repair reactions of ALKBH2 and other AlkB family enzymes under various conditions, including reactions under ss- and ds-DNA conditions and with the presence of different inhibitors, such as copper ion and D-/L-2-hydroxy-glutarate.12–14 Although ALKBH2 prefers ds-DNA repair, we did not find a big difference between ss- and ds-DNA reaction conditions. Because there are many factors for an enzymatic reaction (such as αKG, Fe(II) ion, protein and DNA), we chose to use ss- DNA for all inhibition reactions on ALKBH2 to simplify the analysis. To test the inhibitory effects of hydrolyzable tannins on ALKBH2, 16mer oligonucleotides bearing a site-specific m3C/m1A adduct, the primary substrate of ALKBH2, were synthesized by using solid-phase phosphoramidite chemistry.12,15 The recombinant human ALKBH2 protein was expressed and purified according to previous studies.13,14 Then, the repair efficiency of ALKBH2 on m3C/m1A was assessed. For the enzymatic reactions, the m3C/m1A-containing oligonucleotides were incubated with ALKBH2 in the presence of two cofactors (αKG and Fe(II) ion) and other necessary reagents and buffers (see Experimental Procedures for details), and the repair ratio of m3C/m1A was quantified by HPLC after reaction. We used 5.0 μM DNA and 1.25 μM ALKBH2 for a typical enzymatic reaction to test the inhibition of different compounds; these concentrations are similar to the conditions previously reported.14
The IC50 values of the six hydrolyzable tannins on inhibition of ALKBH2 repairing m3C and m1A were determined. These compounds were either purchased or purified and characterized as reported.9–11 Rhein (compound 1, Figure 1b), a well-known inhibitor to the AlkB family enzymes was served as a positive control.16 The concentrations of rhein varied from 0 to 150 μM and the IC50 values were found as 52.7 μM for m3C and 71.1 μM for m1A (Figure S1 and S8), which are comparable to the values on inhibiting AlkB and FTO reported by Li et al.16 For the hydrolyzable tannins, the concentrations were tested between 0 and 80 μM (Figures S2–S7 and S9–S14). The IC50 values of compounds 2–7 were found from 1.8 to 38.2 μM (Table 1 and Figures S2–S7 and S9–S14).
Table 1.
IC50 Values of Hydrolyzable Tannins Inhibiting the ALKBH2 Enzyme Repairing m3C and m1A
| compound | IC50 (μM) of m3C | IC50 (μM) of m1A | galloyl units |
|---|---|---|---|
| 2 | 38.2 | 30.9 | 1 |
| 3 | 25.5 | 28.3 | 2 |
| 4 | 22.6 | 10.6 | 2 |
| 5 | 18.9 | 6.9 | 5 |
| 6 | 8.1 | 2.5 | 6 |
| 7 | 3.9 | 1.8 | 10 |
Interestingly, based on the structures of these hydrolyzable tannins, we found a correlation of their structural features to their IC50 values: as the number of galloyl groups in the molecule increases, the IC50 value decreases (Table 1). For example, the one galloyl unit containing compound 2 on m3C has an IC50 of 38.2 μM, the five galloyl unit containing compound 5 has an IC50 of 18.9 μM, and the ten galloyl unit containing compound 7 has an IC50 of 3.9 μM. Both compounds 3 and 4 have two galloyl units, and they have similar IC50 values (25.5 and 22.6 μM, correspondingly). A similar trend was observed for m1A repair. These results indicate that the basic galloyl unit of these derivatives are potentially the structural motif responsible for the inhibitory effects.
We further selectively explored the inhibition mechanism of representative compounds on the adduct repair by ALKBH2. Previously, rhein was reported as an inhibitor to AlkB and FTO by competitively replacing αKG in the active site.16,17 And hydrolyzable tannins, including tannic acid (compound 7), have been reported to ameliorate Fe(II) ion and reactive oxygen species-induced neurological diseases.18 In addition, tannic acid has been reported to mitigate iron overload by its chelating ability through the neighboring phenolic hydroxyl groups.4,19 We wanted to investigate the inhibitory mechanisms of the hydrolyzable tannins and chose tannic acid because of its reported activities and also because it has the most galloyl groups10 in the molecules tested here. In addition to compound 7, we also selected compounds 2 (1 galloyl unit) and 5 (5 galloyl units) to study the inhibitory mechanism of hydrolyzable tannins.
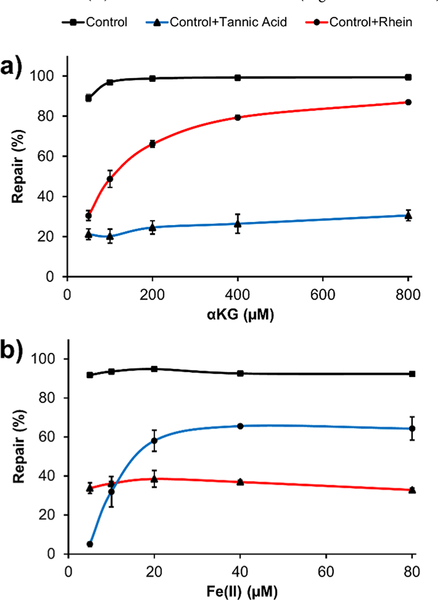
We varied the concentration of αKG and the Fe(II) ion in the reactions without any inhibitor, and with rhein or hydrolyzable tannins. For the reactions without the inhibitor, small variations on the reactivity were found with αKG changing from 50 to 800 μM (89% to 99%, control in Figure 2a) and Fe(II) ion changing from 5 to 80 μM (staying around 93%, control in Figure 2b). First, we tested the reactivity change with different concentrations of αKG with a fixed Fe(II) ion concentration of 10 μM. When 50 μM of rhein was added to the reaction medium, we found the repair ratio decreased to 30% with 50 μM αKG. As the amount of αKG increased up to 800 μM, the repair efficiency was recovered in a concentration-dependent manner (Figure 2a). Then, we tested the reactivity change with different concentrations of Fe(II) ion (from 5 to 80 μM) with a fixed αKG concentration of 93 μM. The reactions with 50 μM rhein all had a repair efficiency around 36% and showed no significant change even with the increment of Fe(II) ion to 80 μM (Figure 2b). These observations are consistent with previously reported data of rhein and show that it is a competitive inhibitor to αKG but not an Fe(II) ion chelator.16,17 For compound 7 under variable concentrations of αKG, all of the reactions with 2 μM of it had a decreased repair efficiency down to ~25% and did not vary significantly with the increment of αKG even to 800 μM (Figure 2a, Fe(II) ion kept at 10 μM). For the reactions with varying concentrations of Fe(II) ion, the reactivity was decreased to 5% with 5 μM Fe(II) ion and was restored in a concentration-dependent manner as Fe(II) ions stepwisely increased up to 80 μM (Figure 2b, αKG kept at 93 μM). These data indicate that compound 7 is an Fe(II) ion chelator but not a competitive inhibitor to αKG. In addition to compound 7, we also tested compounds 2 and 5 under similar conditions to expand the generality of the iron chelating mechanism of hydrolyzable tannins. The results showed the repair efficiencies were also recovered in a concentration-dependent manner as Fe(II) ion concentration elevated (Figures S15 and S16).
Figure 2.
Inhibition of ALKBH2 repair reactions by tannic acid and rhein (control: no inhibitor added). All of the tests were carried out in triplicate, and the error bars represent standard deviation. (a) Tests of compounds on the competition to αKG and (b) tests of compounds on the chelation to the Fe(II) ion.
Rhein has been reported as an inhibitor to αKG/Fe(II)- dependent enzymes, including AlkB and FTO, through different mechanisms, such as a competitor to either αKG or DNA substrate.16,17 The hydrolyzable tannins tested in this work are metal chelators and thus could inhibit ALKBH2 and other αKG/Fe(II)-dependent proteins. Besides chelating the metal ions, there might be additional inhibitory mechanisms for the hydrolyzable tannins. For example, the repair efficiency of ALKBH2 treated with compound 7 reached up to 60% even with the increment of Fe(II) ion concentration up to 80 μM and could not be recovered to the level of the control (~95% repair, Figure 2b). These results indicate the inhibition may also come from the binding of compound 7 to the ALKBH2 protein, possibly leading the protein to a fully or partially inactive conformation.
In this study, we investigated hydrolyzable tannins as a new class of natural product inhibitors for ALKBH2, the major human DNA repair enzyme in the AlkB family. Hydrolyzable tannins have been identified as antioxidative, antiproliferative, and neuroprotective agents. Here, we discovered that these compounds are Fe(II) ion chelators that can decrease the level of free Fe(II) ion, which is critical for the catalytic activity of the AlkB family enzymes. If the chelation of the Fe(II) ion by the hydrolyzable tannins decreases the iron level too much, the reactivity of the AlkB enzymes might be inhibited. It is possible that the overconsumption of hydrolyzable tannins may affect iron homeostasis in the body, leading to the dysfunction of iron-dependent proteins, such as the AlkB homologues and other Fe(II)/αKG-dependent enzymes. The results reported in this paper are from in vitro experiments. Further cellular or animal studies are needed to investigate the adverse effect demonstrated in this work.
Supplementary Material
ACKNOWLEDGMENTS
The authors want to thank the RI-INBRE program, its director Prof. Bongsup Cho, and staff Dr. Al Bach, Kim Andrews, and Patricia Murray for their kind help.
Funding
This work was supported by an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103430. This work was also supported by National Institutes of Health under grant numbers R15 CA213042 and R01 ES028865 (to D.L.).
ABBREVIATIONS
- ss
single stranded
- ds
double stranded
- αKG
alpha-ketoglutarate
- m3C
3-methylcytosine
- m1A
1-methlyadenine
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemres-tox.8b00398.
REFERENCES
- (1).Okuda T, and Ito H (2011) Tannins of Constant Structure in Medicinal and Food Plants—Hydrolyzable Tannins and Polyphenols Related to Tannins. Molecules 16, 2191–2217. [Google Scholar]
- (2).Arapitsas P (2012) Hydrolyzable tannin analysis in food. Food Chem. 135, 1708–1717. [DOI] [PubMed] [Google Scholar]
- (3).Chung KT, Wong TY, Wei CI, Huang YW, and Lin Y (1998) Tannins and human health: a review. Crit. Rev. Food Sci. Nutr. 38, 421–464. [DOI] [PubMed] [Google Scholar]
- (4).Basu T, Panja S, Shendge AK, Das A, and Mandal N (2018) A natural antioxidant, tannic acid mitigates iron-overload induced hepatotoxicity in Swiss albino mice through ROS regulation. Environ. Toxicol. 33, 603–618. [DOI] [PubMed] [Google Scholar]
- (5).Hausinger RP, and Schofield CJ (2015) 2-Oxoglutarate- dependent oxygenases, Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- (6).Fedeles BI, Singh V, Delaney JC, Li D, and Essigmann JM (2015) The AlkB family of Fe(II)/a-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yi C, and He C (2013) DNA repair by reversal of DNA damage. Cold Spring Harbor Perspect. Biol. 5, a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sedgwick B (2004) Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5, 148–157. [DOI] [PubMed] [Google Scholar]
- (9).Yuan T, Ma H, Liu W, Niesen DB, Shah N, Crews R, Rose KN, Vattem DA, and Seeram NP (2016) Pomegranate’s neuroprotective effects against Alzheimer’s disease are mediated by urolithins, its ellagitannin-gut microbial derived metabolites. ACS Chem. Neurosci. 7, 26–33. [DOI] [PubMed] [Google Scholar]
- (10).Ma H, Liu W, Frost L, Kirschenbaum LJ, Dain JA, and Seeram NP (2016) Glucitol-core containing gallotannins inhibit the formation of advanced glycation end-products mediated by their antioxidant potential. Food Funct. 7, 2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ma H, Liu W, Frost L, Wang L, Kong L, Dain JA, and Seeram NP (2015) The hydrolyzable gallotannin, penta-O-galloyl- β-D-glucopyranoside, inhibits the formation of advanced glycation endproducts by protecting protein structure. Mol. BioSyst. 11, 1338–1347. [DOI] [PubMed] [Google Scholar]
- (12).Chen F, Tang Q, Bian K, Humulock ZT, Yang X, Jost M, Drennan CL, Essigmann JM, and Li D (2016) Adaptive response enzyme AlkB preferentially repairs 1-methylguanine and 3- methylthymine adducts in double-stranded DNA. Chem. Res. Toxicol. 29, 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chen F, Bian K, Tang Q, Fedeles BI, Singh V, Humulock ZT, Essigmann JM, and Li D (2017) Oncometabolites D- and L-2-hydroxyglutarate inhibit the AlkB family DNA repair enzymes under physiological conditions. Chem. Res. Toxicol. 30, 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bian K, Chen F, Humulock ZT, Tang Q, and Li D (2017) Copper inhibits the AlkB family DNA repair enzymes under Wilson’s disease condition. Chem. Res. Toxicol. 30, 1794–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tang Q, Cai A, Bian K, Chen F, Delaney JC, Adusumalli S, Bach AC, Akhlaghi F, Cho BP, and Li D (2017) Characterization of byproducts from chemical syntheses of oligonucleotides containing 1-methyladenine and 3-methylcytosine. ACS Omega 2, 8205–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li Q, Huang Y, Liu X, Gan J, Chen H, and Yang C-G (2016) Rhein inhibits AlkB repair enzymes and sensitizes cells to methylated DNA damage. J. Biol. Chem. 291, 11083–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, Peng S, Chen K, Wang M, Gong S, et al. (2012) Development of cell- active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134, 17963–17971. [DOI] [PubMed] [Google Scholar]
- (18).Chan S, Kantham S, Rao VM, Palanivelu MK, Pham HL, Shaw PN, McGeary RP, and Ross BP (2016) Metal chelation, radical scavenging and inhibition of Aβ42 fibrillation by food constituents in relation to Alzheimer’s disease. Food Chem. 199, 185–194. [DOI] [PubMed] [Google Scholar]
- (19).Phiwchai I, Yuensook W, Sawaengsiriphon N, Krungchanuchat S, and Pilapong C (2018) Tannic acid (TA): A molecular tool for chelating and imaging labile iron. Eur. J. Pharm. Sci. 114, 64–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.