Abstract
Background and aim
Long non-coding RNA nuclear-enriched abundant transcript 1 (NEAT1) is abnormally expressed in various human malignancies, including hepatocellular carcinoma (HCC). Let-7b is a miRNA with the effect of a tumor suppressor gene, and its expression level in various tumor tissues is lower than that in normal tissues. Studies have found that IGF-1R can be abnormally activated in the process of hepatocyte deterioration, and the expression level of IGF-1R in HCC is significantly up-regulated. The aim of this study was to investigate the functional mechanism of NEAT1/let-7b-IGF-1R axis in HCC.
Methods
The expressions of NEAT1 and microRNA (miR)-let-7b in HCC tissues and cell lines were quantified by quantitative real-time PCR (qRT-PCR). The effect of NEAT1 on tumor growth was observed in a mice model of transplanted hepatoma. The effects of down-regulation or up-regulation of NEAT1 expression in HCC cell lines were analysed from the perspectives of cell viability and apoptosis. The binding sites of NEAT1 and miR-let-7b were predicted by biological software. The expression of the miR-let-7b target molecules IGF-1R was detected by Western blotting.
Results
The results showed that the expressions of NEAT1 were significantly increased, while the expressions of miR-let-7b were decreased in the HCC tissues and cell lines. Additionally, it was found that the expressions of NEAT1 and miR-let-7b showed a negative correlation in HCC tissues. The mouse model experiments confirmed that the interference with NEAT1 expression inhibited the tumor growth. Meanwhile, the cell viability of HepG2/Huh7 cell lines was significantly decreased via the downregulation of NEAT1, whereas the corresponding rates of apoptosis were significantly increased. It was further proven that there was a certain negative regulatory mechanism between NEAT1 and miR-1et-7b, which was related to the expression of IGF-1R.
Conclusion
The over-expression of NEAT1 could promote the proliferation of HCC cells by inhibiting the expression of the miR-let-7b regulated by IGF-1R.
Keywords: long non-coding RNA NEAT1, microRNA-let-7b, hepatocellular carcinoma, IGF-1R, transplanted hepatoma mice model
Background
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors, which has the characteristics of high morbidity, poor prognosis and high mortality.1,2 By the time symptoms appear, hepatocellular carcinoma has entered the middle and advanced stage, so the cure rate is very low.3 Currently, hepatectomy and selective liver transplantation are effective treatments, but most patients have missed the best time for surgery. Therefore, finding markers for early diagnosis of HCC is the key to current studies. It has been found that tumor is a complex genetic disease, and various genetic or environmental carcinogenic factors may synergistically or sequentially activate proto-oncogenes and/or inactivate tumor suppressor genes, thus causing abnormal gene expression and ultimately carcinogenesis of target cells.4 In the process of tumor development, tumor suppressor gene expression deletion or oncogene over-expression are involved.5,6
Micro RNA (miRNA) is a kind of non-coding single-stranded small molecular RNA, which is involved in physiological processes such as growth, metabolism, and apoptosis in vivo.7 miRNA can regulate the expressions of target genes, mainly by binding to the 3 ‘UTR of target gene mRNA, so as to degrade the target gene mRNA or inhibit the protein synthesis at the transcription level.8,9 Recent studies have shown that miRNA are abnormally expressed in liver cancer tissues and can promote the proliferation of HCC cells by regulating target genes, which are miRNA with the role of oncogenes.10–12 On the contrary, miRNA with anti-oncogene effect can induce apoptosis of cancer cells.13,14 RNA interference (RNAi) refers to the degradation of a specific gene induced by double-stranded RNA (dsRNA) homologous to the target gene sequence during evolution.15 RNAi technology is an effective means to specifically inhibit gene expression and has been applied in gene-targeted therapy for major diseases such as tumor.16 It was found that the vector plasmid constructed by lentivirus could stabilize the transfected cells and achieve the purpose of gene silencing.17
The NEAT1, the core structure of the neat site, is involved in the transcriptional regulation of various genes.18 The NEAT1 is abnormally expressed in various human malignancies, such as glioma, ovarian cancer, leukemia and breast cancer.19–21 Studies have found that the NEAT1 interacts with various RNA-binding proteins located in the neat site, and regulates the mRNA removal from the nucleus, thus promoting the occurrence and development of the tumor.22 In 2018, Zheng et al demonstrated that HIF-2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition.23 Inspired by this research, we aim to investigate the deep role and mechanism of NEAT1 in HCC.
As a member of the let-7 family, let-7b is a miRNA with the effect of tumor suppressor gene, and its expression level in various tumor tissues is lower than that in normal tissues.24,25 In addition, human insulin-like growth factor type 1 receptor (IGF-1R), as a new target of tumor therapy, has become a research hotspot of anti-tumor targeted drugs.26 Studies have found that IGF-1R can be abnormally activated in the process of hepatocyte deterioration, and the expression level of IGF-1R in HCC is significantly up-regulated.27 Then, during the occurrence and development of HCC, what will be the interaction mechanism of NEAT1, let-7b, and IGF-1R. In the present study, the functional mechanism of long non-coding RNA NEAT1 in HCC was investigated, so as to provide a certain theoretical basis for clinical studies.
Materials and Methods
Materials
Animals
24 BALB/c athymic nude mouse, 4–5 weeks of age, were provided by Shandong Provincial Third Hospital. This study was performed with the approval of the local ethical committee and all the experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Patients with HCC
The HCC tissues (n=25) and corresponding para-cancer tissues (n=25) of patients pathologically diagnosed as HCC in Shandong Provincial Third Hospital were used for the experiment. The patients did not receive local or systemic treatment before operation. All tissue samples were immediately stored in a refrigerator at −80 °C after collection. All the patients who participated in this experiment were informed in writing of the purpose of this study.
HCC Cell Lines
The human HCC cell lines (HepG2, SMMC 7721, Bel 7402 and Huh7) and an immortalized normal liver cell line (L02) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Beijing, China), and were maintained in a Dulbecco’s modified Eagle’s medium (DMEM, Gibco Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco Life Technologies, Grand Island, NY, USA), 100 U·mL−1 penicillin and 0.1 mg·mL−1 streptomycin in a humidified chamber at 37°C with 95% air and 5% CO2 atmosphere.
Quantitative Real-Time PCR (qRT-PCR)
The miRNeasy Mini Kit (Qiagen) and Trizol reagent (Invitrogen) were used to extract the microRNAs and total RNA of HCC tissues or cell lines, respectively. Absorbance was measured at 260 nm using a NanoDrop ND-2000 for RNA quantification. RNA was reversely transcribed into cDNA using Super Script VILO cDNA Synthesis Kit (Thermo Fisher Scientific). The forward primer of NEAT1 was 5ʹ-TTGTTCCAGAGCCCATGAT-3ʹ, and the reverse primer was 5ʹ-TGAAAACCTTTACCCCAGGA-3ʹ. qRT-PCR used Super Script III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen) and the reaction conditions were 95°C for 10 min, 95°C for 30 s, 60°C for 30 s, and 74°C for 25 s (38 cycles). The mRNA expressions of NEAT1 and miR-let-7b were quantified by the TaqMan Non-coding RNA/MicroRNA Assays (Thermo Fisher Scientific).
The Assay of Tumor-Bearing Mice
24 BALB/c athymic nude mouse was randomly divided into the lentiviral vector carrying negative control (Lv-si-NC) group and the lentiviral vector carrying si-NEAT1 (Lv-si-NEAT1) group. In the Lv-si-NC group, the HepG2 cells transfected with an Lv-si-NC were suspended in 80 mL PBS and then injected subcutaneously into either side of mice. In the Lv-si-NEAT1 group, an Lv-si-NEAT1 was used to transfect into the HepG2 cells. The mouse was maintained in an SPF grade environment with free access to standard pellet diet and water. The growth of tumor in mice was monitored weekly and the tumor volume (V) was calculated as described in the literature.28
The Transfection of miR/siRNA
The siRNAs were transfected into the HepG2 and Huh7 cells using Lipofectamine RNAiMAX reagent (Invitrogen). The cells were re-transfected after the first transfection and cultured for 48 h. The interference efficiency of siRNA in HepG2/Huh7 was detected by qRT-PCR (Table 1).
Table 1.
Primers of qRT-PCR Genes
| Gene Name | NEAT1 siRNA | Control siRNA |
|---|---|---|
| Forward primer | 5ʹ-phosGUGAGAAGUUGCUUAGAAACUUUdCdC-3’ | 5ʹ-GUACCUGACUAGUCGCAGAAG-3’ |
| Reverse primer | 5ʹ -GGAAAGUUUCUAAGCAACUUCUCACUU-3’ | 5ʹ UCUGCGACUAGUCAGGUACGG-3’ |
The synthetic oligonucleotides (Beijing AuGCT DNA-SYN Biotechnology Co. Ltd) was used to construct the pre-let-7b expression plasmid. The HCC cells were adhered to the wall for 24 hrs in the growth medium without antibiotics.
According to the manufacturer’s instructions, the Roche (No. 04476093001, 06366236001) was used to transiently transfect the siRNAs against IGF-1R, pre-let-7b expression plasmid or pcDNA6.2-GW vector into the HCC cells. The cells were transfected with the pre-let-7b control (pcDNA6.2-GW vector), scramble siRNA and anti-let-7b control as negative controls. The primer of small interfering RNA against IGF-1R (si-IGF-1R#1, No. 2779; si-IGF-1R#2, No. 2122) was purchased from Gene Pharma. All the miRNA/siRNA were transfected for 24 h.
Detection of Cell Viability and Apoptosis
The cell viability was quantified by WST-1 reagent (Roche), which was added directly into the HepG2 or Huh7 cells, and then the cells were incubated for 30 min. The absorbance value at 630 nm was determined, and the absorbance value at 450 nm was used as reference. The amounts of living cells were calculated according to absorbance value.
The TUNEL Apoptosis Assay Kit (Roche) was used to detect the apoptosis of cells, and the specific steps were carried out in strict accordance with the manufacturer’s instructions.
The Assay of Luciferase
In the present study, all the DNA constructs were derived from pGL3 luciferase reporter vectors (Promega). The genomic DNA of human HCC cells were used to amplify the human 3=UTR sequences by PCR. The vectors cloned with PCR products were used for the transfection of HCC cells. The transfected cells were lysed with a lysis solution for 15 min, and the Dual-Luciferase Reporter Assay System (Promega) was used to detect the luciferase activity.
Western Blot Analysis
A total protein extraction kit (Sigma-Aldrich) was used to extract the total protein of HCC cells. The protein concentration was determined by BCA kit (Beyotime Institute of Biotechnology). The SDS-PAGE electrophoresis was used for the separation of samples. After electrophoresis, the proteins on the gel were transferred to a polyvinylidene difluoride (PVDF) membrane. Then, 5% bovine serum albumin was used to closed of PVDF membrane at 4 °C to save up for the night. After incubation with primary antibody for 2 h, the PVDF membrane was washed with Tris-buffered saline-Tween 20 (TBST, BioTNT) for three times. Then, the PVDF membrane was incubated by horseradish peroxidase-labeled secondary antibody dilution (BD Biosciences) for 1 h. After washed with TBST three times, the PVDF membrane was detected by Western Blotting Substrate ECL (Invitrogen).
The Assay of 5-Ethynyl-20-Deoxyuridine (EDU)
A Cell Light EDU DNA imaging kit (Guangzhou RiboBio) was used to the assay of EDU. Briefly, after the cells transfected for 24 h on 96-well plates, the 100 mM of EDU has used to culture the cells for 2 h. The specific steps of cell staining were strictly followed in accordance with the manufacturer’s instructions. A BD Pathway 855 high-content analysis system (BD, San Jose, CA, USA) was used to image and analyze the cells. The percentage of EDU-positive cells was calculated as follows:
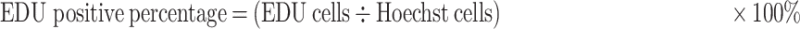 |
(1) |
EDU cells: The number of EDU-strained cells;
Hoechst cells: The number of Hoechst-strained cells.
Statistical Analysis
The data were presented as mean± SD. The significance analysis was analyzed by Bonferroni’s posttest and one way ANOVA (P <0.05). GraphPad Prism (Graph-Pad Software, La Jolla, CA) was used to process all statistical analyses and graphing.
Results
The Expressions of NEAT1 and miR-Let7b in HCC Tissues and Cell Lines
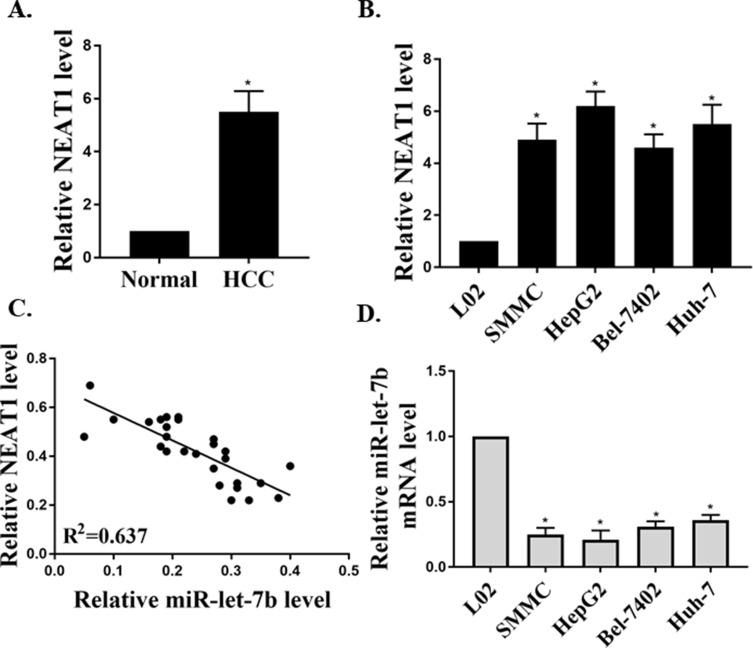
Studies have shown that the NEAT1 abnormal expression in vivo is related to various human cancers. In the present study, the NEAT1 expression in HCC tissues and its correlation with the expression of miR-let-7b were investigated. As shown in Figure 1A, the expression of NEAT1 in HCC tissues was significantly higher than that in the adjacent non-tumor tissues. The result of linear regression (Figure 1C) suggested that the mRNA expression of NEAT1 was negatively correlated with that of miR-let-7b in the HCC tissues. In addition, the expressions of NEAT1 and let-7b in the HCC cell lines (HepG2, SMMC, Bel-7402 and Huh-7) were measured, and the results were consistent with the trend of research results in HCC tissues. The results suggested that NEAT1 expression in HCC cell lines was significantly increased, compared to the L02 (normal liver) cells (Figure 1B). Compared with the L02 cells, the expression of miR-let-7b in HCC cell lines was lower (Figure 1D). Therefore, these results indicated that NEAT1 expression was abnormal in HCC tissues/cells, and there was a highly negative correlation between the mRNA expression NEAT1 and miR-let-7 in HCC tissues.
Figure 1.
The expressions of NEAT1 and miR-let-7b in HCC tissues/cell lines. (A) The relative expression of NEAT1 in HCC tissues (n=25) and adjacent non-tumor tissues (n=25, GAPDH as an internal normalizer). (B) The relative expression of NEAT1 in cells (HepG2, SMMC, Bel-7402, Huh-7 and L02). (C) The linear regression analysis of the expressions of NEAT1 and miR-let-7b (U6 as an internal normalizer) in HCC tissues (n=25). (D) The relative expression of miR-let-7b in cells. *P <0.05.
Effect of NEAT1 Expression on Tumor Growth
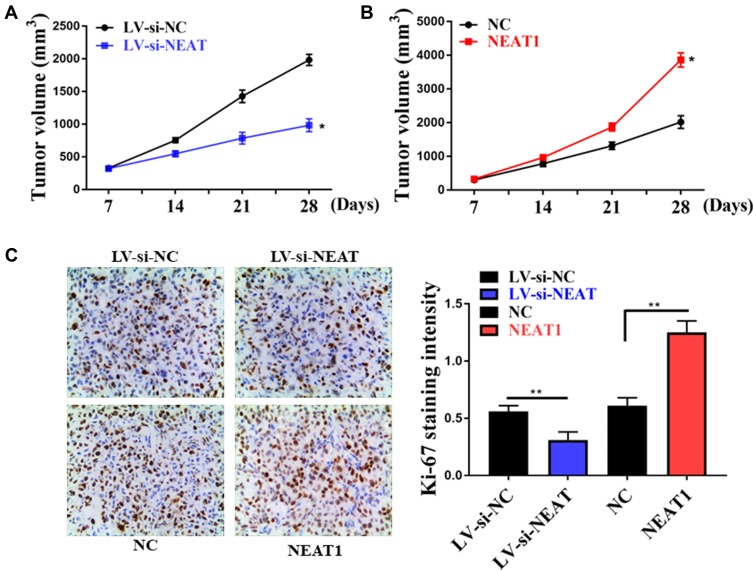
In the present study, a mice model of transplanted hepatoma was used to observe the role of NEAT1 expression in tumor growth. After subcutaneous injection of HepG2 with NEAT1 plasmids or NEAT1 silencing, the tumor volume in mouse was continuously observed for 28 days. The results showed that the tumor volume was markedly decreased in the si-NEAT1 group (Figure 2A), and NEAT1 plasmid group was increased (Figure 2B). Furthermore, HCC cell proliferation was inhibited by NEAT1 silencing and were promoted by NEAT1 over-expression (Figure 2C).
Figure 2.
Effect of interfering with NEAT1 expression on tumor growth. (A) Effect of NEAT1 silencing on tumor growth in mice. (B) Effect of NEAT1 over-expression on tumor growth in mice. (C) Immunohistochemical staining of Ki-67. *P <0.05, **P <0.05.
Role of NEAT1 Expression in the Proliferation and Apoptosis of HCC Cell Lines
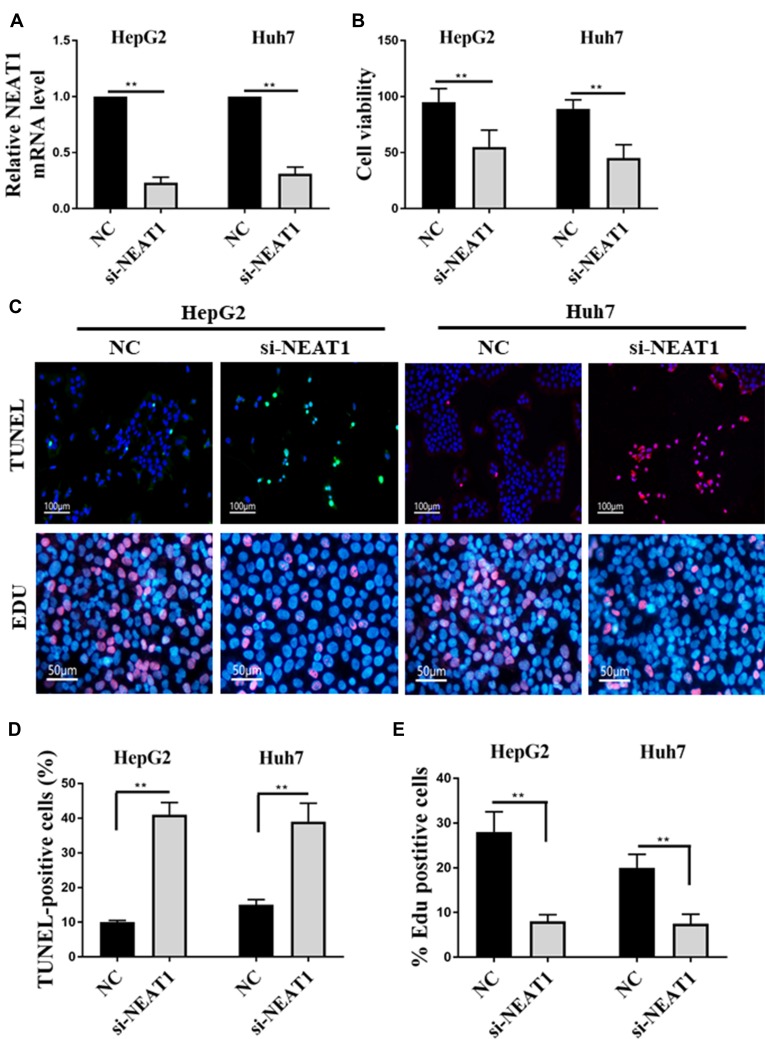
In this study, the expression of NEAT1 was downregulated by the RNA interference technique to investigate the effect of NEAT1 expression on the physiological activity of HCC cell lines (HepG2 and Huh7). As shown in Figure 3A, the results of qRT-PCR showed that the HCC cell lines with low NEAT1 expression (si-HepG2 and si-Huh7) were successfully obtained. In addition, the cell viability of si-HepG2 and si-Huh7 were significantly decreased (Figure 3B), and the apoptosis rates of si-HepG2 and si-Huh7 were remarkably increased (Figure 3C–E). These results indicated that the down-regulation of NEAT1 expression could inhibit the proliferation and induce the apoptosis of HepG2 and Huh7 cells.
Figure 3.
Effect of NEAT1 expression on the proliferation and apoptosis of HepG2/Huh7 cells. (A) Detection of NEAT1 expression in HepG2/Huh7 cells by qRT-PCR. (B) Effect of NEAT1 expression on the proliferation of HepG2/Huh7 cells. (C) The TUNEL (terminal deoxynucleotidyl transferase dUTP nickend labeling) and EDU labeling of HepG2/Huh7 cells. (D) Effect of NEAT1 expression on the percentage of TUNEL positive cells. (E) Effect of NEAT1 expression on the percentage of EDU positive cells. **P <0.05.
Role of Let-7b on NEAT1-Induced the Proliferation in HCC Cells
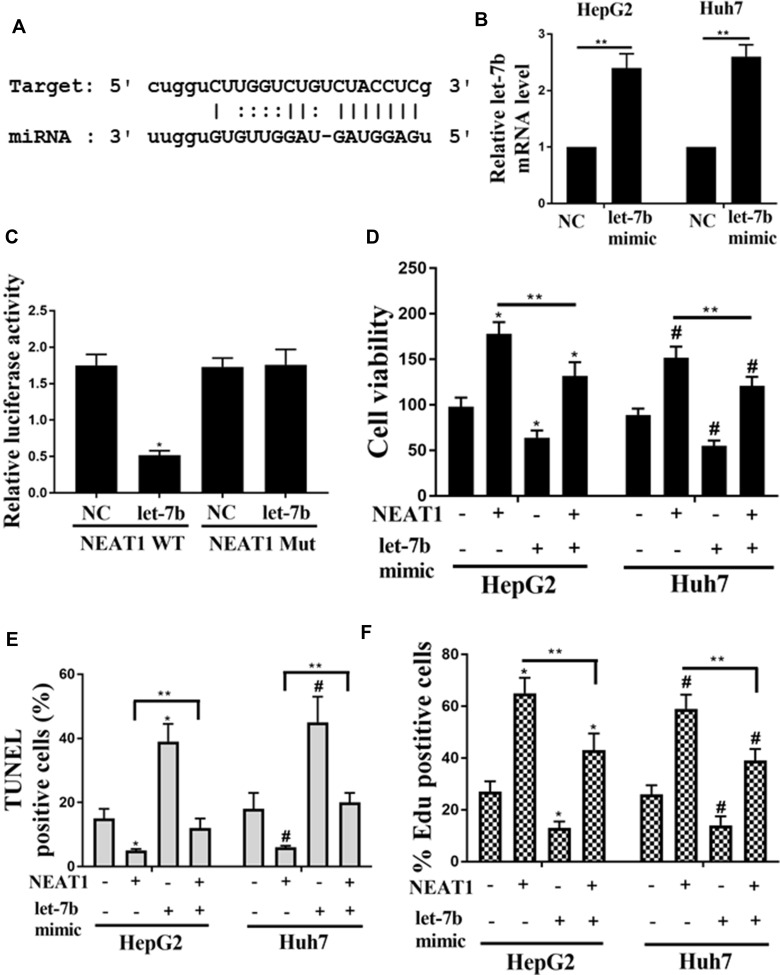
The potential targeted microRNAs of NEAT1, such as Targetscan, Starbase, and microRNA.org, were searched by bioinformatics analysis. Figure 4A shows the bioinformatics results, and we found that the NEAT1 was the predictive gene of let-7b, as a let-7b binding site was found in the NEAT1 transcript. In addition, the result of dual-luciferase reporter assay suggested that, by up-regulating the expression of Let-7b, the relative luciferase activity in HEK293 cells transfected with wild type (WT) vector was decreased, and the Mut vector could recover this inhibition (Figure 4B). In order to investigate the effect of let-7b over-expression on the NEAT1-regulated proliferation and apoptosis of cells, the let-7b mimic was used to transfect the HepG2 and Huh7 cells. And the result of let-7b mRNA levels in HepG2 and Huh7 cells were shown in Figure 4C. It was obvious that NEAT1 WT could significantly lower the luciferase activity of let-7b. Figure 4D shows that the cell viabilities of NEAT1 group were dramatically reversed by the co-transfection of let-7b mimics in both HepG2 and Huh7 cells. Figure 4E and F demonstrate that the cell apoptosis of HepG2 and Huh7 cells were dramatically reversed by co-transfection of let-7b mimics.
Figure 4.
Role of let-7b on NEAT1-induced the proliferation in HCC cells. (A) Bioinformatic analysis showed the common binding sequence between let-7b and NEAT1. (B) Effect of Let-7b expression on the luciferase activity in HEK293 cells transfected with NEAT1 or shNEAT1. (C) Detection of the mRNA levels of let-7b in HepG2 and Huh7 cells by qRT-PCR. (D) Detection of the cell viability of HepG2 and Huh7 cells. (E) Effect of let-7b expression on the percentage of TUNEL positive cells. (F) Effect of let-7b expression on the percentage of EDU positive cells. *P <0.05 vs control in HepG2, #P <0.05 vs control in Huh7, **P <0.05.
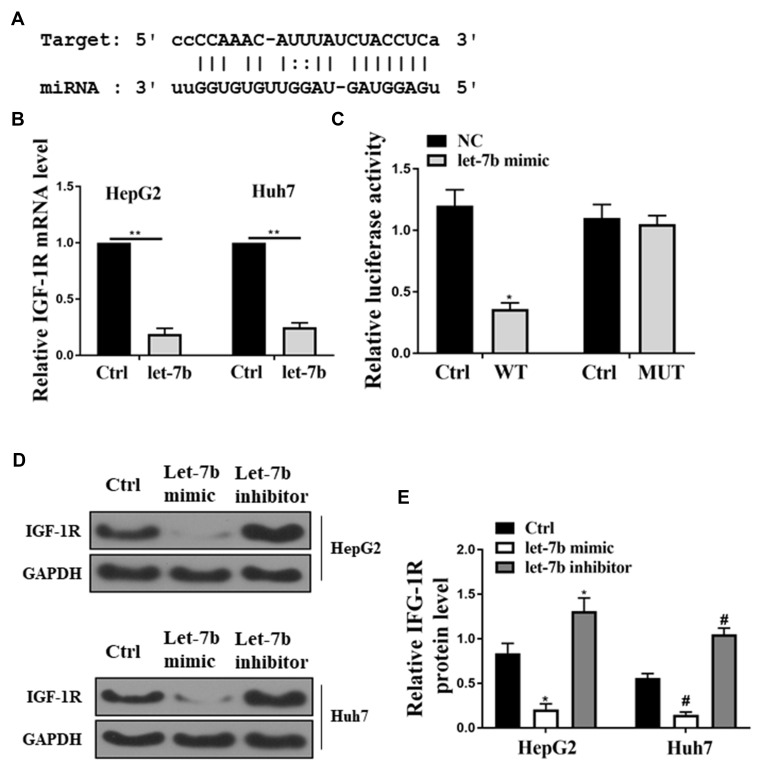
Identification of IGF-1R as A Let-7b Target Binding Site
In the present study, the computational programs TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/cgi-bin) and miRanda (www.microrna.org/) were used to predict the target let-7b binding site. Figure 5A demonstrates that IGF-1R had a shared binding sequence with let-7b. Since IGF-1R was considered to be involved in the regulation of HCC, the interaction between let-7b and IGF-1R was investigated. A dual-luciferase reporter assay was performed using vectors containing let-7b precursors (PRE) and mutant let-7b binding site (MUT) to determine target specificity between let-7b and IGF-1R. The result of dual-luciferase reporter assay suggested that the relative luciferase activity in HEK293 cells co-transfected with et-7b precursors and IGF-1R WT was significantly lower than HEK293T cells transfected with MUT (Figure 5C). In addition, the HepG2 and Huh7 cells were transfected with the let-7b precursor. And the results showed that the mRNA and protein expressions of IGF-1R in the HepG2 and Huh7 cells were downregulated by transfected with the let-7b precursor, compared with the control group that was transfected with the empty vector (Figure 5B, D and E). However, after transfection of let-7b inhibitor into the HepG2 and Huh7 cells, the opposite results were detected.
Figure 5.
Identification of IGF-1R as a let-7b target binding site. (A) Bioinformatic analysis showed the common binding sequence between IGF-1R and let-7b. (B) Detection of the mRNA expression of IGF-1R in the HepG2 and Huh7 cells. (C) Analysis of the relative luciferase activity in HEK-293T cells co-transfected with et-7b precursors and IGF-1R WT. (D) Detection of the protein expression of IGF-1R in HCC cells by Western blot. *P <0.05 vs control in HepG2, #P <0.05 vs control in Huh7, **P <0.05.
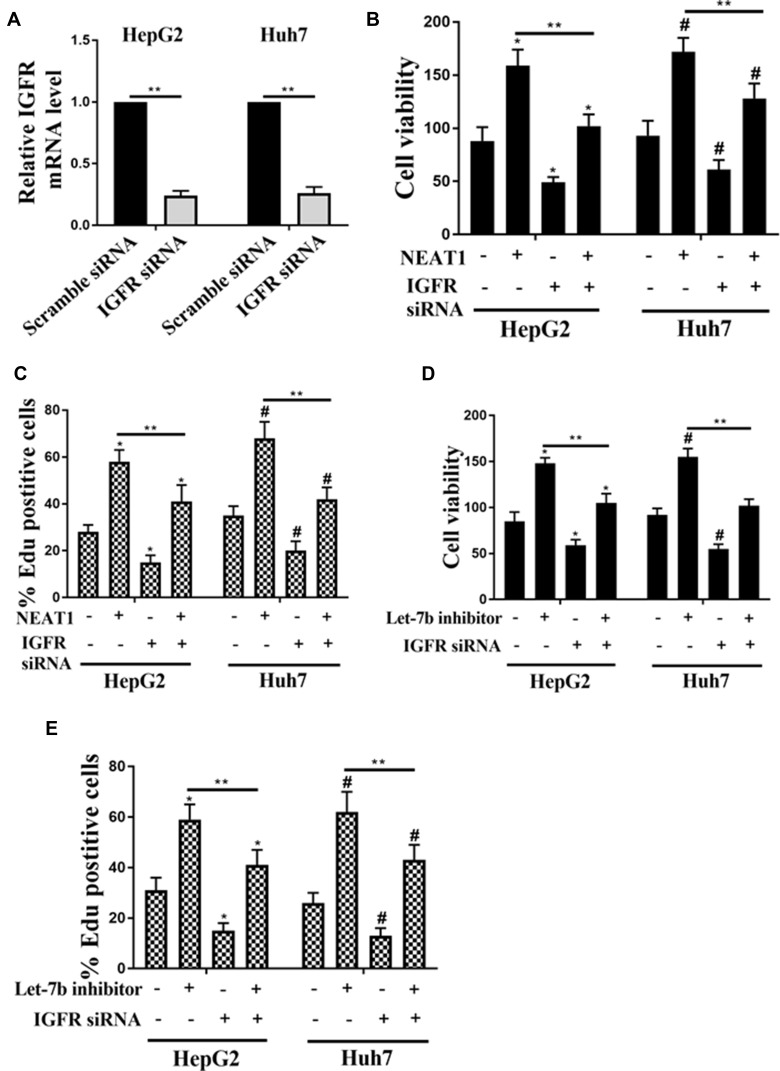
Role of IGF-1R on NEAT1- and Let-7b-Induced the Cell Viability and Proliferation in HCC Cells
In the present study, the HepG2 and Huh7 cells were transfected with IGF-1R siRNA and scramble siRNA. As shown in Figure 6A, the mRNA expression of IGF-1R in the HepG2 and Huh7 cells were significantly decreased in the IGF-1R siRNA group, compared with the scramble siRNA group. In addition, the promotion effects of NEAT1 on cell viability and proliferation were inhibited by IGF-1R interfering (Figure 6B and C). Then the effect of IGF-1R on Let-7b inhibitor-induced cell proliferation was investigated. As shown in Figure 6D and E, the cell viability and the percentage of Edu positive cells were increased by let-7b inhibitor, while these effects were attenuated by IGF-1R siRNA transfection.
Figure 6.
Role of IGF-1R on NEAT1- and let-7b-induced the cell viability and proliferation in HCC cells. (A) Detection of mRNA expression of IGF-1R in HepG2 and Huh7 cells transfected with scramble siRNA or IGF-1R siRNA by qRT-PCR. The cell viability (B) and the percentage of EDU positive cells (C) of HepG2 and Huh7 cells transfected with NEAT1 or IGF-1R siRNA alone or co-transfection NEAT1 and IGF-1R siRNA. The cell viability (D) and the percentage of EDU positive cells (E) of HepG2 and Huh7 cells transfected with let-7b inhibitor or IGF-1R siRNA alone or co-transfection with let-7b inhibitor and IGF-1R siRNA. *P <0.05 vs control in HepG2, #P <0.05 vs control in Huh7, **P <0.05.
Discussion
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in clinical practice, which involves abnormal activation of multiple signaling pathways, including gene mutation, chromosome aberration, abnormal secretion of growth factors and epigenetic changes.29–31 Early diagnosis and effective treatment of HCC is still a worldwide medical problem.32,33 Therefore, finding markers for early diagnosis of HCC is the key to current studies. In this study, the mouse model of liver cancer transplantation, HCC tissues, and cell lines were selected as the study objects. And the results indicated that the NEAT1 could promote the proliferation of HCC cells by inhibiting the expression of the miR-let-7b regulated by IGF-1R.
Guo et al found that, compared to the adjacent normal liver tissues, NEAT1 was highly expressed in the HCC tissues, which associated with the expression levels of, MALAT1, MDTH, and NM23.34 Mang et al revealed that the NEAT1 expression was upregulated in HCC tissues, which promoted the proliferation, invasion, and migration of HCC cells via the NEAT1-hnRNP A2.35 In this study, NEAT1 was abnormally expressed in HCC tissues/cells, and the NEAT1 down-regulation expression could inhibit the proliferation and induce the apoptosis of HepG2 and Huh7 cells, which was consistent with the results reported in the literature. In recent years, RNA interference technology has been successfully applied in tumor gene therapy.36,37 In this study, the lentiviral vector carrying si-NEAT1 was transfected into the HepG2 cells to establish a mouse model of liver cancer transplantation, and the results showed that the NEAT1 expression down-regulation could inhibit the growth rate of tumor and the proliferation of HCC cells.
Studies have confirmed that let-7b is a miRNA with the effect of tumor suppressor gene, and its expression level in various tumor tissues is lower than that in normal tissues.24,38–40 In the present study, the bioinformatics analysis showed that the NEAT1 was the predictive gene of let-7b, as a let-7b binding site was found in the NEAT1 transcript. On the basis of the target gene prediction, the result of dual-luciferase reporter assay suggested that the NEAT1 gene was neatly regulated by the let-7b over-expression vector transfected into HEK293 cells. To further investigate the effect of miRNA let-7b on the proliferation and apoptosis of HCC cells, let-7b mimic was transfected into HepG2 and Huh7 cells. And the functional experiments showed that the roles of NEAT1 in the proliferation and apoptosis of HepG2 and Huh7 cells were dramatically reversed by co-transfection of let-7b mimics. In addition, there was a highly negative correlation between the mRNA expression NEAT1 and miR-let-7 in HCC tissues. Therefore, it was inferred that the NEAT1 gene expression was regulated by let-7b in HCC tissues or cells.
Conclusion
Studies have shown that IGF-1R is involved in regulating the growth, proliferation, apoptosis and other physiological processes of HCC cells.27,41,42 In vivo experiments confirmed that IGF-1R could inhibit the growth and metastasis of HCC cells.43 Since IGF-1R was considered to be involved in the regulation of HCC, the interaction between let-7b and IGF-1R was investigated. The present study indicated that IGF-1R was a let-7b target binding site, and transfecting with the let-7b precursor could inhibit the mRNA and protein expressions of IGF-1R in the HepG2 and Huh7 cells. In addition, IGF-1R could inhibit the promotion effects of NEAT1 on the cell viability and proliferation in HepG2 and Huh7 cells. As mentioned above, the NEAT1 could promote the proliferation of HCC cells by inhibiting the expression of the miR-let-7b regulated by IGF-1R. It was speculated that this might be the NEAT1’s specific mechanism for promoting the development of HCC. The present study provided a new theoretical basis for the pathogenesis of HCC and was conducive to the diagnosis and treatment of HCC.
Abbreviations
HCC, Hepatocellular carcinoma; miRNA, Micro RNA; RNAi, RNA interference; dsRNA, double-stranded RNA; IGF-1R, insulin-like growth factor type 1 receptor.
Availability of Data and Materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Shandong Provincial Third Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients and healthy volunteers provided written informed consent prior to their inclusion within the study.
This study was performed with the approval of the Ethics Committee of Shandong Provincial Third Hospital and all the experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020. doi: 10.1002/hep.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han D, Li J, Wang H, et al. Circular RNA MTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 3.Yong T, Jue R, Shiou-Hwei Y, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci U S A. 2011;108(29):12042–12047. doi: 10.1073/pnas.1108715108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balmain A. Cancer as a complex genetic trait: tumor susceptibility in humans and mouse models. Cell. 2002;108(2):145–152. doi: 10.1016/S0092-8674(02)00622-0 [DOI] [PubMed] [Google Scholar]
- 5.Nishiyama H, Takahashi T, Kakehi Y, et al. Homozygous deletion at the 9q32–33 candidate tumor suppressor locus in primary human bladder cancer. Genes Chromosomes Cancer. 2015;26(2):171–175. doi: [DOI] [PubMed] [Google Scholar]
- 6.Hsieh HY, Jou YC, Tung CL, et al. Epigenetic silencing of the dual-role signal mediator, ANGPTL4 in tumor tissues and its overexpression in the urothelial carcinoma microenvironment. Oncogene. 2017;37(5):673–686. [DOI] [PubMed] [Google Scholar]
- 7.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15(1):1–6. doi: 10.1186/s12935-015-0185-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo JW, Wang X, Yang Y, et al. Role of micro-RNA (miRNA) in pathogenesis of glioblastoma. Eur Rev Med Pharmacol Sci. 2015;19(9):1630. [PubMed] [Google Scholar]
- 9.Paydas S, Acikalin A, Ergin M, et al. Micro-RNA (miRNA) profile in Hodgkin lymphoma: association between clinical and pathological variables. Med Oncol. 2016;33(4):34. doi: 10.1007/s12032-016-0749-5 [DOI] [PubMed] [Google Scholar]
- 10.Yanchun L, Hongmei Y, Zhihong C, et al. MicroRNA-192-5p promote the proliferation and metastasis of hepatocellular carcinoma cell by targeting SEMA3A. Appl Immunohistochem Mol Morphol. 2015;25(4):1. [DOI] [PubMed] [Google Scholar]
- 11.Hu S, Tao R, Wang S, et al. MicroRNA-21 promotes cell proliferation in human hepatocellular carcinoma partly by targeting HEPN1. Tumor Biol. 2015;36(7):5467–5472. doi: 10.1007/s13277-015-3213-9 [DOI] [PubMed] [Google Scholar]
- 12.Chang RM, Yang H, Fang F, et al. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology. 2015;60(4):1251–1263. doi: 10.1002/hep.27221 [DOI] [PubMed] [Google Scholar]
- 13.Yuan C, Liu C, Zou N. MicroRNA-340 targets NF-κB1 to inhibit cell proliferation, migration and invasion in hepatocellular carcinoma cell line. J Biomater Tissue Eng. 2016;6(8):649–658. doi: 10.1166/jbt.2016.1481 [DOI] [Google Scholar]
- 14.Wang P, Cao J, Liu S, et al. Upregulated microRNA-429 inhibits the migration of HCC cells by targeting TRAF6 through the NF-κB pathway. Oncol Rep. 2017;37(5):2883. doi: 10.3892/or.2017.5507 [DOI] [PubMed] [Google Scholar]
- 15.Plasterk RHA. RNA interference (RNAi). Encycl Entomol. 2008;7(5):3195–3196. [Google Scholar]
- 16.Mansoori B, Sandoghchian SS, Baradaran B. RNA interference and its role in cancer therapy. Adv Pharm Bull. 2014;4(4):313–321. doi: 10.5681/apb.2014.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng C, Chen Z, Bai E, et al. [Construction and identification of a lentiviral vector for RNA interference of human GLUT3 gene]. J Central South Uni. 2016;41(5):455–462. [DOI] [PubMed] [Google Scholar]
- 18.West JA, Davis CP, Sunwoo H, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhen L, Yun-Hui L, Hong-Yu D, et al. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2016;37(1):673. doi: 10.1007/s13277-015-3843-y [DOI] [PubMed] [Google Scholar]
- 20.Chai Y, Liu J, Zhang Z, et al. HuR‐regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016;5(7):1588–1598. doi: 10.1002/cam4.2016.5.issue-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian K, Liu G, Tang Z, et al. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch Biochem Biophys. 2017;615:1. doi: 10.1016/j.abb.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 22.Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34(34):4482–4490. doi: 10.1038/onc.2014.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng X, Zhang Y, Liu Y, et al. HIF‐2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial‐mesenchymal transition. J Cell Biochem. 2018;119(4):3247–3256. doi: 10.1002/jcb.v119.4 [DOI] [PubMed] [Google Scholar]
- 24.Jusufović E, Rijavec M, Keser D, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non–small–cell lung cancer. PLoS One. 2012;7(9):e45577. doi: 10.1371/journal.pone.0045577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z, Gan J, Long Z, et al. Targeted delivery of let-7b to reprogramme tumor-associated macrophages and tumor infiltrating dendritic cells for tumor rejection. Biomaterials. 2016;90:72–84. doi: 10.1016/j.biomaterials.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Liu K, Liu S, et al. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumor Biol. 2015;36(12):9779–9788. doi: 10.1007/s13277-015-3749-8 [DOI] [PubMed] [Google Scholar]
- 27.Refolo MG, D’Alessandro R, Lippolis C, et al. IGF-1R tyrosine kinase inhibitors and Vitamin K1 enhance the antitumor effects of Regorafenib in HCC cell lines. Oncotarget. 2017;8(61):103465–103476. doi: 10.18632/oncotarget.v8i61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jean-Fran Ois D, Thierry D, Birgit W, et al. Tumor volume in pharyngolaryngeal squamous cell carcinoma: comparison at CT, MR imaging, and FDG PET and validation with surgical specimen. Radiology. 2004;233(1):93. doi: 10.1148/radiol.2331030660 [DOI] [PubMed] [Google Scholar]
- 29.Yao Y, Mao W, Dong M, et al. Serum Insulin-Like Growth Factor-1 (IGF-1): a novel prognostic factor for early recurrence of Hepatocellular Carcinoma (HCC). Clin Lab. 2017;63(2):261. doi: 10.7754/Clin.Lab.2016.160732 [DOI] [PubMed] [Google Scholar]
- 30.Jun T, Yang JD, Yeh ML, et al. 904 – significantly lower rates of cirrhosis and lower long-term survival in cryptogenic Hepatocellular Carcinoma (HCC) compared to viral etiologies: results of a multi-center international cohort. Gastroenterology. 2017;152(5):S1071. doi: 10.1016/S0016-5085(17)33616-8 [DOI] [Google Scholar]
- 31.Chen X, Ren Z, Yin S, et al. The local liver ablation with pulsed electric field stimulate systemic immune reaction against hepatocellular carcinoma (HCC) with time-dependent cytokine profile. Cytokine. 2017;93:44–50. doi: 10.1016/j.cyto.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 32.Bolondi L, Sofia S, Siringo S, et al. Surveillance programme of cirrhotic patients for early diagnosis and treatment of hepatocellular carcinoma: a cost effectiveness analysis. Gut. 2001;48(2):251–259. doi: 10.1136/gut.48.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamimura K, Kobayashi Y, Takahashi Y, et al. Tumor markers for early diagnosis for brain metastasis of hepatocellular carcinoma: a case series and literature review for effective loco-regional treatment. Cancer Biol Ther. 2017;18(2):79–84. doi: 10.1080/15384047.2016.1276134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo S, Chen W, Luo Y, et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8(5):5395. [PMC free article] [PubMed] [Google Scholar]
- 35.Mang Y, Li L, Ran J, et al. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther. 2017;10:1003–1016. doi: 10.2147/OTT.S116319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han J, Zhang F, Yu M, et al. RNA interference-mediated silencing of NANOG reduces cell proliferation and induces G0/G1 cell cycle arrest in breast cancer cells. Cancer Lett. 2012;321(1):80–88. doi: 10.1016/j.canlet.2012.02.021 [DOI] [PubMed] [Google Scholar]
- 37.Pai SI, Lin YY, Macaes B, et al. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13(6):464–477. doi: 10.1038/sj.gt.3302694 [DOI] [PubMed] [Google Scholar]
- 38.Wu S, Wang X, Chen JX, et al. Predictive factors for the sensitivity of radiotherapy and prognosis of esophageal squamous cell carcinoma. Int J Radiat Biol. 2014;90(5):407–413. doi: 10.3109/09553002.2014.894649 [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Feng J, Zhi X, et al. Let-7b inhibits cell proliferation, migration, and invasion through targeting Cthrc1 in gastric cancer. Tumour Biol. 2015;36(5):3221–3229. doi: 10.1007/s13277-014-2950-5 [DOI] [PubMed] [Google Scholar]
- 40.Lu PW, Li L, Wang F, et al. Effects of long non-coding RNA HOST2 on cell migration and invasion by regulating MicroRNA Let-7b in breast cancer. J Cell Biochem. 2017;19(6):4570–4580. [DOI] [PubMed] [Google Scholar]
- 41.Han X, Wang X, Zhao B, et al. MicroRNA-187 inhibits tumor growth and metastasis via targeting of IGF-1R in hepatocellular carcinoma. Mol Med Rep. 2017;16(2):2241–2246. doi: 10.3892/mmr.2017.6788 [DOI] [PubMed] [Google Scholar]
- 42.Rahmoon MA, Youness RA, Gomaa AI, et al. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017;35(2–3):1–12. doi: 10.1080/08977194.2017.1354859 [DOI] [PubMed] [Google Scholar]
- 43.Chen CL, Wu JC, Chen GY, et al. Baculovirus-mediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol Ther. 2015;23(1):79–88. doi: 10.1038/mt.2014.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.