Aberrant feeding patterns whereby people eat more frequently throughout the day and with a bias toward late-night eating are prevalent in society today. However, whether restriction of food to daytime in comparison to nighttime, coupled with restricted calorie intake, can influence gut microbiota, metabolism, and overall health requires further investigation. We surveyed the effects of the shift in feeding time on gut microbiota and metabolic phenotype in calorie-restricted mice and found that avoiding eating during the rest period may generate more beneficial effects in mice. This work strengthens the evidence for using “when to eat” as an intervention to improve health during calorie restriction.
KEYWORDS: calorie restriction, shift of feeding time, gut microbiota, anti-inflammation
ABSTRACT
Calorie restriction (CR) is accompanied by self-imposed daily restriction of food intake and an extended fasting period between meals. The impact of restricting feeding to the dark or light phase on the effects of CR remains elusive. Here, light-fed CR mice showed physiological changes, such as muscle loss, concomitant with changes in the gut microbiota structure and composition. After switching to ad libitum access to food, light-fed mice had a period of food-craving behavior and short-lived physiological changes, while dark-fed mice displayed lasting changes in fat accumulation, glucose metabolism, intestinal barrier function, and systemic inflammatory markers. Moreover, the gut microbiota was modulated by when the food was consumed, and the most abundant Lactobacillus operational taxonomic unit (OTU) promoted by CR was enhanced in dark-fed mice. After switching to ad libitum feeding, the gut microbiota of dark-fed mice returned to the state resembling that of mice fed normal chow ad libitum, but that of light-fed mice was still significantly different from the other two groups. Together, these data indicate that for CR, restricting food consumption to the active phase brought better metabolic phenotype associated with potentially beneficial structural shifts in the gut microbiota.
IMPORTANCE Aberrant feeding patterns whereby people eat more frequently throughout the day and with a bias toward late-night eating are prevalent in society today. However, whether restriction of food to daytime in comparison to nighttime, coupled with restricted calorie intake, can influence gut microbiota, metabolism, and overall health requires further investigation. We surveyed the effects of the shift in feeding time on gut microbiota and metabolic phenotype in calorie-restricted mice and found that avoiding eating during the rest period may generate more beneficial effects in mice. This work strengthens the evidence for using “when to eat” as an intervention to improve health during calorie restriction.
INTRODUCTION
Calorie restriction (CR) has been shown to extend life span and improve health in various animal models (1, 2). Long-term calorie restriction of mice, with either a normal or high-fat diet, alleviates early life metabolic phenotypes and leads to changes in liver gene expression, which could effectively reflect causal factors critical for life span-extending effects (3). Calorie-restricted mice are considered a healthier group than their ad libitum companions. An important way in which CR improves health during aging is through attenuation of chronic and systemic inflammation (4–6). Prior studies have reported that age-associated microbial dysbiosis aggravates intestinal permeability and systemic inflammation (7). Furthermore, results from our previous studies in mice showed that CR changed gut microbiota into a Lactobacillus-dominated structure, and a strain of Lactobacillus murinus isolated from CR mice contributed to the alleviation of aging-associated inflammation, thereby demonstrating a microbiota-dependent mechanism for the metabolic improvement by CR (8–10).
The gut microbiota also plays a pivotal role in clock-nutrition interplay (11). First, the composition of gut microbiota exhibits diurnal oscillations, which are mostly driven by quality of the diet and food consumption rhythmicity (12–14). Second, the feeding rhythm is critical, but host circadian factors also affect the microbial oscillations (15). Disruption of the host circadian clock by mutation of clock genes or by jetlag blunts the diurnal oscillation in gut microbiota composition, while timed feeding can restore microbiota oscillations in mice deficient in circadian clock genes or a functional circadian clock (12, 13, 16). Third, depletion of the microbiota disrupts circadian rhythmicity of gene expression, not only locally in the intestinal epithelium (17), but also in distal organs such as the liver (18).
It is important to note that CR is also accompanied by self-imposed feeding behavior changes, tending to cause gorging instead of slower-paced intake in mice (1, 19, 20). Seventy percent of ad libitum intake for the CR mice is provided once daily, which means that they have 24 h in which to consume their food allotment. However, CR mice tend to consume food within the first 2 h in which it is available, and the consequence of this behavior is fasting for 22 h until food is next available (19). Food composition (such as the proportion of fat in the diet) and food quantity (through the control of calorie intake) profoundly affect gut microbiota and health outcomes (9, 15). Moreover, an emerging area of research is the investigation of feeding time and thus changed feeding parameters, such as feeding duration and timing of food access (1, 19, 20). Aberrant time-restricted feeding may be involved in the development of disease and the acceleration of aging (21–24). In light of the mainly nocturnal feeding habits of mice, the self-imposed 2-h/22-h feeding-fasting cycles under the CR regimen and the nutrition-microbiota-clock interplay, when the food is made available (dark or light phase), may impact the benefits of CR on the animals.
In this study, to evaluate the influence of shifting feeding time on the effectiveness of CR and the gut microbiota, mice were placed on a light-fed or dark-fed CR protocol, and their metabolic phenotype and gut microbiota were investigated. The lasting effects of these two types of CR were explored by switching the mice to an ad libitum feeding schedule after the 4-week CR period. Light-fed CR mice displayed physiological changes such as muscle loss and changes in the structure and composition of gut microbiota. Compared to their dark-fed CR companions, light-fed mice showed a period of food-craving behavior after shifting to ad libitum feeding. Moreover, the gut microbiota of light-fed mice was still significantly different from dark-fed mice and the mice fed ad libitum, concomitant with short-lived physiological changes. We describe the changes in the microbiota and metabolic phenotype under different timings of CR and concluded that synchronizing light-dark cycles with feeding-fasting cycles might generate a better metabolic phenotype associated with an improved gut microbiota.
RESULTS
Calorie-restricted mice exhibited altered metabolic outcomes and reduced inflammatory markers regardless of feeding time.
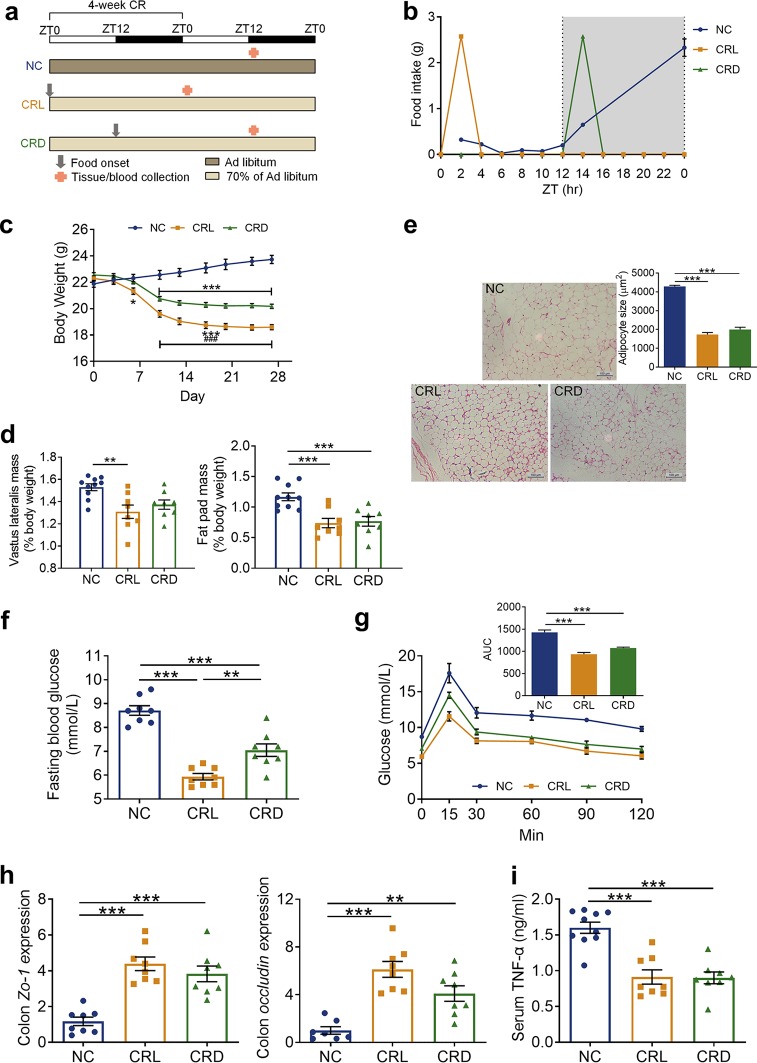
To evaluate how a shift in feeding time impacted physiological and metabolic consequences under the CR regimen, 8-week-old male C57BL/6 mice were randomly assigned to one of the following three groups (Fig. 1a): (i) the normal chow (NC) group received ad libitum access to the normal chow diet, (ii) the CRL group was fed 70% of the ad libitum chow that was provided at the beginning of the light phase (Zeitgeber time 0 [ZT0]), and (iii) the CRD group was fed 70% of the ad libitum chow provided at the beginning of the dark phase (ZT12). At the end of the 4-week CR, the schedule of tissue collection in relation to the light-dark cycle and food availability was designed to ensure the same fasting duration in the CRL and CRD groups (Fig. 1a) (19).
FIG 1.
Calorie-restricted mice exhibited altered metabolic outcomes and reduced inflammatory markers regardless of feeding time. (a) Schematic of the experimental design during 4-week CR and the schedule of tissue collection in relation to the light-dark cycle and food availability after 4-week CR. The light-dark cycle is shown in the bar across the top. (b) Monitored every-2-h food intake after 4-week CR. (c) Body weight curve of each group during 4-week CR. *, P < 0.05; ***, P < 0.001 (compared to the NC mice); ###, P < 0.001 (compared to CRD). (d) Relative weights of vastus lateralis and fat pad by the end of 4-week CR. **, P < 0.01; ***, P < 0.001. (e) Representative H&E-stained histological sections of epipdidymal adipose tissue (eAT) from NC, CRL, and CRD mice after 4-week CR (Scale bar = 100 μm) and calculated mean cell area of adipocytes in eAT. ***, P < 0.001. (f) Fasting blood glucose. **, P < 0.01; ***, P < 0.001. (g) Oral glucose tolerance test (OGTT) after 4-week CR and the calculated area under the curve (AUC) of blood glucose. ***, P < 0.001. (h) Relative mRNA expression of Zo-1 and occludin genes in the colon after 4-week CR. **, P < 0.01; ***, P < 0.001. (i) Serum level of TNF-α after 4-week CR. ***, P < 0.001. NC, normal chow diet; CRL, light-phase-fed with 30% calorie restriction; CRD, dark-phase-fed with 30% calorie restriction. For NC, n = 10; for CRL and CRD, n = 8. (a and c to i) Data are presented as the mean ± SEM and analyzed using one-way ANOVA (d, e, f, h, and i) and two-way repeated-measures ANOVA (c and g), followed by a Tukey post hoc test.
The daily food intake of NC mice was recorded as 3.9 ± 0.52 g or 13.7 ± 1.8 kcal (mean ± standard deviation [SD]), of which 77.5% ± 5.7% was consumed during the dark/active phase (Fig. 1b). It has previously been described that calorie-restricted mice needed approximately 4 weeks to adapt to externally imposed feeding schedules and reach a relatively stable state (19); therefore, we measured the feeding behavior of our mice every 2 h after 4 weeks of CR. CRL and CRD mice consumed all their food within 2 h as soon as it was made available (Fig. 1b). This type of self-imposed feeding behavior in response to reduced calorie intake, no matter the time that the food was made available to the mice (i.e., ZT0 for CRL or ZT12 for CRD), was in agreement with results from a recently published study (19).
During the 4-week CR period, the CRL and CRD mice exhibited a decrease in body weight compared to the NC mice, but the CRL mice lost significantly more weight than did the CRD mice (Fig. 1c). By the end of the 4-week CR, the CRL mice exhibited a significant decrease in vastus lateralis mass compared to NC mice, but this was not observed in the CRD mice (Fig. 1d). The CRL and CRD mice both showed a significant decrease in fat pad weight compared to the NC mice (Fig. 1d), and hematoxylin and eosin (H&E)-stained epididymal fat of CRL and CRD mice revealed smaller adipocytes (Fig. 1e). Furthermore, the CRL and CRD mice both showed a reduction in fasting blood glucose levels compared to the NC mice (Fig. 1f) and an improvement in glucose tolerance in the oral glucose tolerance test (OGTT) compared to the NC mice (Fig. 1g). A previous study identified that hyperglycemia correlated with a loss of intestinal barrier permeability (25). To determine if intestinal permeability was also impacted by light versus dark CR, we measured the expression of two epithelial tight junction proteins zonula occludens 1 (Zo-1) and occludin at the mRNA and protein levels (26, 27). CRL and CRD mice upregulated expression of the Zo-1 and occludin genes in the colon (Fig. 1h). The integrated optical densities (IOD) of Zo-1 and occludin determined by immunohistochemistry were also significantly higher in the CRL and CRD mice than in the NC mice after the 4-week CR period (see Fig. S1 in the supplemental material), implying an improvement in intestinal barrier function in CRL and CRD mice after the 4-week CR. Moreover, the serum level of tumor necrosis factor alpha (TNF-α), which plays an essential role in gut microbiota-induced aging-associated systemic inflammation (7), was significantly reduced in the CRL and CRD mice (Fig. 1i). This reflects an attenuation of inflammation after the 4-week CR. In short, CR profoundly improved the metabolic phenotype of mice regardless of the feeding time.
Immunohistochemical staining of colon and integrated optical density (IOD) analysis for Zo-1 (upper) and occludin (lower) from NC mice (left), CRL mice (middle), and CRD mice (right) after a 4-week CR. Scale bar = 50 μm. Data are presented as the mean ± standard error of the mean (SEM) and analyzed using one-way ANOVA, followed by a Tukey post hoc test. Download FIG S1, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Dark-fed CR mice demonstrated lasting effects on metabolic phenotype and systemic inflammatory marker after switching to ad libitum feeding.
To investigate the lasting effects of CR with light or dark feeding, the CRL and CRD mice were switched to ad libitum access for 4 weeks. During the first light-dark cycle, food intake was assessed every 2 h (Fig. 2a). Both the CRL and CRD mice consumed more food than did the NC mice in the first 2 h, which might be due to a deficit of satiety during the 4-week CR. Notably, food intake in the CRL mice in the first 2 h was significantly higher than in the CRD mice (Fig. 2a). The food intake pattern of the CRD mice resembled that of the NC mice after the first 2 h, but the food intake of the CRL mice in the first light-dark cycle was significantly greater than that of the NC mice and was predominantly consumed during the light phase (Fig. 2a). Interestingly, expression of the neuropeptide Y gene (NPY), which is involved in appetite regulation, was significantly increased in the hypothalamus of CRL mice compared to that of the NC mice after the 4-week CR period (Fig. 2b). From the third light-dark cycle onwards, the feeding behavior of CRD mice was not significantly different from that of the NC mice, whereas it took 10 days for the CRL mice to fully recover to a normal feeding pattern (Fig. 2c and S2). Overall, these results suggest that eating during the rest period might drive a period of food craving behavior when switched to ad libitum feeding.
FIG 2.
Light-fed CR mice displayed a period of food craving behavior after switching to ad libitum feeding. (a) Every-2-h food intake of the 1st light-dark cycle after ad libitum feeding. (b) Relative mRNA expression of the NPY gene in hypothalamus after 4-week CR. (c) Light-phase and dark-phase food intake of each group over the course of 11 consecutive light-dark cycles after ad libitum feeding. For the NC group, n = 10; for CRL and CRD, n = 8. Data are presented as the mean ± SEM and analyzed using one-way ANOVA (b) and two-way repeated-measures ANOVA (a and c), followed by a Tukey post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared to the NC mice); #, P < 0.05; ##, P < 0.01; ###, P < 0.001 (compared to CRD).
Every 2-hr food intake of the 10th light-dark cycle after ad libitum. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. Download FIG S2, TIF file, 0.2 MB (215.9KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
By the end of the 4-week ad libitum period, the body weights of CRL and CRD mice rebounded to a level identical to that of the NC mice (Fig. 3a and b). The vastus lateralis mass of CRL exhibited no significant difference with that of the NC mice, but it was significantly lower than that in the CRD mice (Fig. 3c). Although the fat pad weight of the CRD mice only showed a decrease in trend compared to the NC and CRL mice (Fig. 3d), H&E-stained epididymal fat revealed that the CRD mice still had smaller adipocytes (Fig. 3e). OGTTs were performed at 12 and 24 days after the switch to ad libitum feeding. Interestingly, the improvement in glucose tolerance (measured by the OGTT) in the CRD mice during the CR phase was maintained until day 12 compared to the NC mice. However, in the CRL mice, there was no improvement in glucose tolerance at day 12 compared to the NC mice, and on day 24, the CRL mice showed a deterioration in OGTT response (Fig. 3f). In addition, the CRD mice continued to maintain the significant increase in Zo-1 and occludin expression at both mRNA and protein levels in the colon (Fig. 3g and S3), as well as the lower serum TNF-α level compared to that in the NC and CRL mice even after 4 weeks of ad libitum feeding (Fig. 3h). These observations suggest that the beneficial effects of improved intestinal barrier function and alleviation of the systemic inflammatory marker induced by CR persisted in the CRD mice after the 4-week ad libitum period.
FIG 3.
Dark-fed CR mice demonstrated lasting effects on the metabolic phenotype and systemic inflammatory marker after switching to ad libitum feeding. (a) Schematic of the experimental design during 4-week ad libitum (AL) feeding and the schedule of final tissue collection in relation to the light-dark cycle and food availability after 4-week AL feeding. The light-dark cycle is shown in the bar across the top. (b) Body weight curve of each group during 4-week ad libitum feeding. (c) Relative weight of vastus lateralis by the end of 4-week ad libitum feeding. (d) Relative weight of fat pad by the end of 4-week ad libitum feeding. (e) Representative H&E-stained histological sections of eAT from NC, CRL, and CRD mice after 4-week ad libitum feeding (scale bar = 100 μm), and the calculated mean cell area of adipocytes in eAT. (f) OGTTs performed at 12 and 24 days after the ad libitum feeding (day 12 AL and day 24 AL) and the calculated area under the curve (AUC) of blood glucose. *, P < 0.05, CRL-day 24 compared to CRD-day 24; **, P < 0.01, CRL-day 12 compared to CRD-day 12. (g) Relative mRNA expression of Zo-1 and occludin genes in the colon after 4-week ad libitum feeding. (h) Serum level of TNF-α after 4-week ad libitum feeding. For the NC mice group, n = 10; for CRL and CRD mice, n = 8. Data are presented as the mean ± SEM and analyzed using one-way ANOVA (c to e, g, and h) and two-way repeated-measures ANOVA (b and f), followed by a Tukey post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Immunohistochemical staining of colon and integrated optical density (IOD) analysis for Zo-1 (upper) and occludin (lower) from NC mice (left), CRL mice (middle), and CRD mice (right) after a 4-week ad libitum period. Scale bar = 50 μm. Data are presented as the mean ± SEM and analyzed using one-way ANOVA, followed by a Tukey post hoc test. Download FIG S3, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taken together, these data indicate that protective effects on fat accumulation, glucose metabolism, and intestinal barrier function and on attenuation of the systemic inflammatory marker were maintained in the CRD mice over prolonged times after shifting from CR to ad libitum access. On the other hand, disrupted feeding behavior in the CRL mice was observed after switching to ad libitum access, and the physiological changes of CR were short-lived in the CRL mice.
Light-fed CR mice showed different fecal microbiota structural changes from those of dark-fed mice.
Microbial oscillations are mainly driven by the food consumption rhythmicity (12–14); therefore, we collected fecal samples on the day before treatment and at two time points (ZT0 and ZT12) of the day after the 4-week CR and of days 2, 11, 19, and 26 after switching to ad libitum feeding and profiled the gut microbiota by sequencing the V3-V4 regions in the 16S rRNA gene. Since CRL mice consumed all their food in the light phase during the 4-week CR period (Fig. 1b), we compared the gut microbiota of the CRL mice at ZT0 with that of the NC and CRD mice at ZT12, or that of the CRL mice at ZT12 with that of the NC and CRD mice at ZT0 after 4-week CR, in order to keep the same time before or after the onset of meal time. The same plan was followed for analyzing the gut microbiota for day 2 AL (day 2 after switching to ad libitum feeding) considering that most of the food in CRL mice was still being consumed during the light phase in the first light-dark cycle after switching to ad libitum feeding (Fig. 2a and c). For days 11, 19, and 26 AL, we compared the gut microbiota between the groups at the same time points, as the CRL mice had changed their feeding pattern to one similar to that of the NC mice after 10 days of ad libitum access.
Before the treatment, no significant difference in gut microbiota structure was observed among the three groups (Fig. S4). Four-week CR had no significant effect on the diversity and richness of the gut microbiota among the three groups (Fig. S5a). Principal-coordinate analysis (PCoA) and permutational multivariate analysis of variance (PERMANOVA) based on the Bray-Curtis distance show that after 4 weeks of CR, the gut microbiota structure of CRL and CRD significantly diverged from that of the NC mice mainly along the axis representing the first principal component (PC1), irrespective of sample collection time (ZT0 or ZT12), reflecting that the amount of food intake was the predominant factor in shifting the gut microbiota (Fig. 4a and b). However, the shift in feeding time also exerted a significant change in gut microbiota structure, as shown by the distancing in clustering of gut microbiota isolated from CRL mice compared to CRD mice (Fig. 4b). On the other hand, the variation in the gut microbiota at ZT0 and ZT12 was more pronounced in both the CRL and CRD mice than in the NC mice (Fig. 4b), which may be due to the self-imposed 2-h daily restriction of food intake and the extended fasting period between meals. Interestingly, it was noted that CRL-ZT0 and CRD-ZT12 were most similar in gut microbiota structure, as shown by the PCoA plot and the comparison on distance (Fig. 4a and S6).
FIG 4.
Light-fed CR mice showed different fecal microbiota structural changes compared to dark-fed mice. (a) Principal-coordinate analysis (PCoA) based on Bray-Curtis distance of all the samples collected at two time points (ZT0 and ZT12) of the day after the 4-week calorie restriction (CR) period and of days 2, 11, 19, and 26 after ad libitum feeding. (b) Top, PCoA based on Bray-Curtis distance. Bottom, heatmap of Bray-Curtis distances between each group and permutational multivariate analysis of variance (PERMANOVA; 9,999 permutations) were shown separately on the day after 4-week CR and days 2, 11, 19, and 26 AL (from left to right). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
16S rRNA profiles of baseline samples. Top, principal-coordinate analysis (PCoA) based on Bray-Curtis distance. Bottom, heatmap of Bray-Curtis distances between each group and permutational multivariate analysis of variance (PERMANOVA; 9,999 permutations). Download FIG S4, TIF file, 0.8 MB (826.2KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alpha diversity of the gut microbiota. (a to e) The observed OTUs (left), Shannon diversity index (middle), and Faith’s phylogenetic diversity values (right) after 4-week calorie restriction (CR) (a) and on days 2 (b), 11 (c), 19 (d), and 26 (e) after ad libitum (AL). The sampling level is 12,300. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. **, P < 0.01. Download FIG S5, TIF file, 1.3 MB (1.4MB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bray-Curtis distance between CRL and CRD mice compared at two time points after a 4-week CR period. Data are presented as the mean ± SEM and analyzed using one-way ANOVA, followed by a Tukey post hoc test. ***, P < 0.001. Download FIG S6, TIF file, 0.2 MB (190.8KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
At day 2 AL, alpha diversity was significantly reduced in CRL-ZT0 mice compared to that in the NC-ZT12 and CRD-ZT12 mice (Fig. S5b), which might be associated with the increased food intake in CRL mice after switching to ad libitum feeding (Fig. 2a). From the score plot of the PCoA, the gut microbiota of the CRL and CRD mice was still significantly different from that of the NC mice, regardless of the collection time (ZT0 or ZT12) (Fig. 4a and b). We also noticed significant differences between the CRL-ZT0 and CRD-ZT12 mice, as well as between the CRL-ZT12 and CRD-ZT0 mice (Fig. 4b). Interestingly, the significant variation at ZT0 and ZT12 in the gut microbiota in both CRL and CRD mice disappeared from day 2 onwards (Fig. 4a and b). Furthermore, the gut microbiota of the CRD mice moved close to that of the NC mice in the PCoA plot and showed no significant difference from that of the NC mice from day 11 onwards (Fig. 4a and b). However, the gut microbiota of the CRL mice at days 11 and 19 AL moved to the left part of the PC1 axis, which changed in a direction opposite from that of the CR stage (Fig. 4a). Significant differences in the gut microbiota were still observed between the CRL and NC mice and/or the CRD mice from day 11 onwards, and from the PCoA plot, the gut microbiota of CRL mice still separated from that of the NC and CRD mice along PC4 at day 26 AL (Fig. 4b).
Shifting feeding time impacts fecal microbiota composition.
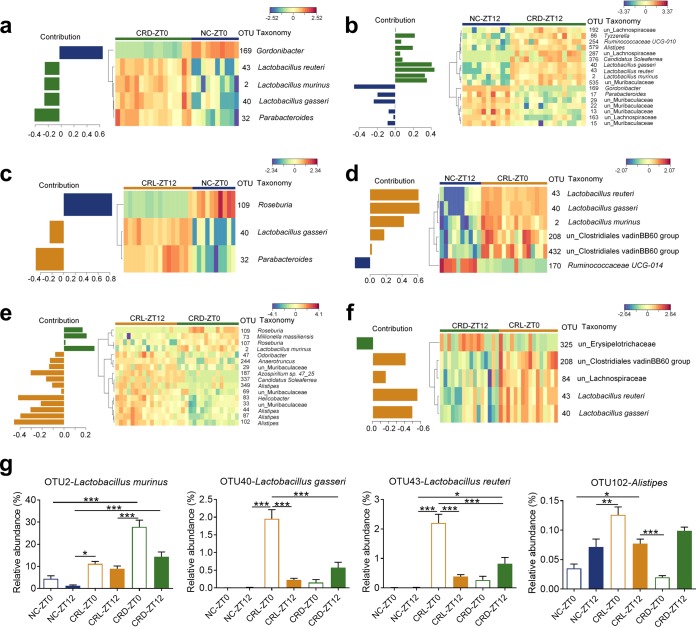
The sparse partial least squares discriminant analysis (sPLS-DA) (28) was utilized to identify members of the gut microbiota that were affected by CR and/or the shift in feeding time after the 4-week CR. First, the NC and CRD mice were compared to investigate the effects of CR on gut microbiota composition (Fig. 5a and b and S7a and b). Four OTUs changed in a similar way at ZT0 and ZT12, including three Lactobacillus OTUs (OTU2, OTU40, and OTU43) which were significantly enriched in the CRD mice, while Gordonibacter (OTU169) was depleted in CRD mice (Fig. 5a and b). Parabacteroides (OTU32) was enriched in the CRD-ZT0 mice compared to the NC-ZT0 mice (Fig. 5a), but OTU17, belonging to the same genus, was reduced in CRD-ZT12 mice compared to NC-ZT12 mice (Fig. 5b). These results substantiate our previous findings that bacteria belonging to Lactobacillus become prevalent phylotypes in CR mice (8, 9). Second, the NC and CRL mice were compared to identify OTUs affected by CR with reversal of feeding time (Fig. 5c and d and S7c and d). Lactobacillus gasseri (OTU40) was enriched in CRL mice irrespective of sample collection time (ZT0 or ZT12), whereas two other Lactobacillus OTUs (OTU2 and OTU43) were significantly increased in CRL-ZT0 mice compared to NC-ZT12 mice (Fig. 5c and d). This enrichment of Lactobacillus spp. was observed in both the CRL and CRD mice. It was noted that some OTUs only responded to either CRL or CRD in comparison to the NC mice, such as Roseburia spp. (OTU109), which were decreased in CRL-ZT12 mice compared to NC-ZT0 mice (Fig. 5c) but were not found in the CRD mice compared to the NC mice. This indicated that gut microbiota is not only affected by how much is eaten but also by when the food is consumed. Third, we compared the CRL and CRD mice, which were both fed 70% of the ad libitum intake to determine the effects of shifting feeding time on gut microbiota composition (Fig. 5e and f and S7e and f). The relative abundance of L. murinus (OTU2) was higher in the CRD-ZT0 than in the CRL-ZT12 mice (Fig. 5e), while the relative abundances of L. gasseri (OTU40) and Lactobacillus reuteri (OTU 43) were lower in CRD-ZT12 mice than in CRL-ZT0 mice (Fig. 5f). Notably, the relative abundance of OTU2 was elevated to an average of 28% ± 3% in CRD-ZT0 mice (Fig. 5g). Also, the relative abundances of two Roseburia OTUs (OTU107 and OTU109) were higher in the CRD-ZT0 mice than in the CRL-ZT12 mice (Fig. 5e), while the relative abundances of Alistipes (OTU44, OTU87, OTU102, and OTU349), Odoribacter (OTU47), and Helicobacter (OTU83) spp. were higher in the CRL-ZT12 mice than in the CRD-ZT0 mice (Fig. 5e).
FIG 5.
Shifting feeding time impacts fecal microbiota composition. (a to f) Contribution plots and heatmaps of key OTUs for separating the gut microbiota among different groups after 4-week CR are shown in NC-ZT0 and CRD-ZT0 (a), NC-ZT12 and CRD-ZT12 (b), NC-ZT0 and CRL-ZT12 (c), NC-ZT12 and CRL-ZT0 (d), CRL-ZT12 and CRD-ZT0 (e), and CRL-ZT0 and CRD-ZT12 (f). The color of the spots represented the relative abundance (normalized and log transformed) of the OTUs in each group. OTUs were arranged according to clusters based on the complete linkage method. The taxonomy of each OTU is shown on the right. See Fig. S7 for sPLS-DA models. (g) Relative abundances of three Lactobacillus OTUs and one Alistipes OTU after 4-week CR. For NC group, n = 10; for CRL and CRD groups, n = 8. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
The optimal classification performance of the sPLS-DA model related to FIG 5. (a to f) The plots of the classification error rate of different components averaged across the leave-one-out cross-validation for maximum distances (left side of each panel), and the sPLS-DA plots based on OTU table (right side of each panel) for separating the gut microbiota of NC-ZT0 and CRD-ZT0 (a), NC-ZT12 and CRD-ZT12 (b), NC-ZT0 and CRL-ZT12 (c), NC-ZT12 and CRL-ZT0 (d), CRL-ZT12 and CRD-ZT0 (e), and CRL-ZT0 and CRD-ZT12 (f). Download FIG S7, TIF file, 0.6 MB (586.7KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
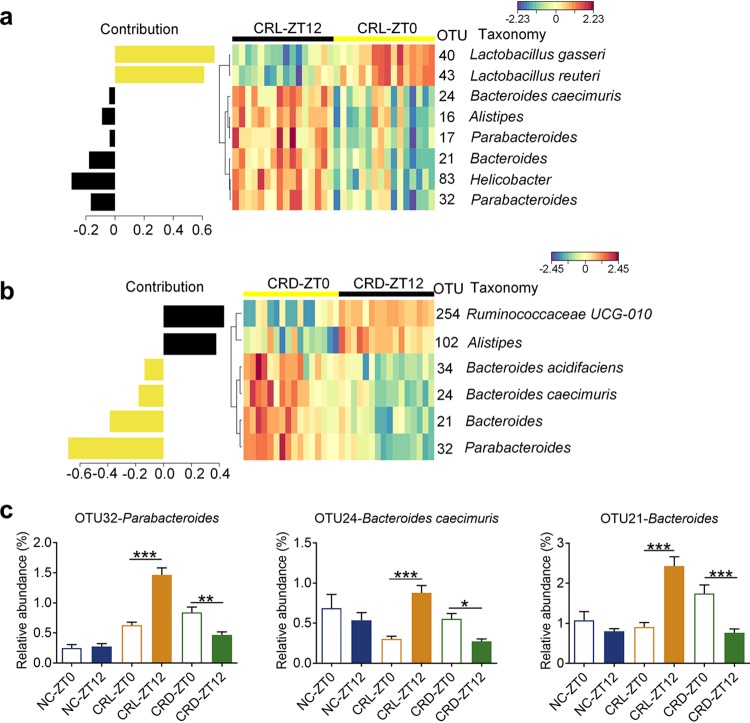
Additionally, we compared the variation of ZT0 and ZT12, respectively, in CRL and CRD mice to identify OTUs that were affected by the self-imposed feeding-fasting cycles (Fig. 6 and S8). OTU40 (L. gasseri) and OTU43 (L. reuteri) were significantly higher in the CRL-ZT0 mice than in the CRL-ZT12 mice, and a distinct trend could be observed in CRD mice (Fig. 6a and 5g), which might suggest that they were at higher levels after longer fasting duration. The abundances of OTU32 (Parabacteroides), OTU24 (Bacteroides caecimuris), and OTU21 (Bacteroides) were significantly higher in CRD-ZT0 mice than in CRD-ZT12 mice, in line with the trends seen in the NC mice, while the CRL mice showed adverse variation at ZT0 and ZT12 (Fig. 6). This might suggest that they were at lower levels after longer fasting durations. The CRL-ZT0 and CRD-ZT12 mice were most similar in terms of both the compositional analysis and the comparison on distance (Fig. S6), suggesting that it is the fasting state that is driving the differences in gut microbiota.
FIG 6.
The variation of gut microbiota at ZT0 and ZT12 in two types of CR mice. (a and b) The contribution plot of OTUs and heat maps of key OTUs for separating the gut microbiota of CRL-ZT0 and CRL-ZT12 (a) and CRD-ZT0 and CRD-ZT12 (b). See Fig. S8 for sPLS-DA models. (c) Relative abundances of 3 OTUs (OTU32, OTU24, and OTU21). For the NC group, n = 10; for the CRL and CRD groups, n = 16. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The optimal classification performance of the sPLS-DA model related to FIG 6. (a and b) The plots of the classification error rate of different components averaged across the leave-one-out cross-validation for maximum distances (left), and the sPLS-DA plots based on OTU table (right) for separating the gut microbiota of CRL-ZT0 and CRL-ZT12 (a) and CRD-ZT0 and CRD-ZT12 (b). Download FIG S8, TIF file, 0.1 MB (145.4KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Furthermore, as mentioned earlier, the gut microbiota structures of the CRL and NC mice were significantly different during the 4-week ad libitum feeding, and particularly at day 19 AL (Fig. 4a). Next, we also used sPLS-DA model to identify the phylotypes that showed differences between CRL and the NC mice at day 19 AL (Fig. S9). The compositional difference between the CRL and NC mice was that the relative abundances of Alistipes spp. (OTU102) and L. gasseri (OTU40) were higher in CRL mice than in the NC mice (Fig. S9). The enrichment of Alistipes spp. (OTU102) was also observed in CRL after 4-week CR and at day 2 AL (Fig. 5g and S10a). Interestingly, three Lactobacillus OTUs (OTU2, OTU40, and OTU43) showed different responses in CRL and CRD mice, not only after the 4-week CR, but also during the subsequent ad libitum period. The relative abundance of L. murinus (OTU2) was rapidly reduced in CRD at day 2 AL and did not differ among the three groups from day 11 onwards (Fig. 5g and S10), suggesting a strong correlation with CR. However, L. gasseri (OTU40) and L. reuteri (OTU 43) were elevated in the CRD mice at day 2 AL compared to the preceding CR period and were higher in both the CRL and CRD mice than in the NC mice at day 2 AL (Fig. 5g and S10). These OTUs remained significantly higher in the CRL mice than in the NC mice at day 11 AL and day 19 AL, but this was not observed in the CRD mice (Fig. S10).
The difference of gut microbiota between CRL and NC at ZT0 on day 19 after ad libitum. The plot of the classification error rate of different components averaged across the leave-one-out cross validation for maximum distances and sPLS-DA plot based on OTU table (top) and contribution plot of OTUs and heat map of key OTUs (bottom) for separating the gut microbiota of CRL-ZT0 and NC-ZT0. Download FIG S9, TIF file, 0.2 MB (201.7KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of three Lactobacillus OTUs and one Alistipes OTU. (a to d) OTU2 (left), OTU40 (middle-left), OTU43 (middle-right), and OTU102 (right) in days 2 (a), 11 (b), 19 (c), and 26 (d) after ad libitum. For the NC group, n = 10; for CRL and CRD, n = 8. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. ***, P < 0.001; **, P < 0.01; *, P < 0.05. Download FIG S10, TIF file, 0.9 MB (950.7KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Collectively, 16S rRNA amplicon sequencing analysis from CRL and CRD mouse gut microbiota revealed that CR alone elicited a substantial influence on microbiota composition; however, the time of feeding also contributed.
DISCUSSION
Our study found that the shift in feeding time impacts CR-induced effects, including physiological changes, modulation of gut microbiota, and anti-inflammation changes, all of which may contribute to improved health and extended longevity (8, 9, 19, 20). Two major characteristics of the CR regimen are (i) the reduction in total calorie intake without malnutrition, and (ii) periodicity in availability of food (e.g., self-imposed 2-h daily restriction of food intake in mice). Previous studies paid much attention to the former but ignored the latter. Feeding parameters (amount, duration, and timing of food access) strongly and differently impact the beneficial health outcomes (19), but the timing of food access in many CR studies is not even stated or specified (3, 29, 30). The current work clarified that when the food is made available (dark or light phase) was essential and influenced the outcomes of CR.
Previous studies showed that limiting the daily food intake of an isocaloric diet to a smaller time window of 8 to 12 h per day was sufficient to improve health and longevity in mice, and restricting both feeding duration and caloric intake (i.e., CR) exhibited much more beneficial effects (1, 11, 20). In the current work, light-fed mice lost significantly more weight than did their dark-fed companions and exhibited muscle loss compared to the controls after the 4-week CR. This difference in body weight and muscle mass under the two CR conditions reinforces the idea that feeding time is critical, since these mice consumed the same amount of food each day. Previous work showed that the clock in skeletal muscle appeared to be more sensitive to feeding patterns, resulting in large changes in gene and protein expression (31). Future work on the timing of food access and changes in muscle might provide insights into its molecular mechanisms of action. During the subsequent ad libitum feeding period, mice that had previously been light fed demonstrated disrupted feeding behavior, short-lived physiological changes, and even a deterioration of glucose tolerance. This suggests that in addition to the amount of food and feeding duration, when to eat (light phase or dark phase) also had an important influence on metabolic health. In future studies, it would be interesting to investigate whether the timing of CR can have effects on life span and aging through lifelong CR experiments.
CR profoundly improves metabolic phenotype through its potential anti-inflammatory effects (32, 33). In agreement with this, a significant reduction in inflammatory markers and improvement in intestinal barrier function were observed in our CR mice. A recent study on the anti-inflammatory effects of CR showed that CR-induced microbiota alterations dictated the tone of the immune response and contributed to the improvements by the reduction of endotoxin in mice (5), but which member of the gut microbiota acted as a determinant of the beneficial effects was not identified. Here, we observed that the timing of CR impacts the structure and composition of fecal microbiota in mice, and a higher abundance of L. murinus was noted in dark-fed CR mice than in their light-fed companions. Previously, we isolated a strain designated L. murinus CR147, which could represent the most abundant Lactobacillus OTU enriched by CR. The similarity of the representative sequence of OTU2 in the current work and the V3-V4 regions of the 16S rRNA gene sequence of L. murinus CR147 was 100%. Since this strain has been shown to protect the gut barrier and alleviate aging-associated inflammation (9), promoting this OTU might be informative for the better metabolic consequences after switching to ad libitum feeding. Furthermore, members of the dominant genus Lactobacillus might take advantage of the resource and make the gut environment unfavorable to pathogenic or detrimental bacteria (34, 35). Moreover, other bacteria might also contribute to the beneficial effects of dark-fed CR. Roseburia spp. were also enriched in dark-fed CR mice, and a member of this genus was reported to produce butyrate, decrease levels of inflammatory markers, and improve gut barrier function (36). Members of the gut microbiota that increased in light-fed mice, such as Alistipes, Odoribacter, and Helicobacter spp., were reported to be correlated with some diseases. Members of the genus Alistipes were associated with the pain in patients with irritable bowel syndrome (37). Higher abundances of Alistipes and Odoribacter spp. were found in grid floor stress-induced BALB/c mice and mice fed a high-fat diet (38, 39). Additionally, Helicobacter infection is known to cause gastric cancer (40). Light-fed CR may promote detrimental shifts in the microbiota that could elicit negative consequences on metabolic outcomes.
The genus Lactobacillus, which is widely used as probiotics, has been shown in many studies to protect against pathogen-induced gut barrier disruption and to alleviate inflammation (9, 41–45). However, two recent clinical studies reported that Lactobacillus rhamnosus GG did not bring better outcomes (46, 47), and L. reuteri was even reported to be obesogenic in humans (48). In the current work, three Lactobacillus OTUs were enriched by CR but showed different responses in light-fed and dark-fed mice, not only after the 4-week CR, but also during the follow-up ad libitum period. Specifically, L. gasseri (OTU40) and L. reuteri (OTU43) still presented a higher abundance in CRL mice after ad libitum feeding, concomitant with the short-lived physiological changes. The similarities of the representative sequences of OTU40 and OTU43 with the V3-V4 regions of the 16S rRNA gene sequence of L. murinus CR147 were 88% and 92%, respectively. In this regard, future work on probiotics is required to explore the strain-specific protective mechanisms for the development of targeted therapies (45, 49, 50).
Comparing the two time points of the fecal microbiota in the two CR groups provides an opportunity to find that some OTUs, such as Lactobacillus, Parabacteroides, and Bacteroides, featured distinct phases between dark- and light-fed groups, all of which were highly robust oscillations reported previously (12, 18). Prolonged fasting duration might drive the differences in gut microbiota, as it influences the nutrient availability to bacteria (16). Our microbial analysis was based on the stool 16S rRNA gene sequencing as a proxy of gastrointestinal composition, which may yield limited conclusions (51). For future studies, it would be of utmost significance to profile the microbiota along the entirety of the intestinal tract to accomplish a comprehensive assessment of CR effects on mucosal and luminal microbiota and the mechanism of blooming of some bacteria.
Many people who diet to lose weight find it logistically difficult to implement CR over their lifetime and thereby undergo excessive weight regain cycles. Research previously investigated the “yo-yo effect,” based on the nutrition quality changing from a high-fat diet to normal chow diet, and demonstrated that the microbiome plays a key role in this phenomenon (52). The CR regimen focuses on the quantity of diet, and the current work shows that when to eat might act as a modulator of gut microbiota, impacting the lasting effects of CR. The causal role of CR-induced microbiota modulation and metabolic health has been established through microbiota transplantations (5). In addition, the role of the time-shifted microbiota in the metabolic imbalances observed in jet-lagged mice was corroborated upon fecal transplantation (12). Further work is needed to build upon the existing findings to identify the causative linkage between timed CR-induced gut microbes and metabolic outcomes.
Timing of food access should be taken into careful consideration in a CR regimen. Erratic daily eating patterns are prevalent in modern society, with people eating more frequently throughout the day and with a bias toward late-night eating (53). In light of the fact that humans naturally spend the light phase in the active/feeding state and the dark phase in the resting/fasting state, avoiding eating close to or during the rest period when people are on a diet might generate better metabolic outcomes. Our work strengthens the evidence for using “when to eat” as an intervention to improve health during calorie restriction.
MATERIALS AND METHODS
Ethical approval.
All animal experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the School of Life Sciences and Biotechnology of Shanghai Jiao Tong University (no. 2014005).
Animals, housing, and experimental design.
Specific-pathogen-free (SPF) 4-week-old male C57BL/6J mice were purchased from SLAC, Inc. (Shanghai, China). The mice arrived at the animal center of Shanghai Jiao Tong University at the same time and were single housed and acclimatized to the animal facility environment for 4 weeks before experimentation; the mice were therefore 8 weeks old at the beginning of the experiments. Mice were randomly assigned to one of the following three groups and individually caged for an 8-week trial: (i) normal chow diet (NC, n = 10), with 8-week ad libitum access to food; (ii) normal chow diet with 30% calorie restriction, with food provided at the beginning of the light phase (CRL, n = 16) and 4-week calorie restriction followed by 4-week ad libitum access; (iii) normal chow diet with 30% calorie restriction, with food provided at the beginning of the dark phase (CRD, n = 16) and 4-week calorie restriction followed by 4-week ad libitum access (Fig. 1a and 3a). For the NC group, food was available in excess of normal consumption at all time; for the two CR groups, 70% of the ad libitum intake was provided once daily at either the beginning of the night or the day. The normal chow diet (22.1% protein, 5.3% fat, 3% fiber, 3.52 total kcal/g; product number P1101F) was purchased from SLAC, Inc. The daily food consumption of the NC group was recorded over 1 week and averaged to determine the amount of food for the two CR groups in the following week. All mice were single housed during the study to avoid any competition effects, ensuring that each mouse in the two CR groups had access to 70% of the intake of the NC group (3, 9). All mice were kept under a strict 24-h light-dark cycle, with lights being turned on from 7 a.m. to 7 p.m. (Zeitgeber time 0 [ZT0], 7 a.m., lights on; ZT12, 7 p.m., lights off).
Sample collection.
During the 4-week CR, body weight was measured twice weekly. The Zeitgeber time of when the body weight was measured, when the OGTT was performed, when blood samples were taken, and when the mice were dissected was before the onset of food availability for each group (Fig. 1a), as previously described (19). At the end of the 4-week CR, half of the animals were sacrificed, and adipose tissues (epididymal, mesenteric, subcutaneous inguinal, and retroperitoneal), vastus lateralis muscles, colon, and hypothalamus, etc., were collected and weighed. Blood samples were collected from the orbital vascular plexus, and the serum was isolated for TNF-α assays. After a 4-week ad libitum feeding period, as the two CR groups had recovered to normal feeding patterns after 10 days of ad libitum feeding, the rest of the animals were sacrificed at the same Zeitgeber time after fasting for 6 h, and tissue and blood samples were collected (Fig. 3a). The animals did not receive food on the day of dissection. All samples were stored at –80°C until analysis.
Histopathology.
Fresh epididymal fat pads were fixed with 4% paraformaldehyde and then embedded in paraffin. Subsequently, 4-μm sections were stained by hematoxylin and eosin (H&E). Digital images of sections were acquired with a Leica DMRBE microscope. Adipocyte size (cross-sectional area) was counted using Image Pro Plus 6.0. For each mouse, areas of adipocytes were determined in at least three histological sections and 300 total adipocytes.
Oral glucose tolerance test.
The Zeitgeber time of when the OGTT was performed after the 4-week CR was before the onset of food availability for each group, while that after 4-week ad libitum feeding was after 6 h fasting for all groups (54). Glucose was administered to mice by oral gavage at a dose of 2.0 g/kg of body weight. Blood glucose concentrations, in samples collected from the tip of the tail vein, were determined at 0, 15, 30, 60, 90, and 120 min after glucose challenge (Accu-Chek 381 Performa; Roche, USA).
RNA isolation and RT-qPCR.
Total RNA was extracted from the colon and hypothalamus using an RNeasy lipid tissue minikit (catalog no. 74804; Qiagen, Germany), according to the manufacturer’s protocol. Subsequent to the verification of RNA quality (gel electrophoresis), 1 μg of each total RNA sample (determined spectrophotometrically) was treated with RNase-free DNase I (catalog no. 18068-015; Invitrogen, USA). First-strand cDNA was synthesized using random hexanucleotide primers and SuperScript III reverse transcriptase (catalog no. 18080-051; Invitrogen, USA). Forward and reverse primers were added to SYBR green I PCR supermix (Bio-Rad) to amplify the Zo-1, occludin, and NPY genes. Real-time quantitative PCR (RT-qPCR) was performed on a LightCycler 96 system (Roche Applied Science). The PCR conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 20 s, 56°C for 30 s, and 72°C for 30 s, and plate reads for 5 s. Gene expression levels were determined using the comparative ΔΔCT method (2−ΔΔCT method), with the β-actin gene serving as the reference gene. Forward (F) and reverse (R) primer sequences are as follows: Zo-1 gene, F-ACCCGAAACTGATGCTGTGGATAG, and R-AAATGGCCGGGCAGAACTTGTGTA; occludin gene, F-ATGTCCGGCCGATGCTCTC and R-TTTGGCTGCTCTTGGGTCTGTAT; NPY, F-ATGCTAGGTAACAAGCGAATGG and R-TGTCGCAGAGCGGAGTAGTAT; and β-actin gene, F-GGCTGTATTCCCCTCCATCG and R-CCAGTTGGTAACAATGCCATGT.
Immunohistochemistry.
After deparaffinization and processing for antigen retrieval, the blank colon sections were incubated overnight at room temperature with anti-rabbit Zo-1 antibody (1:500, catalog no. GB11195; Servicebio, China) and occludin antibody (1:200, catalog no. GB11149; Servicebio). Sections were then incubated with anti-rabbit IgG secondary antibody (KPL, MA, USA) for 50 min at room temperature. After washing with phosphate-buffered saline (PBS), the DAB Envision kit (catalog no. K5007; Dako, Copenhagen, Denmark) was used to develop color; Zo-1 and occludin appear brown, while nuclei are blue. Images of each colon section were obtained in tripartitely by experienced staff who were blind to the experiment under ×400 magnification using a Leica DMRBE microscope and were analyzed using Image Pro Plus 6.0, according to the method previously described (55). The integrated optical density (IOD) values were log10 transformed.
Serum TNF-α measurements.
Blood samples were centrifuged at 4,000 rpm and 4°C for 15 min, and the serum was stored at –80°C until further analyses. Enzyme-linked immunosorbent assays (ELISAs) for tumor necrosis factor-alpha (TNF-α) were purchased from R&D Systems, Inc. (MHSTA50; Minneapolis, MN, USA), and were conducted according to the manufacturer’s instructions.
Fecal DNA extraction and 16S rRNA gene V3-V4 region sequencing.
Microbial DNA was extracted, as previously described (56), from fecal samples collected the day before treatment (for baseline samples, NC, n = 10; CRL, n = 16; and CRD, n = 16) and at two time points (ZT0 and ZT12) of the day after 4-week CR (NC, n = 10; CRL, n = 16; and CRD, n = 16) and of days 2, 11, 19, and 26 after switching to ad libitum feeding (NC, n = 10; CRL, n = 8; and CRD, n = 8). A total of 334 samples were sequenced on the MiSeq system (Illumina, Inc., USA) with the MiSeq reagent kit v3 (600 cycles, catalog no. MS-102-3033; Illumina). A sequencing library of the V3-V4 regions of the 16S rRNA gene was constructed according to the manufacturer’s instructions (Part # 15044223 Rev. B; Illumina Inc., USA) with some modifications as previously published (57).
Analysis of 16S rRNA V3-V4 sequencing data.
High-quality sequence alignments, sequence clustering, and operational taxonomic unit (OTU) delineation were performed as previously described (57). The OTU table was finalized by mapping all unique sequences to the obtained OTUs with the USEARCH (58) global alignment algorithm at a 97% cutoff. A total of 7,968,290 high-quality reads (27,288 ± 6,254 reads per sample) were clustered into 736 OTUs at a threshold of 97% identity.
The sequences of all samples were downsized to 12,300 (1,000 permutations) to standardize sequencing depth. All subsequent analyses were performed on the QIIME platform (version 1.9.1). The alpha diversity of each sample was calculated with observed OTUs, Shannon index, and Faith’s phylogenetic diversity (PD whole tree). Representative sequences for each OTU were built into a phylogenetic tree by FastTree and assigned taxonomic classifications using the SILVA rRNA database project. Bray-Curtis matrices for 16S rRNA gene sequencing samples were computed using QIIME via the beta_diversity script (59). The statistical significance of gut microbiota among different groups was assessed by permutational multivariate analysis of variance (PERMANOVA; 9,999 permutations, P < 0.001) in R (3.4.2). PCoA and PERMANOVA were based on the data matrix of the Bray-Curtis distances. To identify the most discriminative OTUs among different groups, sparse partial least-squares discriminant analysis (sPLS-DA) was performed using the mixOmics v6.3.1 (60) R package. Centered log ratio (CLR) transformations were implemented in sPLS-DA to circumvent spurious results. The optimal classification performance of the sPLS-DA model was assessed with the perf function using leave-one-out cross-validation with the smallest error rate.
Statistical analysis.
Statistical analysis was carried out using Prism version 7 (GraphPad Software, Inc.). One-way analysis of variance (ANOVA) or two-way repeated-measures ANOVA, followed by a Tukey post hoc test, was used to analyze variation among the three groups. Differences were considered statistically significant when the P value was <0.05.
Data availability.
Raw Illumina sequence data of the 16S rRNA gene generated in this study are available in the NCBI Sequence Read Archive (SRA) under accession number SRP166705.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 81871091, 81401141, and 31330005).
C. Zhang conceived and designed the study. L. Zhang, X. Xue, R. Zhai, and X. Yang conducted the animal trial and sample collection. L. Zhang conducted the RNA isolation and physiological data analysis. L. Zhang prepared the DNA and conducted the sequencing of 16S rRNA gene. L. Zhang and H. Li conducted the sequencing data analysis. L. Zhang, C. Zhang, and L. Zhao wrote and revised the manuscript. All authors read and approved the final manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Di Francesco A, Di Germanio C, Bernier M, de Cabo R. 2018. A time to fast. Science 362:770–775. doi: 10.1126/science.aau2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontana L, Partridge L. 2015. Promoting health and longevity through diet: from model organisms to humans. Cell 161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou B, Yang L, Li S, Huang J, Chen H, Hou L, Wang J, Green CD, Yan Z, Huang X, Kaeberlein M, Zhu L, Xiao H, Liu Y, Han JD. 2012. Midlife gene expressions identify modulators of aging through dietary interventions. Proc Natl Acad Sci U S A 109:E1201–E1209. doi: 10.1073/pnas.1119304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung HY, Kim HJ, Kim KW, Choi JS, Yu BP. 2002. Molecular inflammation hypothesis of aging based on the anti-aging mechanism of calorie restriction. Microsc Res Tech 59:264–272. doi: 10.1002/jemt.10203. [DOI] [PubMed] [Google Scholar]
- 5.Fabbiano S, Suarez-Zamorano N, Chevalier C, Lazarevic V, Kieser S, Rigo D, Leo S, Veyrat-Durebex C, Gaia N, Maresca M, Merkler D, Gomez de Aguero M, Macpherson A, Schrenzel J, Trajkovski M. 2018. Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab 28:907–921.e7. doi: 10.1016/j.cmet.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omodei D, Fontana L. 2011. Calorie restriction and prevention of age-associated chronic disease. FEBS Lett 585:1537–1542. doi: 10.1016/j.febslet.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, Schertzer JD, Larche MJ, Davidson DJ, Verdu EF, Surette MG, Bowdish D. 2017. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 21:455–466.e4. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Li S, Yang L, Huang P, Li W, Wang S, Zhao G, Zhang M, Pang X, Yan Z, Liu Y, Zhao L. 2013. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun 4:2163. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan F, Zhang L, Li M, Hu Y, Zeng B, Yuan H, Zhao L, Zhang C. 2018. Predominant gut Lactobacillus murinus strain mediates anti-inflammaging effects in calorie-restricted mice. Microbiome 6:54. doi: 10.1186/s40168-018-0440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T, Wu Y, Wang L, Pang X, Zhao L, Yuan H, Zhang C. 2019. A more robust gut microbiota in calorie-restricted mice is associated with attenuated intestinal injury caused by the chemotherapy drug cyclophosphamide. mBio 10:e02903-19. doi: 10.1128/mBio.02903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asher G, Sassone-Corsi P. 2015. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. 2014. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 13.Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. 2014. Circadian disorganization alters intestinal microbiota. PLoS One 9:e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrinpar A, Chaix A, Yooseph S, Panda S. 2014. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. 2015. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, Bushman FD, FitzGerald GA. 2015. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci U S A 112:10479–10484. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherji A, Kobiita A, Ye T, Chambon P. 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, Federici S, Zmora N, Zeevi D, Dori-Bachash M, Pevsner-Fischer M, Kartvelishvily E, Brandis A, Harmelin A, Shibolet O, Halpern Z, Honda K, Amit I, Segal E, Elinav E. 2016. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167:1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Acosta-Rodríguez VA, de Groot MHM, Rijo-Ferreira F, Green CB, Takahashi JS. 2017. Mice under caloric restriction self-impose a temporal restriction of food intake as revealed by an automated feeder system. Cell Metab 26:267–277.e2. doi: 10.1016/j.cmet.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell SJ, Bernier M, Mattison JA, Aon MA, Kaiser TA, Anson RM, Ikeno Y, Anderson RM, Ingram DK, de Cabo R. 2018. Daily fasting improves health and survival in male mice independent of diet composition and calories. Cell Metab 29:221–228.e3. doi: 10.1016/j.cmet.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froy O. 2011. Circadian rhythms, aging, and life span in mammals. Physiology (Bethesda) 26:225–235. doi: 10.1152/physiol.00012.2011. [DOI] [PubMed] [Google Scholar]
- 22.Paschos GK, FitzGerald GA. 2017. Circadian clocks and metabolism: implications for microbiome and aging. Trends Genet 33:760–769. doi: 10.1016/j.tig.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longo VD, Panda S. 2016. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab 23:1048–1059. doi: 10.1016/j.cmet.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Froy O, Miskin R. 2010. Effect of feeding regimens on circadian rhythms: implications for aging and longevity. Aging (Albany NY) 2:7–27. doi: 10.18632/aging.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thaiss CA, Levy M, Grosheva I, Zheng D, Soffer E, Blacher E, Braverman S, Tengeler AC, Barak O, Elazar M, Ben-Zeev R, Lehavi-Regev D, Katz MN, Pevsner-Fischer M, Gertler A, Halpern Z, Harmelin A, Aamar S, Serradas P, Grosfeld A, Shapiro H, Geiger B, Elinav E. 2018. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science 359:1376–1383. doi: 10.1126/science.aar3318. [DOI] [PubMed] [Google Scholar]
- 26.Volynets V, Louis S, Pretz D, Lang L, Ostaff MJ, Wehkamp J, Bischoff SC. 2017. Intestinal barrier function and the gut microbiome are differentially affected in mice fed a Western-style diet or drinking water supplemented with fructose. J Nutr 147:770–780. doi: 10.3945/jn.116.242859. [DOI] [PubMed] [Google Scholar]
- 27.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 28.Lê Cao KA, Boitard S, Besse P. 2011. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. 2010. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. 2010. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest 120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinouchi K, Magnan C, Ceglia N, Liu Y, Cervantes M, Pastore N, Huynh T, Ballabio A, Baldi P, Masri S, Sassone-Corsi P. 2018. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep 25:3299–3314.e6. doi: 10.1016/j.celrep.2018.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogan S, Ray A, Cleary MP. 2017. The influence of different calorie restriction protocols on serum pro-inflammatory cytokines, adipokines and IGF-I levels in female C57BL6 mice: short term and long term diet effects. Meta Gene 12:22–32. doi: 10.1016/j.mgene.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meydani SN, Das SK, Pieper CF, Lewis MR, Klein S, Dixit VD, Gupta AK, Villareal DT, Bhapkar M, Huang M, Fuss PJ, Roberts SB, Holloszy JO, Fontana L. 2016. Long-term moderate calorie restriction inhibits inflammation without impairing cell-mediated immunity: a randomized controlled trial in non-obese humans. Aging (Albany NY) 8:1416–1431. doi: 10.18632/aging.100994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, Yu L, Xu C, Ren Z, Xu Y, Xu S, Shen H, Zhu X, Shi Y, Shen Q, Dong W, Liu R, Ling Y, Zeng Y, Wang X, Zhang Q, Wang J, Wang L, Wu Y, Zeng B, Wei H, Zhang M, Peng Y, Zhang C. 2018. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, Zhang M, Wang L, Hou Y, Ouyang H, Zhang Y, Zheng Y, Wang J, Lv X, Wang Y, Zhang F, Zeng B, Li W, Yan F, Zhao Y, Pang X, Zhang X, Fu H, Chen F, Zhao N, Hamaker BR, Bridgewater LC, Weinkove D, Clement K, Dore J, Holmes E, Xiao H, Zhao G, Yang S, Bork P, Nicholson JK, Wei H, Tang H, Zhang X, Zhao L. 2015. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine 2:968–984. doi: 10.1016/j.ebiom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, Mehrabian M, Denu JM, BäCkhed F, Lusis AJ, Rey FE. 2018. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol 3:1461. doi: 10.1038/s41564-018-0272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. 2011. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bangsgaard Bendtsen KM, Krych L, Sorensen DB, Pang W, Nielsen DS, Josefsen K, Hansen LH, Sorensen SJ, Hansen AK. 2012. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One 7:e46231. doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker A, Pfitzner B, Harir M, Schaubeck M, Calasan J, Heinzmann SS, Turaev D, Rattei T, Endesfelder D, Castell WZ, Haller D, Schmid M, Hartmann A, Schmitt-Kopplin P. 2017. Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci Rep 7:11047. doi: 10.1038/s41598-017-10369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goh KL, Chan WK, Shiota S, Yamaoka Y. 2011. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter 16(Suppl 1):1–9. doi: 10.1111/j.1523-5378.2011.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zareie M, Johnson-Henry K, Jury J, Yang PC, Ngan BY, McKay DM, Soderholm JD, Perdue MH, Sherman PM. 2006. Probiotics prevent bacterial translocation and improve intestinal barrier function in rats following chronic psychological stress. Gut 55:1553–1560. doi: 10.1136/gut.2005.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernardeau M, Guguen M, Vernoux JP. 2006. Beneficial lactobacilli in food and feed: long-term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol Rev 30:487–513. doi: 10.1111/j.1574-6976.2006.00020.x. [DOI] [PubMed] [Google Scholar]
- 43.Johnson-Henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. 2007. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol 9:356–367. doi: 10.1111/j.1462-5822.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- 44.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Frätzer C, Krannich A, Gollasch M, Grohme DA, Côrte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, Müller DN. 2017. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551:585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Schillde M-A, Hörmannsperger G, Weiher M, Alpert C-A, Hahne H, Bäuerl C, van Huynegem K, Steidler L, Hrncir T, Pérez-Martínez G, Kuster B, Haller D. 2012. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11:387–396. doi: 10.1016/j.chom.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Schnadower D, Tarr PI, Casper TC, Gorelick MH, Dean JM, O'Connell KJ, Mahajan P, Levine AC, Bhatt SR, Roskind CG, Powell EC, Rogers AJ, Vance C, Sapien RE, Olsen CS, Metheney M, Dickey VP, Hall-Moore C, Freedman SB. 2018. Lactobacillus rhamnosus GG versus placebo for acute gastroenteritis in children. N Engl J Med 379:2002–2014. doi: 10.1056/NEJMoa1802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freedman SB, Williamson-Urquhart S, Farion KJ, Gouin S, Willan AR, Poonai N, Hurley K, Sherman PM, Finkelstein Y, Lee BE, Pang XL, Chui L, Schnadower D, Xie J, Gorelick M, Schuh S. 2018. Multicenter trial of a combination probiotic for children with gastroenteritis. N Engl J Med 379:2015–2026. doi: 10.1056/NEJMoa1802597. [DOI] [PubMed] [Google Scholar]
- 48.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. 2012. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond) 36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Zhang C, Zhao L. 2016. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med 8:41. doi: 10.1186/s13073-016-0304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao L. 2013. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol 11:639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 51.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB, Federici S, Cohen Y, Linevsky R, Rothschild D, Moor AE, Ben-Moshe S, Harmelin A, Itzkovitz S, Maharshak N, Shibolet O, Shapiro H, Pevsner-Fischer M, Sharon I, Halpern Z, Segal E, Elinav E. 2018. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 52.Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, Dohnalova L, Braverman S, Rozin S, Malitsky S, Dori-Bachash M, Kuperman Y, Biton I, Gertler A, Harmelin A, Shapiro H, Halpern Z, Aharoni A, Segal E, Elinav E. 2016. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540:544–551. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 53.Gill S, Panda S. 2015. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab 22:789–798. doi: 10.1016/j.cmet.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. 2008. Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab 295:E1323–E1332. doi: 10.1152/ajpendo.90617.2008. [DOI] [PubMed] [Google Scholar]
- 55.Wang CJ, Zhou ZG, Holmqvist A, Zhang H, Li Y, Adell G, Sun XF. 2009. Survivin expression quantified by Image Pro-Plus compared with visual assessment. Appl Immunohistochem Mol Morphol 17:530–535. doi: 10.1097/PAI.0b013e3181a13bf2. [DOI] [PubMed] [Google Scholar]
- 56.Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol 63:2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Wu Y, Wang J, Wu G, Long W, Xue Z, Wang L, Zhang X, Pang X, Zhao Y, Zhao L, Zhang C. 2016. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci Rep 6:27572. doi: 10.1038/srep27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 59.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lê Cao K-A, Rohart F, Gonzalez I, Dejean S, Abadi A, Gautier B, Bartolo F, Monget P, Coquery J, Yao F, Liquet B. 2016. mixOmics: omics data integration project. R package version 6.1.1. doi: 10.18129/B9.bioc.mixOmics. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunohistochemical staining of colon and integrated optical density (IOD) analysis for Zo-1 (upper) and occludin (lower) from NC mice (left), CRL mice (middle), and CRD mice (right) after a 4-week CR. Scale bar = 50 μm. Data are presented as the mean ± standard error of the mean (SEM) and analyzed using one-way ANOVA, followed by a Tukey post hoc test. Download FIG S1, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Every 2-hr food intake of the 10th light-dark cycle after ad libitum. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. Download FIG S2, TIF file, 0.2 MB (215.9KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Immunohistochemical staining of colon and integrated optical density (IOD) analysis for Zo-1 (upper) and occludin (lower) from NC mice (left), CRL mice (middle), and CRD mice (right) after a 4-week ad libitum period. Scale bar = 50 μm. Data are presented as the mean ± SEM and analyzed using one-way ANOVA, followed by a Tukey post hoc test. Download FIG S3, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA profiles of baseline samples. Top, principal-coordinate analysis (PCoA) based on Bray-Curtis distance. Bottom, heatmap of Bray-Curtis distances between each group and permutational multivariate analysis of variance (PERMANOVA; 9,999 permutations). Download FIG S4, TIF file, 0.8 MB (826.2KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alpha diversity of the gut microbiota. (a to e) The observed OTUs (left), Shannon diversity index (middle), and Faith’s phylogenetic diversity values (right) after 4-week calorie restriction (CR) (a) and on days 2 (b), 11 (c), 19 (d), and 26 (e) after ad libitum (AL). The sampling level is 12,300. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. **, P < 0.01. Download FIG S5, TIF file, 1.3 MB (1.4MB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bray-Curtis distance between CRL and CRD mice compared at two time points after a 4-week CR period. Data are presented as the mean ± SEM and analyzed using one-way ANOVA, followed by a Tukey post hoc test. ***, P < 0.001. Download FIG S6, TIF file, 0.2 MB (190.8KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The optimal classification performance of the sPLS-DA model related to FIG 5. (a to f) The plots of the classification error rate of different components averaged across the leave-one-out cross-validation for maximum distances (left side of each panel), and the sPLS-DA plots based on OTU table (right side of each panel) for separating the gut microbiota of NC-ZT0 and CRD-ZT0 (a), NC-ZT12 and CRD-ZT12 (b), NC-ZT0 and CRL-ZT12 (c), NC-ZT12 and CRL-ZT0 (d), CRL-ZT12 and CRD-ZT0 (e), and CRL-ZT0 and CRD-ZT12 (f). Download FIG S7, TIF file, 0.6 MB (586.7KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The optimal classification performance of the sPLS-DA model related to FIG 6. (a and b) The plots of the classification error rate of different components averaged across the leave-one-out cross-validation for maximum distances (left), and the sPLS-DA plots based on OTU table (right) for separating the gut microbiota of CRL-ZT0 and CRL-ZT12 (a) and CRD-ZT0 and CRD-ZT12 (b). Download FIG S8, TIF file, 0.1 MB (145.4KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The difference of gut microbiota between CRL and NC at ZT0 on day 19 after ad libitum. The plot of the classification error rate of different components averaged across the leave-one-out cross validation for maximum distances and sPLS-DA plot based on OTU table (top) and contribution plot of OTUs and heat map of key OTUs (bottom) for separating the gut microbiota of CRL-ZT0 and NC-ZT0. Download FIG S9, TIF file, 0.2 MB (201.7KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of three Lactobacillus OTUs and one Alistipes OTU. (a to d) OTU2 (left), OTU40 (middle-left), OTU43 (middle-right), and OTU102 (right) in days 2 (a), 11 (b), 19 (c), and 26 (d) after ad libitum. For the NC group, n = 10; for CRL and CRD, n = 8. Data are presented as the mean ± SEM and analyzed using two-way repeated-measures ANOVA, followed by a Tukey post hoc test. ***, P < 0.001; **, P < 0.01; *, P < 0.05. Download FIG S10, TIF file, 0.9 MB (950.7KB, tif) .
Copyright © 2019 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw Illumina sequence data of the 16S rRNA gene generated in this study are available in the NCBI Sequence Read Archive (SRA) under accession number SRP166705.