Abstract
The Myxococcales order consist of eleven families comprising30 genera, and are featured by the formation of the highest level of differential structure aggregations called fruiting bodies. These multicellular structures are essential for their resistance in ecosystems and is used in the primitive identification of these bacteria while their accurate taxonomic position is confirmed by the nucleotide sequence of 16SrRNA gene. Phenotypic classification of these structures is currently performed based on the stereomicroscopic observations that demand personal experience. The detailed phenotypic features of the genera with similar fruiting bodies are not readily distinctive by not particularly experienced researchers. The human examination of the fruiting bodies requires high skill and is error-prone. An image pattern analysis of schematic images of these structures conducted us to the construction of a database, which led to an extractable recognition of the unknown fruiting bodies. In this paper, Convolutional Neural Network (CNN) was considered as a baseline for recognition of fruiting bodies. In addition, to enhance the result the classifier, part of CNN is replaced with other classifiers. By employing the introduced model, all 30 genera of this order could be recognized based on stereomicroscopic images of the fruiting bodies at the genus level that not only does not urge us to amplify and sequence gene but also can be attained without preparation of microscopic slides of the vegetative cells or myxospores. The accuracy of 77.24% in recognition of genera and accuracy of 88.92% in recognition of suborders illustrate the applicability property of the proposed machine learning model.
Subject terms: Imaging, Data mining
Introduction
Myxobacteria are nonpathogenic, free-living bacteria that mainly thrive in terrestrial excosystems as well as marine habitats. They are aerobic and mesophilic bacteria that mainly exist near the surface of the soil. Myxobacteria are Gram-negative with high genomic GC content bacteria that exhibit outstanding characteristics such as the formation of the multicellular fruiting body, gliding motility, predation of microorganisms and cellulolysis. They are considered as a taxonomically distinct group due to their elaborated life cycle that is uncommon in the prokaryotic domain1. They are able to lyse other bacteria and yeast cells by predatory behavior. In this process, hydrolyzing enzymes and secondary metabolite molecules secrete to the medium and hydrolyze the prey to consumable nutrients2.
During the intricate life cycle, Myxobacteria can produce a resistant and dormant form of cells called myxospore, which is generated, forms vegetative cells inside the fruiting body structures during their complex life cycle. Fruiting bodies of Myxobacteria vary in colour, shape, and size. The size of the fruiting body ranges from 10 to 1000 μm depending on genus and species1. The shape of fruiting bodies emanates as spherical, cylindrical, mounds and either hybride form of two or more morphologies like fruiting body of the Chondromyces species3. Most of the genera produce distinctive and colored fruiting bodies on the surface of the medium that often can be observed through naked eyes. Predominantly, fruiting bodies comprise sporangioles that enclose myxospores in the form of single or clusters. Identification and classification of Myxobacteria extensively dependent on the morphology detail of fruiting bodies, swarming pattern on solid media and shape of the vegetative cells and myxospores.
The correlation between morphology and phylogeny of Myxobacteria investigated by Sproer et al. (1999) has shown that the phenotypic classification can provide a consistent basis for the description of neotype species. Accordingly, the findings suggested the classification at the genus level based on morphology is consistent for most of the Myxobacterial genera.
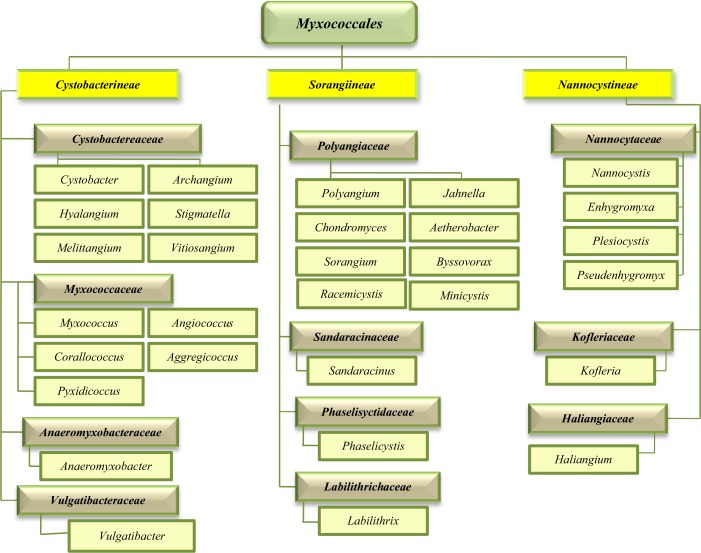
The order Myxococcales is placed in δ class of Proteobacteria phylum and consists of three suborders, 11 families, 30 genera and 58 species4. The taxonomic classification and phylogeny of the order Myxococcales updated on September 2018 is presented in Fig. 1.
Figure 1.
Phylogeny of the order Myxococcales updated on September 2018.
There are only few reports on image analysis of Myxobacteria, which have been focused on their swarming motility patterns. Fruiting body formation in Myxobacteria requires swarming by the sensing of signals under starvation conditions. A-signal is involved in the detection of nutrient deficiency in the environment. By entering the cells to a developmental pathway, a large number of cells convert to myxospores and a limited population of cells forms a layer around of myxospores called peripheral cells. On the other hand, C-signal is essential for rippling and differentiation.
It is repoted that 49% of drugs approved between 1981 and 2014 originated from natural products or their semisynthetic derivatives5. In the last decade, myxobacteria were recognized to be a valuable source of bioactive secondary metabolites producing novel structure skeletons. Thus, they have received notable attention in drug discovery plans due to the diversity and unique modes of action of their metabolites6,7. A large number of myxobacterial metabolites have been reported having antibacterial activity such as myxovalargin8, sorangicin9, saframycin10, sorangiolid11, chondrochlorens12, and thuggacins13 while metabolites like rhizopodin14 and chondramides15 have shown anticancer activity through interactions with microtubule assembly in the eukaryotic cell lines. In addition, these bacteria represent other rare bioactivities such as anti-malarial, insecticidal, immunosuppressive, and anti-viral activities, etc.7.
To screen Myxobacteria for their bioactive metabolites, samples collected from different environments are cultured on several media and differentiated manually. This often makes the whole process of screening labour intensive, time-consuming, and experienced-technicians-oriented16. In addition to the 16S rRNA gene sequencing, other genomic approaches such as full genome sequencing, multi-locus sequencing, and metagenomics have been used in their identification17 which are time-consuming18 and are not needed for the primary identification at the genus level. Therefore, for impressive deduction of labour work during strains screening programs, a high throughput computer-based isolation instead of the experience of an expert is highly appreciated. Artificial pattern recognition or machine learning systems was applied in this study to provide the identification of Myxobacteria at the genus level based on self-experience approach. Consequently, pattern recognition systems implicate algorithms, which their performance will be improved through experience19.
No study aimed at characterizing the Myxobacterial genera based on their automated image analysis is reported so far.
In the current study, the development and the application of user-aided image acquisition and automated processing pipeline for the identification of myxobacterial strains based on their descriptive features were achieved. The method proved its robustness toward size and shape variations of the fruiting bodies due to their different maturity state; and changing qualities of images, different technical alterations, and different fields of view. The provided method is fast and accurate, while requires inexpensive assembly of the instrument to assemble the system. In addition, learning of their pattern is active, which results in auto-updating the database. Finally, the application of the method is simple, and there is no need for complex knowledge or years of experience or courses.
Convolutional Neural Network is employed for discrimination between different myxobacterial genera. Furthermore, to increase the accuracy of recognition, the fully connected part of DCNN was swapped with other classifiers. Experimental results illustrate that the proposed model can identify the genera with the accuracy of 77.24% and the higher taxa of suborder with the accuracy of 88.92%.
The structure of the paper is as the following. Section 2 introduces preliminary work includes Extreme Learning Machine (ELM) classifier, On-Line Sequential ELM (OSELM)20, Constraint ELM (CELM)21, and Convolutional Neural Network. Section 3 describes materials and methods. Experiments and validation are explained in Section 5 and Section 6 concludes the results.
Preliminary Work
ELM model
Suppose a set of samples available for building a model is {(xi, ti)|xi ∈ RD, ti ∈ RM, i = 1, 2, … N}. Also, suppose l as the number of hidden nodes and g(x) as the activation function in a multi-layer perceptron neural network. With these assumptions, input weights W and the hidden biases b can be specified randomly. In this regard, the hidden layer output of ELM can be obtained by Eq. (1):
| 1 |
where wi ∈ RD, bi ∈ R, i = 1, 2, …. L.
Considering β as the output weights, based on the proof presented by Huang et al. (Huang et al., 2017), the norm of β is smaller, and the generalization performance of ELM is more suitable. Consequently, by finding the least square solution of the problem the output weights can be obtained by Eq. (2):
| 2 |
where h(xi) is the i th output vector of the hidden layer, ti is the i th label vector
Based on the Karush–Kuhn–Tucker22 theory, Eq. (2) can be expressed by the following Lagrange function:
| 3 |
where each αi Lagrange multiplier relates to an instance xi. The following set of equations can be calculated by the partial derivative of Eq. (3):
| 4 |
| 5 |
| 6 |
where α = [α1, …, αN]T and the least square solution of β is attained by computing the three equations. The answer is as Eq. (7):
| 7 |
Accordingly, the output function of ELM is as Eq. (8):
| 8 |
The calculation of dot product in ELM can be replaced by introducing the kernel function k(xi . xj) as Eq. (9). To decrease the computational complexity of high dimensional dot product, it is essential to make sure that k(xi . xj) is simply a mapping method of the relative location of two input examples23.
| 9 |
OSELM
In the first phase of OSELM which is boosting phase the Single Layer Feed forward Network (SLFNs) is trained using the primitive ELM method with some batch of training data in the initialization stage and these boosting training data will be discarded as soon as this phase is completed. The required batch of training data is very small, which can be equal to the number of hidden neurons (e.g. for 10 neurons, 10 training samples may be needed to boost the learning).
In the second phase, the OSELM learns the train data one-by-one or chunk-by-chunk and all the training data will be discarded after the learning process on these data is finished22.
CELM
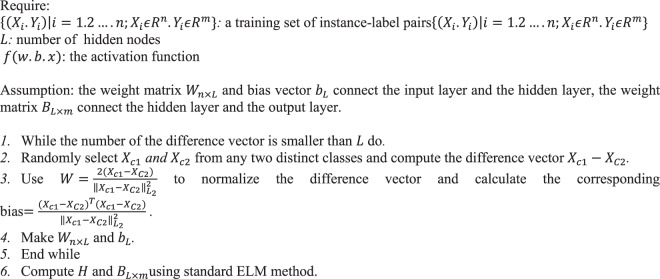
The algorithm of CELM was proposed for constraining the weight vectors {Wj|j = 1 .…. L} from the input layer to the hidden layer by drawing from the closed set of difference vectors of between-class instances, which are the set of vectors correlating the instances of one class with instances of a different class24. The pseudo code of CELM training process is illustrated in Algorithm 1. It can be seen that except that the CELM constrains the input connection weights of the hidden neuron, the CELM is similar to the ELM. Experiment results are shown that CELM has a performance with higher efficiency compared to ELM24.
Algorithm 1.
Training process of CELM.
Convolutional neural network
On a general overview, after feeding images to convolution neural network, which includes several layers of types convolutional, nonlinear, and pooling, the transformed images are delivered to the output layer that can predict a class in classification problems or a single number in regression problems. Typically, the convolutional neural network includes the following layers:
Input layer
Usually values for raw pixels of the input image are incorporated into the input layer.
Convolution layer
The convolutional layer is the main layer of a CNN. Neurons in this layer are connected to the regions of the image or the previous layer. These areas called the filters to move vertically and horizontally and extract features from the image of the previous layer. For each part, a dot product of the weights and the input is computed, and a bias value is added to it. The step size of filter shift is named stride. The number of weights applied for a filter is obtained by Eq. (10) 25:
| 10 |
where h and w is the height and the width of the filter, and c is considered the number of channels in the input. By using Eq. (11), the number of parameters in a convolution layer is calculated.
| 11 |
where 1 is for the bias and n is the number of filters. The output size of the convolutional layer is calculated using Eq. (12):
| 12 |
where I is input size of layer, F is filter size, P is padding dimension and S is stride number.
The layer’s parameters consist of a set of learnable filters. Through the forward flow, each filter is convolved with the input data. Convolution is simply the result of the dot product between the elements of the filter and the input. Accordingly, in the training process of the network, the filters are learnt and are activated when it faced with some specific features at some spatial positions in the input.
A neuron in CNN investigates a small region in the input and shares parameters with neurons in the same activation map.
Batch normalization layer
This layer normalizes its inputs xi by estimating the mean μB and variance of a mini-batch and on each input channel. Afterward, it normalizes the activations using Eq. (13):
| 13 |
when the mini-batch variance is very small, improves numerical stability. To allow for the possibility that inputs with zero mean and unit variance are not optimal for the layer that follows the batch normalization layer, the batch normalization layer further shifts and scales the activations Eq. (14):
| 14 |
Here, the offset β and scale factor γ are learnable parameters that are updated during network training. At the end of the learning process, batch normalization layer calculates mean and variance over the full training set and stores them in sequence in trained mean and trained variance properties25.
Pooling layer
Pooling layer does a form of down sampling. There are several non-linear functions to implement pooling among which max-pooling is the most common. It partitions the input image into a set of non-overlapping rectangles and, for each such sub-region, outputs the maximum. The intuition is that the exact location of a feature is less important than its rough location relative to other features. The pooling layer serves to progressively reduce the spatial size of the representation, to reduce the number of parameters and amount of computation in the network, and hence to also control overfitting. It is common to periodically insert a pooling layer between successive convolutional layers in CNN architecture. Pooling layers provide a form of translation invariance. Specially max-pooling across rotated/scaled database images gains rotation/scale invariance. The Poolsize property determines the size of the rectangular regions. The output size of a pooling layer with input size InputSize is as Eq. (15):
| 15 |
Activation layer
This layer applies the non-saturating activation function. It increases the nonlinear properties of the decision function and of the overall network without affecting the receptive fields of the convolution layer. ReLU is the abbreviation of Rectified Linear Units. ReLU is preferable to other functions, because it trains the neural network several times faster without a significant penalty to generalization accuracy26. Other functions are also used to increase nonlinearity, for example, the saturating hyperbolic tangent and the sigmoid function.
Fully connected layer
The fully connected layer is a traditional Multi-Layer Perceptron that uses a Softmax activation function in the output layer. The term “Fully Connected” implies that every neuron in the previous layer is connected to every neuron on the next layer. The result of this layer is a vector of 1 × 1 × n, where n is the number of classes. After several convolutional and pooling layers, the classification in the neural network is done via fully connected layers. This layer(s) is the part that learns supervisory in contrast to the convolutional, pooling and activation layers that learn nonsupervisory.
Softmax layer
This layer receives the output of the previous (fully connected) layer and converts it to a probability distribution on the classes. This is done through Eq. (16):
| 16 |
where 0 < p(Cr|x.θ) ≤ 1 and . Also , p(x. θ. cr) is the conditional probability of the sample given class r and p(cr) is the class prior probability27.
Materials and Methods
The details of preparing MYXO.DB and the proposed method are described in this section.
Classification of Myxobacteria based on the appearance of the fruiting bodies
The appearance of the fruiting body that harbors the myxobacterial spores (myxospores) can be categorized as Table 1. These morphological characteristics are currently used by expert researchers to distinguish the genera from each other. Therefore, this morphological identification key demands the deep understanding and visualization skill of the researchers in order to lead them to the right genus.
Table 1.
The macro-morphological specification of Myxobacterial genera used in observational identification.
| Family | Genus | Sporangiole¥ | Characteristics of the Fruiting Body | Characteristics of the Swarm | |||||
|---|---|---|---|---|---|---|---|---|---|
| Stalk | Shape of the fruiting body | Size (μm) | Texture | Color and shape of the swarm | Swarmedge | Agar corrosion | |||
| Cystobactereaceae | Archangium | − | − | Variable in size and shape, strings of myxospores in hardened slime | 50–1000 | Hard | Branched radial veins | Flame like | − |
| Cystobacter | + | − | Rounded, elongate, or coiled singly or in groups | 50–180 | Hard | Tough slime sheet with veins | Flame like | − | |
| Hyalangium | + | − | Small spherical sporangioles with glassy shape | 35–45 | Glassy | Yellow or brown/Thin, tough slime sheet with very fine veins | Fine veins | − | |
| Melittangium | + | + | Semispherical sporangiole like a mushroom cap | 50–100 | Soft | Bright yellow/Slime sheet and radial veins | Flare- to flame-like | − | |
| Stigmatella | + | + | Spherical sporangioles singly or in clusters | 300–350 | Hard | Yellow/Tough slime sheet with oscillating waves | Flare like | − | |
| Vitiosangium | − | − | Oval to Bean shaped, solitary mounds | 20–200 | Soft | Coherent swarm with scattered ripples | Flare like | — | |
| Myxococcaceae | Corallococcus | − | − | Coral, hornlike, often solitary | 20–1,000 | Hard | Colorless/thin and transparent | Flares, flames | − |
| Myxococcus | − | +| | Rounded to oval, often solitary | 50–200 | Soft | Colorless to shade of orange and yellow/thin, film-like | Flares, flames | − | |
| Pyxidicoccus | + | − | Ovoid clusters | 30–80 | Hard | Colorless/thin, film-like | Flares, flames | − | |
| Aggregicoccus | − | − | Spherical fruiting body-like aggregates | ND | Soft | Transparent swarms, wavy, rippling structures | Intricate veins on edges | − | |
| Angiococcus | + | − | Spheroidal sporangioles | 30–40 | Soft | Thin, spreading swarm of gliding cells | Flare-like | − | |
| Anaeromyxobacteraceae | Anaeromyxobacter | + | − | Polyhedral or spherical solitary or cluster | ND | Soft | — | — | − |
| Vulgatibacteraceae | Vulgatibacter | − | − | Fruiting body-like aggregates | ND | Soft | No swarming | unstructured | − |
| Polyangiaceae | Byssovora | + | − | Polyhedral sporangioles in sorus | 220–560 | Soft | Pseudoplasmodial thin layer | Fanlike | + |
| Chondromyces | + | + | Sessile, spectacular, complex, and elegant miniature tree- or flowerlike fruiting body | 1000 | Hard | light orange and burrow/Thin, filmlike, transparent | Fanlike | + | |
| Jahnella | + | − | Coils shape sporangioles in cluster | 60–90 × 80–120 | Tough | Orange/Scattered long veins | Bands in agar | + | |
| Polyangium | + | − | Oval to polyhedral sessile sporangioles, arranged in a cluster or solitary | 50–400 | Soft | Pseudoplasmodial swarm | Fan-shaped | + | |
| Sorangium | + | − | Ovoid to polyhedral sporangioles in cluster and chain | 20–30 | Hard | Yellow or orange/Soft radial veins | Curtain-like | + | |
| Aetherobacter | + | − | Fascicles in chains or rolls in aggregates | 50–3000 | Soft | Swarm forms ring- or halo-like colonies | Coherent migrating cells | + | |
| Minicystis | + | − | Small fruiting bodies, ovoid sporangioles | 4.0–12.0 | Soft | Swarm appears film-like, thin and transparent | unstructured, with loose migrating cells sometimes with tiny flares | − | |
| Racemicystis | + | — | Varying size | 200–800 | Tough | Orange to beige | Sweep like | − | |
| Sandaracinaceae | Sandaracinus | − | − | Fruiting body-like aggregates sessile and irregular | 50–150 | Soft | Orange/shallow wave depressions | Cell mounds at the end | − |
| Phaselicystidaceae | Phaselicystis | + | − | Bean, sausage, or ovoid shaped sporangioles | 20 × 25, 49 × 56 | Tough | Tough, slimy net-like veins | Flame- or flare-like | + |
| Labilithrichaceae | Labilithrix | − | − | Raised colonies instead of the Fruiting body | ND | Soft | Slimy | Hairy-like | − |
| Nanocystacea | Enhygromyxa | − | − | Fruiting body like aggregates (Rounded, hump, globular) | 100–150 | Soft | Colorless/light orange to red/delicate slimy veins | Flare to pseudoplasmodium | + |
| Nannocystis | + | − | Spherical, oval to short sausage-shaped sporangiole | 6 × 3.5–110 × 0 | Hard | Excavated/deep tunnels | Trails or fine wave | + | |
| Plesiocystis | − | − | Fruiting-like body aggregates | 100–500 | Soft | Thin, transparent, pseudoplasmodium | Flare-like | + | |
| Pseudenhygromyxa | − | − | Fruiting-like body aggregates | 50–800 | Soft | Colorless to pale peach/slimy veins | Flare-like | + | |
| Kofleriaceae | Kofleria | − | − | Yellow knobs in agar or on surface | ND | Soft | Yellow/film-like with radial veins | Lateral rim | − |
| Haliangiaceae | Haliangium | + | − | Fruiting-like aggregate or sessile oval-shaped sporangioles | 15–150 | Soft | Colorless to yellow shades/thin, film-like | Flare- to flame-like | + |
¥Sporangiole: Packages of myxospores.
+Indicate the presence.
−Indicate the absence.
ND: Not Distinguishable.
Selection of the stereomicroscope images of the typical fruiting bodies
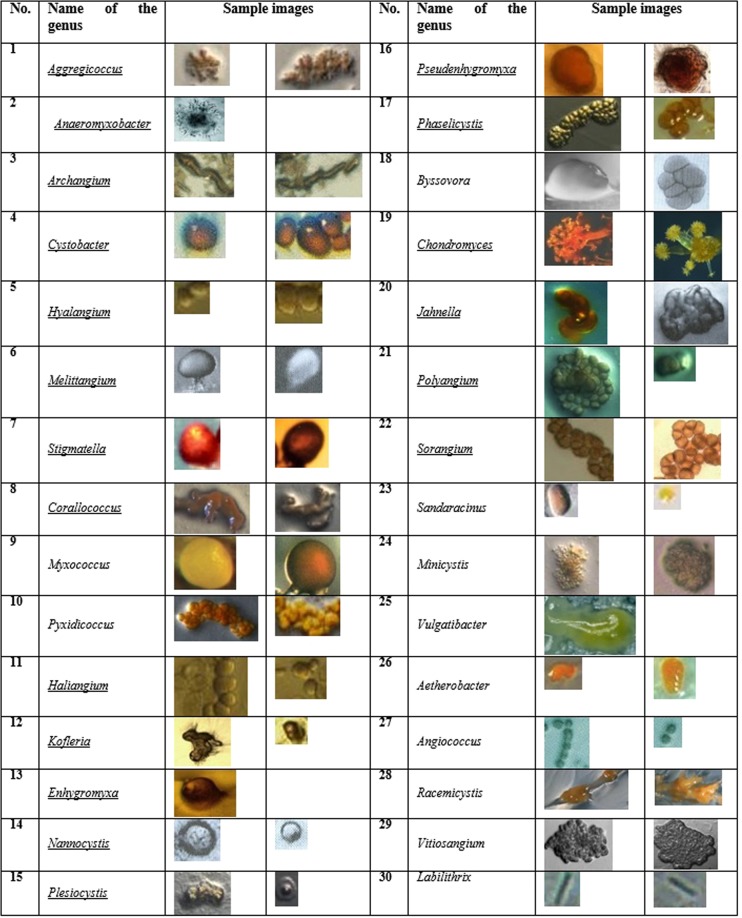
Morphological features of the spore producing structures called fruiting bodies were selected based on their typical descriptive morphology from the valid publications. The images were selected based on the descriptive literature and the number of investigated genera was 30 as exists in the order of Myxobacteria at the time of conducting the experiment.
During the process of pattern recognition, the features that describe the fruiting bodies including shape, size, intensity, and texture were extracted from the typical fruiting bodies of all 30 genera of this order.
Preparing the dataset of MYXO.DB
Since the source of images were quite different in terms of the parameters including contrast, resolution, size, illumination, having noise and capturing camera aspects, designing an automatic image processing method for automatic segmentation of samples from images was complex. Therefore, to form a dataset for automatic recognition of Myxobacteria, the single samples or compact fruiting bodies were cropped manually. The genera, which produce single fruiting body, were considered individually, while the genera that does not form concrete fruiting bodies are not produced, their swarm pattern was considered.
As mentioned before, identification and classification of Myxobacteria dependents principally on the morphology of fruiting bodies, swarming pattern on medium, shape of vegetative cells and myxospores shape. In preparing the dataset of MYXO.DB, morphology of fruiting bodies was analyzed. Furthermore, the colony morphology for some Myxobacteria was considered. Table 2 represents the number of single fruity bodies, which could be extracted from each genus of the 30 classes. Some sample derived images of each genus of MYXO.DB are illustrated in Fig. 2.
Table 2.
The quantity of each class in MYXO.DB, which contains 322 samples and 30 classes.
| No. | Name of the genus | No. of images | No. | Name of the genus | No. of images |
|---|---|---|---|---|---|
| 1 | Aggregicoccus | 11 | 16 | Pseudenhygromyxa | 4 |
| 2 | Anaeromyxobacter | 1 | 17 | Phaselicystis | 13 |
| 3 | Archangium | 8 | 18 | Byssovora | 3 |
| 4 | Cystobacter | 10 | 19 | Chondromyces | 17 |
| 5 | Hyalangium | 9 | 20 | Jahnella | 11 |
| 6 | Melittangium | 9 | 21 | Polyangium | 9 |
| 7 | Stigmatella | 14 | 22 | Sorangium | 21 |
| 8 | Corallococcus | 15 | 23 | Sandaracinus | 14 |
| 9 | Myxococcus | 38 | 24 | Minicystis | 20 |
| 10 | Pyxidicoccus | 13 | 25 | Vulgatibacter | 1 |
| 11 | Haliangium | 5 | 26 | Aetherobacter | 14 |
| 12 | Kofleria | 10 | 27 | Angiococcus | 9 |
| 13 | Enhygromyxa | 1 | 28 | Racemicystis | 11 |
| 14 | Nannocystis | 14 | 29 | Vitiosangium | 6 |
| 15 | Plesiocystis | 8 | 30 | Labilithrix | 3 |
Figure 2.
The representative images of each genus in MYXO.DB.
As mentioned in the previous section for learning the difference between distinct types of Myxobacteria and the similarities within the samples of a certain genus, an adequate number of samples should be available. In this regard, some classes with a small number of instances have not been considered in some experiments. In this regard, two datasets with a smaller number of classes were constructed.
MY25 dataset is a subset of MYXO.DB, which contains 313 images, which are categorized into 25 classes. In this dataset, the classes with less than four samples have been removed from the MYXO.DB.
In MY22 dataset, the MYXO.DB dataset is augmented and the number of instances increased by cropping some sub images from each image sample. The augmented dataset contains 629 images, which are categorized into 22 classes. In this dataset, the classes with less than eight samples have been omitted.
To recognize the suborder of each strain, MYCategories dataset is built which contains 319 images samples of MYCategories were categorized into three classes includes Cystobacterineae, Sorangiineae and Nannocystineae.
By viewing the samples, it can be concluded that some classes are similar to each other for example Stigmatella and Myxococcus in terms of the morphology of fruiting body. Such similarities can encounter the automatic recognition systems with a notable challenge. On the other hand, a few number of samples for some classes like Anaeromyxobacter, Myxococcus and Vulgatibacter prevents a pattern recognition system from automatic learning. Like learning of a human being child, for an efficient machine learning process, there should be sufficient training samples or experiences28.
Further, the color of the fruiting body and the background, and the shape and orientation of colonies are diverse in different samples of a certain class.
Preprocessing
The discriminating features of Myxobacteria are their shape and texture. Their color can be altered by some culture conditions; therefore, color is not a reliable distinguishing feature for recognizing different types of Myxobacteria.
Some descriptors of the object morphology like area, convex area, roundness etc. denote the size and shape approximately. Consequently, before imposing the sample images to any pattern recognition system, initially, all the images should be converted to gray scale image to drop the color information.
In addition, all the images have been normalized to the size of 100 × 100 pixels because of the below reasons:
Capturing device for each image of Myxobacteria has a certain property.
The resolution and the amount of the noise in images are dissimilar.
Different level of maturity causes diverse size of individuals or colonies.
The magnification magnitude of the microscope in image capturing may be disparate.
The input of the proposed method is normalized gray scale images. Equation (17) is used for converting a colored image I in RGB color space to gray scale image Ig 29 where IR, IG and IB are the image I in Red, Green and Blue color plate and Ig is the resulted gray level image.
| 17 |
Feature extraction by convolution neural network
In this paper, for automated recognition of different fruiting bodies a CNN classification model is designed. By the emerging application of deep learning in computer vision applications, extracting the features through the CNNs is beneficial for producing general image descriptors. CNNs resulted in high efficiency in many image classification applications30. CNNs were inspired by biological processes in which the connectivity pattern between neurons is deduced by the organization of the animal visual cortex31. The major ascendancy of CNNs is partial independence from previous information and human effort in feature design.
A CNN includes a stack of different types of layers that convert the input data into an output value or label. After a brief description of the applied algorithm in this study, the structure of the designed CNN for automatic recognition of Myxobacteria fruiting bodies is described as the following.
Input layer
The input of this layer is images with equal size and labeled. In our modeling, raw pixels of images with the size of 28 × 28 are fed into the input layer.
Convolutional layer
Our model comprised four convolution layers. In the first convolution layer, 16 filters, in the second layer, 32 filters, in the third layer, 64 filters, and in the last layer 128 filters are used. The size of filters is considered 3 × 3 with the padding size of one.
Batch normalization layer
In order to normalize the extracted features using the convolution layer, a batch normalization layer is considered following each convolution layer. The applied model contains four convolutional layers and one batch normalization layer after each one.
Rectified linear unit (ReLU) layer
This layer activates max (0, x) activation function on each neuron, that causes the negative values to be converted to zero. The proposed model comprises four ReLU layers.
Pooling layer
Four max-pooling layers are considered in the model of this study. The pool size in these layers as well as the stride size of each layer, is [2 2]. Therefore, the size of the output from the first, the second, the third, and the fourth pooling layers are 14, 7, 3, and 1, respectively.
Fully connected layer
The proposed model encompassed one fully connected layer.
Softmax Layer
The ultimate layer in the model is Softmax layer. This layer is placed after fully connected layer to replicate the outputs of fully connected layers to a probability distribution on the defined classes.
The features extracted from the convolutional and pooling layers comprise descriptors reflecting data on the acquired inside shape of the fruiting bodies (e.g., area of the convolutional mask relative to the colony size, mask area in the center and border of the colony, object sizes, number of objects in the mask, and deviations).
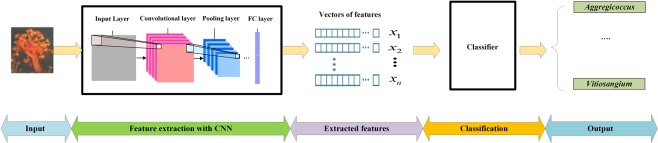
The combined feature set obtained from the last layer of unsupervised part of CNN assists as a quantitative signature of the phenotype of the Fruiting bodied. The images from the same genus or belonging to the same phenotypic class share some matching characteristics from the various existing features. The structure of the designed CNN for automatic recognition of the genera is displayed in Fig. 3.
Figure 3.
The structure of the proposed CNN for automatic identification of the genera.
In each convolutional layer, the input convolves with some different filters. In the next pooling layer the size of convolved images reduced to smaller images. This routine is continued to the end of the first phase of CNN.
The first part of a CNN is a feature extraction phase, which also called unsupervised phase. In this phase, the features, which are corresponding to texture and shape, are learned. The output of this phase is diverted to the second part. The second part is a classifier also called supervised phase.
The learning algorithm of the second part is stochastic gradient descent with momentum32.
Extreme convolution neural network (E-CNN)
In this algorithm, in the first step, the features of images are extracted using the convolution neural network and then these features are sent as inputs for Extreme learning machines algorithm.
Feature extraction
As we see in the second method, convolution neural networks are one of the best methods for feature extraction. The first step in this method is feature extraction. We carried out this using a convolution neural network. The features that are extracted by this method are mostly related to the edges, colors, and textures of the images. We use the model defined in the second method to extract the features at this stage. The best features that can be selected for this are features that are extracted from the fully connected layer. Our defined model for convolution neural network extracts three features for each image. Then these features are sent as inputs to the next step of the algorithm
Classification
At this stage, the features extracted by CNN are classified by different classifiers. Considering that CNN in its late layer uses back propagation algorithms, its speed and accuracy are low. Therefore, by putting this layer in place with a better learning algorithm such as SVM, the speed, and accuracy of this algorithm can be increased. In this paper, we will use different algorithms to examine classification accuracy.
Experiments and Validation
The accuracy of our model was assessed by evaluating it to differentiate the complex and not very typical phenotypes of the fruiting bodies.
All the experiments were run by 10-fold cross-validation strategy. One round of cross-validation involves partitioning the dataset into complementary subsets, performing the analysis on one subset (i.e. training set), and validating the analysis on the other subset (i.e. validation set or testing set). To reduce variability, 10 rounds of cross-validation were performed using different partitions, and the validation results were averaged over the rounds33. The used performance measure is classification accuracy, which is obtained by Eq. (18):
| 18 |
where C is the number of classes which in our case is 22. TDi denotes true detection of instances in class i and Ti is the total number of instances in class i.
The accuracy of our model in the detection of distinct classes is reported by TP and FP values that indicate True Positive Rate and False Positive Rate, respectively.
TP denotes the number of instances, which belong to a class and recognized truly by the proposed modeling as the members of that class. FP denotes the number of instances, which are wrongly recognized as the members of a genus, but they truly belong to other genera. The classification models are trained by the training set and evaluated by the test set which has not been encountered during modeling. This evaluation strategy confirms the ability of the models to predict unseen samples.
Here, we describe an automated image analysis tool that facilitates the identification of Myxobacterial genera independent of need for microscopic or nucleotide sequencing In addition, this can increase the efficiency of the isolation process by optimization of the isolation condition from early on towards retrieving more of the diverse genera instead of compiling the strains that are member of a few limited genera.
Preparing a dataset that includes images with different size and variation in the shape of the fruiting body, results robustness of the proposed method toward shape variations and size of the fruiting bodies due to their maturity, using various culture media or different fields of view.
Configuration of CNN on myxobacterial pictures-MY22
In Table 3, three different configurations of baseline CNN executed on 22 classes were compared. As one can see, the third configuration provides the best accuracy. Thus, this configuration is used for all of the experiments in our study. However, the best accuracy of CNN is 77.24%.
Table 3.
Configuration of CNN on Myxobacterial pictures in MY22.
| Structure | Feature Extraction Layers | Fully Connected | Accuracy |
|---|---|---|---|
| 1 | Convolution Layer (fi (2,16), Padding (3,3)) | F (O(22)) | 65.46% |
| Max-Pooling (Pol (2,2), Stir(2,2)) | |||
| Convolution Layer (fi (2,32), Padding (3,3)) | |||
| Max-Pooling (Pol (2,2), Stir(2,2)) | |||
| Convolution Layer (fi (2,64), Padding (3,3)) | |||
| Max-Pooling (Pol (2,2), Stir(2,2)) | |||
| Convolution Layer (fi (2,128), Padding (3,3)) | |||
| 2 | Convolution Layer (fi (3,16), Padding (1,1)) | F (O(22)) | 74.246% |
| Max-Pooling (Pol (2,2)), Stir(2,2)) | |||
| Convolution Layer (fi (3,32), Padding (1,1)) | |||
| Max-Pooling (Pol (2,2), Stir(2,2)) | |||
| Convolution Layer (fi (3,64), Padding (1,1)) | |||
| 3 | Convolution Layer (fi (3,16), Padding (1,1)) | F (O(22)) | 77.24% |
| Max-Pooling (Pol (2,2)), Stir(2,2)) | |||
| Convolution Layer (fi (3,32), Padding (1,1)) | |||
| Max-Pooling (Pol (2,2), Stir(2,2)) | |||
| Convolution Layer (fi (3,64), Padding (1,1)) | |||
| Max-Pooling (Pol (2,2), Stir(2,2)) | |||
| Convolution Layer (fi (3,128), Padding (1,1)) |
fi: Filter.
M: Max-Pooling Layer.
Pol: Poll-size.
Stir: Stride.
O: Output.
Results on myxobacterial pictures- MY25
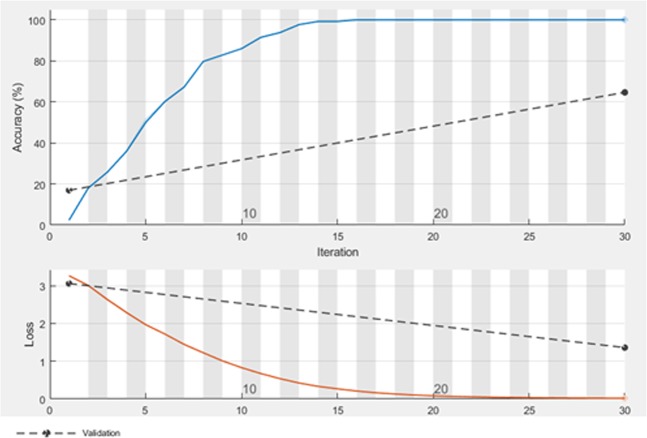
The results of MY25 dataset are shown in Table 4. The size of the extracted feature vector for this dataset is 1 × 25. As it can be seen, in this case CNN-MLP has the best result among other algorithms. The accuracy and loss of training and validation set of MY25 when the number of iterations is increased is presented in Fig. 4.
Table 4.
Configuration of CNN on Myxobacterial pictures in MY25.
| Feature extraction | Classifier | Accuracy (%) | Precision (%) | Recall (%) | Parameters |
|---|---|---|---|---|---|
| CNN | MLP | 80.7 | 100 | 100 | Learning Rate: 0.3 Hidden Layer: 1 No. of Nodes: 3 |
| RBF | 25.4 | 46.2 | 100 | No. of Batches: 100 | |
| SVM | 76.84 | 100 | 100 | Kernel Function: Linear | |
| XGBoost | 74.91 | 100 | 100 | ||
| ELM | 73.85 | 75.6 | 100 | Activation Function: Sigmoid No. of Nodes: 100 | |
| CELM | 21.54 | 35.6 | 100 | Activation Function: Sigmoid No. of Nodes: 20 | |
| OSELM | 70.77 | 75.3 | 78.4 | No. of Nodes: 50 No. of train samples: 50 No. of Blocks: 20 | |
| KELM | 76.92 | 100 | 100 | Kernel Function: RBF No. of Nodes: 500 | |
| CNN baseline | 64.62 | 100 | 100 | Epochs: 30 Learning rate: 0.01 Iteration per epoch: 1 |
Figure 4.
Accuracy and loss of training and validation set of MY25.
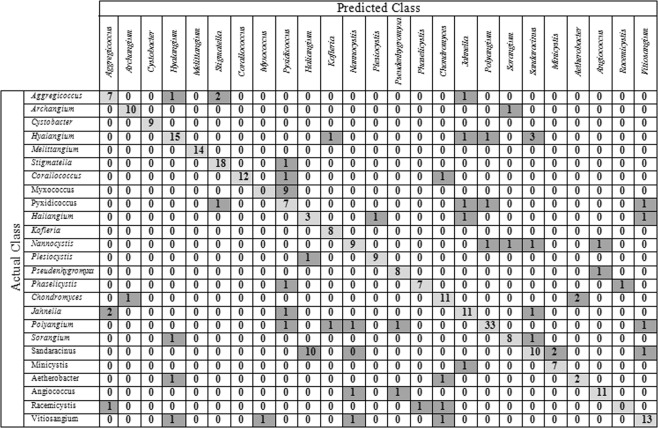
Figure 5 shows the confusion matrix for CNN-SVM on MY25 dataset. Light gray cells show the number of correct classified samples and the dark gray cells related to the number of misclassified samples. As can be seen in the figure, the number of correct classified samples from Myxococcus genus is zero and all the samples in this class were classified as Pyxidicoccus wrongly. The samples from Myxococcus genus include single bodies but the samples from Pyxidicoccus genus are colonies. The structure of their fruiting body is similar to each other as both belong to the same family. Although it seems that, the CNN features should be discriminative enough to separate these two classes but in practice, it is not successful. Finding other structures for CNN may reduce the misclassification error.
Figure 5.
Confusion matrix for CNN-SVM on MY25.
Results on myxobacterial pictures-MY22
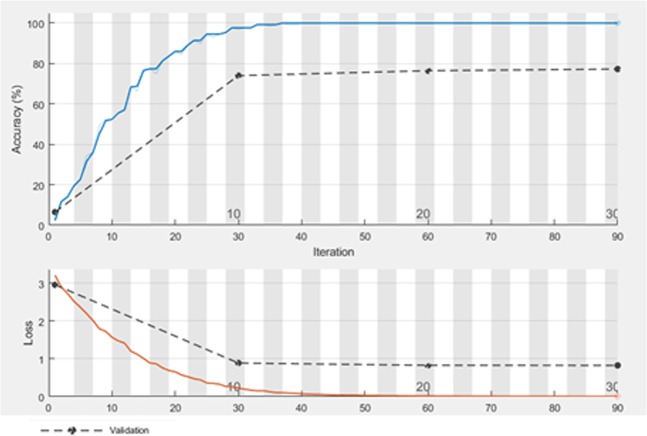
The results of MY22 dataset are listed in Table 5. The size of the extracted feature vector for this dataset is 1 × 22. As can been seen, CNN-SVM has the optimum result among other algorithms. The accuracy and loss of training and validation set of MY22 when the number of iterations is increased is shown in Fig. 6.
Table 5.
Configuration of CNN on Myxobacterial pictures in MY22 dataset.
| Feature extraction | Classifier | Accuracy (%) | Recall (%) | Precision (%) | Parameters |
|---|---|---|---|---|---|
| CNN | MLP | 89.72 | 96.7 | 1 | Learning Rate: 0.3 Hidden Layer: 1 No. of Nodes: 3 |
| RBF | 31.81 | 97.3 | 50.7 | No. of Batches: 100 | |
| SVM | 89.88 | 1 | 1 | Kernel Function: Linear | |
| XGBoost | 86.94 | 97.3 | 1 | ||
| ELM | 77.24 | 80.4 | 79.6 | Activation Function: Sigmoid No. of Nodes: 100 | |
| CELM | 9.7 | 24.6 | 1 | Activation Function: Sigmoid No. of Nodes: 20 | |
| OSELM | 78.86 | 82.2 | 79.8 | No. of Nodes: 180 No. of train samples: 300 No. of Blocks: 10 | |
| KELM | 85.37 | 87.3 | 1 | Kernel Function: RBF No. of Nodes: 20 | |
| CNN baseline | 77.24 | 1 | 1 | Epochs: 30 Learning rate: 0.01 Iteration per epoch: 3 |
Figure 6.
Accuracy and loss on training and validation set of MY22.
Results on myxobacterial MYCategories
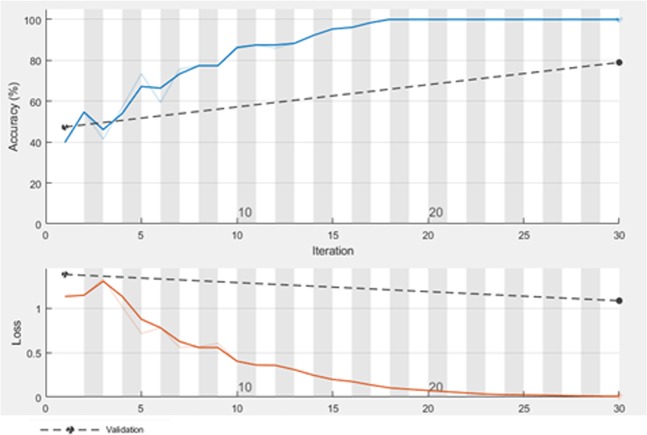
The results of MYCategories dataset are listed in Table 6. The size of the extracted feature vector for this dataset is 1 × 3. As you see in Table 6 where CNN-SVM has the distinguished result among other algorithms. The accuracy and loss of training and validation set of MYCategories when the number of iterations is increased is illustrated in Fig. 7.
Table 6.
Performance of different methods on MYCategories.
| Feature extraction | Classifier | Accuracy (%) | Precision (%) | Recall (%) | Parameters |
|---|---|---|---|---|---|
| CNN | MLP | 88.23 | 92.9 | 89.6 | Learning Rate: 0.3 Hidden Layer: 1 No. of Nodes: 3 |
| RBF | 88.58 | 92.3 | 88.6 | No. of Batches: 100 | |
| SVM | 88.92 | 91.7 | 92.4 | Kernel Function: Linear | |
| XGBoost | 86.15 | 90.8 | 89.4 | ||
| ELM | 77.19 | 88.4 | 82 | Activation Function: Sigmoid No. of Nodes: 10 | |
| CELM | 61.4 | 70.4 | 65.3 | Activation Function: Sigmoid No. of Nodes: 20 | |
| OSELM | 80.7 | 75.2 | 82.3 | No. of Nodes: 180 No. of train samples: 10 No. of Blocks: 20 | |
| KELM | 78.95 | 79.8 | 80.4 | Kernel Function: RBF No. of Nodes: 10 | |
| CNN base line | 78.95 | 86.9 | 86.9 | Epochs: 30 Learning rate: 0.01 Iteration per epoch: 1 |
Figure 7.
Accuracy and loss on training and validation set of MYCategories.
Figure 8 shows the confusion matrix for CNN-SVM on MYCategories. Row 1 shows that from 135 samples belong to Cystobacterineae suborder. The number of 116 samples were been recognized correctly, 11 samples were wrongly recognized in the suborder Sorangiineae and eight samples were displaced as Nannocystineae suborder. The results of the Fig. 8 show that the probability of wrong recognition between Cystobacterineae and Sorangiineae is higher compared to others.
Figure 8.
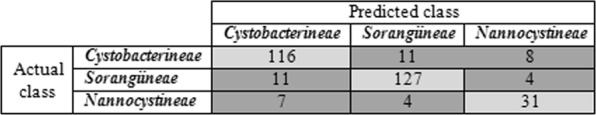
Confusion matrix for CNN-SVM on MYCategories.
Conclusion
Application of pattern analysis in microbiology and biotechnology is accelerating the speed and accuracy of the procedures and practices. By increasing the number of Myxobacterial genera especially with similar fruiting bodies, the challenge in their instant recognition has emerged recently. The conventional techniques that involve observation of the macromorphology and its characteristics such as color, shape, and pigment is considered acceptable criteria for bacterial identification in some groups of bacteria. However, morphology oriented methods have some impediment, for instance, they usually need proficient personnel who have a solidified knowledge on the morphology and taxonomy of Myxobacteria.
In recent decades, automated methods have been applied in laboratories for rapid identification of bacteria. Using machine learning methods, a combination of experience and technology in the task of bacterial identification will be provided. It is anticipated that in the near future the number of new genera in all microbial taxa will dramatically be increased. This fact has been observed in the last five years with the introduction of six new genera that do not have the peculiar characteristic shape of fruiting bodies that are even named as pseudo fruiting bodies. However identification of some myxobacterial genera such as Stigmatella, Chondromyces, Corallococcus, Myxococcus, Cystobacter, and Archangium is rather easy by observation, there are challenges in case of some genera that produce a amorphousand disorded forms of the fruiting body like Kofleria, Jahnella, Enhygromyxa, Plesiocystis, Pseudenhygromyxa and Haliangium, etc.
Automated imaging and analysis have the potential to improve the duration and accuracy of identification of Myxobacteria required for a variety of ecological and biotechnological projects.
The introduced automated recognition can enable an analysis of individual fruiting bodies, taken over time or all presented at a single image. Additionally, the classification of distinct fruiting body shapes based on image-derived features was independent of whether pictures are colored or on a grey scale. Phenotypic changes in the morphology of fruiting body due to being in variant maturity stage can be expressed as minor changes in feature space, which can be corrected and attributed to the respected suborder.
In this study, a platform that uses stereomicroscopic image analysis and pattern recognition to differentiate between 30 genera of Myxobacteria was developed based on the phenotypic signatures. The images of the newly discovered genera of Myxobacteria can be added to the dataset and the proposed structure can be retrained to discriminate between genera automatically.
In this work, the images were gathered from different resources, captured by various cameras with disparate resolution and lightening environment. Image capturing with the similar situation can enhance the accuracy of the recognition result and hence the efficiency of automated identificatio system.
Author contributions
H. Sajedi and F. Mohammadipanah wrote the main manuscript text. H. Sajedi designed the experiments; F. Mohamadipanah collected the images and Ali Pashaie executed ELM experiments.
Data availability
The analysis data of this study are available from the corresponding authors for follow up studies.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hedieh Sajedi, Email: hhsajedi@ut.ac.ir.
Fatemeh Mohammadipanah, Email: fmohammadipanah@ut.ac.ir.
References
- 1.Shimkets, L. J., Dworkin, M., Reichenbach, H. The myxobacteria. The prokaryotes. Springer, 31–115 (2006).
- 2.Xiao, Y., Wei, X., Ebright, R., Wall, D. Antibiotic Production by Myxobacteria Plays a Role in Predation. J. Bacteriol. 4626–4633 (2011). [DOI] [PMC free article] [PubMed]
- 3.Feldbauer, R. Machine Learning for Microbial Phenotype Prediction. BestMasters. Springer Fachmedien Wiesbaden (2016).
- 4.Parte AC. LPSN – List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- 5.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 6.Dale K. Signaling in myxobacteria. Annu. Rev. Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 7.Weissman KJ, Müller R. A brief tour of myxobacterial secondary metabolism. Bioorg. Med. Chem. 2009;17(6):2121–2136. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Irschik H, Gerth K, Kemmer T, Steinmetz H, Reichenbach H. The myxovalargins, new peptide antibiotics from Myxococcus fulvus (myxobacterales). I. Cultivation, isolation, and some chemical and biological properties. J. Antibiot. 1983;36(1):6–12. doi: 10.7164/antibiotics.36.6. [DOI] [PubMed] [Google Scholar]
- 9.Irschik H, Jansen R, Gerth K, Hofle G, Reichenbach H. The sorangicins, novel and powerful inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J. Antibiot. 1987;40(1):7–13. doi: 10.7164/antibiotics.40.7. [DOI] [PubMed] [Google Scholar]
- 10.Irschik H, Trowitzsch-kienast, Gerth W, Hofle K, Reichenbach G. Saframycin H. Mx1, a new natural saframycin isolated from a myxobacterium. J. Antibiot. 1988;41(8):993–998. doi: 10.7164/antibiotics.41.993. [DOI] [PubMed] [Google Scholar]
- 11.Irschik H, Jansen R, Gerth K, Hofle G, Reichenbach H. Antibiotics from gilding bacteria. No. 67. Sorangiolid A, a New Antibiotic Isolated from the Myxobacterium Sorangium cellulosum So ce 12. J. Antibiot. 1995;48(8):886–887. doi: 10.7164/antibiotics.48.886. [DOI] [PubMed] [Google Scholar]
- 12.Jansen R, et al. β‐Amino Styrenes from Chondromyces crocatus (Myxobacteria) Eur. J. Org. Chem. 2003;14:2684–2689. doi: 10.1002/ejoc.200200699. [DOI] [Google Scholar]
- 13.Irschik H, Reichenbach H, Höfle G, Jansen R. The thuggacins, novel antibacterial macrolides from Sorangium cellulosum acting against selected Gram-positive bacteria. J. Antibiot. 2007;60(12):733. doi: 10.1038/ja.2007.95. [DOI] [PubMed] [Google Scholar]
- 14.Kunze B, Jansen R, Sasse F, Höfle G, Reichenbach H. Chondramides A~D, new antifungal and cytostatic depsipeptides from Chondromyces crocatus (Myxobacteria). Production, physico-chemical and biological properties. J Antibiot. 1995;48:1262–1266. doi: 10.7164/antibiotics.48.1262. [DOI] [PubMed] [Google Scholar]
- 15.Gronewold TM, Sasse F, Lunsdorf H, Reichenbach H. Cell Tissue Res. 1999;295:121–129. doi: 10.1007/s004410051218. [DOI] [PubMed] [Google Scholar]
- 16.Luo, Y., Cobb, R. E. & Zhao, H. Recent Advances in Natural Product Discovery. Curr Opin Biotechnol. 230–237 (2014). [DOI] [PMC free article] [PubMed]
- 17.Salipant SJ, et al. Application of Whole-Genome Sequencing for Bacterial Strain Typing in Molecular Epidemiology. J. Clin. Microbiol. 2015;53(4):1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sentausa E, Fournier P-E. Advantages and limitations of genomics in prokaryotic taxonomy. Clin. Microbiol. Infect. 2013;19:790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 19.Spröer C, Reichenbach H, Stackebrandt E. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Bacteriol. 1999;49(3):1255–62. doi: 10.1099/00207713-49-3-1255. [DOI] [PubMed] [Google Scholar]
- 20.Razavi SF, Sajedi H, Shiri ME. Integration of colour and uniform interlaced derivative patterns for object tracking. IET Image Process. 2016;10(5):381–390. doi: 10.1049/iet-ipr.2015.0610. [DOI] [Google Scholar]
- 21.Zhu, W., Miao, J. & Qing, L. Constrained Extreme Learning Machine: A novel highly discriminative random feedforward neural network, International Joint Conference on Neural Networks (IJCNN), China (2015).
- 22.Huang, G.-B., Liang, N.-Y., Rong, H.-J., Saratchandran, P., Sundararajan, N. On-line sequential extreme learning machine, Computational Intelligence, 232–237 (2005).
- 23.Wang, J., Song, Y.-F., Ma, T.-L. Mexican Hat Wavelet Kernel ELM for Multiclass Classification, Computational Intelligence and Neuroscience, Article ID 7479140, 8 pages (2017). [DOI] [PMC free article] [PubMed]
- 24.Zhu, W., Miao, J. & Qing, L. Constrained extreme learning machine: a novel highly discriminative random feedforward neural network, International Joint Conference on Neural Networks (IJCNN), 800–807: IEEE (2014).
- 25.Nair, V. Hinton G. E. Rectified linear units improve restricted boltzmann machines. 27th international conference on Machine Learning (ICML-10), 807–814 (2010).
- 26.Krizhevsky A, Sutskever I, Hinton G. Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process Syst. 2012;1:1097–1105. [Google Scholar]
- 27.Bishop, C. M. Pattern recognition and machine learning. springer (2006).
- 28.Sajedi H, Mohammadipanah F, Rahimi SAH. Actinobacterial strains recognition by Machine learning methods. Multimed. Tools Appl. 2019;78(14):20285–20307. doi: 10.1007/s11042-019-7379-9. [DOI] [Google Scholar]
- 29.Aboura K. Pseudo Bayesian and Linear Regression Global Thresholding. AEU-Int. J. Electron. C. 2010;56(1):63–72. [Google Scholar]
- 30.Bahrami M, Sajedi H. Image Concept Detection in Imbalanced Datasets with Ensemble of Convolutional Neural Networks. Intelligent Data Analysis. 2019;28:426–439. [Google Scholar]
- 31.Masakazu M, Mori K, Mitari Y, Kaneda Y. Subject independent facial expression recognition with robust face detection using a convolutional neural network. Neural Networks. 2003;16(5):555–559. doi: 10.1016/S0893-6080(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 32.Rahimi SA, Sajedi H. Monitoring air pollution by deep features and extreme learning machine. J Exp Theor Artif Intell. 2019;31(4):517–531. doi: 10.1080/0952813X.2019.1572658. [DOI] [Google Scholar]
- 33.Grossman, R., Giovanni S., Elder, J., Agarwal N., Liu H. Ensemble Methods in Data Mining: Improving Accuracy Through Combining Predictions. Morgan & Claypool Publishers (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analysis data of this study are available from the corresponding authors for follow up studies.