Immunocompromised individuals often become symptomatic with infections which are normally fairly asymptomatic in healthy individuals. The particular mechanisms that underlie susceptibility to coinfections in human immunodeficiency virus (HIV)-infected individuals are multifaceted. ZIKV and other flaviviruses are sensitive to neutralizing antibodies, whose production can be limited in HIV-infected individuals but are also sensitive to type I interferons, which are expressed at high levels in HIV-infected individuals. Data in this study highlight how individual components of the innate and adaptive immune responses which become perturbed in HIV-infected individuals influence ZIKV infection.
KEYWORDS: SIV, ZIKV, innate immunity, type I interferon
ABSTRACT
Flaviviruses are controlled by adaptive immune responses but are exquisitely sensitive to interferon-stimulated genes (ISGs). How coinfections, particularly simian immunodeficiency viruses (SIVs), that induce robust ISG signatures influence flavivirus clearance and pathogenesis is unclear. Here, we studied how Zika virus (ZIKV) infection is modulated in SIV-infected nonhuman primates. We measured ZIKV replication, cellular ZIKV RNA levels, and immune responses in non-SIV-infected and SIV-infected rhesus macaques (RMs), which we infected with ZIKV. Coinfected animals had a 1- to 2-day delay in peak ZIKV viremia, which was 30% of that in non-SIV-infected animals. However, ZIKV viremia was significantly prolonged in SIV-positive (SIV+) RMs. ISG levels at the time of ZIKV infection were predictive for lower ZIKV viremia in the SIV+ RMs, while prolonged ZIKV viremia was associated with muted and delayed adaptive responses in SIV+ RMs.
INTRODUCTION
The high prevalence of Zika virus (ZIKV) in northeast Brazil and its spread in the Americas in 2015 to 2016 (1, 2) were declared a public health emergency following the linkage between infection and microcephaly in fetuses of ZIKV-infected mothers (3–5). Since ZIKV and other flaviviruses are transmitted by arthropods, flavivirus epidemics are often restricted to geographies where mosquitoes flourish, and these areas overlap areas with a high incidence of HIV infection (6, 7).
ZIKV and other flaviviruses are sensitive to type I interferons (IFNs). Indeed, ZIKV replicates poorly in immunocompetent mice (8–10), in part due to an inability of the ZIKV nonstructural 5 (NS5) protein to degrade mouse STAT2 (11–13). STAT2 activation occurs after type I IFN signaling, which facilitates the transcription of over 300 IFN-stimulated genes (ISGs) (13). Mouse models of ZIKV infection, therefore, generally require type I IFN signaling to be abrogated or altered to escape IFN-mediated control (14, 15). The IFN-mediated control of flaviviruses is, in part, attributed to the interferon-inducible transmembrane proteins (IFITMs) (16–18).
Adaptive immune responses are also clearly important for the clearance of ZIKV and other flaviviruses (19–21). Human immunodeficiency virus (HIV) infection of humans and simian immunodeficiency virus (SIV) infection of Asian macaques result in a loss of memory CD4+ T cells, which provide critical immunological defense functions. Most HIV/SIV-infected individuals who are not treated with combination antiretroviral therapy will lose sufficient numbers of these CD4+ T cells to render them susceptible to opportunistic infections (22, 23). While this sensitivity to opportunistic infections generally occurs very late after the acquisition of infection, several studies have shown that HIV-infected individuals and SIV-infected rhesus macaques (RMs) mount suboptimal adaptive immune responses to vaccination and viral infections (24–27). The decreased humoral responses to vaccination, in particular, have been attributed to preferential infection of lymph node follicle-resident T follicular cells, which provide critical help to B cells during the germinal center reaction (27–33).
While HIV/SIV-infected individuals become immunocompromised and mount suboptimal adaptive immune responses, it is also important to note that another hallmark of these progressive immunodeficiency lentiviral infections is chronic immune activation (34–36). This immune activation is very commonly associated with increased expression of ISGs (37–40). Indeed, transcriptional analysis of cell types of multiple lineages from HIV/SIV-infected individuals indicates recent type I IFN signaling (41). Thus, it is also reasonable to predict that the type I IFN production which occurs in HIV infection might limit ZIKV replication.
Both ZIKV and SIV have individually been isolated from wild nonhuman primates (NHPs) and, when experimentally introduced into captive NHPs, have been shown to mirror aspects of human disease progression (42–45). To explore how HIV infection might impact ZIKV antiviral responses and disease progression, we followed ZIKV viremia, ZIKV RNA levels within cells targeted by ZIKV in vivo, and innate and adaptive immune responses in peripheral blood and lymph nodes (LNs) in non-SIV-infected and SIV-infected RMs that were infected with ZIKV. We found that ZIKV viremia peaked later and was prolonged in SIV-coinfected RMs. These data provide insight into how preexisting lentiviral infection modulates ZIKV viremia in a relevant NHP model for both infections.
RESULTS
Experimental sampling and infection establishment.
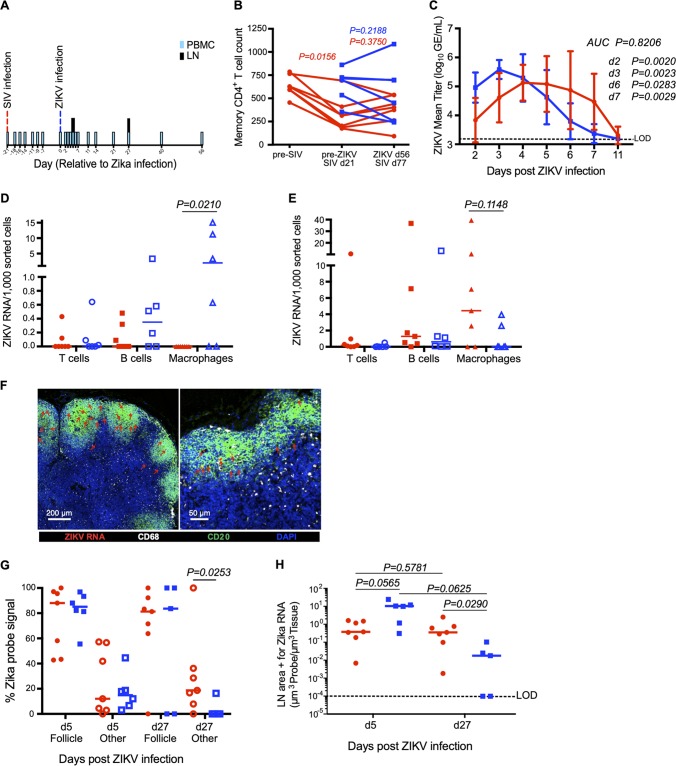
To determine how SIV infection might influence viremia and the host response to ZIKV infection, we infected 7 Asian RMs with 3,000 50% tissue culture infective doses (TCID50) of SIVmac239 intravenously (Table 1). Three weeks thereafter, we subcutaneously infected these animals with 104 PFU of ZIKV isolate Nicaragua/2016 (SIV/ZIKV animals) (Table 1). For a control, we also infected 12 rhesus macaques (RMs) with ZIKV only (Table 1). The time point 3 weeks post-SIV infection was chosen to correspond to the peak inflammation and expression of ISGs during acute SIV infection in rhesus macaques but prior to the animals becoming dramatically immunocompromised (38, 46). We sampled peripheral blood and lymph nodes at numerous time points both pre- and post-SIV and/or ZIKV infection (Fig. 1A) to study viremia and innate and adaptive immune responses. To measure SIV progression, we measured SIV RNA levels in plasma (Table 1) as well as counted peripheral blood CD4+ T cells (the number of cells per microliter of blood) (Fig. 1B). We found that the SIV/ZIKV cohort had a significant loss of memory CD4+ T cells prior to ZIKV challenge (Fig. 1B) (P = 0.0156), consistent with these animals being progressively SIV infected, but ZIKV infection did not exacerbate the loss of CD4+ T cells in the peripheral blood of either the SIV-infected or the uninfected group of animals (Fig. 1B).
TABLE 1.
Study animalsa
| Animal | Gender | Wt (kg) | Age (yr) | Group | SIV plasma viremiab |
Phenotype |
|---|---|---|---|---|---|---|
| RH37033 | Male | 11.9 | 11 | SIV/ZIKV | 200,000 | A01+ B08– B17– |
| RH37034 | Male | 12.4 | 11 | SIV/ZIKV | 670,000 | A01– B08– B17– |
| RHDE8Z | Male | 11.9 | 8 | SIV/ZIKV | 12,000 | A01– B08– B17– |
| RHDFiV | Male | 10.3 | 6 | SIV/ZIKV | 800,000 | A01– B08– B17– |
| RHDF31 | Male | 7.7 | 6 | SIV/ZIKV | 830,000 | A01– B08– B17– |
| RH4345 | Male | 10.8 | 7 | SIV/ZIKV | 260,000 | A01– B08– B17– |
| RH08D030 | Male | 12.9 | 12 | SIV/ZIKV | 2,300,000 | A01+ B08– B17– |
| RH37073 | Female | 10.2 | 12 | ZIKV | NA | A01+ B08– B17+ |
| RH788 | Male | 16.5 | 13 | ZIKV | NA | A01– B08– B17– |
| RHDF86 | Male | 13.2 | 6 | ZIKV | NA | A01– B08– B17– |
| RHDBC4 | Male | 14.9 | 13 | ZIKV | NA | A01– B08– B17– |
| RHA0P003 | Male | 13.3 | 18 | ZIKV | NA | ND |
| RH734 | Male | 12.0 | 16 | ZIKV | NA | A01– B08– B17– |
| RHZJ31 | Male | 7.2 | 7 | ZIKV | NA | ND |
| RHZG41 | Female | 5.6 | 10 | ZIKV | NA | A01– B08– B17– |
| RHDF0B | Male | 8.6 | 4 | ZIKV | NA | ND |
| RHDF2H | Male | 4.7 | 4 | ZIKV | NA | ND |
| RHDFWF | Male | 4.9 | 4 | ZIKV | NA | ND |
| RHDFWL | Male | 4.7 | 5 | ZIKV | NA | ND |
NA, not applicable; ND, not determined.
The number of copies of viral SIV RNA per milliliter of plasma at day 0 of ZIKV infection.
FIG 1.
Effect of SIV coinfection on ZIKV viremia and cellular infection in peripheral lymph nodes. (A) Study design depicting longitudinal time points for SIV infection, ZIKV infection, and PBMC and peripheral LN sampling. (B) Memory CD4+ T cell counts for ZIKV (blue squares; n = 6) and SIV/ZIKV animals (red circles; n = 7). Each line represents an individual animal. (C) ZIKV mean viral titer in plasma (ZIKV, n = 12; SIV/ZIKV, n = 7). AUC was calculated from day 2 (d2) to day 7. (D and E) ZIKV RNA per 1,000 sorted cells in peripheral LN T cells (circles), B cells (squares), and macrophages (triangles) at 5 days post-ZIKV infection (D) and 27 days post-ZIKV infection (E) in SIV/ZIKV-coinfected animals (red) and ZIKV animals (blue). (F) Representative lymph node immunohistochemistry images at day 27 post-ZIKV infection of ZIKV vRNA (RNAscope; red), CD20 (B cell follicle; green), CD68 (white), and DAPI (blue). (G) Percentages of the ZIKV probe signal that are found within the B cell follicle region (filled symbols) and all other regions of the LNs (Other; open symbols) at day 5 and day 27 post-ZIKV infection. (H) Quantification of the LN area positive for ZIKV vRNA probe per total LN area quantified at day 5 and day 27 post-ZIKV infection. The dashed line represents the limit of detection (LOD). For this figure, all P values between groups represent Mann-Whitney t test results (red compared to blue), while P values between cell types or days within the same group represent Wilcoxon matched-pair results.
Peak plasma ZIKV viremia is delayed in SIV-infected RMs.
We next measured plasma ZIKV viremia between animals that were and were not SIV infected (Fig. 1C). Consistently with the results of previous reports, in non-SIV-infected RMs, ZIKV RNA was detectable in plasma by 2 days postinfection (dpi), peaked to approximately 500,000 copies/ml at 2 to 3 dpi, and then declined to undetectable levels by 7 dpi (47–53). In contrast, the dynamics of ZIKV viremia were delayed in SIV-infected animals (Fig. 1C; see also the ZIKV viremia in individual animals discussed in Fig. S1 in the supplemental material). Peak ZIKV RNA in SIV-infected animals occurred at approximately 4 days postinfection and was 30% of that in non-SIV-infected animals. Furthermore, the SIV-infected RMs had significantly lower ZIKV plasma RNA at both day 2 (Fig. 1C) ( P = 0.0020) and day 3 (Fig. 1C) ( P = 0.0023) post-ZIKV infection. Likely as a consequence of this delayed peak, ZIKV RNA levels were higher in SIV+ animals than in SIV-negative animals at day 7 after ZIKV infection, a time when ZIKV had been cleared from most non-SIV-infected RMs (Fig. 1C) ( P = 0.0029), suggesting that lentiviral coinfection might prolong the flavivirus transmission period.
ZIKV plasma viral titer. Dotted lines represent titers for individual animals, while solid lines represent the mean viral titer for the ZIKV cohort (blue; n = 12) and the SIV/ZIKV cohort (red; n = 7). Download FIG S1, PDF file, 0.1 MB (76.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Peripheral lymph node macrophages are a source of ZIKV replication.
To understand ZIKV tropism in our coinfection model, we used flow cytometry to sort T cells, B cells, and macrophages from LNs of animals from our cohort at day 5 and day 27 post-ZIKV infection (our flow cytometric sorting strategy is shown in Fig. S2) and measured ZIKV RNA by quantitative reverse transcription-PCR (qRT-PCR). We found ZIKV RNA in lymph node-resident macrophages and, to a lesser extent, in B cells, with higher levels of ZIKV RNA in macrophages isolated from non-SIV-infected animals at day 5 post-ZIKV infection (Fig. 1D) than in SIV-infected animals ( P = 0.021). Interestingly, we also found ZIKV RNA in macrophages isolated from LNs of animals at day 27 post-ZIKV infection, a time point when plasma ZIKV RNA was undetectable in plasma (Fig. 1E). This is consistent with previous reports of the persistence of ZIKV in the lymph nodes of RMs, with virus being detected in peripheral LNs for up to 72 days (47).
Representative flow cytometry gating for cell sorting of peripheral lymph nodes. Parent gating strategy for the flow cytometric sorting of day 5 and day 27 post-ZIKV infection LN samples for analysis of cellular infection status by RT-PCR. A representative staining profile from the ZIKV cohort is shown. Live, singlet leukocytes were sorted into T cell (CD3+), B cell (CD20+), or myeloid cell (CD11b+) populations. Download FIG S2, PDF file, 0.1 MB (157.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Fluorescence in situ hybridization for ZIKV RNA in lymph node sections verified that ZIKV RNA could be detected both within B cell follicles and in the paracortex (Fig. 1F, where ZIKV RNA is indicated with red arrows), with the majority of the ZIKV RNA at days 5 and day 27 post-ZIKV infection found within the B cell follicle (Fig. 1G). To understand how lymph node levels of ZIKV RNA compared between SIV+ and non-SIV-infected animals, we performed quantitative image analysis (Fig. 1G and H). Consistently with our plasma ZIKV RNA analysis, we observed modestly higher levels of ZIKV RNA within lymph nodes of non-SIV-infected animals at day 5 post-ZIKV infection than in SIV+ animals (P = 0.056) (Fig. 1H) but significantly higher levels of ZIKV RNA in lymph nodes of SIV+ animals at day 27 post-ZIKV infection (P = 0.029) (Fig. 1H). These data are consistent with delayed clearance of ZIKV in SIV+ animals and further demonstrate that the LN serves as a prolonged ZIKV reservoir in RMs.
SIV coinfection reduces immune responses to ZIKV.
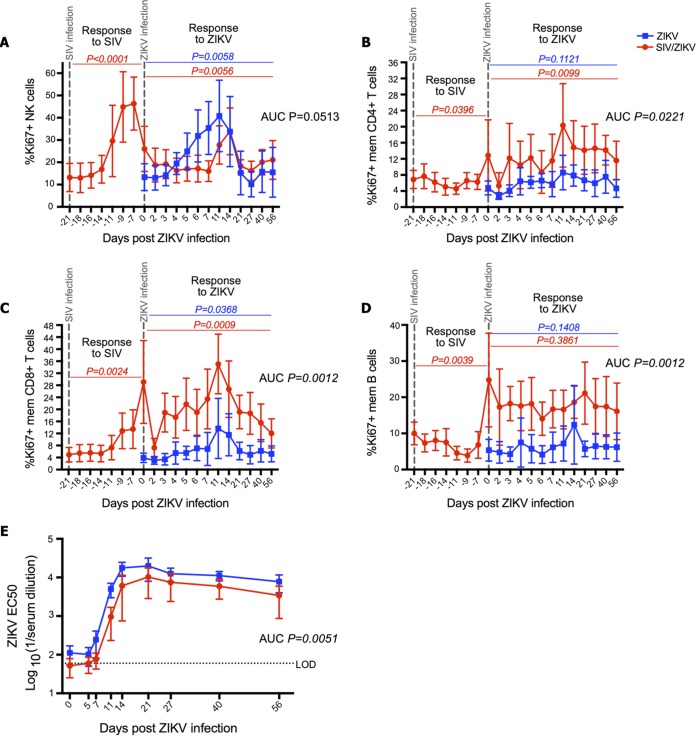
Using flow cytometric analysis, we next measured innate and adaptive immunological responses during ZIKV infection of the animals in our cohort (the flow cytometric gating strategy is shown in Fig. S3). We found that RMs mounted robust proliferative responses to SIV and ZIKV infections in several cellular populations as measured by Ki67 expression (Fig. 2). The SIV-infected animals mounted NK cell proliferative responses to both SIV infection and ZIKV infection (Fig. 2A). However, the NK cell proliferative response during ZIKV replication among the SIV+ animals was restricted compared to that of the non-SIV-infected animals, showing a delayed, dampened, and shorter duration of proliferation (Fig. 2A) (area under the curve [AUC], P = 0.05). We also measured the proliferation of memory CD4+ T cells (Fig. 2B), memory CD8+ T cells (Fig. 2C), and memory B cells (Fig. 2D) in our longitudinal peripheral blood samples. These proliferative responses tended to peak around day 11 post-ZIKV infection, with a significant response for both cohorts in the Ki67+ memory CD8+ T cell population (P = 0.0368 in the ZIKV cohort and P = 0.0009 in the SIV/ZIKV cohort). We did not observe any proliferative response to ZIKV among B cells (Fig. 2D). SIV infection was also associated with robust proliferative responses from NK cells, T cells, and B cells (Fig. 2). Proliferative responses of T and B cells to ZIKV infection among SIV+ animals were, thus, less clear given their already-heightened proliferation (Fig. 2B to D).
FIG 2.
SIV coinfection reduces immune responses to ZIKV. (A to D) Percentages of Ki67+ NK cells (A), memory CD4+ T cells (B), memory CD8+ T cells (C), and memory (mem) B cells (D) longitudinally pre- and post-ZIKV infection. ZIKV cohort mean percentages are illustrated by blue squares (n = 6), while SIV/ZIKV cohort mean percentages are illustrated by red circles (n = 7). P values represent repeated-measures one-way ANOVA. AUCs were calculated from day 2 to day 21 post-ZIKV infection, followed by a Mann-Whitney U test comparison between groups. Error bars represent standard deviations for each time point. (E) Zika virus-neutralizing antibody titers (EC50 values) post-ZIKV infection in ZIKV RMs (blue squares; n = 5) and SIV/ZIKV RMs (red circles; n = 7). AUCs were calculated from day 0 to day 56. The dashed line represents the limit of detection (LOD).
Representative flow cytometry gating for phenotypic cell characterization. Parent gating strategy for the analysis of PBMCs. A representative staining profile from the ZIKV cohort is shown. Live, singlet lymphocytes were phenotypically characterized as memory B cells (CD27+ CD20+ CD3−), memory CD4+ T cells (CD28+/− CD95+ CD4+ CD3+), memory CD8+ T cells (CD28+/− CD95+ CD8+ CD3+), or NK cells (NKG2a+). Proliferation was assessed for each cell type by the expression of Ki67. Download FIG S3, PDF file, 0.2 MB (240.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The humoral immune response plays a significant role in controlling flavivirus infection (20, 21). We next measured titers of ZIKV-specific neutralizing antibody (NAb) responses in longitudinal plasma samples (Fig. 2E). In non-SIV-infected animals, NAbs to ZIKV became detectable by day 7 post-ZIKV infection and then peaked at days 14 to 21, after which, titers plateaued until at least day 56 (Fig. 2E). In SIV/ZIKV-coinfected RMs, the NAb response to ZIKV was slightly delayed and peaked at a lower titer than that in non-SIV-infected animals (Fig. 2E) (AUC, P = 0.0051). The SIV/ZIKV-coinfected RMs had a 5.3-fold-lower average EC50 NAb titer at day 11 and a 2.9-fold-lower titer at day 14. Taken together, these data suggest that SIV-infected RMs have reduced innate and adaptive immune responses to ZIKV infection.
ZIKV has no significant effect on CD4 T cell functionality in SIV/ZIKV-coinfected RM.
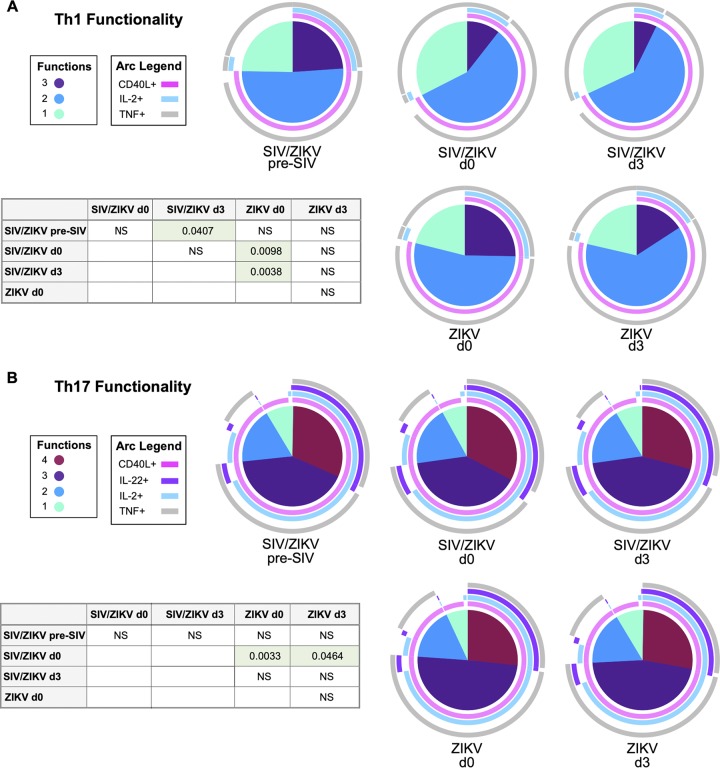
HIV and SIV infection are associated with decreased T cell functionality and loss of CD4+ T cells, particularly Th17 cells from mucosal tissues, supporting the delayed and decreased immune responses observed in our ZIKV/SIV-coinfected animals (54–56). This immunological perturbation is thought to be due, in part, to alterations to antigen-presenting cells, which result in an environment conducive to the maintenance and development of Th1 cells (57). Given that ZIKV is also an RNA virus which induces Th1 cells (58), we sought to explore whether ZIKV coinfection further exacerbates immune dysfunction. Thus, we assessed whether SIV/ZIKV coinfection changed the functional profile of memory CD4+ T cells. We, therefore, measured the expression of CD40L, interleukin 22 (IL-22), IL-17, IL-2, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) from mitogenically stimulated memory CD4+ T cells by flow cytometry. We then determined the overall functional capacity of Th1 (IFN-γ+ IL-17–) and Th17 (IL-17+ IFN-γ–) CD4+ T cells using a simplified presentation of incredibly complex evaluations (SPICE) analysis (Fig. 3). SIV infection decreased the polyfunctional capacity (defined here as the ability to simultaneously express CD40L, TNF-α, and IL-2) of Th1 cells (Fig. 3A). Consistently with previous reports (54, 56), SIV infection resulted in decreased polyfunctionality (simultaneous expression of CD40L, IL-22, IL-2, and TNF-α) of Th17 cells (Fig. 3B). ZIKV infection did not influence the functional capacity of memory CD4+ T cells (either Th1 or Th17 cells) (Fig. 3).
FIG 3.
ZIKV, on its own, does not affect CD4 T cell polyfunctionality. (A and B) Pie charts depicting Th1 functionality and the ability of IFN-γ+ IL-17– CD4+ T cells to simultaneously produce CD40L, TNF-α, and IL-2 (A) and Th17 functionality and the ability of IL-17+ IFN-γ– CD4+ T cells to simultaneously produce CD40L, IL-22, IL-2, and TNF-α (B) in SIV/ZIKV (top pie charts)- and ZIKV (bottom pie charts)-infected RMs prior to and after infection with SIV and ZIKV. P values are based on SPICE permutation tests and reflect differences between T cell polyfunctionality based on the expression of different combinations of cytokines (as illustrated by arcs surrounding each pie). P values of >0.05 are listed as not significant (NS).
Interferon signaling predicts peak ZIKV plasma viremia.
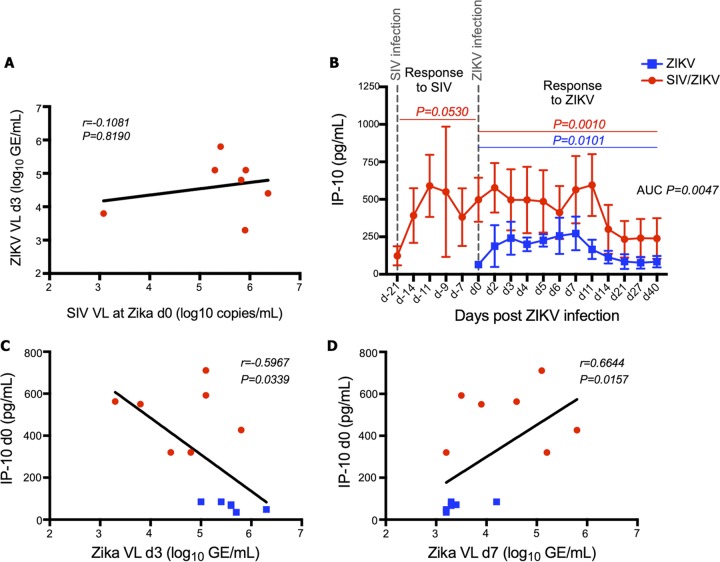
The observed delays in both ZIKV viremia and immunological responses in coinfected RMs suggested that SIV viremia might, itself, either directly or indirectly limit ZIKV replication. However, when we compared SIV plasma viral loads at day 0 of ZIKV infection with ZIKV plasma viral loads at day 3 post-ZIKV infection (peak ZIKV viremia in non-SIV-infected animals) we found no clear evidence that SIV viremia correlates with the extent of subsequent ZIKV replication (Fig. 4A) (r = –0.1081, P = 0.8190). Considering that both HIV and SIV infections induce significant type I IFN production and signaling (37–40) and that flaviviruses replicate poorly in IFN-α-primed environments (59–62), we next sought to examine whether there was an association of ISGs with levels of ZIKV viremia in our SIV/ZIKV animals. To measure systemic levels of IFN signaling, we assayed plasma levels of IP-10/CXCL10 in our animals longitudinally after SIV and ZIKV infections (Fig. 4B). This IFN-induced protein has been shown to be high in acutely ZIKV-infected individuals and strongly associated with exanthema (63). In non-SIV-infected animals, ZIKV infection led to increased plasma levels of IP-10 that were maintained until day 14 post-ZIKV infection (Fig. 4B). In SIV/ZIKV-coinfected animals, SIV itself (pre-ZIKV) induced and maintained high plasma levels of IP-10 (higher than those observed in the animals infected only with ZIKV) that did not further increase following coinfection (Fig. 4B). We, thus, questioned whether the IFN response elicited by SIV infection might predict lower levels of peak ZIKV viremia and compared the plasma levels of IP-10 prior to ZIKV infection with the levels of ZIKV viremia at day 3 (Fig. 4C). Higher levels of IP-10 prior to ZIKV infection, produced in response to SIV infection, were predictive of lower levels of ZIKV viremia. Moreover, the prolonged ZIKV viremia observed in SIV/ZIKV-coinfected RMs at day 7 post-ZIKV infection was associated with plasma levels of IP-10 prior to ZIKV infection (Fig. 4D). Thus, the IFN-stimulated response mounted against SIV correlated with a delayed peak and prolonged clearance of ZIKV viremia.
FIG 4.
IFN-stimulated IP-10 gene levels are predictive of peak ZIKV plasma viremia. (A) Association between ZIKV plasma viremia (log10 GE/ml) at day 3 and SIV plasma viremia at day 0 of ZIKV infection (7 RMs in the SIV/ZIKV cohort). VL, viral load. (B) IP-10 (picograms per milliliter) measured by ELISA in longitudinal plasma samples pre- and post-ZIKV infection. Mean IP-10 levels with standard deviation error bars are represented for the ZIKV cohort (n = 6) in blue squares and for the SIV/ZIKV cohort in red circles. P values represent repeated-measures one-way ANOVA. AUCs were calculated from day 2 to day 21 post-ZIKV infection, followed by Mann-Whitney comparisons between groups. (C to D) Associations between IP-10 plasma levels (picograms per milliliter) at day 0 of ZIKV infection and ZIKV plasma viremia (log10 GE/ml) at day 3 (C) and day 7 (D) post-ZIKV-infection. SIV/ZIKV RM data points are shown as red circles, and ZIKV RM data points are shown as blue squares. Spearman correlations with linear regression were utilized to determine statistical significance for associations in panels A, C, and D.
Transcriptional analysis of ZIKV and SIV immune responses.
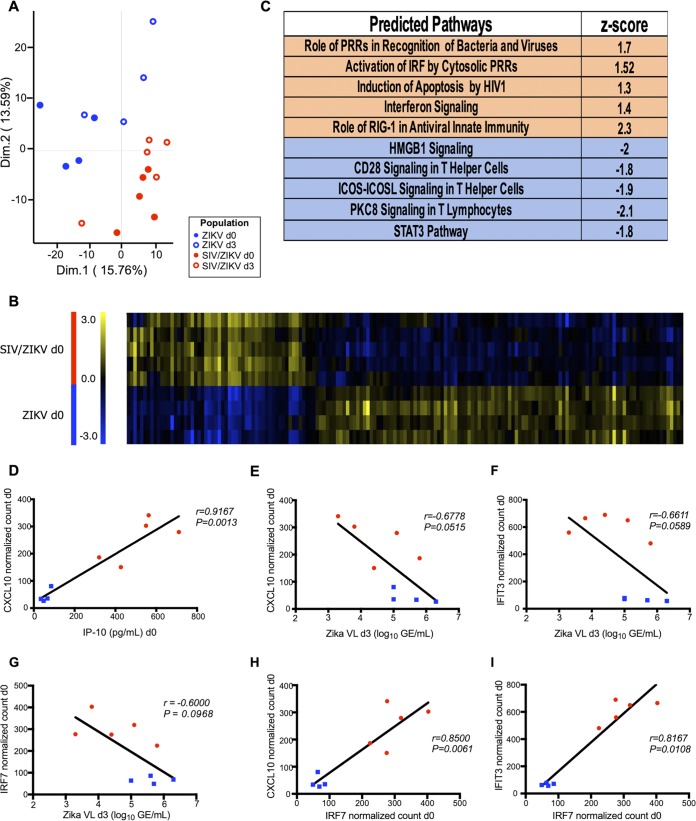
Since we observed associations between increased IP-10 levels and lower peak ZIKV viremia at day 3 post-Zika infection in our coinfected RMs, we next sought to explore global transcriptional responses that coincide with SIV and ZIKV infection. Gene expression was measured from mRNAs extracted from peripheral blood mononuclear cells (PBMCs) by NanoString nCounter analysis. We studied samples corresponding to pre-ZIKV infection (day 0) and day 3 post-ZIKV infection, with and without SIV coinfection. Principal-component analysis revealed that transcript profiles of PBMCs from the ZIKV RMs were distinct from those of the SIV/ZIKV RMs (Fig. 5A). Most of the variation within the gene expression data was related to SIV or ZIKV infection, most of which was attributable to SIV infection. While ZIKV coinfection did not dramatically alter the transcriptional profile of PBMCs from SIV-infected animals, ZIKV did alter the transcriptional profile of circulating leukocytes (Fig. S4A). Many of the pathways which were induced by SIV infection were also induced by ZIKV infection, albeit to lower levels. Indeed, when we compared non-SIV-infected and SIV-infected RMs prior to ZIKV infection (ZIKV day 0), 163 genes were significantly differentially expressed between the two cohorts (Fig. 5B and Table S1).
FIG 5.
Transcriptional gene profiling of expression changes in acute ZIKV infection. Immunology panel gene expression profiling in the ZIKV RM cohort (n = 4) at day 0 (filled blue circles) or day 3 (open blue circles) and SIV/ZIKV RM cohort (n = 5) at day 0 (filled red circles) or day 3 (open red circles). (A) Principal-component analysis comparing the four sample groups. The plot represents clustering of the data in a 2-dimensional (Dim.1 and Dim.2) matrix. Each dot represents one RM. (B) Heatmap of 163 differentially expressed genes (P > 0.05) in ZIKV and SIV/ZIKV RM cohorts (n = 4 and n = 5, respectively) at day 0 post-Zika infection. Blue indicates low relative gene expression, and yellow indicates high relative gene expression. Each row represents a normalized expression value for a single gene, and each row represents one animal from the corresponding cohort. (C) Pathway activation determined by z-scores. The top five pathways predicted to be activated (orange) or deactivated (blue) are displayed. PRRs, pseudoresponse regulators; IRF, interferon regulatory factor; HIV1, HIV type 1; ICOS, inducible T cell costimulator; ICOSL, inducible T cell costimulator ligand. (D) Association between the CXCL10-normalized count at day 0 post-Zika infection and IP-10 levels (picograms per milliliter) in plasma at day 0. (E to G) Associations between ZIKV plasma viremia (log10 number of GE per milliliter) at day 3 post-Zika infection and CXCL10 (E)-, IFIT3 (F)-, or IRF7 (G)-normalized counts at day 0 post-ZIKV infection. (H and I) Associations between normalized counts at day 0 post-ZIKV infection of IRF7 and CXCL10 (H) or IFIT3 (I). (D to I) Blue squares represent data from the ZIKV cohort, and red circles represent data from the SIV/ZIKV cohort. Correlations in panels D to I were determined using Spearman’s rank analysis.
Transcriptional gene profiling of expression changes in acute ZIKV infection. Immunology panel gene expression profiling in the ZIKV RM cohort (n = 4) at day 0 (filled blue circles) or day 3 (open blue circles). (A) Principal-component analysis comparing the two sample groups. The plot represents clustering of the data in a 2-dimensional matrix. Each dot represents one RM. (B) Pathway activation determined by z-score. The top five pathways predicted to be activated (orange) or deactivated (blue) are displayed. Download FIG S4, PDF file, 0.1 MB (134.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcriptional gene expression differences between SIV/ZIKV and ZIKV cohorts at day 0. Download Table S1, PDF file, 0.1 MB (65.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Ingenuity pathway analysis (IPA) of the gene expression profiles revealed a robust type I IFN response to SIV (Fig. 5C and Fig. S4B, ZIKV monoinfection). We compared levels of mRNA for IP-10/CXCL10 in peripheral blood mononuclear cells (from normalized counts from nanostring analysis) to protein levels in plasma (from ELISA) and found a strong, positive correlation (Fig. 5D) (r = 0.9167, P = 0.0013). We further observed an association between levels of IP-10 mRNA at day 0 and ZIKV viremia at day 3 (Fig. 5E), as we did with IP-10 protein levels (Fig. 4C). This association was not limited to IP-10/CXCL10, as we also identified message levels of interferon-stimulated IFIT3 as a predictor of subsequent ZIKV peak viremia (Fig. 5F). To further understand the robustness of these findings, we also examined and found a relationship between ZIKV viremia and levels of IRF7, a central ISG (Fig. 5G). Indeed, IRF7 correlated positively with both CXCL10 and IFIT3 (Fig. 5H) (P = 0.0061 (Fig. 5I) ( P = 0.0108). Taken together, our findings demonstrate that the SIV-induced type I IFN response may lead to diminished ZIKV viremia during coinfection.
DISCUSSION
NHPs are often used as models to study viral vaccine efficacy and antiviral therapeutic safety and efficacy and to understand aspects of viral pathogenesis. Owing to >90% genetic coding conservation between humans and nonhuman primates, viral infections in NHP models result from infections of the same cell types and induce nearly identical pathologies (44). Although NHPs are well-known models for lentiviral and flaviviral infections, to date, their use in coinfection models has been limited. Here, we studied how preexisting SIV infection influences subsequent ZIKV infection. We find that the type I IFN responses elicited in SIV-infected RMs correlates with reduced ZIKV replication, that these coinfected animals mount inferior humoral responses to ZIKV, and that these animals have delayed ZIKV clearance. A recent study, using a small cohort of simian-human immunodeficiency virus (SHIV)- and SIV-infected RMs also found that ZIKV viremia was uncharacteristically low during coinfection; however, the authors concluded that SHIV/SIV infections did not dramatically influence the course of ZIKV infection (64). This small study of only 4 Asian macaques used historical controls and did not study immune responses to ZIKV; thus, the study is somewhat difficult to interpret.
Decreased adaptive immune responses are a hallmark of progressive lentiviral infection (27–33). Although the underlying mechanisms are multifactorial, preferential HIV/SIV infection of follicle-resident CD4+ Tfh cells (reviewed in reference 65), loss of pathogen-specific memory CD4+ T cells (66, 67), exhaustion of adaptive lymphocytes (68, 69), and aberrant inflammation (70) are all described contributors. In HIV and SIV infection, antibody/virus complex trapping by follicular dendritic cell (FDC) networks promotes the increased HIV/SIV exposure and subsequent infection of Tfh cells, and this is exacerbated by an inability of virus-specific CD8+ T cells to enter the follicle (65). Indeed, replication-competent HIV can be recovered from the FDC network for years after HIV-infected individuals are treated with antiretroviral therapy (71). That we also find higher levels of ZIKV RNA in the lymph node follicles than in the paracortex (irrespective of coinfection) (Fig. 1G) coupled with delayed ZIKV clearance (Fig. 1C) in SIV-coinfected animals suggests that replication-competent ZIKV may persist in lymphoid follicles and suggests that similar mechanisms may be operative for other flaviviruses. However, it is unclear which particular types of cells become ZIKV RNA+ in lymphoid follicles or whether FDCs can become infected themselves. Additional work is required.
Consistently with previous reports, our data clearly demonstrate that SIV infection leads to a robust production of ISGs (37–41). While infection with ZIKV was sufficient to induce expression of ISGs, the response generated in acute SIV infection far exceeded that induced by ZIKV (Fig. 4B). The differences in ISG induction may be due to distinct differences between the duration of viremia and magnitude of ZIKV infection, which is typically cleared from plasma within 7 days, and those of HIV/SIV, which is persistent in plasma at a typical set point viral load of 1 million copies per milliliter of plasma. Although we further demonstrate that the level of IP-10/CXCL10 at the time of ZIKV infection predicts subsequent ZIKV replication (Fig. 4C and D), the SIV-generated IFN response was insufficient to completely prevent ZIKV replication. Moreover, recent work suggests that particular ISGs may competitively bind to flavivirus receptors (72–74), which may explain the delayed ZIKV viremia that we observed in SIV+ animals. Paradoxically, this may result from the interferon-responsive genes themselves, in that reduced antigenemia may lead to decreased B cell stimulation and in turn limit the anti-ZIKV antibody responses (75–77), as observed in Fig. 2E. The delayed antibody responses, in turn, may lead to moderately prolonged ZIKV viremia in SIV+ animals. Interestingly, levels of IP-10 have also been shown to differentiate between patients with and without neurological complications after ZIKV infection in Brazil and between infants with and without congenital deformities born to ZIKV-infected mothers, consistent with the idea that IFN signaling is associated with ZIKV pathogenesis (78).
Tfh cells are an important reservoir for HIV/SIV (27–33), and their predicted infection by SIV in our coinfection model may compromise the generation of ZIKV antibodies. A combination of Tfh loss and ISG-mediated hypoantigenemia may be cumulatively operative in decreasing ZIKV-specific NAbs. Although the degree to which lower levels of antibodies influence susceptibility to flaviviral coinfection is unclear, these data coupled with the prolonged ZIKV viremia demonstrate that SIV infection shapes the resultant anti-ZIKV response.
Importantly, we coinfected our RMs during the early phase of SIV infection, before they were clinically symptomatic and prior to the onset of immunodeficiency. Although vaccine immunogenicity is known to be compromised in chronically HIV-infected individuals, our data further suggest that vaccine efficacy may be similarly compromised or biased during hyperacute infection.
Delayed clearance of ZIKV in SIV-infected RMs coupled with the higher levels of virus within the lymphoid follicle might result in prolonged ZIKV transmission risks. This may be related to antibody complex interactions with follicular dendritic cells, as in previous reports of FDC trapping of HIV/SIV virions (71).
Taken together, this study provides mechanistic insights into how simultaneous viral infections of primates can influence one another and highlights the importance of the innate arm of the immune system in limiting RNA virus replication in vivo. These data also highlight that perturbations to immune responses to secondary infections can be influenced by immunodeficiency virus infection, even before progression to AIDS.
MATERIALS AND METHODS
Sample processing.
Whole blood was centrifuged for plasma collection, and subsequently, peripheral blood mononuclear cells (PBMCs) were isolated by standard density centrifugation and cryopreserved. LN biopsy samples were collected from axillary or inguinal sites and then either processed into single-cell suspensions for flow cytometry analysis or fixed in 4% paraformaldehyde and paraffin embedded for immunohistochemistry (79). For flow cytometry experiments, all time points from a specific animal were stained and run together after their collection on day 56. For all other experiments, assayed time points were run within the same experiment.
Plasma viral-load assessment.
SIV viral RNA (vRNA) levels in plasma were determined by real-time RT-PCR using the ABI Prism 7900 sequence detection system (Applied Biosystems) as previously described (79). Primer pairs for the assay corresponded to the forward nucleotides 1181 to 1208 and reverse nucleotides1338 to 1317 of the SIVmac239 gag gene (80).
To assess ZIKV RNA levels in the plasma of our study RMs, viral RNA was extracted using EZ1 virus minikit v2.0 (Qiagen), and cDNA was synthesized with the SuperScript III first-strand synthesis system (Life Technologies). Absolute quantification of virus genome equivalents (GE; log10 per milliliter) was determined by real-time PCR using a TaqMan probe (Integrated DNA Technologies) targeting the ZIKV E protein. ZIKV RNA levels were measured in all animals to at least day 11 post-ZIKV infection. All animals, except RHDFiV (SIV/ZIKV cohort) measured below the limit of detection (LOD) of 3.2 log10 GE/ml at 11 days postinfection (dpi).
Immunophenotyping and cellular functionality profiling.
Isolated PBMCs were used for multicolor flow analysis and cellular sorting using monoclonal antibodies (MAbs) with specific cross-reactivities to RM antigens. All PBMC samples at various time points from a specific animal were stained and run at one time. Cellular viability was assessed by Live/Dead Aqua (Invitrogen) fixable dead-cell staining, followed by staining with fluorescently conjugated MAbs against CCR5 phycoerythrin (PE) (clone 3A9), CD20 PE-Cy7 (L27), CD27 V450 (M-T271), CD3 Alx700 (SP34-2), HLA-DR allophycocyanin (APC)-H7 (L243), CD11b/MAC-1 BV605 (ICRF44), CD16 BV711 (3G8), and CD14 BV786 (M5E2) from BD Biosciences, CD28 ECD (28.2) and NKG2a APC (Z199) from Beckman Coulter, CD95 PE-Cy5 (DX2) and CD8a peridinin chlorophyll protein (PerCP)-Cyanine5.5 (RPA-T8) from eBioscience/Invitrogen, and CD4 BV650 from BioLegend to determine immunophenotype. Cells were then permeabilized (Cytofix/Cytoperm; BD Biosciences), and their proliferation was assessed via Ki-67 fluorescein isothiocyanate (FITC) (clone B56; BD Biosciences) staining. Cellular phenotypes were defined as outlined in Fig. S3 in the supplemental material.
Cellular functionality was assessed after overnight stimulation at 37°C with phorbol myristate acetate (PMA; 2.5 ng/ml) and ionomycin (1 μM) in the presence of brefeldin A (1 μg/ml; Sigma-Aldrich) using the following MAbs (in addition to or intermixed with the above panel): IFN-γ V450 (B27) from BD Biosciences, NKG2a PE (Z199) from Beckman Coulter, TNF-α BV605 (MAb11) and IL-2 BV785 (MQ1-17H12) from BioLegend, and IL-22 APC (IL22JOP), IL-17 Alx488 (eBio64DEC17), and CD154 APC-eFluor780 (24-31) from eBioscience/Invitrogen. All samples were run on an LSRFortessa apparatus (BD Biosciences) and analyzed using FlowJo software version 9.9.5 (TreeStar, Ashland, OR). Any cell subset analysis that was based on fewer than 200 cells of the parent population was not included.
RVP production (for neutralization assays).
Reporter virus particles (RVPs) incorporating the structural proteins of ZIKV (strain H/PF/2013) (81), dengue virus 2 (DENV2; strain 16681) (82), or West Nile virus (WNV) lineage I (strain NY99) (83) were produced by complementation of a subgenomic green fluorescent protein (GFP)-expressing replicon derived from a lineage II strain of WNV as previously described (81–84). To determine virus titer, 2-fold dilutions of RVPs were used to infect Raji cells that express the flavivirus attachment factor DC-SIGNR (Raji-DCSIGNR cells) (85) in duplicate technical replicates at 37°C. GFP-positive infected cells were detected by flow cytometry 2 days later. In subsequent neutralization assays, RVPs were sufficiently diluted to within the linear range of the virus infectivity dose-response curve to ensure antibody excess at informative points.
Neutralization assays.
For neutralization studies, ZIKV RVPs were mixed with serial dilutions of heat-inactivated macaque plasma for 1 h at 37°C, followed by infection of Raji-DCSIGNR cells in duplicate technical replicates. Infections were carried out at 37°C, and GFP-positive infected cells were quantified by flow cytometry 2 days later. Results were analyzed by nonlinear regression analysis to estimate the concentration of plasma required to inhibit 50% of infection (IC50). For neutralization assays, all plasma samples were initially tested at a starting dilution of 1:60 (based on the final volume of cells, virus, and plasma per well), which was designated the limit of detection.
ZIKV infection assessment in LN cells.
Lymph node cells from day 5 and day 27 post-ZIKV infection were sorted into T cell, B cell, and macrophage populations (Fig. S2) for assessment of ZIKV viral RNA via qRT-PCR. RNA was extracted from sorted cells using the Qiagen RNeasy kit (Qiagen) with on-column DNase I treatment per the manufacturer’s instructions. ZIKV positive-strand RNA was measured by qRT-PCR using the primers and probe described by Lanciotti et al., with a slightly modified probe to more closely correspond to the Nicaragua ZIKV sequence (86). The following primer and probe sequences were used: forward, 5′-CCGCTGCCCAACACAAG-3′; reverse, 5′-CCACTAACGTTCTTTTGCAGACAT-3′; and probe, 5′-6-carboxyfluorescein (6FAM)-AGCCTACCTTGACAAGCAATCAGACACTCAA-MGBNFQ-3′. The qRT-PCR was performed in triplicate using Applied Biosystems TaqMan fast virus 1-step master mix and a QuantStudio 6 instrument (ThermoFisher). A standard curve was generated using in vitro-transcribed ZIKV RNA using an Ambion MEGAscript kit (ThermoFisher) and a pCRII-TOPO plasmid containing French Polynesia ZIKV bases 700 to 1500 (kindly provided by Thomas C. Friedrich, University of Wisconsin, Madison, WI). The standard curve was used to extrapolate the number of copies of ZIKV RNA per 1,000 cells. Samples were considered positive if at least one of the triplicate wells had a threshold cycle (CT) value of less than 38. If not all of the wells of the triplicate sample were positive, then any positive CTs from that sample were averaged and divided by 3 to calculate the copy number.
IFN response assessment.
IP-10 levels were determined in plasma using the CXCL10/IP-10 human Quantikine enzyme-linked immunosorbent assay (ELISA; R&D Systems). Plasma from day −14 to day 40 (SIV/ZIKV cohort) and day 0 to day 40 (ZIKV cohort) were run at a 1:2 dilution in duplicate.
In situ hybridization.
Rhesus macaque LNs were biopsied 5 or 27 days post-ZIKV infection in both the ZIKV and SIV/ZIKV cohorts. LNs were then fixed overnight in 4% paraformaldehyde at 4°C. LNs were then dehydrated by washing them in sequentially increasing concentrations of ethanol, embedded in paraffin, and allowed to sit overnight at 4°C. LNs were sectioned at 5 μm (Histoserv). Sectioned slides were deparaffinized in a dry oven for 1 h at 60°C, followed by sequential washes in 100% xylene, ethanol of decreasing concentrations, and distilled water (dH2O). Antigen retrieval was performed by steaming slides immersed in a 0.5% citraconic acid solution (pH 7.4) at 95°C for 20 min, followed by two 10-min rinses in dH2O. Sections were treated with Protease Plus pretreatment solution (Advanced Cell Diagnostics) diluted 1:5 in cold phosphate-buffered saline (PBS) for 20 min at 40°C, rinsed twice in dH2O, and then treated with hydrogen peroxide for 10 min at room temperature and rinsed twice in dH2O. Fluorescent in situ hybridization was conducted using an RNAscope 2.5 HD Brown detection reagent kit (Advanced Cell Diagnostics) according to the manufacturer’s suggestions. ZIKV RNA was detected using the V-ZIKA-pp RNAscope probe, and SIV RNA was detected using the SIVmac239 RNAscope probe (Advanced Cell Diagnostics). Following the final amplification step, tissue was stained with Trypticase soy agar (TSA) plus cyanine 3.5 amplification reagent (Perkin Elmer).
After RNA-fluorescent in situ hybridization (FISH), sections were blocked overnight at 4°C in washing/blocking solution (0.1 M Tris-HCl [Quality Biological], 0.1%, vol/vol, Tween 20 [Sigma-Aldrich], 0.5%, vol/vol, cold water fish skin gelatin [Sigma-Aldrich]). Sections were stained with anti-CD68 (KP1; BioLegend) or anti-CD20 (L26; Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI) as a counterstain.
Primary-antibody staining was detected with Alexa Fluor 568 or 647 goat anti-mouse IgG1 (Invitrogen). Some sections were stained with anti-ZIKV envelope (BioFront Technologies) for comparative analysis. Stained tissue was treated with 10 mM CuSO4-50 mM NH4Cl (pH 5.0), TrueBlack lipofuscin quencher (Biotium), and TrueVIEW (Vector Laboratories) to quench autofluorescence. Slides were then mounted using Fluoromount G (Electron Microscopy Sciences).
Confocal imaging and analysis.
Stained LN sections were imaged on an inverted SP5 confocal microscope (Leica Microsystems), and images were analyzed using Imaris software (Bitplane). Three-dimensional (3D) surfaces were constructed according to an absolute intensity threshold for fluorescent channels detecting either CD20 (B cell follicle), DAPI (all tissue), or the RNA-FISH probe (ZIKV/SIV RNA). Summed volumes of constructed 3D surfaces were then calculated. 3D surface volume overlap was determined manually. ZIKV/SIV RNA quantifications were normalized as a function of their total tissue volume (e.g., cubic micrometers of the RNA-FISH probe per cubic micrometer of DAPI).
RNA expression profiling.
RNA was isolated from 3 million PBMCs for each animal at day 0 and day 3 post-ZIKV infection using the MagMAX-96 total RNA isolation kit (Thermo Fisher Scientific). Each RNA sample was normalized to 50 ng/μl. Preparation, hybridization, and detection of RNA samples were carried out by following the NanoString manufacturer’s instructions (NanoString Technologies). Subsequent analyses were performed using the nCounter analysis system (NanoString Technologies) and TM4 MeV microarray software suite (http://mev.tm4.org). Principal-component analysis was performed with the R package FactoMineR at default settings. Reads from NanoString were normalized to internal positive and negative controls and housekeeping genes. For ingenuity pathway analysis (IPA), log2 fold change values (SIV+ day 3/SIV– day 0) were uploaded to the IPA software (Qiagen) to determine potential upstream regulators.
Quantification and statistical analysis.
All analyses were performed using Prism v7.0 software (GraphPad). Mann-Whitney U tests were used when Zika mean titers and infection cell frequencies were compared between cohorts. The Wilcoxon matched-pair signed-rank test was used for cell subset comparisons between time points within the same cohort (i.e., 1B, P = 0.0156; memory CD4+ T cell count preinfection to postinfection). AUC analysis for phenotypic proliferation analysis was carried out from day 2 to day 21 post-ZIKV infection for each animal, and then group cohort comparisons were made using the Mann-Whitney U test (Fig. 2). AUC analysis for Zika mean titers was carried out similarly, except that it was limited to the curve defined by day 2 to day 7 of ZIKV infection (Fig. 1C). Comparisons of ZIKV 50% effective concentrations (EC50s) were analyzed via 2-way analysis of variance (ANOVA) of log-transformed data (Fig. 2E). Responses to SIV and ZIKV were determined by repeated-measures one-way ANOVA analysis (Fig. 2A to D and 4B). Pie charts and polyfunctional capability comparisons were assessed using SPICE v5.3 (National Institute of Allergy and Infectious Diseases), with the permutations set at 10,000 (Fig. 3). The P values included within the SPICE analysis reflect the differences between T cell polyfunctionality based on the expression of different combinations of cytokines and not differences in the total number of functions (e.g., 1+ function, 2+ functions). Spearman’s rank-transformed correlation analysis was utilized to evaluate any associations between immune parameters (Fig. 4A, C, and D and Fig. 5D to I).
Contact for reagent and resource sharing.
Further information and requests for resources and reagents should be directed to Jason Brenchley.
Experimental model and subject details.
Nineteen healthy RMs (Macaca mulatta) were randomly divided into 2 cohorts, consisting of 7 RMs that were first infected with SIVmac239 (3,000 TCID50 intravenously) (87) and then subsequently challenged subcutaneously on day 21 post-SIV infection with Zika virus isolate Nicaragua/2016 (UCB 7420) at 104 PFU (SIV/ZIKV cohort); 12 RMs were infected with the Zika virus isolate Nicaragua/2016 alone (ZIKV cohort) (Table 1). This ZIKV challenge virus was chosen due to its relatively high viral replication in RMs. The original isolate (UCB 7420) was from patient serum collected at the University of California, Berkley (kindly provided by Eva Harris, GenBank accession number MN577550). The cohort sizes were based on previous studies of experimental manipulations of SIV progression in Asian macaques (45, 79, 88). All animals for this coinfection study were between 4.7 and 17 kg and 4 and 18 years of age (Table 1).
All study macaques were housed in certified facilities and cared for in accordance with standards recommended by the American Association for the Accreditation of Laboratory Animal Care (AAALAC). All procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the National Institute of Allergy and Infectious Diseases (animal protocol number LVD26). All procedures were carried out under ketamine anesthesia by trained personnel with veterinary staff supervision. This study followed recommended guidelines outlined in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (89) as well as the 2006 Weatherall report (90). RMs were fed twice daily with standard commercial monkey chow and produce and provided with continuous access to water. Each of the animals was singly housed in adjoining cages to allow for noncontact social interaction. Environmental enrichment was provided in the form of primate puzzle feeders, mirrors, and other appropriate toys.
Peripheral blood and lymph node biopsy specimens were collected pre- and post-ZIKV infection as shown in Fig. 1A. These sampling time points were chosen to focus extensively on early ZIKV infection prior to clearance from plasma circulation, as well as to include later time points in order to determine viral immune response dynamics and persistence. Study animals were prospectively screened and, with the exception of RHA0P003, free of the presence of antibodies for West Nile virus, dengue virus, and ZIKV prior to study infection. Neutralizing antibody titers from RHA0P003 were left out of the analysis in Fig. 2E due to clear evidence of prior WNV infection detected in a prospective antibody-dependent enhancement WNV reporter virus particle assay. The NAb titers from this animal were high enough to drive significant differences at day 7 (a 17.8-fold higher average NAb titer in non-SIV-infected RMs), which was no longer significant when the RHA0P003 titers were left out.
ACKNOWLEDGMENTS
We thank Heather Kendall, JoAnne Swerczek, Richard Herbert, and all the exceptional veterinary staff at the National Institutes of Health Animal Center. We thank Shurjo Sen, Giorgio Trinchieri, and the Genomics Laboratory/Leidos Biomedical Research, Inc., for technical assistance with the NanoString NHP assay.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Funding for this study was provided in part by the Division of Intramural Research/NIAID/NIH.
J.M.B. conceived the project. C.L.V. and J.M.B. designed the project and wrote the manuscript. C.L.V., K.A.D., S.M.B., S.S.W., T.C.P., H.D.H., and J.M.B. provided input for study methodology/assays. C.L.V., S.J.M., K.A.D., E.A.-C., E.P.K., S.J.R., A.M.O., and C.E.S. performed experiments. C.L.V., S.J.M., K.A.D., E.A.-C., E.P.K., S.J.R., J.C.M., and J.M.B. analyzed the data.
We declare no competing interests.
Footnotes
This article is a direct contribution from Jason M. Brenchley, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Ann Chahroudi, Emory University School of Medicine, and Nick Mannes, Tulane.
Citation Vinton CL, Magaziner SJ, Dowd KA, Robertson SJ, Amaro-Carambot E, Karmele EP, Ortiz AM, Starke CE, Mudd JC, Whitehead SS, Best SM, Pierson TC, Hickman HD, Brenchley JM. 2019. Simian immunodeficiency virus infection of rhesus macaques results in delayed Zika virus clearance. mBio 10:e02790-19. https://doi.org/10.1128/mBio.02790-19.
REFERENCES
- 1.Faria NR, Azevedo RDSDS, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami FGDS, Macedo FLDL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, Medeiros DBDA, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, Simith DDB, Messina JP, Abade L, Lourenço J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, Lemos PDS, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Júnior JLDSG, Mir D, Bello G, Delatorre E, Khan K, et al. 2016. Zika virus in the Americas: early epidemiological and genetic findings. Science 352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q, Sun K, Chinazzi M, Pastore YPA, Dean NE, Rojas DP, Merler S, Mistry D, Poletti P, Rossi L, Bray M, Halloran ME, Longini IM Jr, Vespignani A. 2017. Spread of Zika virus in the Americas. Proc Natl Acad Sci U S A 114:E4334–E4343. doi: 10.1073/pnas.1620161114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbi L, Coelho AVC, Alencar LCA, Crovella S. 2018. Prevalence of Guillain-Barre syndrome among Zika virus infected cases: a systematic review and meta-analysis. Braz J Infect Dis 22:137–141. doi: 10.1016/j.bjid.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Oliveira WK, de Franca GVA, Carmo EH, Duncan BB, de Souza Kuchenbecker R, Schmidt MI. 2017. Infection-related microcephaly after the 2015 and 2016 Zika virus outbreaks in Brazil: a surveillance-based analysis. Lancet 390:861–870. doi: 10.1016/S0140-6736(17)31368-5. [DOI] [PubMed] [Google Scholar]
- 5.Nascimento OJM, da Silva I. 2017. Guillain-Barre syndrome and Zika virus outbreaks. Curr Opin Neurol 30:500–507. doi: 10.1097/WCO.0000000000000471. [DOI] [PubMed] [Google Scholar]
- 6.Samy AM, Thomas SM, Wahed AA, Cohoon KP, Peterson AT. 2016. Mapping the global geographic potential of Zika virus spread. Mem Inst Oswaldo Cruz 111:559–560. doi: 10.1590/0074-02760160149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anonymous. 2017. Prevalence of HIV among adults aged 15 to 49, 2017 by WHO region. (IER) WHO, Geneva, Switzerland: http://apps.who.int/gho/data/view.main.22500. [Google Scholar]
- 8.Larocca RA, Abbink P, Peron JPS, Zanotto PMDA, Iampietro MJ, Badamchi-Zadeh A, Boyd M, Ng'ang'a D, Kirilova M, Nityanandam R, Mercado NB, Li Z, Moseley ET, Bricault CA, Borducchi EN, Giglio PB, Jetton D, Neubauer G, Nkolola JP, Maxfield LF, De La Barrera RA, Jarman RG, Eckels KH, Michael NL, Thomas SJ, Barouch DH. 2016. Vaccine protection against Zika virus from Brazil. Nature 536:474–478. doi: 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. 2016. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg 94:1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep 17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thurmond S, Wang B, Song J, Hai R. 2018. Suppression of type I interferon signaling by flavivirus NS5. Viruses 10:E712. doi: 10.3390/v10120712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alves Dos Santos E, Fink K. 2018. Animal models for dengue and Zika vaccine development. Adv Exp Med Biol 1062:215–239. doi: 10.1007/978-981-10-8727-1_16. [DOI] [PubMed] [Google Scholar]
- 15.Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH III, Ruckert C, Young MC, Zhu Z, Robertson SJ, McNally KL, Ye J, Cao B, Mysorekar IU, Ebel GD, Baric RS, Best SM, Artyomov MN, Garcia-Sastre A, Diamond MS. 2018. An immunocompetent mouse model of Zika virus infection. Cell Host Microbe 23:672–685.e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John SP, Chin CR, Perreira JM, Feeley EM, Aker AM, Savidis G, Smith SE, Elia AE, Everitt AR, Vora M, Pertel T, Elledge SJ, Kellam P, Brass AL. 2013. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J Virol 87:7837–7852. doi: 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, Brass AL. 2016. The IFITMs inhibit Zika virus replication. Cell Rep 15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 19.Abbink P, Stephenson KE, Barouch DH. 2018. Zika virus vaccines. Nat Rev Microbiol 16:594–600. doi: 10.1038/s41579-018-0039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richner JM, Diamond MS. 2018. Zika virus vaccines: immune response, current status, and future challenges. Curr Opin Immunol 53:130–136. doi: 10.1016/j.coi.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond MS. 2003. Evasion of innate and adaptive immunity by flaviviruses. Immunol Cell Biol 81:196–206. doi: 10.1046/j.1440-1711.2003.01157.x. [DOI] [PubMed] [Google Scholar]
- 22.Crowe SM, Carlin JB, Stewart KI, Lucas CR, Hoy JF. 1991. Predictive value of CD4 lymphocyte numbers for the development of opportunistic infections and malignancies in HIV-infected persons. J Acquir Immune Defic Syndr 4:770–776. [PubMed] [Google Scholar]
- 23.Ruegg CL, Engleman EG. 1990. Impaired immunity in AIDS. The mechanisms responsible and their potential reversal by antiviral therapy. Ann N Y Acad Sci 616:307–317. doi: 10.1111/j.1749-6632.1990.tb17851.x. [DOI] [PubMed] [Google Scholar]
- 24.Ortona L, Tamburrini E, Tumbarello M, Ventura G, Cauda R. 1988. Natural killer (NK) activity in patients with HIV infection. Boll Ist Sieroter Milan 67:135–141. (In Italian.) [PubMed] [Google Scholar]
- 25.Overton ET, Nurutdinova D, Sungkanuparph S, Seyfried W, Groger RK, Powderly WG. 2007. Predictors of immunity after hepatitis A vaccination in HIV-infected persons. J Viral Hepat 14:189–193. doi: 10.1111/j.1365-2893.2006.00822.x. [DOI] [PubMed] [Google Scholar]
- 26.Malaspina A, Moir S, Orsega SM, Vasquez J, Miller NJ, Donoghue ET, Kottilil S, Gezmu M, Follmann D, Vodeiko GM, Levandowski RA, Mican JM, Fauci AS. 2005. Compromised B cell responses to influenza vaccination in HIV-infected individuals. J Infect Dis 191:1442–1450. doi: 10.1086/429298. [DOI] [PubMed] [Google Scholar]
- 27.Klatt NR, Vinton CL, Lynch RM, Canary LA, Ho J, Darrah PA, Estes JD, Seder RA, Moir SL, Brenchley JM. 2011. SIV infection of rhesus macaques results in dysfunctional T- and B-cell responses to neo and recall Leishmania major vaccination. Blood 118:5803–5812. doi: 10.1182/blood-2011-07-365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moysi E, Pallikkuth S, De Armas LR, Gonzalez LE, Ambrozak D, George V, Huddleston D, Pahwa R, Koup RA, Petrovas C, Pahwa S. 2018. Altered immune cell follicular dynamics in HIV infection following influenza vaccination. J Clin Invest 128:3171–3185. doi: 10.1172/JCI99884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moysi E, Petrovas C, Koup RA. 2018. The role of follicular helper CD4 T cells in the development of HIV-1 specific broadly neutralizing antibody responses. Retrovirology 15:54. doi: 10.1186/s12977-018-0437-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovas C, Koup RA. 2014. T follicular helper cells and HIV/SIV-specific antibody responses. Curr Opin HIV AIDS 9:235–241. doi: 10.1097/COH.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 32.Poultsidi A, Dimopoulos Y, He TF, Chavakis T, Saloustros E, Lee PP, Petrovas C. 2018. Lymph node cellular dynamics in cancer and HIV: what can we learn for the follicular CD4 (Tfh) cells? Front Immunol 9:2233. doi: 10.3389/fimmu.2018.02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, Ollerton MT, Connick E. 2018. Follicular T-cell subsets in HIV infection: recent advances in pathogenesis research. Curr Opin HIV AIDS 14:71–76. doi: 10.1097/COH.0000000000000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenchley JM. 2013. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol 6:657–665. doi: 10.1038/mi.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klatt NR, Chomont N, Douek DC, Deeks SG. 2013. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev 254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klatt NR, Funderburg NT, Brenchley JM. 2013. Microbial translocation, immune activation, and HIV disease. Trends Microbiol 21:6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle T, Goujon C, Malim MH. 2015. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol 13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris LD, Tabb B, Sodora DL, Paiardini M, Klatt NR, Douek DC, Silvestri G, Muller-Trutwin M, Vasile-Pandrea I, Apetrei C, Hirsch V, Lifson J, Brenchley JM, Estes JD. 2010. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol 84:7886–7891. doi: 10.1128/JVI.02612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roff SR, Noon-Song EN, Yamamoto JK. 2014. The significance of interferon-gamma in HIV-1 pathogenesis, therapy, and prophylaxis. Front Immunol 4:498. doi: 10.3389/fimmu.2013.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, Hill BJ, Timmer JK, Reiss E, Yarden G, Darko S, Contijoch E, Todd JP, Silvestri G, Nason M, Norgren RB Jr, Keele BF, Rao S, Langer JA, Lifson JD, Schreiber G, Douek DC. 2014. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosinger SE, Utay NS. 2015. Type I interferon: understanding its role in HIV pathogenesis and therapy. Curr HIV/AIDS Rep 12:41–53. doi: 10.1007/s11904-014-0244-6. [DOI] [PubMed] [Google Scholar]
- 42.Bradley MP, Nagamine CM. 2017. Animal models of Zika virus. Comp Med 67:242–252. [PMC free article] [PubMed] [Google Scholar]
- 43.Dudley DM, Aliota MT, Mohr EL, Weiler AM, Lehrer-Brey G, Weisgrau KL, Mohns MS, Breitbach ME, Rasheed MN, Newman CM, Gellerup DD, Moncla LH, Post J, Schultz-Darken N, Schotzko ML, Hayes JM, Eudailey JA, Moody MA, Permar SR, O'Connor SL, Rakasz EG, Simmons HA, Capuano S, Golos TG, Osorio JE, Friedrich TC, O'Connor DH. 2016. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun 7:12204. doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estes JD, Wong SW, Brenchley JM. 2018. Nonhuman primate models of human viral infections. Nat Rev Immunol 18:390–404. doi: 10.1038/s41577-018-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klatt NR, Canary LA, Vanderford TH, Vinton CL, Engram JC, Dunham RM, Cronise HE, Swerczek JM, Lafont BA, Picker LJ, Silvestri G, Brenchley JM. 2012. Dynamics of simian immunodeficiency virus SIVmac239 infection in pigtail macaques. J Virol 86:1203–1213. doi: 10.1128/JVI.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, Zeng M, Masopust D, Carlis JV, Ran L, Vanderford TH, Paiardini M, Isett RB, Baldwin DA, Else JG, Staprans SI, Silvestri G, Haase AT, Kelvin DJ. 2009. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest 119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aid M, Abbink P, Larocca RA, Boyd M, Nityanandam R, Nanayakkara O, Martinot AJ, Moseley ET, Blass E, Borducchi EN, Chandrashekar A, Brinkman AL, Molloy K, Jetton D, Tartaglia LJ, Liu J, Best K, Perelson AS, De La Barrera RA, Lewis MG, Barouch DH. 2017. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell 169:610–620.e14. doi: 10.1016/j.cell.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coffey LL, Pesavento PA, Keesler RI, Singapuri A, Watanabe J, Watanabe R, Yee J, Bliss-Moreau E, Cruzen C, Christe KL, Reader JR, von Morgenland W, Gibbons AM, Allen AM, Linnen J, Gao K, Delwart E, Simmons G, Stone M, Lanteri M, Bakkour S, Busch M, Morrison J, Van Rompay KK. 2017. Zika virus tissue and blood compartmentalization in acute infection of rhesus macaques. PLoS One 12:e0171148. doi: 10.1371/journal.pone.0171148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirsch AJ, Smith JL, Haese NN, Broeckel RM, Parkins CJ, Kreklywich C, DeFilippis VR, Denton M, Smith PP, Messer WB, Colgin LM, Ducore RM, Grigsby PL, Hennebold JD, Swanson T, Legasse AW, Axthelm MK, MacAllister R, Wiley CA, Nelson JA, Streblow DN. 2017. Zika virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLoS Pathog 13:e1006219. doi: 10.1371/journal.ppat.1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li XF, Dong HL, Huang XY, Qiu YF, Wang HJ, Deng YQ, Zhang NN, Ye Q, Zhao H, Liu ZY, Fan H, An XP, Sun SH, Gao B, Fa YZ, Tong YG, Zhang FC, Gao GF, Cao WC, Shi PY, Qin CF. 2016. Characterization of a 2016 clinical isolate of Zika virus in non-human primates. EBioMedicine 12:170–177. doi: 10.1016/j.ebiom.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Connor MA, Tisoncik-Go J, Lewis TB, Miller CJ, Bratt D, Moats CR, Edlefsen PT, Smedley J, Klatt NR, Gale M, Fuller DH. 2018. Early cellular innate immune responses drive Zika viral persistence and tissue tropism in pigtail macaques. Nat Commun 9:3371. doi: 10.1038/s41467-018-05826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osuna CE, Lim SY, Deleage C, Griffin BD, Stein D, Schroeder LT, Omange RW, Best K, Luo M, Hraber PT, Andersen-Elyard H, Ojeda EF, Huang S, Vanlandingham DL, Higgs S, Perelson AS, Estes JD, Safronetz D, Lewis MG, Whitney JB. 2016. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat Med 22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silveira ELV, Rogers KA, Gumber S, Amancha P, Xiao P, Woollard SM, Byrareddy SN, Teixeira MM, Villinger F. 2017. Immune cell dynamics in rhesus macaques infected with a Brazilian strain of Zika virus. J Immunol 199:1003–1011. doi: 10.4049/jimmunol.1700256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klatt NR, Brenchley JM. 2010. Th17 cell dynamics in HIV infection. Curr Opin HIV AIDS 5:135–140. doi: 10.1097/COH.0b013e3283364846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryan ES, Micci L, Fromentin R, Paganini S, McGary CS, Easley K, Chomont N, Paiardini M. 2016. Loss of function of intestinal IL-17 and IL-22 producing cells contributes to inflammation and viral persistence in SIV-infected rhesus macaques. PLoS Pathog 12:e1005412. doi: 10.1371/journal.ppat.1005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, Tabb B, Canary LA, Dang Q, Hirsch VM, Alter G, Belkaid Y, Lifson JD, Silvestri G, Milner JD, Paiardini M, Haddad EK, Brenchley JM. 2012. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol 5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abbink P, Larocca RA, Visitsunthorn K, Boyd M, De La Barrera RA, Gromowski GD, Kirilova M, Peterson R, Li Z, Nanayakkara O, Nityanandam R, Mercado NB, Borducchi EN, Chandrashekar A, Jetton D, Mojta S, Gandhi P, LeSuer J, Khatiwada S, Lewis MG, Modjarrad K, Jarman RG, Eckels KH, Thomas SJ, Michael NL, Barouch DH. 2017. Durability and correlates of vaccine protection against Zika virus in rhesus monkeys. Sci Transl Med 9:eaao4163. doi: 10.1126/scitranslmed.aao4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson JF, Rahal JJ. 2002. Efficacy of interferon alpha-2b and ribavirin against West Nile virus in vitro. Emerg Infect Dis 8:107–108. doi: 10.3201/eid0801.010252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crance JM, Scaramozzino N, Jouan A, Garin D. 2003. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antiviral Res 58:73–79. doi: 10.1016/s0166-3542(02)00185-7. [DOI] [PubMed] [Google Scholar]
- 61.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. 2000. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol 74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snyder B, Goebel S, Koide F, Ptak R, Kalkeri R. 2018. Synergistic antiviral activity of Sofosbuvir and type-I interferons (alpha and beta) against Zika virus. J Med Virol 90:8–12. doi: 10.1002/jmv.24932. [DOI] [PubMed] [Google Scholar]
- 63.Barros JBS, da Silva PAN, Koga RCR, Gonzalez-Dias P, Carmo Filho JR, Nagib PRA, Coelho V, Nakaya HI, Fonseca SG, Pfrimer I. 2018. Acute Zika virus infection in an endemic area shows modest proinflammatory systemic immunoactivation and cytokine-symptom associations. Front Immunol 9:821. doi: 10.3389/fimmu.2018.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bidokhti MRM, Dutta D, Madduri LSV, Woollard SM, Norgren R Jr, Giavedoni L, Byrareddy SN. 2018. SIV/SHIV-Zika co-infection does not alter disease pathogenesis in adult non-pregnant rhesus macaque model. PLoS Negl Trop Dis 12:e0006811. doi: 10.1371/journal.pntd.0006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pallikkuth S, de Armas L, Rinaldi S, Pahwa S. 2017. T follicular helper cells and B cell dysfunction in aging and HIV-1 infection. Front Immunol 8:1380. doi: 10.3389/fimmu.2017.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 67.Geldmacher C, Schuetz A, Ngwenyama N, Casazza JP, Sanga E, Saathoff E, Boehme C, Geis S, Maboko L, Singh M, Minja F, Meyerhans A, Koup RA, Hoelscher M. 2008. Early depletion of Mycobacterium tuberculosis-specific T helper 1 cell responses after HIV-1 infection. J Infect Dis 198:1590–1598. doi: 10.1086/593017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Appay V, Sauce D. 2017. Assessing immune aging in HIV-infected patients. Virulence 8:529–538. doi: 10.1080/21505594.2016.1195536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saeidi A, Zandi K, Cheok YY, Saeidi H, Wong WF, Lee CYQ, Cheong HC, Yong YK, Larsson M, Shankar EM. 2018. T-cell exhaustion in chronic infections: reversing the state of exhaustion and reinvigorating optimal protective immune responses. Front Immunol 9:2569. doi: 10.3389/fimmu.2018.02569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overton ET. 2007. An overview of vaccinations in HIV. Curr HIV/AIDS Rep 4:105–113. doi: 10.1007/s11904-007-0016-7. [DOI] [PubMed] [Google Scholar]
- 71.Keele BF, Tazi L, Gartner S, Liu Y, Burgon TB, Estes JD, Thacker TC, Crandall KA, McArthur JC, Burton GF. 2008. Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J Virol 82:5548–5561. doi: 10.1128/JVI.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen JP, Lu HL, Lai SL, Campanella GS, Sung JM, Lu MY, Wu-Hsieh BA, Lin YL, Lane TE, Luster AD, Liao F. 2006. Dengue virus induces expression of CXC chemokine ligand 10/IFN-gamma-inducible protein 10, which competitively inhibits viral binding to cell surface heparan sulfate. J Immunol 177:3185–3192. doi: 10.4049/jimmunol.177.5.3185. [DOI] [PubMed] [Google Scholar]
- 73.Kim SY, Zhao J, Liu X, Fraser K, Lin L, Zhang X, Zhang F, Dordick JS, Linhardt RJ. 2017. Interaction of Zika virus envelope protein with glycosaminoglycans. Biochemistry 56:1151–1162. doi: 10.1021/acs.biochem.6b01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med 3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 75.Dowd KA, Ko SY, Morabito KM, Yang ES, Pelc RS, DeMaso CR, Castilho LR, Abbink P, Boyd M, Nityanandam R, Gordon DN, Gallagher JR, Chen X, Todd JP, Tsybovsky Y, Harris A, Huang YS, Higgs S, Vanlandingham DL, Andersen H, Lewis MG, De La Barrera R, Eckels KH, Jarman RG, Nason MC, Barouch DH, Roederer M, Kong WP, Mascola JR, Pierson TC, Graham BS. 2016. Rapid development of a DNA vaccine for Zika virus. Science 354:237–240. doi: 10.1126/science.aai9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaudinski MR, Houser KV, Morabito KM, Hu Z, Yamshchikov G, Rothwell RS, Berkowitz N, Mendoza F, Saunders JG, Novik L, Hendel CS, Holman LA, Gordon IJ, Cox JH, Edupuganti S, McArthur MA, Rouphael NG, Lyke KE, Cummings GE, Sitar S, Bailer RT, Foreman BM, Burgomaster K, Pelc RS, Gordon DN, DeMaso CR, Dowd KA, Laurencot C, Schwartz RM, Mascola JR, Graham BS, Pierson TC, Ledgerwood JE, Chen GL, VRC 319, VRC 320 study teams. 2018. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet 391:552–562. doi: 10.1016/S0140-6736(17)33105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pierson TC, Graham BS. 2016. Zika virus: immunity and vaccine development. Cell 167:625–631. doi: 10.1016/j.cell.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kam YW, Leite JA, Lum FM, Tan JJL, Lee B, Judice CC, Teixeira DAT, Andreata-Santos R, Vinolo MA, Angerami R, Resende MR, Freitas ARR, Amaral E, Junior RP, Costa ML, Guida JP, Arns CW, Ferreira LCS, Renia L, Proenca-Modena JL, Ng LFP, Costa FTM, Zika-Unicamp N. 2017. Specific biomarkers associated with neurological complications and congenital central nervous system abnormalities from Zika virus-infected patients in Brazil. J Infect Dis 216:172–181. doi: 10.1093/infdis/jix261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ortiz AM, Flynn JK, DiNapoli SR, Vujkovic-Cvijin I, Starke CE, Lai SH, Long ME, Sortino O, Vinton CL, Mudd JC, Johnston L, Busman-Sahay K, Belkaid Y, Estes JD, Brenchley JM. 2018. Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat Med 24:1313–1316. doi: 10.1038/s41591-018-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hansen SG, Piatak M, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Fruh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. 2017. Addendum: immune clearance of highly pathogenic SIV infection. Nature 547:123–124. doi: 10.1038/nature22984. [DOI] [PubMed] [Google Scholar]
- 81.Dowd KA, DeMaso CR, Pelc RS, Speer SD, Smith ARY, Goo L, Platt DJ, Mascola JR, Graham BS, Mulligan MJ, Diamond MS, Ledgerwood JE, Pierson TC. 2016. Broadly neutralizing activity of Zika virus-immune sera identifies a single viral serotype. Cell Rep 16:1485–1491. doi: 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ansarah-Sobrinho C, Nelson S, Jost CA, Whitehead SS, Pierson TC. 2008. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology 381:67–74. doi: 10.1016/j.virol.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pierson TC, Sanchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, Altamura LA, Diamond MS, Doms RW. 2006. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology 346:53–65. doi: 10.1016/j.virol.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 84.Mukherjee S, Pierson TC, Dowd KA. 2014. Pseudo-infectious reporter virus particles for measuring antibody-mediated neutralization and enhancement of dengue virus infection. Methods Mol Biol 1138:75–97. doi: 10.1007/978-1-4939-0348-1_6. [DOI] [PubMed] [Google Scholar]
- 85.Davis CW, Nguyen HY, Hanna SL, Sanchez MD, Doms RW, Pierson TC. 2006. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J Virol 80:1290–1301. doi: 10.1128/JVI.80.3.1290-1301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burns DP, Desrosiers RC. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol 65:1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, Engram JC, Little DM, Ryzhova E, Gonzalez-Scarano F, Paiardini M, Ansari AA, Ratcliffe S, Else JG, Brenchley JM, Collman RG, Estes JD, Derdeyn CA, Silvestri G. 2011. Depletion of CD4(+) T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J Clin Invest 121:4433–4445. doi: 10.1172/JCI46023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 90.Weatherall D, et al. 2006. The use of non-human primates in research. The Royal Society, London, United Kingdom. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ZIKV plasma viral titer. Dotted lines represent titers for individual animals, while solid lines represent the mean viral titer for the ZIKV cohort (blue; n = 12) and the SIV/ZIKV cohort (red; n = 7). Download FIG S1, PDF file, 0.1 MB (76.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative flow cytometry gating for cell sorting of peripheral lymph nodes. Parent gating strategy for the flow cytometric sorting of day 5 and day 27 post-ZIKV infection LN samples for analysis of cellular infection status by RT-PCR. A representative staining profile from the ZIKV cohort is shown. Live, singlet leukocytes were sorted into T cell (CD3+), B cell (CD20+), or myeloid cell (CD11b+) populations. Download FIG S2, PDF file, 0.1 MB (157.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative flow cytometry gating for phenotypic cell characterization. Parent gating strategy for the analysis of PBMCs. A representative staining profile from the ZIKV cohort is shown. Live, singlet lymphocytes were phenotypically characterized as memory B cells (CD27+ CD20+ CD3−), memory CD4+ T cells (CD28+/− CD95+ CD4+ CD3+), memory CD8+ T cells (CD28+/− CD95+ CD8+ CD3+), or NK cells (NKG2a+). Proliferation was assessed for each cell type by the expression of Ki67. Download FIG S3, PDF file, 0.2 MB (240.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcriptional gene profiling of expression changes in acute ZIKV infection. Immunology panel gene expression profiling in the ZIKV RM cohort (n = 4) at day 0 (filled blue circles) or day 3 (open blue circles). (A) Principal-component analysis comparing the two sample groups. The plot represents clustering of the data in a 2-dimensional matrix. Each dot represents one RM. (B) Pathway activation determined by z-score. The top five pathways predicted to be activated (orange) or deactivated (blue) are displayed. Download FIG S4, PDF file, 0.1 MB (134.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcriptional gene expression differences between SIV/ZIKV and ZIKV cohorts at day 0. Download Table S1, PDF file, 0.1 MB (65.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.