Wang et al. identify Rab7 as a novel regulator of primary cilia disassembly. Their findings demonstrate that Rab7 localization to primary cilia is required for intraciliary F-actin polymerization, which is indispensable for the regulation of cilia ectocytosis and disassembly.
Abstract
The primary cilium is a sensory organelle that protrudes from the cell surface. Primary cilia undergo dynamic transitions between assembly and disassembly to exert their function in cell signaling. In this study, we identify the small GTPase Rab7 as a novel regulator of cilia disassembly. Depletion of Rab7 potently induced spontaneous ciliogenesis in proliferating cells and promoted cilia elongation during quiescence. Moreover, Rab7 performs an essential role in cilia disassembly; knockdown of Rab7 blocked serum-induced ciliary resorption, and active Rab7 was required for this process. Further, we demonstrate that Rab7 depletion significantly suppresses cilia tip excision, referred to as cilia ectocytosis, which has been identified as required for cilia disassembly. Mechanically, the failure of F-actin polymerization at the site of excision of cilia tips caused suppression of cilia ectocytosis on Rab7 depletion. Overall, our results suggest a novel function for Rab7 in regulating cilia ectocytosis and cilia disassembly via control of intraciliary F-actin polymerization.
Introduction
The primary cilium is an antenna-like, microtubule-based organelle that extends from the cell surface to sense and transduce extracellular signals (Singla and Reiter, 2006; Berbari et al., 2009; Goetz and Anderson, 2010; Satir et al., 2010; Ishikawa and Marshall, 2011; Sung and Leroux, 2013). Dysfunctions of the primary cilium underlie a group of human diseases referred to as ciliopathies, including polycystic kidney disease, retinal dystrophy, and Bardet-Biedl syndrome (Fliegauf et al., 2007; Gerdes et al., 2009; Nigg and Raff, 2009; Veland et al., 2009; Hildebrandt et al., 2011). Primary cilia are dynamically expressed in cycling cells (Rieder et al., 1979; Tucker et al., 1979; Pan and Snell, 2007; Pugacheva et al., 2007; Kim et al., 2011; Kim and Dynlacht, 2013; Pan et al., 2013; Sánchez and Dynlacht, 2016). Briefly, they are formed in quiescent cells and resorbed during cell cycle reentry. Alongside discoveries indicating diverse roles for ciliogenesis, emerging evidence suggests that ciliary resorption is associated with cellular functions, including stress responses (Iomini et al., 2004; McGlashan et al., 2010; Prodromou et al., 2012; Luo et al., 2014), cell cycle progression (Rieder et al., 1979; Pugacheva et al., 2007; Kim et al., 2011; Li et al., 2011; Inoko et al., 2012; Plotnikova et al., 2012; Spalluto et al., 2013), and cell differentiation (Marion et al., 2009; Plaisant et al., 2009; Forcioli-Conti et al., 2015).
Recently, researchers have made increasing efforts to uncover the mechanisms underlying cilia disassembly. Initial studies showed that Aurora A (AurA) kinase induces disassembly of cilia by phosphorylation and activation of the tubulin deacetylase HDAC6, deacetylating tubulin molecules within the axoneme and resulting in the destabilization of axonemal microtubules to facilitate ciliary resorption (Pugacheva et al., 2007). Similarly, two microtubule depolymerizing kinesins, Kif2a (Miyamoto et al., 2015) and Kif24 (Kobayashi et al., 2011; Kim et al., 2015b), were found to directly promote the depolymerization and destabilization of ciliary microtubules and postulated to facilitate cilia disassembly independently of AurA. In addition, cilia disassembly also requires the participation of actin dynamics, since inhibition of actin polymerization induces cilia assembly and prevents cilia disassembly through orchestration of intracellular trafficking and transcription regulation (Kim et al., 2010, 2015a; Pitaval et al., 2010; Cao et al., 2012; Yan and Zhu, 2013; Saito et al., 2017). Interestingly, a recent study provided more direct evidence of the role of actin dynamics in cilia disassembly, demonstrating that F-actin can polymerize in primary cilia to excise cilia tips for cilia ectocytosis (also called cilia decapitation; Nager et al., 2017; Phua et al., 2017), thus triggering disassembly of cilia and entry into the cell cycle (Phua et al., 2017).
Rab GTPases are key regulators of membrane trafficking in the endomembrane system (Barr and Lambright, 2010; Hutagalung and Novick, 2011; Itzen and Goody, 2011; Barr, 2013). Their activity is strictly controlled through cycling between inactive GDP-bound and active GTP-bound forms. Rab8 and Rab11 have been reported to be involved in various steps during ciliogenesis, including vesicle docking, ciliary membrane formation, and intraflagellar transport (Nachury et al., 2007; Omori et al., 2008; Knödler et al., 2010; Westlake et al., 2011); however, it remains unclear whether Rab GTPases participate in the process of cilia disassembly. Here, we report that the small GTPase Rab7, a key regulator of endosomal biogenesis and maturation (Bucci et al., 2000; Rink et al., 2005; Hyttinen et al., 2013), is also an essential regulator of primary cilium disassembly, which depends on its active state. Further, we found that cilia ectocytosis, a newly defined process required for cilia disassembly, is suppressed by Rab7 depletion because of a failure of F-actin polymerization at the site of cilia tip excision. Overall, our results suggest that Rab7 is required for intraciliary F-actin polymerization and is responsible for regulation of cilia ectocytosis and disassembly.
Results and discussion
Depletion of Rab7 can promote ciliogenesis by increasing both the number and length of primary cilia
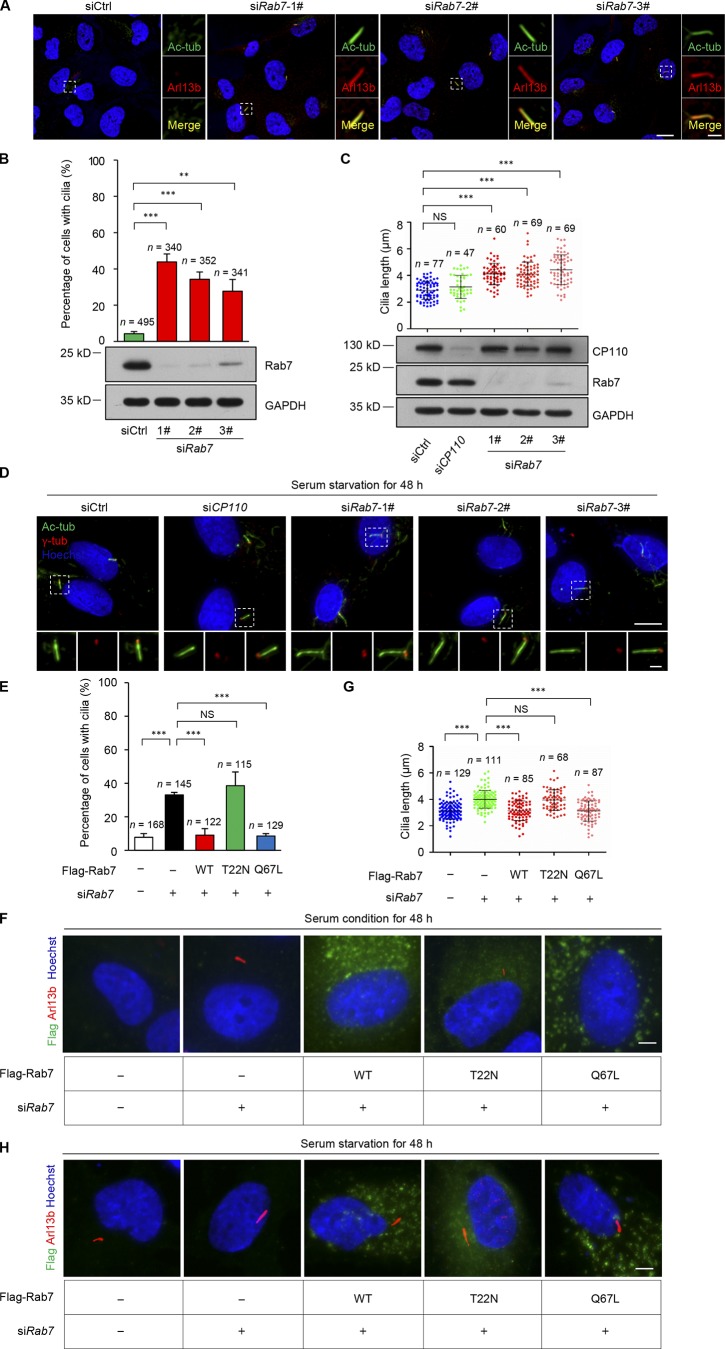
Our data from a separate study indicate that Rab7 knockdown can promote ciliogenesis. To elucidate its function in cilia expression, we knocked down Rab7 in RPE-1 cells using three individual siRNA molecules with nonoverlapping sequences. Spontaneous ciliogenesis was observed in Rab7 knockdown cells, even in the presence of serum, with percentages as high as 28–44%, compared with 4% in control cells (Fig. 1, A and B). Notably, the variations in levels of ciliogenesis induced by the different siRNAs were consistent with their efficiency in knocking down Rab7, as determined by immunoblotting (Fig. 1 B). To further investigate its effect on primary cilia, we measured cilia length in serum-starved cells and found that Rab7 depletion significantly increased the length of primary cilia; however, silencing of CP110, a well-known ciliogenesis suppressor that must be removed from the mother centriole for ciliogenesis (Spektor et al., 2007; Schmidt et al., 2009; Kobayashi et al., 2011), had no such effects (Fig. 1, C and D). Moreover, immunoblotting results showed that Rab7 knockdown did not affect CP110 expression (Fig. 1 C).
Figure 1.
Depletion of Rab7 promotes ciliogenesis. (A) Rab7 knockdown using three individual siRNAs induced spontaneous ciliogenesis in cycling cells (+serum). RPE-1 cells transfected with the indicated siRNAs were stained with acetylated α-tubulin (Ac-tub, green) and ADP ribosylation factor–like GTPase 13B (Arl13b, red). Nuclei were stained with Hoechst (blue). Scale bars, 10 µm (main image) and 2 µm (magnified regions). (B) The percentage of cells with primary cilia in A were quantified. Data are means ± SD of four independent experiments. Student’s t test was performed. **P < 0.01, ***P < 0.001. Cell lysates were immunoblotted with the indicated antibodies. GAPDH was used as a loading control. (C) Rab7 knockdown promoted cilia elongation in quiescent (−serum) RPE-1 cells. The length of primary cilia was measured. CP110 was used as a negative control. Data are means ± SD. Mann-Whitney rank sum test was performed. ***P < 0.001. Cell lysates were immunoblotted with the indicated antibodies. GAPDH was used as a loading control. (D) Representative images of C. RPE-1 cells transfected with the indicated siRNAs were stained with acetylated α-tubulin (green) and γ-tubulin (red). Acetylated α-tubulin labels cilia and γ-tubulin labels centrosomes. Nuclei were stained with Hoechst (blue). Scale bars, 10 µm (main image) and 2 µm (magnified regions). (E and F) Spontaneous ciliogenesis induced by Rab7 knockdown in RPE-1 cells were rescued by Flag-Rab7-WT and the Q67L mutant, but not by the T22N mutant. RPE-1 cells transfected with the indicated siRNAs and plasmids were cultured in serum medium for 48 h. WT, wild-type Rab7; T22N, dominant negative mutant of Rab7; Q67L, dominant active mutant of Rab7. All constructs were RNAi resistant. Cells were stained with Flag (green) and Arl13b (red). Nuclei were stained with Hoechst (blue). Scale bar, 5 µm. Data are means ± SD of three independent experiments. ***P < 0.001. (G and H) Prolonged cilia length induced by Rab7 knockdown in RPE-1 cells was rescued by Flag-Rab7-WT and the Q67L mutant, but not by the T22N mutant. RPE-1 cells transfected with the indicated siRNAs and plasmids were cultured in serum-free medium for 48 h. All constructs were RNAi resistant. Cells were stained with Flag (green) and Arl13b (red). Nuclei were stained with Hoechst (blue). Scale bar, 5 µm. Data are means ± SD. Student’s t test was performed. ***P < 0.001.
As a small GTPase, Rab7 can hydrolyze GTP to GDP to transform from an active to an inactive state. We found that both wild-type and the dominant active mutant (Q67L) of Rab7 could efficiently rescue the spontaneous ciliogenesis and prolonged cilia length in Rab7 knockdown cells, while the dominant negative Rab7 mutant, T22N, failed to rescue these phenotypes (Fig. 1, E–H). Together, these data indicate a negative regulatory role of Rab7 in ciliogenesis and demonstrate that depletion of Rab7 promotes ciliogenesis, likely in a different way than CP110.
Active Rab7 is required for serum-induced disassembly of primary cilia
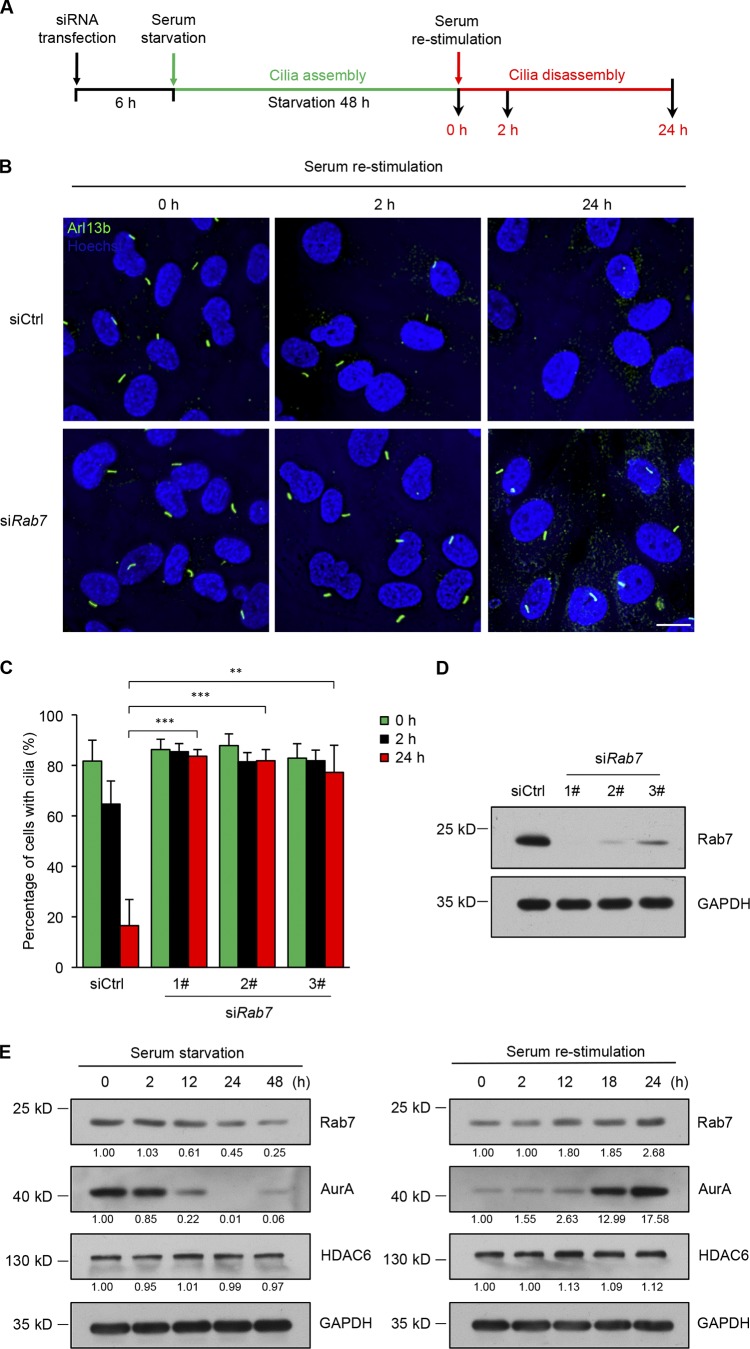
Cilia dynamics reflect the equilibrium between cilia assembly and disassembly (Marshall and Rosenbaum, 2001; Liang et al., 2016; Hsu et al., 2017). It has been shown experimentally that serum starvation of cells induces cilia assembly, while culture in serum-containing medium induces cilia disassembly (Fig. 2 A; Pugacheva et al., 2007; Li et al., 2011; Kim et al., 2015b). Since Rab7 negatively regulates ciliogenesis in a different way from CP110, which suppresses cilia assembly, we next examined whether Rab7 plays a role in cilia disassembly. The results showed that cilia disassembly was induced upon serum stimulation in control cells. However, cilia disassembly was blocked in Rab7 knockdown conditions (Fig. 2, B–D), suggesting an essential role for Rab7 in cilia disassembly. Accordingly, the expression profile of Rab7 is consistent with a role in cilia dynamics in that the level of endogenous Rab7 decreased continuously during ciliogenesis and increased moderately during cilia disassembly, resembling the profile of the established cilia disassembly regulator, AurA (Fig. 2 E).
Figure 2.
Rab7 is required for serum-induced disassembly of primary cilia. (A) Schematic diagram summarizing the strategy used for the cilia disassembly experiments. (B) Quiescent RPE-1 cells transfected with control or Rab7 siRNA were subjected to serum restimulation for the indicated times. Primary cilia were scored at 0, 2, and 24 h after serum readdition. Cilia were stained with anti-Arl13b (green), and nuclei were stained with Hoechst (blue). Scale bar, 10 µm. (C) RPE-1 cells transfected with the indicated siRNAs were subjected to cilia disassembly assay. The percentage of cells with primary cilia was quantified. Data are means ± SD of three independent experiments. Student’s t test was performed. **P < 0.01, ***P < 0.001. From left to right, cell n = 426, 402, 395; 489, 450, 389; 407, 372, 390; and 407, 382, 324. (D) Immunoblots of RPE-1 cell lysates in B with the indicated antibodies. GAPDH was used as a loading control. (E) Western analysis of Rab7 in RPE-1 cells after serum starvation and serum restimulation for the indicated times. AurA, HDAC6, and GAPDH were used as positive, negative, and loading controls, respectively. Quantification of relative amounts of Rab7, AurA, and HDAC6, normalized to respective GAPDH levels, is shown at the bottom of each immunoblot.
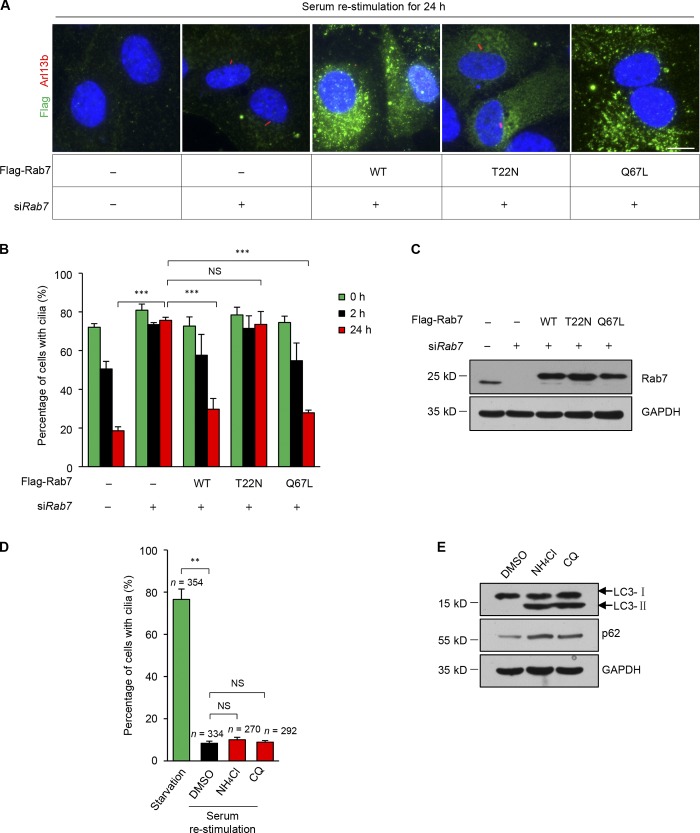
Next, we determined whether active Rab7 is required for cilia disassembly. A rescue experiment showed that expression of wild-type or the dominant active mutant (Q67L) of Rab7 could efficiently reverse the inhibition of cilia disassembly caused by Rab7 silencing, whereas the dominant negative mutant (T22N) of Rab7 had no effect (Fig. 3, A–C). To confirm the activity of the Rab7 constructs, GST pull-down assays were performed, and only the wild-type and Q67L mutant could be pulled down using GST-RILP, a well-known effector of Rab7 (Cantalupo et al., 2001; Jordens et al., 2001; Carroll et al., 2013), whereas the T22N mutant protein was not pulled down (Fig. S1 A). Thus, these results suggest that the activity of Rab7 is indeed required for cilia disassembly.
Figure 3.
The activity of Rab7 is required for the disassembly of primary cilia. (A) Quiescent RPE-1 cells transfected with the indicated siRNAs and plasmids were subjected to serum restimulation for 24 h. Cells were stained with Flag (green) and Arl13b (red). Nuclei were stained with Hoechst (blue). WT, wild-type Rab7; T22N, dominant negative mutant of Rab7; Q67L, dominant active mutant of Rab7. All constructs were RNAi resistant. Scale bar, 10 µm. (B) Cells were collected at the indicated times after serum restimulation, and the percentages of transfected cells with cilia were quantified. Cilia disassembly defects induced by Rab7 knockdown in RPE-1 cells were rescued by Flag-Rab7-WT and the Q67L mutant, but not by the T22N mutant. Data are means ± SD of three independent experiments. ***P < 0.001. From left to right, cell n = 365, 295, 264; 270, 245, 242; 169, 147, 129; 137, 132, 122; and 153, 155, 162. (C) Immunoblots of RPE-1 cell lysates in B with the indicated antibodies. GAPDH was used as a loading control. (D) Quiescent RPE-1 cells were treated with DMSO, 10 mM NH4Cl, or 20 µM CQ in serum-containing media for cilia disassembly. The percentages of cells with primary cilia were quantified after the serum was restimulated for 24 h. Data are means ± SD of three independent experiments. Student’s t test was performed. **P < 0.01. (E) Western blotting was used to detect the inhibition effect of NH4Cl and CQ on lysosome. The accumulation of p62 and LC3-II indicates the inhibition of lysosome activity.
Rab7 has an established role in endo-lysosomal maturation. To verify whether lysosome function is also required for cilia disassembly, we applied two lysosome inhibitors, NH4Cl and chloroquine (CQ), to inhibit the lysosome activity. Lysosome inhibition was confirmed by the accumulation of p62 and LC3-II in the presence of NH4Cl and CQ (Fig. 3 E); however, lysosomal inhibition had no effect on cilia disassembly (Fig. 3 D). These results suggest that the role of Rab7 in cilia disassembly is unlikely to involve regulation of lysosome activity. Rab7 is also reported to localize to ER and mitochondrial membranes. To verify whether intracellular localization of Rab7 is required for cilia disassembly, we knocked down the retromer subunit VPS35 to inhibit Rab7 localization on ER and mitochondrial membranes (Fig. S1, B–D; Jimenez-Orgaz et al., 2018). However, VPS35 knockdown had no effect on cilia disassembly, suggesting it is unlikely that the role of Rab7 in cilia disassembly depends on its localization to ER or mitochondrial membranes.
Rab7 is required for cilia ectocytosis during serum-induced cilia disassembly
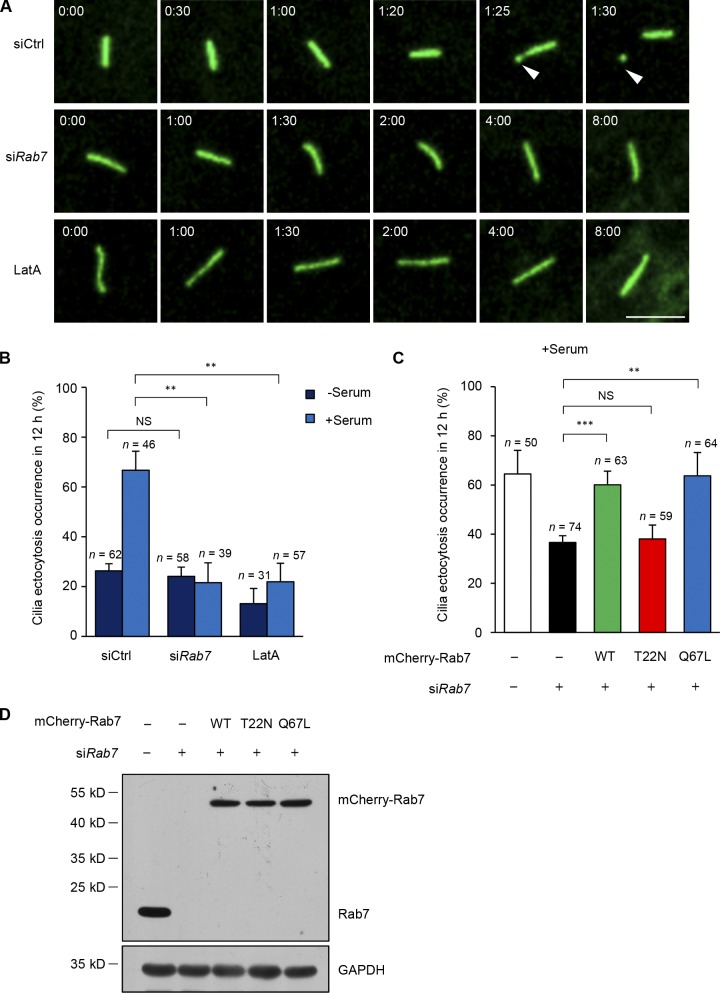
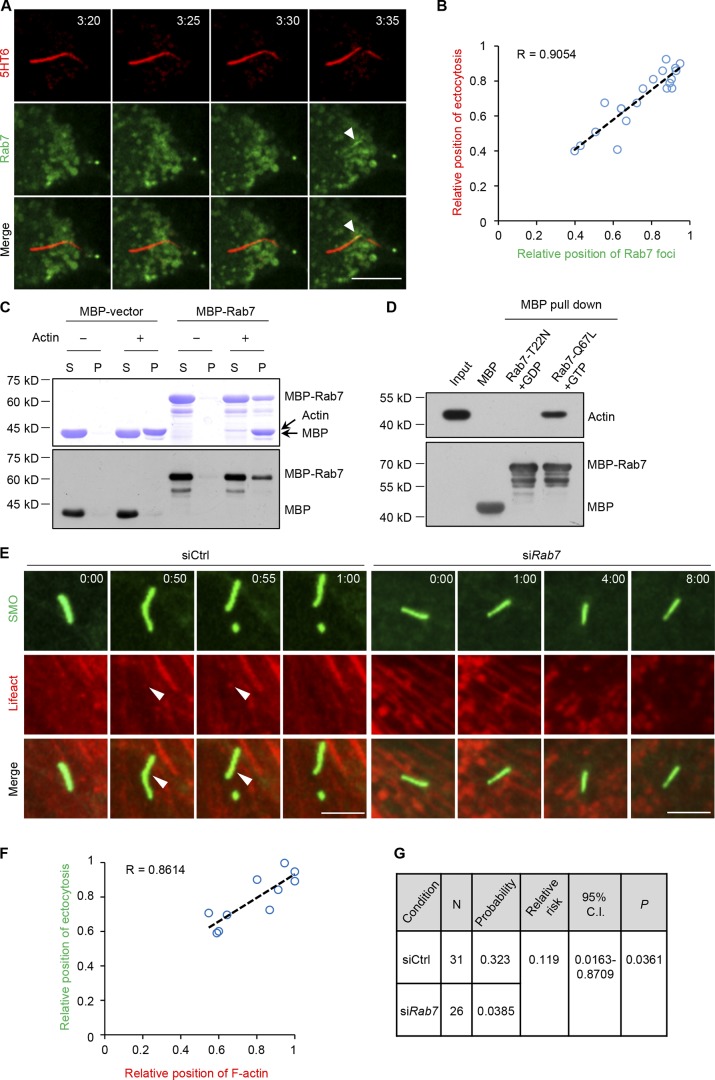
It has been recently reported that serum induction triggers the excision of cilia tips, referred to as cilia ectocytosis (also called cilia decapitation), which is required for cilia disassembly (Phua et al., 2017). To test whether Rab7 could function in cilia ectocytosis, we adopted RPE-1 cells expressing fluorescent protein-tagged ciliary membrane markers (GFP-SMO or 5HT6-mCherry; Corbit et al., 2005; Kim et al., 2010; Lu et al., 2015; Phua et al., 2017) to visualize cilia ectocytosis in live cells. Primary cilia were first induced by starving the cells for 48 h, and then cilia ectocytosis was monitored within 12 h after serum restimulation. The results demonstrated that 66% of control cells underwent cilia ectocytosis within 12 h; however, Rab7 knockdown significantly suppressed the occurrence of ciliary excision after serum stimulation (Fig. 4, A and B). Meanwhile, Latrunculin A (LatA), an inhibitor of actin polymerization, could restrict cilia ectocytosis after serum treatment (Fig. 4, A and B). A similar result was also observed in a previous study (Phua et al., 2017). Notably, some ectocytosis also occurred under serum-free conditions and did not decrease further in Rab7 knockdown cells, suggesting that Rab7 is not involved in cilia ectocytosis under serum-starved conditions. Further, in accordance with the finding that Rab7 activity is required for cilia disassembly, wild-type Rab7 and the Q67L mutant efficiently rescued the reduction of ectocytosis in Rab7 knockdown cells, while the dominant negative Rab7 mutant, T22N, failed to rescue the phenotype (Fig. 4, C and D). These results indicate that active Rab7 is required for serum-stimulated cilia ectocytosis during cilia disassembly. Phua et al. (2017) found that ciliary ectocytosis was an initiating step during cilia-loss process in RPE-1 and IMCD3 cells. However, Mirvis et al. (2019) reported that whole-cilium shedding in IMCD3 cells mainly occurs during cilia loss. We speculate that the difference might be a result of the different culture or imaging conditions.
Figure 4.
Depletion of Rab7 inhibits cilia ectocytosis during cilia disassembly. (A) Knockdown of Rab7 inhibits cilia ectocytosis. RPE-1 cells stably expressing GFP-SMO were transfected with the indicated siRNA or treated with 200 nM LatA (positive control) during serum restimulation for live-cell imaging. Arrowheads, excised cilia tips. Time in hour:minute. Scale bar, 5 µm. (B) Scoring of the percentage of cells that underwent cilia ectocytosis under quiescent (−serum) or growth-stimulated (+serum) conditions. Data are means ± SD of three independent experiments. Student’s t test was performed. **P < 0.01. (C) The percentages of transfected cells that underwent cilia ectocytosis under growth-stimulated (+serum) conditions were quantified. Cilia ectocytosis defects induced by Rab7 knockdown in RPE-1 cells were rescued by mCherry-Rab7-WT or mCherry-Rab7-Q67L, but not by the T22N mutant of Rab7. Data are means ± SD of three independent experiments. Student’s t test was performed. **P < 0.01, ***P < 0.001. (D) Immunoblots of RPE-1 cell lysates in C with the indicated antibodies. GAPDH was used as a loading control.
Role of Rab7 in regulation of actin polymerization during cilia ectocytosis
To better understand the role of Rab7 in cilia ectocytosis, we performed live-cell imaging to monitor Rab7 localization during cilia ectocytosis by serum stimulation. Interestingly, we found that Rab7 transiently localized at the cilia excision site and the position of cilia excision correlated with Rab7 accumulation during ectocytosis (Fig. 5, A and B; and Fig. S2, A and B). To see whether the relative position of Rab7 is near the base or tip of the cilium, we applied both base and ciliary membrane markers to orient cilia and found that Rab7 localized at the tip of primary cilia (Fig. S2 C). We also found that the dominant active mutant of Rab7 (Q67L) could localize at the cilia excision site, which was not the case for the dominant negative mutant of Rab7 (T22N; Fig. S2, D and E).
Figure 5.
Rab7 regulates F-actin assembly to induce cilia ectocytosis. (A) Rab7 localization during cilia ectocytosis. Time-lapse images of RPE-1 cells expressing 5HT6-mCherry and GFP-Rab7 during serum restimulation. Arrowheads mark GFP-Rab7 foci at the site of cilia excision. Time is in hour:minute. Scale bar, 10 µm. (B) Correlation plot of the intraciliary Rab7 position with position of excision, each provided as a relative ratio of cilia length. The position of Rab7 was measured at the estimated centroid position of GFP-Rab7 foci at excision. Linear regression is indicated by the dashed line, with the Pearson correlation coefficient R value indicated. n = 20 cells from four independent experiments. (C) Rab7 binds to F-actin. Purified MBP protein and MBP-tagged Rab7 protein were incubated with (+) or without (−) polymerized F-actin filaments and subjected to ultracentrifugation. The supernatant (S) or pellet (P) samples were subjected to SDS-PAGE, followed by Coomassie blue staining. The samples that we used in Western blot amount to 2% of those in Coomassie blue staining, and indicated antibodies were used in Western blot. (D) Rab7 binds to F-actin in a GTP-dependent manner. Purified MBP and MBP-Rab7-Q67L, and MBP-Rab7-T22N immobilized on amylose-resin beads were incubated with GTP, or with GDP, respectively, and then incubated with polymerized F-actin. After extensive washing, bound proteins were resolved by SDS-PAGE and Western blotting using indicated antibodies. (E) Ciliary F-actin polymerization was inhibited by siRab7. RPE-1 cells stably expressing GFP-SMO were transfected with the indicated siRNAs and mCherry-Lifeact during serum restimulation and subjected to live-cell imaging. Arrowheads mark mCherry-Lifeact foci at the site of cilia excision. Time is in hour:minute. Scale bars, 5 µm. (F) Correlation plot of the intraciliary F-actin position against the excision position in control cells in the left panel of E, each provided as a relative ratio of cilia length. The position of F-actin was measured at the estimated centroid position of F-actin foci at excision. Linear regression is indicated by the dashed line, with the Pearson correlation coefficient R value indicated. n = 10 cells from three independent experiments. (G) Relative risk ratio analyses of the effect of siRab7 on intraciliary F-actin incidence in E.
F-actin polymerization at the ciliary tip is the key step during cilia ectocytosis (Phua et al., 2017). We then investigated whether Rab7 could regulate F-actin dynamics in primary cilia. We found that purified Rab7 recombinant protein could interact with polymerized F-actin in a GTP-dependent manner (Fig. 5, C and D; and Fig. S2 F), which suggests a direct binding of Rab7 to F-actin. Furthermore, we investigated the involvement of Rab7 in intraciliary actin polymerization using live-cell visualization of F-actin dynamics. mCherry-Lifeact was used as an F-actin biosensor. As reported previously (Phua et al., 2017), F-actin was polymerized at the site of cilia excision, and the position of cilia excision correlated with F-actin accumulation during ectocytosis (Fig. 5, E and F), consistent with the transient localization of Rab7. Importantly, Rab7 knockdown significantly diminished actin accumulation at the cilia ectocytosis sites (Fig. 5, E and G). According to previous reports (Phua et al., 2017), PI(4)P/PI(4,5)P2 switching is reported to be involved in cilia ectocytosis. Rab7 is also known to play roles in controlling phosphoinositide switching in intracellular compartments, such as PI (phosphatidylinositol), PI(3)P, and PI(3,5)P2. To exclude that Rab7 regulates ciliary ectocytosis indirectly through PIs, we next examined whether Rab7 could regulate the ciliary localization of Inpp5e and PI(4,5)P2 during serum stimulation. The results showed that Rab7 knockdown did not abolish the ciliary reduction of Inpp5e and ciliary accumulation of PI(4,5)P2 (Fig. S3, A–C). Taken together, our results suggest that Rab7 regulates intraciliary actin polymerization and promotes cilia ectocytosis, potentially through its direct binding to F-actin.
The primary cilium is a microtubule-based organelle protruding from the cell surface. The important functions of primary cilia in signal transduction, cell proliferation and differentiation, and embryo development are well understood. In recent years, great progress has been made in uncovering how regulatory proteins control ciliogenesis; however, our knowledge of cilia disassembly lags somewhat behind that of ciliogenesis. A recent study reported that cilia ectocytosis, in which polymerized F-actin excises cilia tips, is required to trigger cilia disassembly (Phua et al., 2017). In our study, we define a novel function of the small GTPase Rab7 in regulating F-actin polymerization at cilia tips to control cilia ectocytosis as well as cilia disassembly.
Emerging evidence indicates that actin dynamics is involved in the regulation of ciliogenesis and cilia disassembly. Depletion of the actin filament nucleator ARP2/3 significantly promotes both the number and length of cilia (Bershteyn et al., 2010; Kim et al., 2010; Cao et al., 2012). Low-dose treatment with the actin polymerization inhibitors cytochalasin D or LatA not only promotes ciliogenesis, but also inhibits cilia disassembly (Li et al., 2011; Kim et al., 2015a; Phua et al., 2017; Saito et al., 2017). Rab7 is reported to be associated with several cytoskeleton-associated factors, including Rac1, vimentin, cortactin, and F-actin (Guerra and Bucci, 2016). Moreover, Rab7 depletion can influence cell spreading and neurite outgrowth (BasuRay et al., 2010; Cogli et al., 2010; Raiborg et al., 2015; Margiotta et al., 2017), likely through influencing cytoskeletal alterations. More importantly, a research group had also identified Rab7 in their cilia proteome (Mick et al., 2015). Our data show that Rab7 could directly bind to F-actin and Rab7 depletion reduces F-actin polymerization at the ciliary excision site, which would directly account for the abolition of cilia ectocytosis that occurs before cilia disassembly. As the activity of Rab7 was found to be required for cilia disassembly and cilia ectocytosis, further research is needed to verify the exact mechanism underlying the coordination of Rab7 activation and intraciliary actin dynamics during cilia ectocytosis.
As inhibition of cilia tip excision suppresses serum-stimulated cilia disassembly, cilia ectocytosis is considered crucial to this process (Phua et al., 2017); however, some cilia ectocytosis also occurs in quiescent cells, with no growth stimulation for cilia disassembly. We assume that the cilia ectocytosis taking place in quiescent cells may maintain the equilibrium between cilia assembly and disassembly to keep length constant (Long et al., 2016). Interestingly, our data show that, unlike serum-induced cilia ectocytosis, cilia ectocytosis in quiescent cells is not regulated by Rab7 (Fig. 4). Moreover, Rab7 protein expression was reduced during serum starvation, which also indicates that Rab7 is not a major regulator in this condition; nevertheless, whether or not cilia ectocytosis requires other regulators remains to be determined.
Abnormalities in cilia can cause developmental disorders, including defects in embryonic pattern formation (Capdevila et al., 2000; Gerdes et al., 2009; Kinzel et al., 2010; Bangs et al., 2015), and Rab7 knockout mice are reported to fail to establish the anterior–posterior axis during gastrulation, exhibiting embryonic lethality at approximately E7–8 (Kawamura et al., 2012). Thus, our research suggests a possible connection between Rab7 and cilia dynamics in early embryonic development. In addition, Rab7 is reported to promote protrusion and neurite outgrowth (BasuRay et al., 2010; Raiborg et al., 2015; Ponomareva et al., 2016), and mutations in Rab7 are well characterized as causing Charcot-Marie-Tooth syndrome type 2B (CMT2B; Cogli et al., 2009; Bucci and De Luca, 2012), a peripheral neuropathy with an unknown pathogenic mechanism. Given the existence of neuronal primary cilia (Lee and Gleeson, 2011; Guemez-Gamboa et al., 2014), further research is needed to investigate whether Rab7-mediated cilia disassembly can account for CMT2B pathogenesis. Thus, our findings relating to the function of Rab7 in cilia disassembly could provide insights into Rab7-associated developmental defects and pathogenesis, which may result from dysregulated cilia dynamics.
Materials and methods
Cells and culture
Human hTERT-RPE-1 cells were a gift from Xueliang Zhu (Chinese Academy of Sciences, Beijing, China), and RPE-1 cell lines stably expressing GFP-SMO (constitutive active W535L mutant of Smoothened, which is constantly present in cilia) were kindly provided by Christopher J. Westlake (National Cancer Institute, Bethesda, MD). RPE-1 cells were cultured in DMEM/F-12 (Life Technologies) supplemented with 10% FBS, 1% penicillin/streptomycin, and 0.01 mg/ml hygromycin B. Cilia assembly and disassembly assays were performed as previously described (Pugacheva et al., 2007; Li et al., 2011; Kim et al., 2015b). Briefly, RPE-1 cells were starved in Opti-MEM reduced-serum media (Life Technologies) for 48 h to induce cilia formation, and then 10% serum was added back to the media for 24 h to induce cilia disassembly. For chemical treatments, cells were treated with 200 nM LatA (428021, Millipore), 10 mM NH4Cl, 20 µM CQ (C6628, Sigma), or DMSO during the last 1 h of starvation, as well as after serum readdition, until the time of harvest.
Plasmids and transfection
Plasmids expressing Rab7, Lifeact, and BFP sequences were obtained from Addgene (#61804, #54610, and #46911, respectively). Plasmids expressing Centrin2 were obtained from Jianguo Chen (Peking University, Beijing, China). The sequence of Rab7 cDNA was subcloned into the pcDNA3.0 or pmCherry-C1 vectors to generate Flag-tagged or mCherry-tagged Rab7, respectively. The Rab7-T22N and Rab7-Q67L mutants were generated by PCR-based site-directed mutagenesis. Both the Centrin2 and BFP cDNA were cloned into pcDNA3.0 to generate BFP-tagged Centrin2. Plasmids of 5HT6-mCherry were obtained from Addgene (#47500). All of the plasmids were transiently transfected into cells using Lipofectamine LTX Reagent (Life Technologies) or X-tremeGENE 9 (Roche) according to the manufacturer’s instructions.
siRNA oligonucleotides and transfection
Synthetic siRNA oligonucleotides were obtained from Life Technologies, with sequences as follows: control siRNA, 5′-UUCUCCGAACGUGUCACGUAA-3′; CP110 siRNA, 5′-GCGGCCAAAUGUUGCGACAAUUUAA-3′; Rab7 siRNA (1#), 5′-CACUCAUGAACCAGUAUGUGAAUAA-3′; Rab7 siRNA (2#), 5′-CCCUAGAUAGCUGGAGAGAUGAGUU-3′; Rab7 siRNA (3#), 5′-GCUGCGUUCUGGUAUUUGA-3′; VPS35 siRNA (1#), 5′-GGUGUAAAUGUGGAACGUUdTdTAACGUUCCACAUUUACACC-3′; VPS35 siRNA (2#), 5′-GAGUUACACCAGAAUGUAAdTdTUUACAUUCUGGUGUAACUC-3′.
siRNAs were transfected using RNAiMAX (Life Technologies) according to the manufacturer’s instructions. Rab7 siRNA #1 was used in the majority of Rab7 depletion experiments, unless indicated otherwise.
Antibodies
The following antibodies were used in this study: mouse anti–Ac-tubulin (1:400; T6793, Sigma), rabbit anti-Arl13b (1:100; 17711–1-AP, Proteintech), rabbit anti–γ-tubulin (1:200; T5192, Sigma), rabbit anti-CP110 (1:200; 12780–1-AP, Proteintech), rabbit anti-AurA (1:1,000; 4718S, CST), rabbit anti-HDAC6 (1:1,000; 7612, CST), mouse anti-calregulin (1:400; sc-11398, Santa Cruz), mouse anti-TOM20 (1:400; sc-17764, Santa Cruz), and rabbit anti-Rab7 (Western blot 1:1,000, immunofluorescence 1:200; ab137029, Abcam).
The activity of Rab7 constructs
HEK293T cells were transfected with different constructs for 24 h, and then lysed with lysis buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 1% NP-40, and 2.5 mM MgCl2) containing 0.5 mM PMSF and complete protease inhibitor cocktail (04693132001, Roche). Lysates were centrifuged at 12,000 rpm for 10 min at 4°C, and supernatants were incubated with GST-RILP beads at 4°C for 1 h. After washing three times with washing buffer (lysis buffer without NP-40), beads were collected, and proteins attached to beads were detected by Western blotting and Ponceau S staining.
Actin-binding assay
The actin co-sedimentation assay was performed according to the manufacturer’s instructions (BK001, Cytoskeleton). Briefly, resuspended actin was kept on ice for 30 min and polymerized in actin polymerization buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 50 mM KCl, 2 mM MgCl2, and 1 mM ATP) at room temperature for 1 h. Purified recombinant proteins were incubated with polymerized F-actin in actin polymerization buffer at room temperature for 30 min. The reaction was then spun down at 150,000× g for 1.5 h at room temperature. Pellets and supernatants were analyzed by SDS-PAGE followed by Coomassie blue staining or immunoblotting.
Maltose-binding protein (MBP) pull-down assay
Purified MBP-tagged proteins (5 µg of each) were immobilized on amylose-resin beads and incubated with polymerized F-actin in binding buffer (PBS with 2 mM MgCl2 and 10 mM GTP or GDP) at 4°C for 2 h. After extensive washing with binding buffer, bound proteins were analyzed by SDS-PAGE and Western blotting using the indicated antibodies.
Immunofluorescence
For detection of primary cilia, RPE-1 cells were fixed in 4% paraformaldehyde for 10 min at room temperature and then permeabilized in 0.5% Triton X-100 in PBS for 10 min. For staining of centrosome proteins, RPE-1 cells were fixed and permeabilized in methanol at −20°C for 5 min, and then blocked with 3% normal goat serum in 0.1% Triton X-100/PBS for 1 h. Primary antibodies were added in blocking buffer for 1 h at room temperature. Secondary antibodies (Alexa Fluor 488– and 546–conjugated donkey anti-mouse and anti-rabbit; Life Technologies) were used at a dilution of 1:400 in blocking buffer for 1 h at room temperature. DNA was stained with Hoechst 33342 (1:1,000; H3570, Invitrogen). Images were taken with a 60×/1.42 (Fig. 1, D, F, and H; and Fig. S1 B) or 40×/1.30 (Figs. 1 A, 2 B, and 3 A) oil objective lens on a DeltaVision Restoration Microscope (Applied Precision Instruments). All acquisition settings were kept constant for experimental and control groups in the same experiment. The representative images acquired by the DeltaVision system were processed by iterative constrained deconvolution (SoftWoRx, Applied Precision Instruments). Maximal intensity projections of the entire Z-stack are shown. Raw images were analyzed using Volocity 6.0 software (PerkinElmer).
Live-cell imaging
RPE-1/GFP–Smo stable cell lines were seeded in 8-chambered cover glasses (Lab-Tek Chambered no. 1.0 Borosilicate Cover Glass System, Thermo). Images were collected every 5 min for 12 h using a 60×/1.40 oil objective lens on an inverted fluorescence microscope (Nikon Eclipse Ti-E) with an UltraView spinning-disc confocal scanner unit (PerkinElmer) at 37°C and 5% CO2. The start points were taken ∼30 min after serum readdition to select appropriate fields. Images were viewed using Volocity 6.0 software, and cell behavior was analyzed manually. All acquisition settings were kept constant for experimental and control groups in the same experiment. Images in Fig. 5 A and Fig. S2, D and E, show single optical sections, and images in Figs. 4 A, 5 E, and Fig. S2, A and C, show projections.
Notably, there were occasions when primary cilia extended from the ventral side of the cell and adhered to the underlying substratum. Subsequent cellular movements could result in significant mechanical pulling and breaking of primary cilia. Care was taken to exclude such instances of passive cilia breaking from all our experiments (Phua et al., 2017).
Statistical analysis
Statistical comparisons between two groups were performed by two-tailed t test or the Mann-Whitney rank sum test when a normal distribution could not be assumed. Statistical analysis was performed using SPSS software. The probability data presented in Fig. 5 G were analyzed using statistical risk ratio analyses: probability was determined by dividing the number of positives by the total number of cells analyzed, and relative risk was determined by dividing the probability value in the test condition by that of the control condition. For all tests, differences were considered statistically significant if P values were <0.05 (as indicated with *; *P < 0.05; **P < 0.01; ***P < 0.001). No statistical methods were used to predetermine sample size. The experiments were not randomized. Investigators were blinded during assessment for all staining and time-lapse assays.
Online supplemental material
Fig. S1 shows that the activity of Rab7 is required for the proper maintenance of ciliogenesis. Fig. S2 illustrates Rab7 localization during cilia ectocytosis. Fig. S3 shows that depletion of Rab7 does not affect ciliary Inpp5e and PI(4,5)P2 distribution during serum stimulation.
Supplementary Material
Acknowledgments
We thank Professor Xueliang Zhu, Christopher J. Westlake, Takanari Inoue (Johns Hopkins University School of Medicine, Baltimore, MD), and Siew Cheng Phua (Johns Hopkins University School of Medicine) for providing cell lines, plasmids, and technical support.
This work was supported by grants from the National Basic Research Program of China (2014CB910603 and 2013CB910302), National Natural Science Foundation of China (81521064, 81790252, 81325014, and 81802764), National Key Research and Development Program (2017YFA0503900, 2017YFC1601100, 2017YFC1601101, 2017YFC1601102, and 2017YFC1601104), International Science and Technology Cooperation Program of China (2015DFA31610 and 2013DFA31710), Beijing Nova Program (Z161100004916147), and National Major Scientific and Technological Special Project for “Significant New Drugs Development.”
The authors declare no competing financial interests.
Author contributions: H-Y. Li and K. He supervised the project; G. Wang and H-B. Hu designed and carried out most of the experiments; Y-C. Zhang, Z-Q. Song, and S-B. Zhou contributed to the preparation of complementary DNA vector constructs; L. Chen, J-F. Yuan, and Y. Huang carried out the statistics; X-M. Zhang, T. Zhou, and A-L. Li provided reagents and suggestions; M. Wu, H-Q. Tu, X. Pan, and N. Wang analyzed the data; G. Wang, Y. Chang, and H-Y. Li wrote the paper. All authors discussed the results and commented on the manuscript.
References
- Bangs F.K., Schrode N., Hadjantonakis A.K., and Anderson K.V.. 2015. Lineage specificity of primary cilia in the mouse embryo. Nat. Cell Biol. 17:113–122. 10.1038/ncb3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A. 2013. Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202:191–199. 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F., and Lambright D.G.. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470. 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BasuRay S., Mukherjee S., Romero E., Wilson M.C., and Wandinger-Ness A.. 2010. Rab7 mutants associated with Charcot-Marie-Tooth disease exhibit enhanced NGF-stimulated signaling. PLoS One. 5:e15351 10.1371/journal.pone.0015351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N.F., O’Connor A.K., Haycraft C.J., and Yoder B.K.. 2009. The primary cilium as a complex signaling center. Curr. Biol. 19:R526–R535. 10.1016/j.cub.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M., Atwood S.X., Woo W.M., Li M., and Oro A.E.. 2010. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev. Cell. 19:270–283. 10.1016/j.devcel.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C., and De Luca M.. 2012. Molecular basis of Charcot-Marie-Tooth type 2B disease. Biochem. Soc. Trans. 40:1368–1372. 10.1042/BST20120197 [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., and van Deurs B.. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480. 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo G., Alifano P., Roberti V., Bruni C.B., and Bucci C.. 2001. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20:683–693. 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Shen Y., Zhu L., Xu Y., Zhou Y., Wu Z., Li Y., Yan X., and Zhu X.. 2012. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat. Cell Biol. 14:697–706. 10.1038/ncb2512 [DOI] [PubMed] [Google Scholar]
- Capdevila J., Vogan K.J., Tabin C.J., and Izpisúa Belmonte J.C.. 2000. Mechanisms of left-right determination in vertebrates. Cell. 101:9–21. 10.1016/S0092-8674(00)80619-4 [DOI] [PubMed] [Google Scholar]
- Carroll B., Mohd-Naim N., Maximiano F., Frasa M.A., McCormack J., Finelli M., Thoresen S.B., Perdios L., Daigaku R., Francis R.E., et al. 2013. The TBC/RabGAP Armus coordinates Rac1 and Rab7 functions during autophagy. Dev. Cell. 25:15–28. 10.1016/j.devcel.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogli L., Piro F., and Bucci C.. 2009. Rab7 and the CMT2B disease. Biochem. Soc. Trans. 37:1027–1031. 10.1042/BST0371027 [DOI] [PubMed] [Google Scholar]
- Cogli L., Progida C., Lecci R., Bramato R., Krüttgen A., and Bucci C.. 2010. CMT2B-associated Rab7 mutants inhibit neurite outgrowth. Acta Neuropathol. 120:491–501. 10.1007/s00401-010-0696-8 [DOI] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., and Omran H.. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893. 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Forcioli-Conti N., Lacas-Gervais S., Dani C., and Peraldi P.. 2015. The primary cilium undergoes dynamic size modifications during adipocyte differentiation of human adipose stem cells. Biochem. Biophys. Res. Commun. 458:117–122. 10.1016/j.bbrc.2015.01.078 [DOI] [PubMed] [Google Scholar]
- Gerdes J.M., Davis E.E., and Katsanis N.. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 137:32–45. 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., and Anderson K.V.. 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11:331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guemez-Gamboa A., Coufal N.G., and Gleeson J.G.. 2014. Primary cilia in the developing and mature brain. Neuron. 82:511–521. 10.1016/j.neuron.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F., and Bucci C.. 2016. Multiple Roles of the Small GTPase Rab7. Cells. 5:. 10.3390/cells5030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Benzing T., and Katsanis N.. 2011. Ciliopathies. N. Engl. J. Med. 364:1533–1543. 10.1056/NEJMra1010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.S., Chuang J.Z., and Sung C.H.. 2017. The Biology of Ciliary Dynamics. Cold Spring Harb. Perspect. Biol. 9:. 10.1101/cshperspect.a027904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen J.M., Niittykoski M., Salminen A., and Kaarniranta K.. 2013. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim. Biophys. Acta. 1833:503–510. 10.1016/j.bbamcr.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Inoko A., Matsuyama M., Goto H., Ohmuro-Matsuyama Y., Hayashi Y., Enomoto M., Ibi M., Urano T., Yonemura S., Kiyono T., et al. 2012. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J. Cell Biol. 197:391–405. 10.1083/jcb.201106101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Tejada K., Mo W., Vaananen H., and Piperno G.. 2004. Primary cilia of human endothelial cells disassemble under laminar shear stress. J. Cell Biol. 164:811–817. 10.1083/jcb.200312133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Itzen A., and Goody R.S.. 2011. GTPases involved in vesicular trafficking: structures and mechanisms. Semin. Cell Dev. Biol. 22:48–56. 10.1016/j.semcdb.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Jimenez-Orgaz A., Kvainickas A., Nägele H., Denner J., Eimer S., Dengjel J., and Steinberg F.. 2018. Control of RAB7 activity and localization through the retromer-TBC1D5 complex enables RAB7-dependent mitophagy. EMBO J. 37:235–254. 10.15252/embj.201797128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kawamura N., Sun-Wada G.H., Aoyama M., Harada A., Takasuga S., Sasaki T., and Wada Y.. 2012. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 3:1071 10.1038/ncomms2069 [DOI] [PubMed] [Google Scholar]
- Kim S., and Dynlacht B.D.. 2013. Assembling a primary cilium. Curr. Opin. Cell Biol. 25:506–511. 10.1016/j.ceb.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J.E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., and Gleeson J.G.. 2010. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 464:1048–1051. 10.1038/nature08895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Jo H., Hong H., Kim M.H., Kim J.M., Lee J.K., Heo W.D., and Kim J.. 2015a Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Nat. Commun. 6:6781 10.1038/ncomms7781 [DOI] [PubMed] [Google Scholar]
- Kim S., Zaghloul N.A., Bubenshchikova E., Oh E.C., Rankin S., Katsanis N., Obara T., and Tsiokas L.. 2011. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat. Cell Biol. 13:351–360. 10.1038/ncb2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee K., Choi J.H., Ringstad N., and Dynlacht B.D.. 2015b Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat. Commun. 6:8087 10.1038/ncomms9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzel D., Boldt K., Davis E.E., Burtscher I., Trümbach D., Diplas B., Attié-Bitach T., Wurst W., Katsanis N., Ueffing M., and Lickert H.. 2010. Pitchfork regulates primary cilia disassembly and left-right asymmetry. Dev. Cell. 19:66–77. 10.1016/j.devcel.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., and Guo W.. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Tsang W.Y., Li J., Lane W., and Dynlacht B.D.. 2011. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 145:914–925. 10.1016/j.cell.2011.04.028 [DOI] [PubMed] [Google Scholar]
- Lee J.E., and Gleeson J.G.. 2011. Cilia in the nervous system: linking cilia function and neurodevelopmental disorders. Curr. Opin. Neurol. 24:98–105. 10.1097/WCO.0b013e3283444d05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Saito M., Chuang J.Z., Tseng Y.Y., Dedesma C., Tomizawa K., Kaitsuka T., and Sung C.H.. 2011. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat. Cell Biol. 13:402–411. 10.1038/ncb2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Meng D., Zhu B., and Pan J.. 2016. Mechanism of ciliary disassembly. Cell. Mol. Life Sci. 73:1787–1802. 10.1007/s00018-016-2148-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Zhang F., Xu N., Liu G., Diener D.R., Rosenbaum J.L., and Huang K.. 2016. Comparative Analysis of Ciliary Membranes and Ectosomes. Curr. Biol. 26:3327–3335. 10.1016/j.cub.2016.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Insinna C., Ott C., Stauffer J., Pintado P.A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.S., et al. 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17:531 10.1038/ncb3155 [DOI] [PubMed] [Google Scholar]
- Luo N., Conwell M.D., Chen X., Kettenhofen C.I., Westlake C.J., Cantor L.B., Wells C.D., Weinreb R.N., Corson T.W., Spandau D.F., et al. 2014. Primary cilia signaling mediates intraocular pressure sensation. Proc. Natl. Acad. Sci. USA. 111:12871–12876. 10.1073/pnas.1323292111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margiotta A., Progida C., Bakke O., and Bucci C.. 2017. Rab7a regulates cell migration through Rac1 and vimentin. Biochim Biophys Acta Mol Cell Res. 1864:367–381. 10.1016/j.bbamcr.2016.11.020 [DOI] [PubMed] [Google Scholar]
- Marion V., Stoetzel C., Schlicht D., Messaddeq N., Koch M., Flori E., Danse J.M., Mandel J.L., and Dollfus H.. 2009. Transient ciliogenesis involving Bardet-Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc. Natl. Acad. Sci. USA. 106:1820–1825. 10.1073/pnas.0812518106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., and Rosenbaum J.L.. 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155:405–414. 10.1083/jcb.200106141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlashan S.R., Knight M.M., Chowdhury T.T., Joshi P., Jensen C.G., Kennedy S., and Poole C.A.. 2010. Mechanical loading modulates chondrocyte primary cilia incidence and length. Cell Biol. Int. 34:441–446. 10.1042/CBI20090094 [DOI] [PubMed] [Google Scholar]
- Mick D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., and Nachury M.V.. 2015. Proteomics of Primary Cilia by Proximity Labeling. Dev. Cell. 35:497–512. 10.1016/j.devcel.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirvis M., Siemers K.A., Nelson W.J., and Stearns T.P.. 2019. Primary cilium loss in mammalian cells occurs predominantly by whole-cilium shedding. PLoS Biol. 17:e3000381 10.1371/journal.pbio.3000381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Hosoba K., Ochiai H., Royba E., Izumi H., Sakuma T., Yamamoto T., Dynlacht B.D., and Matsuura S.. 2015. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Reports. 10:664–673. 10.1016/j.celrep.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., and Jackson P.K.. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nager A.R., Goldstein J.S., Herranz-Pérez V., Portran D., Ye F., Garcia-Verdugo J.M., and Nachury M.V.. 2017. An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell. 168:252–263.e14. 10.1016/j.cell.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Omori Y., Zhao C., Saras A., Mukhopadhyay S., Kim W., Furukawa T., Sengupta P., Veraksa A., and Malicki J.. 2008. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat. Cell Biol. 10:437–444. 10.1038/ncb1706 [DOI] [PubMed] [Google Scholar]
- Pan J., and Snell W.. 2007. The primary cilium: keeper of the key to cell division. Cell. 129:1255–1257. 10.1016/j.cell.2007.06.018 [DOI] [PubMed] [Google Scholar]
- Pan J., Seeger-Nukpezah T., and Golemis E.A.. 2013. The role of the cilium in normal and abnormal cell cycles: emphasis on renal cystic pathologies. Cell. Mol. Life Sci. 70:1849–1874. 10.1007/s00018-012-1052-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua S.C., Chiba S., Suzuki M., Su E., Roberson E.C., Pusapati G.V., Setou M., Rohatgi R., Reiter J.F., Ikegami K., and Inoue T.. 2017. Dynamic Remodeling of Membrane Composition Drives Cell Cycle through Primary Cilia Excision. Cell. 168:264–279.e15. 10.1016/j.cell.2016.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaval A., Tseng Q., Bornens M., and Théry M.. 2010. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J. Cell Biol. 191:303–312. 10.1083/jcb.201004003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisant M., Fontaine C., Cousin W., Rochet N., Dani C., and Peraldi P.. 2009. Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells. 27:703–713. 10.1634/stemcells.2008-0888 [DOI] [PubMed] [Google Scholar]
- Plotnikova O.V., Nikonova A.S., Loskutov Y.V., Kozyulina P.Y., Pugacheva E.N., and Golemis E.A.. 2012. Calmodulin activation of Aurora-A kinase (AURKA) is required during ciliary disassembly and in mitosis. Mol. Biol. Cell. 23:2658–2670. 10.1091/mbc.e11-12-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva O.Y., Eliceiri K.W., and Halloran M.C.. 2016. Charcot-Marie-Tooth 2b associated Rab7 mutations cause axon growth and guidance defects during vertebrate sensory neuron development. Neural Dev. 11:2 10.1186/s13064-016-0058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou N.V., Thompson C.L., Osborn D.P., Cogger K.F., Ashworth R., Knight M.M., Beales P.L., and Chapple J.P.. 2012. Heat shock induces rapid resorption of primary cilia. J. Cell Sci. 125:4297–4305. 10.1242/jcs.100545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., and Golemis E.A.. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 129:1351–1363. 10.1016/j.cell.2007.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., Olsvik H., Schink K.O., Schultz S.W., Vietri M., Nisi V., Bucci C., Brech A., et al. 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520:234–238. 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Jensen C.G., and Jensen L.C.. 1979. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J. Ultrastruct. Res. 68:173–185. 10.1016/S0022-5320(79)90152-7 [DOI] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Saito M., Otsu W., Hsu K.S., Chuang J.Z., Yanagisawa T., Shieh V., Kaitsuka T., Wei F.Y., Tomizawa K., and Sung C.H.. 2017. Tctex-1 controls ciliary resorption by regulating branched actin polymerization and endocytosis. EMBO Rep. 18:1460–1472. 10.15252/embr.201744204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez I., and Dynlacht B.D.. 2016. Cilium assembly and disassembly. Nat. Cell Biol. 18:711–717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P., Pedersen L.B., and Christensen S.T.. 2010. The primary cilium at a glance. J. Cell Sci. 123:499–503. 10.1242/jcs.050377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Singla V., and Reiter J.F.. 2006. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 313:629–633. 10.1126/science.1124534 [DOI] [PubMed] [Google Scholar]
- Spalluto C., Wilson D.I., and Hearn T.. 2013. Evidence for reciliation of RPE1 cells in late G1 phase, and ciliary localisation of cyclin B1. FEBS Open Bio. 3:334–340. 10.1016/j.fob.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., and Dynlacht B.D.. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690. 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Sung C.H., and Leroux M.R.. 2013. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol. 15:1387–1397. 10.1038/ncb2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R.W., Pardee A.B., and Fujiwara K.. 1979. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 17:527–535. 10.1016/0092-8674(79)90261-7 [DOI] [PubMed] [Google Scholar]
- Veland I.R., Awan A., Pedersen L.B., Yoder B.K., and Christensen S.T.. 2009. Primary cilia and signaling pathways in mammalian development, health and disease. Nephron, Physiol. 111:p39–p53. 10.1159/000208212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., and Zhu X.. 2013. Branched F-actin as a negative regulator of cilia formation. Exp. Cell Res. 319:147–151. 10.1016/j.yexcr.2012.08.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.