Abstract
Background
We attempted to train and validate a model of deep learning for the preoperative prediction of the response of patients with intermediate-stage hepatocellular carcinoma (HCC) undergoing transarterial chemoembolization (TACE).
Method
All computed tomography (CT) images were acquired for 562 patients from the Nan Fang Hospital (NFH), 89 patients from Zhu Hai Hospital Affiliated with Jinan University (ZHHAJU), and 138 patients from the Sun Yat-sen University Cancer Center (SYUCC). We built a predictive model from the outputs using the transfer learning techniques of a residual convolutional neural network (ResNet50). The prediction accuracy for each patch was revaluated in two independent validation cohorts.
Results
In the training set (NFH), the deep learning model had an accuracy of 84.3% and areas under curves (AUCs) of 0.97, 0.96, 0.95, and 0.96 for complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD), respectively. In the other two validation sets (ZHHAJU and SYUCC), the deep learning model had accuracies of 85.1% and 82.8% for CR, PR, SD, and PD. The ResNet50 model also had high AUCs for predicting the objective response of TACE therapy in patches and patients of three cohorts. Decision curve analysis (DCA) showed that the ResNet50 model had a high net benefit in the two validation cohorts.
Conclusion
The deep learning model presented a good performance for predicting the response of TACE therapy and could help clinicians in better screening patients with HCC who can benefit from the interventional treatment.
Key Points
• Therapy response of TACE can be predicted by a deep learning model based on CT images.
• The probability value from a trained or validation deep learning model showed significant correlation with different therapy responses.
• Further improvement is necessary before clinical utilization.
Electronic supplementary material
The online version of this article (10.1007/s00330-019-06318-1) contains supplementary material, which is available to authorized users.
Keywords: Hepatocellular carcinoma, Artificial intelligence, Multidetector computed tomography, ROC curve
Introduction
Hepatocellular carcinoma (HCC) ranks second as the major cause of cancer-related deaths globally and is the sixth most common cancer in the world. Its incidence has continuously increased in recent years, and approximately 850,200 new cases of HCC are annually diagnosed worldwide [1, 2]. Less than 30% of patients with HCC are eligible for potentially curative therapies, such as transplantation, resection, or ablation [3, 4]. For selected patients who are not suitable for such interventions, but have liver-confined disease, preserved liver function, and good performance status, transarterial chemoembolization (TACE) is recommended according to international guidelines [5–9].
Although repeated TACE procedures are often needed, the initial response effectively predicts the overall survival (OS) because the best response cannot always be achieved after one session of TACE, especially in large tumors. Moreover, the achievement of a treatment response at an early time point is the robust predictor for favorable outcomes [10]. The texture analysis based on contrast-enhanced magnetic resonance imaging (MRI) before TACE may act as imaging biomarkers to predict an early response from patients with HCC. The highest accuracy for complete response (CR) group and the non-complete response (NCR) was 0.76 [11]. A pretherapeutic dynamic CT texture analysis can also be valuable in predicting the therapy response of HCC to TACE. Higher arterial enhancement and GLCM (gray-level co-occurrence matrix) moments, lower homogeneity, and smaller tumor size are significant predictors of complete response (CR) after TACE [12]. However, based on the traditional statistics and machine learning, the accuracy of this method is limited. On the condition of optimal cutoff values for predicting a CR to TACE in the receiver operating characteristic (ROC) curves, the highest AUC of texture parameters was 0.72. Furthermore, most studies focused on the two classifications (CR or NCR) and the prediction of four classifications (CR, PR, SD, and PD) using CT images is unclear. Therefore, a more effective model to accurately identify patients who would have an initial response after TACE therapy is urgently needed to facilitate individualized treatment strategies.
Deep learning has recently gained attention as a technique for realizing Artificial intelligence (AI) [13–15]. Several types of deep-stacked artificial neural networks, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), have been proposed and judiciously used in various fields. Deep CNNs are especially recognized as demonstrating high performance for image recognition tasks [16, 17]. Some initial successes in applying deep learning to the assessment of radiological images have been witnessed [18–21]. A study used a deep learning algorithm to non-invasively predict the IDH (Isocitrate Dehydrogenase) status within a multi-institutional dataset of low- and high-grade gliomas [22]. Deep learning also shows the potential to stage liver fibrosis based on radiological images [23]. However, there is embarrassed that performing deep learning often faces a shortage of medical data, especially in radiological images of patients undergoing treatment. Transfer learning, which is a feasible deep learning technique for addressing a lack of image data, has been proven a highly effective technique, particularly in the case of limited medical images [24, 25]. The models have been used to distinguish the features of the medical images in a much faster manner and with significantly fewer training medical images [13].
In this study, based on the CT images from three independent centers, we aimed to investigate a deep learning algorithm to precisely and non-invasively evaluate the different therapy response in HCC patients before the TACE treatment.
Materials and methods
Patients
Our retrospective study had been approved by the institutional review board and Ethical Committee (NFEC-201208-K3). This study included patients in Nan Fang Hospital (NFH), Zhu Hai Hospital Affiliated with Jinan University (ZHHAJU), and Sun Yat-sen University Cancer Center (SYUCC), who matched the following criteria for selection: (a) radiologically or pathologically proven HCC; (b) received initial treatment of TACE; (c) availability of hepatic-arterial CT imaging within 7 days before treatment; (d) availability of hepatic-arterial CT imaging within 30 days after treatment; and (e) patients with Barcelona Clinic Liver Cancer (BCLC) stage B. The exclusion criteria were as follows: (a) previous treatments, including loco-regional or whole-body therapies, such as liver transplantation, radiotherapy, radiofrequency ablation, or sorafenib treatment, and (b) other malignant liver tumors. Electronic medical records were used to collect the pretreatment clinical characteristics of patients. Supplemental Figure 1 shows the recruitment pathways for patients in training and two validation cohorts. Based on the radiology evaluation in patients after the first TACE therapy, the different responses on hepatic-arterial CT images were determined by modified RECIST (mRECIST 1.1), including complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). We defined the objective response as CR+PR and the non-response as SD+PD.
CT acquisition and region-of-interest segmentation
Contrast-enhanced computed tomography (CECT) was performed at three hospitals as previously described [26]. Supplemental Table S1 presents the scan characteristics for the three centers. All CT images were downloaded using a picture archiving and communication system (PACS; Nan Fang Hospital, Zhu Hai Hospital Affiliated with Jinan University, and Sun Yat-sen University Cancer Center Network Center, China). The CT images of all patients were input into the ITK-SNAP software (version 3.6). The tumor regions of interest (ROIs) were analyzed by three senior radiologists who had 14-year experience (reader 1, Jing Zhang), 17-year experience (reader 2, Jingxian Shen), and 23-year experience (reader 3, Yikai Xu). The ROIs of CT images from the training cohort (NFH) and two validation cohorts (ZHHAJU and SYUCC) were manually segmented by reader 1 and reader 2, respectively. Then, reader 3 confirmed each ROI and saved as the main file (CT image) and segmentation file (mask image) in the ITK-SNAP software. All radiologists were specifically blinded to the therapy outcome of the patients from three cohorts.
Image preprocessing
The window width and level were transformed into the original one. All CT images were reconstructed using a medium sharp reconstruction algorithm with a thickness of 1 mm. Subsequently, the intensity values of the image were mapped to [0, 1]. This target of the deep learning algorithm is the classified labels of CR, PR, SD, and PD. We saved one CT image and mask of ROI from the largest tumor area for each patient and then saved the other two CT images and two corresponding masks of ROIs from the nearest sequences. Using an in-house algorithm, we extracted an average of approximately three patches with a resize of 224 × 224 × 3 for each patient in the training and two validation cohorts, respectively. Notably, each patch for the training and validation of the network was entirely included in the ROIs of the CT images. Finally, 1687 patches were extracted from 562 patients for training (NFH); 268 patches were extracted from 89 patients for validation 1 (ZHHAJU); and 406 patches were extracted from 138 patients for validation 2 (SYUCC). Data augmentation techniques were introduced before the training procedure considering the potential bias caused by the unbalanced data and big data requirement of deep learning [27]. Specifically, in our study, the patches were equally and randomly distributed across each class by data augmentation, including level flip, vertical flip, level and vertical flip, 90° rotation, and − 90° rotation. Using this method, we built a “new” training data and this augmentation was only performed on the NFH cohort, not on the two external validation cohorts. In order to minimize memory usage, we used data augmentation only in real time.
Transfer learning of the residual neural network
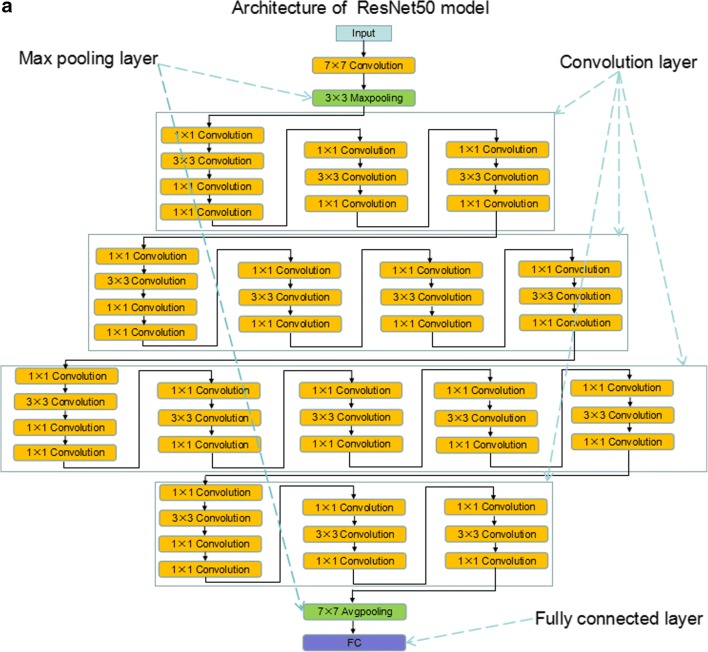
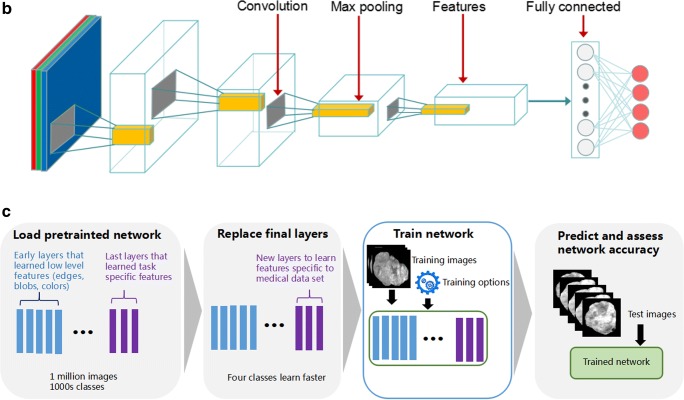
ResNet is a representative deep convolutional neural network integrated with images, auto-encoding, and classification. It won the 2015 ImageNet Large Scale Visual Recognition Challenge (ILSVRC) by using feature transmission to prevent gradient vanishing, such that a much deeper network than those used previously could be effectively trained [28]. Motivated by a previous study that a deeper network is potentially more powerful than shallow networks, our residual network was derived from a 50-layer residual network architecture and has 177 layers in total, showing detailed information in the Supplementary Information (ResNet50). ResNet50 has been trained on a subset of the ImageNet database (http://www.image-net.org) and can classify images into 1000 object categories (e.g., keyboard, mouse, pencil, and many animals). The architecture of ResNet50 and flowchart of deep learning for CT images were shown in Fig. 1a–c. We froze the weights of earlier layers (1 to 174) in the pretrained network. The trained network does not update the parameters of the frozen layers. Freezing the weights of many initial layers can significantly speed up network training and prevent over-fitting to the new medical dataset.
Fig. 1.
The architecture of ResNet50 and deep learning model flowchart. a, b Architecture of ResNet50 is shown and includes convolution layers, max pooling layers, and a fully connected layer. c A ResNet50 model was pretrained on a million images from the ImageNet database and can classify images into 1000 object categories. Based on this new dataset of CT images, a transfer learning model was adapted to significantly shorten the training time and improve the accuracy. The earlier connected layers were frozen (1 to 174), and the final connected layers were replaced (175 to 177). Finally, this model was transferred to a novel network. All patches were augmented in the proposed approach after the ROI patches were determined from the CT images. A transfer learning 50-layer residual convolutional neural network was used to predict the response to TACE therapy
A series of blocks consisting of three convolutional layers (fc1000, fc1000_softmax, and classification layers_fc1000) were replaced by new layers (fc4, fc4_softmax, and classification layers_fc4) to extract deep residual features and transmit features from the front layer to the latter one. At the end of the network, a full-connection layer was used to perform classification. During training, the weights were optimized via the stochastic gradient descent (SGD) optimization algorithm with a mini-batch size 64 [29]. After fine-tuning parameters of deep learning, the learning rate and the number of max epochs were set to 0.0001 and 54, respectively, to ensure covering of the entire data for efficient training. The loss function was identified as binary cross-entropy. We used the sigmoid function to compute the probability before the output layer. The performance of deep learning model was estimated by AUC and accuracy (). The patches from Nan Fang Hospital (NFH) were trained via pretrained ResNet50. Based on the trained model, patches from Zhu Hai Hospital Affiliated with Jinan University (ZHHAJU) and Sun Yat-sen University Cancer Center (SYUCC) were used to be validated, respectively.
Implementation details
Our implementation was based on the Deep Learning Toolbox™ Model for the ResNet50 Network in MATLAB (version 2018a; MathWorks). Our training experiments were performed in a Linux environment on a machine with the following specifications: CPU Intel Xeon Processor E5-2640V3 at 2.60 GHz, GPU NVIDIA Pascal Titan X, and 128-GB RAM.
Statistical analysis
Statistical analyses were performed with R statistical software version 3.5.0 (R Core Team, 2018), GraphPad prism 7.0, and MATLAB 2018a. The receiver operating characteristic (ROC) curves were plotted with the “pROC” package. A confidence interval (CI) of 95% for AUC was calculated using each dataset (1000 bootstrap). The confusion matrices were plotted with MATLAB 2018a in three different cohorts to calculate the accuracies of estimating the response of TACE therapy. Decision curve analysis (DCA) was performed using the “dca.R” package. The Mann–Whitney U test was analyzed by R statistical software. Two-sided p values < 0.05 were considered significant.
Results
Clinical characteristics of patients
Five hundred sixty-two patients with HCC were finally included in the training cohort (NFH), and 89 and 138 patients were allocated to the independent validation cohorts 1 (ZHHAJU) and 2 (SYUCC), respectively. Table 1 summarizes the baseline clinical characteristics of the training and two validation cohorts. In the training cohort, validation cohort 1, and validation cohort 2, the age of 168 (29.90%), 28 (31.46%), and 38 (27.54) patients was more than 60 years, respectively. Sixty (10.68%), 14 (15.73%), and 15 (10.87%) patients were females in the training cohort, validation cohort 1, and cohort 2, respectively. In the training cohort, the patients for CR, PR, SD, and PD were 83 (14.77%), 151 (26.87%), 222 (39.50%), and 106 (18.86%), respectively. In validation cohorts 1 and 2, the patients for CR, PR, SD, and PD were 13 (14.61%) and 21 (15.22%), 24 (26.97%) and 37 (26.81%), 35 (39.33%) and 54 (39.13%), and 17 (19.09%) and 26 (18.84%), respectively. No significant differences were observed between the three cohorts in the clinical database.
Table 1.
Participant characteristics in the training and validation cohorts
| Characteristic | Training cohort (n = 562) | Validation cohort 1 (n = 89) | Validation cohort 2 (n = 138) |
|---|---|---|---|
| Age (years) | |||
| ≤ 60 | 394 (70.10%) | 61 (68.54%) | 100 (72.46%) |
| > 60 | 168 (29.90%) | 28 (31.46%) | 38 (27.54%) |
| Sex | |||
| Male | 502 (89.32%) | 75 (84.27%) | 123 (89.13%) |
| Female | 60 (10.68%) | 14 (15.73%) | 15 (10.87%) |
| HBsAg status | |||
| Positive | 514 (91.46%) | 80 (96.67%) | 125 (90.57%) |
| Negative | 48 (8.54%) | 9 (3.33%) | 13 (9.43%) |
| Child–Pugh classification | |||
| A | 451 (80.25%) | 74 (83.15%) | 111 (80.43%) |
| B | 111 (19.75%) | 15 (16.85%) | 28 (19.57%) |
| ALT (U/mL) | |||
| ≤ 40 | 240 (42.70%) | 45 (50.56%) | 69 (50.00%) |
| > 40 | 322 (57.30%) | 44 (49.44%) | 69 (50.00%) |
| AST (U/mL) | |||
| ≤ 40 | 147 (26.16%) | 27 (30.34%) | 37 (26.81%) |
| > 40 | 415 (73.84%) | 62 (69.66%) | 101 (73.19%) |
| AFP (ng/mL) | |||
| ≤ 20 | 322 (57.30%) | 49 (55.06%) | 83 (60.14%) |
| > 20 | 240 (42.70%) | 40 (44.94%) | 55 (39.86%) |
| Hepatocirrhosis status | |||
| Present | 294 (52.31%) | 40 (44.94%) | 70 (50.73%) |
| Absent | 268 (47.69%) | 49 (55.06%) | 68 (49.27%) |
| Tumor size (cm) | |||
| ≤ 5 | 83 (14.77%) | 12 (13.48%) | 22 (15.94%) |
| > 5, ≤ 10 | 245 (43.59%) | 37 (41.57%) | 52 (37.68%) |
| > 10 | 234 (41.64%) | 40 (44.95%) | 64 (46.38%) |
| Tumor numbers | |||
| ≤ 3 | 463 (82.38%) | 77 (86.51%) | 114 (82.61%) |
| > 3 | 99 (17.62%) | 12 (13.49%) | 24 (17.39%) |
| Response to therapy | |||
| CR | 83 (14.77%) | 13 (14.61%) | 21 (15.22%) |
| PR | 151 (26.87%) | 24 (26.97%) | 37 (26.81%) |
| SD | 222 (39.50%) | 35 (39.33%) | 54 (39.13%) |
| PD | 106 (18.86%) | 17 (19.09%) | 26 (18.84%) |
HBsAg, hepatitis B surface antigen; ALT, alanine aminotransferase; AST, aspartate aminotransferase; AFP, alpha-fetoprotein; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease
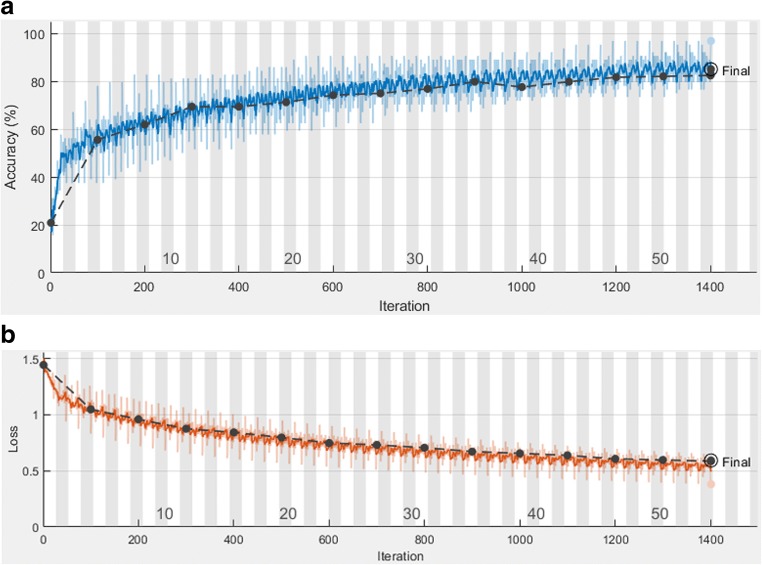
Training and validation of the deep learning model in multi-class classification
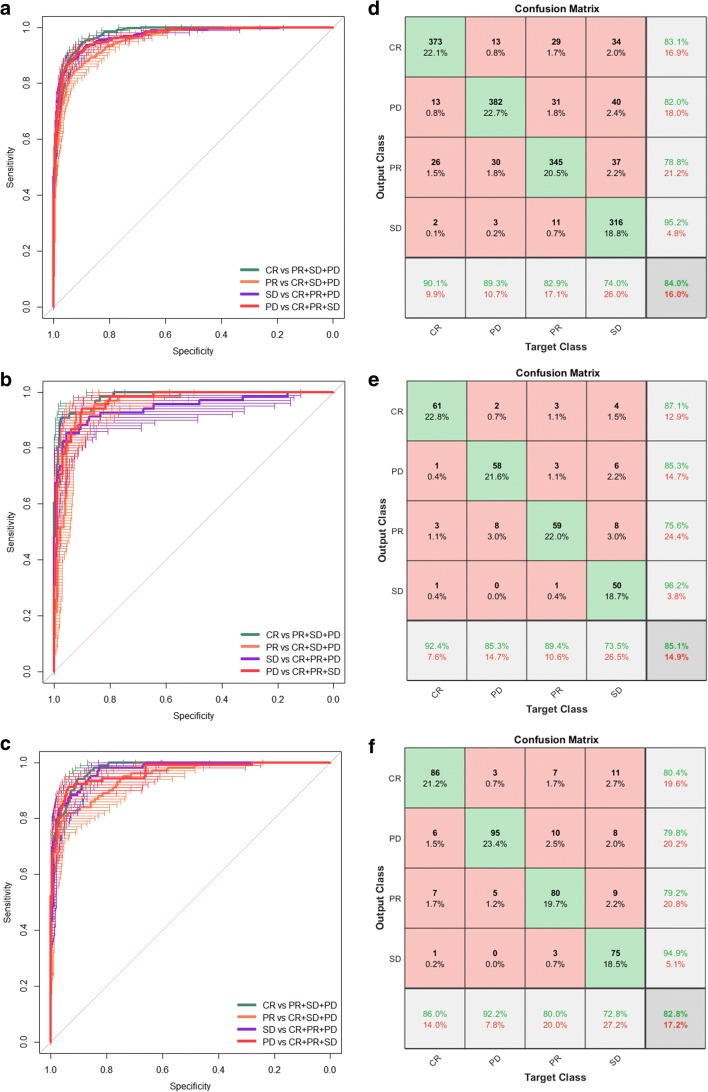
All the patches (n = 1687) were augmented and trained in the training cohort via the residual convolutional neural network (ResNet50) to increase the robustness of the model. Figure 2a shows that the accuracy was above 80% and the cross-entropy loss was close to 0.5 after 54 epochs training (1401 iterations) and 71 min 38 s time (Fig. 2b). The resulting model had an AUC of 0.97 (0.97–0.98), 0.96 (0.96–0.97), 0.95 (0.94–0.96), and 0.96 (0.96–0.97) for CR, PR, SD, and PD, respectively (Fig. 3a). We then tested the patches (n = 268) in validation cohort 1. An AUC of 0.98 (0.97–0.99), 0.96 (0.95–0.98), 0.95 (0.93–0.98), and 0.94 (0.90–0.98) for CR, PR, SD, and PD was observed (Fig. 3b). In validation cohort 2, the AUCs of predicting CR, PR, SD, and PD in TACE treatment were 0.97 (0.96–0.98), 0.96 (0.94–0.98), 0.94 (0.92–0.97), and 0.97 (0.95–0.98), respectively (Fig. 3c). The deep learning model indicated good discrimination of the therapy response using the ROI patches in the two cohorts.
Fig. 2.
Training and validation processes of the deep learning model based on the CT images. Accuracy (a) and cross-entropy loss (b) were plotted against the training step during the length of the training of the four-class classifier over the course of 54 steps. The red and black lines represent the training and validation processes, respectively. The cross-entropy loss was close to 0.5, while the final validation accuracy was 85.07%
Fig. 3.
ROC curve and confusion matrix for predicting the response to TACE therapy. a Each patient for the assessment of the response to TACE therapy is shown via the ROC curve. In the training set, the deep learning model had an AUC of 0.97, 0.96, 0.95, and 0.96 for CR, PR, SD, and PD, respectively. b In validation 1 set, the deep learning model had an AUC of 0.98, 0.96, 0.95, and 0.94 for CR, PR, SD, and PD, respectively. c In validation 2 set, the deep learning model had an AUC of 0.97, 0.96, 0.95, and 0.95 for CR, PR, SD, and PD, respectively. d–f The model exhibited accuracies of 84.3%, 85.1%, and 82.8% in the three cohorts (NFH, ZHHAJU, and SYUCC), respectively
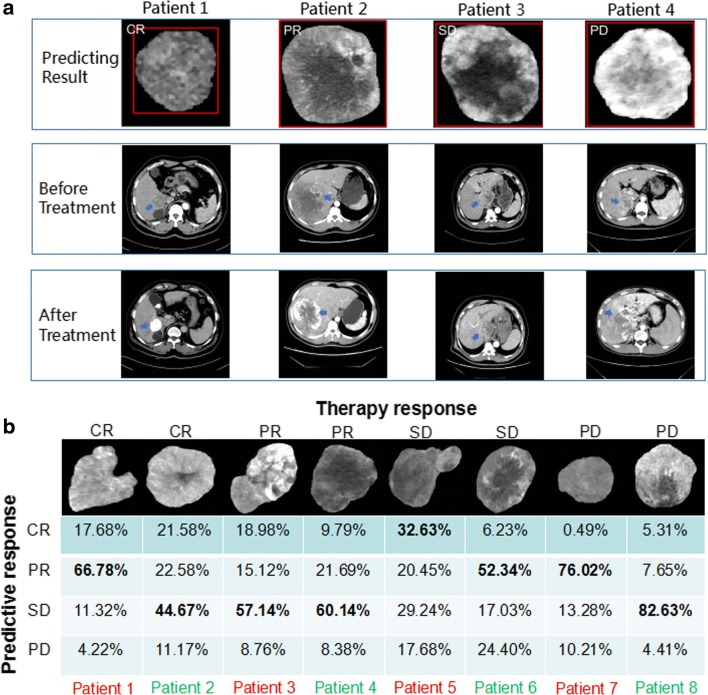
We next focused on the predictive accuracy of the deep learning model in each patch by confusion matrix. The training cohort exhibited an average accuracy of 84.0% and a low error of 16% of predicting CR, PR, SD, and PD in TACE therapy (Fig. 3d). The independent validation cohorts 1 and 2 showed an average accuracies of 85.1% and 82.8% and low errors of 14.9% and 17.2%, respectively (Fig. 3e, f). We further chose four typical patches of different therapy responses from validation cohort 2 and displayed the CT images of the maximum cross-sectional diameter of the liver tumor, including pretreatment and post-treatment of the hepatic-arterial phase images (Fig. 4a). As shown, the deep learning model performed well in the classification of predicting CR, PR, SD, and PD of TACE treatment. There were some difficult cases of these patterns that were misclassified in the ResNet50. We displayed eight misclassified images of ROI from validation cohorts 1 and 2 in Fig. 4b. For example, CR could be incorrectly predicted as SD (44.67%) in patient 2, and PD also could be incorrectly predicted as PR (76.02%) in patient 7.
Fig. 4.
Examples of correctly predicted and misclassified patches by the ResNet50. a Horizontal cross-section CT images through four patients of validation cohort 2 with HCC before and after 1 month of TACE therapy. b Eight patches showed the misclassifications of four therapy responses in validation cohorts 1 and 2, respectively. The output of the deep learning model is presented below each patch. Red and green colors represent the ROI images from validation cohorts 1 and 2, respectively
Evaluation on performance of two-class classification via trained ResNet50 model
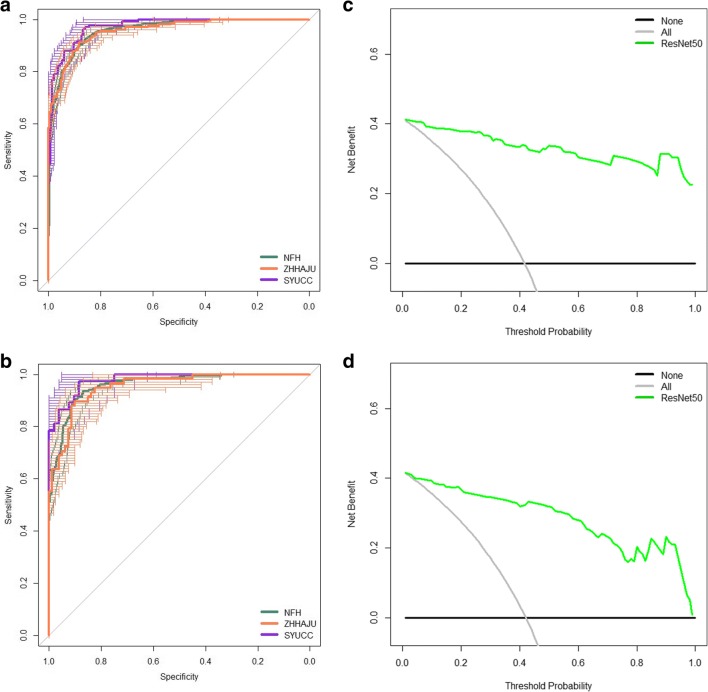
Multiple 2D arrays were output by the convolutional layers of a ResNet50 model. To further estimate the classification performance of our deep network trained on four-category learning, we have considered a clinically important task, including two-class classification of objective response (CR or PR) and non-response (SD or PD). On the task of two-class classification, we have merged the output prediction probability by the trained deep learning model for four-category classification, for adding CR and PR (i.e., yresponse = yCR + yPR). The probability value of therapy response (objective response) was significantly increased in the response HCC patches versus the non-response HCC patches in Supplemental Figure 2A–C (each cohort, p < 0.0001). Throughout this method, the model achieved an AUC of 0.95 (0.95–0.96), 0.96 (0.94–0.97), and 0.97 (0.96–0.98) in the patches from NFH, ZHHAJU, and SYUCC cohorts (Fig. 5a). The probability value of therapy response was calculated bythe average probability values of all patches in each patient of three cohorts. We found the ResNet50 model presented high AUC in the patients of three cohorts (Fig. 5b). To analyze the clinical use of model based on deep learning, the decision curve analysis (DCA) for the predictor of ResNet50 was performed in this study. When the threshold probabilities are ~ 2% and 4%, the model shows stronger benefit in comparison with the treat-all or treat-none strategy in the ZHHAJU (Fig. 5c) and SYUCC (Fig. 5d) cohorts. This similar performance of DCA also has been displayed in the training cohort (Supplemental Figure 3).
Fig. 5.
ROC curve and DCA curve for estimating the objective response to TACE therapy. a In the NFH, ZHHAJU, and SYUCC cohorts, the deep learning model had an AUC of 0.95, 0.96, and 0.97 for predicting therapy response via patches, respectively. b Based on the predictive probability, the model presented an AUC of 0.95, 0.96, and 0.97 for predicting therapy response in all patients from NFH, ZHHAJU, and SYUCC cohorts, respectively. In the ZHHAJU (c) and SYUCC (d) cohorts, the DCA indicated that when the threshold probability was above 2% and 4%, THE use of the deep learning model for predicting TACE response would gain more benefit than the “treat-all” patients or “treat-none” schemes
Discussion
In this study, we demonstrated a novel application of deep learning to predict the response of TACE therapy in a three-institution dataset of HCC. As far as we know, this is the first time that the deep learning model based on radiological images is used to predict the four responses of interventional treatment in liver cancer. This algorithm may facilitate deep learning techniques for the medical field of precise therapy oncology. Based on the pretreatment ROI images of patients with HCC, this utility model of deep learning is a potential method for predicting the response of TACE therapy.
According to the BCLC stage system, TACE is recommended for patients with HCC with BCLC stage B [6, 30–32]. In patients with stage C, TACE therapy is also a frequent and important application treatment, especially in comprehensive treatment [33–35]. Recent studies revealed that the therapy response at first chemoembolization is a good predictor for the favorable outcome in hepatocellular carcinoma [10, 36]. However, no report was associated with the prediction response of TACE therapy in the field of hepatocellular carcinoma via deep learning of CT images. Previous studies showed that different clinical risk factors (e.g., tumor size) perform well in prediction [37–39], but the precise estimation of four therapy responses remains challenging in clinical settings and difficult to implement. A new method involving a radiomics approach based on radiological images (e.g., CT, MR, and PET-CT) is also currently being applied in various tumors. It extracts radiographic features from conventional images and includes the features of tumor shape, texture, intensity, and wavelet transform characteristics [40–46]. However, numerous pre-engineered features are artificial design features. This may lead to poor reproducibility and nonredundant radiomics features (RFs) for CT images because of the variable scan parameters of different types of imaging equipment [47]. The application of radiomics also relies on traditional machine learning techniques. Unlike the above method, the algorithm of deep learning can directly learn predictive features from the images and potentially greatly increase the robust accuracy in these radiological images [48, 49].
Previous studies mostly focused on deep learning based on the small segmentation patches of each ROI image to enhance the size of the sample and frequently showed a significant AUC [50–52]. However, the scenario of predicting the labels of entire ROI images was often ignored. Therefore, we used an algorithm of transfer learning to evaluate the treatment response via the whole CT-ROI patches. Comparing with previous study, high AUCs of predicting CR, PR, SD, and PD in the therapy response of TACE were observed among the three cohorts [12]. This result indicated that our transfer learning model performed well in predicting different therapy responses using CT images from three independent centers. The result of the confusion matrix presented significantly high accuracies of prediction in the NFH, ZHHAJU, and SYUCC cohorts and was distinct from previous report [11]. Interestingly, we found the accuracy for the training cohort was lower than for the validation cohort 1 (84.3% vs. 85.1%). We speculated the phenomenon was correlated with a small sample size of patients in validation cohort 1. Increasing number of patients would potentially reduce the validation accuracy. Misclassified CR patches by the deep learning model were more observed in PR patches than in SD and PD patches in the training cohort (1.5%) and validation cohorts 1 (1.7%) and 2 (1.1%). Meanwhile, misclassified PD patches were more frequently found in PR patches than in SD and CR patches. The precision probability of preoperatively predicting the four therapy responses (i.e., CR, PR, SD, and PD) via each ROI patch was calculated and found useful in individualized clinical treatment. We further investigate the prediction of objective response (response or non- response) in patches or patients and also found high accuracies in the three cohorts. This finding demonstrated that the deep learning model based on CT images may help doctors recognize patients who would acquire well or poor initial response of TACE therapy.
However, our study has several limitations. First, the sample size of patients with HCC was relatively small, and this was a retrospective research. A much larger database of the prospective study would be collected from more centers in the future. Second, we trained and validated all the patches of the 2D CT images from three medical centers. Because of 3D patches’ potential of having more context information, we speculated the 3D CT patches had an accuracy higher and a better model quality than that of the 2D CT patches. The 3D CT patches would be investigated in the next step. Third, the correlation between the biological processes (e.g., differential gene expression and pathway) and the prediction results of deep learning networks in HCC was unknown and should be analyzed in the future. Fourth, the ROIs were drawn manually in our study. Lesions selected from different abdominal radiologists might have various differences, impacting on disease classification. We would use the combination of the algorithm for HCC segmentation and ResNet50 model to automatically predict the outcome of TACE therapy in the following study.
In summary, the deep learning model based on CT images would potentially serve as a new tool for predicting the therapy response of patients undergoing TACE treatment. Our method using transfer learning for predictive classification of radiological images may also be used to determine more precise clinical treatments in other malignant tumors.
Electronic supplementary material
(DOCX 3103 kb)
Acknowledgments
(I) Conception and design: Li Liu and Jie Peng; (II) administrative support: none; (III) provision of study materials or patients: Li Liu and Jie Peng; (IV) collection and assembly of data: Jie Peng, Shuai Kang, Hangxia Deng, Yikai Xu, Jing Zhang, Wuxing Gong, and Jingxian Shen; (V) data analysis and interpretation: Li Liu, Jie Peng, Zhengyuan Ning, and Jinhua Huang; (VI) manuscript writing: all authors; and (VII) final approval of manuscript: all authors.
Abbreviations
- AI
Artificial intelligence
- AUCs
Areas under the curve
- BCLC
Barcelona Clinic Liver Cancer
- CECT
Contrast-enhanced computed tomography
- CI
Confidence interval
- CNNs
Convolutional neural networks
- CR
Complete response
- DCA
Decision curve analysis
- GLCM
Gray-level co-occurrence matrix
- HCC
Hepatocellular carcinoma
- IDH
Isocitrate dehydrogenase
- ILSVRC
ImageNet Large Scale Visual Recognition Challenge
- MRI
Magnetic resonance imaging
- NCR
Non-complete response
- NFH
Nan Fang Hospital
- OS
Overall survival
- PACS
Picture archiving and communication system
- PD
Progressive disease
- PR
Partial response
- RFs
Radiomics features
- ROC
Receiver operating characteristic
- ROIs
Regions of interest
- RNNs
Recurrent neural networks
- SD
Stable disease
- SGD
Stochastic gradient descent
- SYUCC
Sun Yat-sen University Cancer Center
- TACE
Transarterial chemoembolization
- ZHHAJU
Zhu Hai Hospital Affiliated with Jinan University
Funding
This work was supported by the National Nature Science Foundation of China (grant nos. 81372283, 81472711, 81401180, 81672756, and 91540111), Sciences Foundation of Guizhou Province (grant no. 20185579-X), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015), and the Natural Science Foundation of Guangdong Province (grant no. 2014A030311 01 3).
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication is Jie Peng.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Multicenter study
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinhua Huang, Email: huangjh@sysucc.org.cn.
Li Liu, Email: liuli.fimmu@gmail.com.
References
- 1.Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11:317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding J, Wang H. Multiple interactive factors in hepatocarcinogenesis. Cancer Lett. 2014;346:17–23. doi: 10.1016/j.canlet.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Lei Z, Li J, Wu D, et al. Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma within the Milan criteria. JAMA Surg. 2016;151:356–363. doi: 10.1001/jamasurg.2015.4257. [DOI] [PubMed] [Google Scholar]
- 4.Ding XX, Zhu QG, Zhang SM, et al. Precision medicine for hepatocellular carcinoma: driver mutations and targeted therapy. Oncotarget. 2017;8:55715–55730. doi: 10.18632/oncotarget.18382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu K, Huang J, Lai L, et al. Medium or large hepatocellular carcinoma: sorafenib combined with transarterial chemoembolization and radiofrequency ablation. Radiology. 2018;288:300–307. doi: 10.1148/radiol.2018172028. [DOI] [PubMed] [Google Scholar]
- 6.Takayasu K, Arii S, Ikai I, et al. Prospective cohort study of transarterial chemoembolization for unresectable hepatocellular carcinoma in 8510 patients. Gastroenterology. 2006;131:461–469. doi: 10.1053/j.gastro.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 8.Fako V, Wang XW. The status of transarterial chemoembolization treatment in the era of precision oncology. Hepat Oncol. 2017;4:55–63. doi: 10.2217/hep-2017-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biolato M, Gallusi G, Iavarone M, et al. Prognostic ability of BCLC-B subclassification in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. Ann Hepatol. 2018;17:110–118. doi: 10.5604/01.3001.0010.7542. [DOI] [PubMed] [Google Scholar]
- 10.Kim BK, Kim SU, Kim KA, et al. Complete response at first chemoembolization is still the most robust predictor for favorable outcome in hepatocellular carcinoma. J Hepatol. 2015;62:1304–1310. doi: 10.1016/j.jhep.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Yu JY, Zhang HP, Tang ZY, et al. Value of texture analysis based on enhanced MRI for predicting an early therapeutic response to transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma. Clin Radiol. 2018;73:758.e9–758.e18. doi: 10.1016/j.crad.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Park HJ, Kim JH, Choi SY, et al. Prediction of therapeutic response of hepatocellular carcinoma to transcatheter arterial chemoembolization based on Pretherapeutic dynamic CT and textural findings. AJR Am J Roentgenol. 2017;209:W211–W220. doi: 10.2214/AJR.16.17398. [DOI] [PubMed] [Google Scholar]
- 13.Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2017;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 15.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Deep learning for staging liver fibrosis on CT: a pilot study. Eur Radiol 18:514–521 [DOI] [PubMed]
- 17.Wang K, Lu X, Zhou H, et al. Deep learning radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2018;5:136–154. doi: 10.1136/gutjnl-2018-316204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajkomar A, Lingam S, Taylor AG, Blum M, Mongan J (2017) High-throughput classification of radiographs using deep convolutional neural networks. J Digit Imaging 30:95–101 [DOI] [PMC free article] [PubMed]
- 19.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 20.Lee YH. Efficiency improvement in a busy radiology practice: determination of musculoskeletal magnetic resonance imaging protocol using deep-learning convolutional neural networks. J Digit Imaging. 2018;10:107–117. doi: 10.1007/s10278-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamidian S, Sahiner B, Petrick N, Pezeshk A (2017) 3D convolutional neural network for automatic detection of lung nodules in chest CT. Proc SPIE Int Soc Opt Eng 10134 [DOI] [PMC free article] [PubMed]
- 22.Chang K, Bai HX, Zhou H, et al. Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res. 2018;24:1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Liver fibrosis: deep convolutional neural network for staging by using gadoxetic acid-enhanced hepatobiliary phase MR images. Radiology 287:146–155 [DOI] [PubMed]
- 24.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng J, Zhang J, Zhang Q, Xu Y, Zhou J, Liu L (2018) A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn Interv Radiol 24:121–127 [DOI] [PMC free article] [PubMed]
- 27.Ning Z, Luo J, Li Y, et al. Pattern classification for gastrointestinal stromal tumors by integration of radiomics and deep convolutional features. IEEE J Biomed Health Inform. 2018;23(3):1181–1191. doi: 10.1109/JBHI.2018.2841992. [DOI] [PubMed] [Google Scholar]
- 28.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. 2016 IEEE Conf Comput Vis Pattern Recognit 1:770–778
- 29.Wang L, Yang Y, Min R, Chakradhar S (2017) Accelerating deep neural network training with inconsistent stochastic gradient descent. Neural Netw 93:219–229 [DOI] [PubMed]
- 30.Kudo M. Locoregional therapy for hepatocellular carcinoma. Liver Cancer. 2015;4(3):163–164. doi: 10.1159/000367741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiraoka A, Kumagi T, Hirooka M, et al. Prognosis following transcatheter arterial embolization for 121 patients with unresectable hepatocellular carcinoma with or without a history of treatment. World J Gastroenterol. 2006;12(13):2075–2079. doi: 10.3748/wjg.v12.i13.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veloso Gomes F, Oliveira JA, Correia MT, et al. Chemoembolization of hepatocellular carcinoma with drug-eluting polyethylene glycol embolic agents: single-center retrospective analysis in 302 patients. J Vasc Interv Radiol. 2018;29(6):841–849. doi: 10.1016/j.jvir.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Tsurusaki M, Murakami T. Surgical and Locoregional therapy of HCC: TACE. Liver Cancer. 2015;4(3):165–175. doi: 10.1159/000367739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown DB, Geschwind JF, Soulen MC, Millward SF, Sacks D (2006) Society of Interventional Radiology position statement on chemoembolization of hepatic malignancies. J Vasc Interv Radiol 17:217–223 [DOI] [PubMed]
- 36.Gillmore R, Stuart S, Kirkwood A, et al. EASL and mRECIST responses are independent prognostic factors for survival in hepatocellular cancer patients treated with transarterial embolization. J Hepatol. 2011;55:1309–1316. doi: 10.1016/j.jhep.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Vesselle G, Quirier-Leleu C, Velasco S, et al. Predictive factors for complete response of chemoembolization with drug-eluting beads (DEB-TACE) for hepatocellular carcinoma. Eur Radiol. 2016;26(6):1640–1648. doi: 10.1007/s00330-015-3982-y. [DOI] [PubMed] [Google Scholar]
- 38.Park KH, Kwon SH, Lee YS, et al. Predictive factors of contrast-enhanced ultrasonography for the response to transarterial chemoembolization in hepatocellular carcinoma. Clin Mol Hepatol. 2015;21(2):158–164. doi: 10.3350/cmh.2015.21.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JH, Yoon HK, Ko GY, et al. Nonresectable combined hepatocellular carcinoma and cholangiocarcinoma: analysis of the response and prognostic factors after transcatheter arterial chemoembolization. Radiology. 2010;255(1):270–277. doi: 10.1148/radiol.09091076. [DOI] [PubMed] [Google Scholar]
- 40.Xi YB, Guo F, Xu ZL, et al. Radiomics signature: a potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. 2018;47(5):1380–1387. doi: 10.1002/jmri.25860. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer. 2016;17:441–449. doi: 10.1016/j.cllc.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Zheng J, Li Y, et al. A radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin Cancer Res. 2017;23:6904–6911. doi: 10.1158/1078-0432.CCR-17-1510. [DOI] [PubMed] [Google Scholar]
- 43.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Shi Z, Lian Y, et al. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. 2017;27:3509–3522. doi: 10.1007/s00330-016-4653-3. [DOI] [PubMed] [Google Scholar]
- 45.Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung Cancer. Radiology. 2016;281(3):947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Zhu Y, Burnside ES, et al (2016) Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2:16012 [DOI] [PMC free article] [PubMed]
- 47.Berenguer R, Pastor-Juan MDR, Canales-Vazquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288(2):407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 48.Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286(3):887–896 [DOI] [PubMed]
- 49.Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys. 2017;44(10):e360–e375. doi: 10.1002/mp.12344. [DOI] [PubMed] [Google Scholar]
- 50.Shen C, Gonzalez Y, Chen L, Jiang SB, Jia X (2018) Intelligent parameter tuning in optimization-based iterative CT reconstruction via deep reinforcement learning. IEEE Trans Med Imaging 37(6):1430–1439 [DOI] [PMC free article] [PubMed]
- 51.Lam C, Yu C, Huang L, Rubin D (2018) Retinal lesion detection with deep learning using image patches. Invest Ophthalmol Vis Sci 59(1):590–596 [DOI] [PMC free article] [PubMed]
- 52.Gao X, Qian Y. Prediction of multidrug-resistant TB from CT pulmonary images based on deep learning techniques. Mol Pharm. 2018;10:1021. doi: 10.1021/acs.molpharmaceut.7b00875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 3103 kb)