Derrer et al. show that the mRNA export factor Mex67 can perform its essential function when stably tethered to the nuclear pore complex.
Abstract
The RNA export factor Mex67 is essential for the transport of mRNA through the nuclear pore complex (NPC) in yeast, but the molecular mechanism of this export process remains poorly understood. Here, we use quantitative fluorescence microscopy techniques in live budding yeast cells to investigate how Mex67 facilitates mRNA export. We show that Mex67 exhibits little interaction with mRNA in the nucleus and localizes to the NPC independently of mRNA, occupying a set of binding sites offered by FG repeats in the NPC. The ATPase Dbp5, which is thought to remove Mex67 from transcripts, does not affect the interaction of Mex67 with the NPC. Strikingly, we find that the essential function of Mex67 is spatially restricted to the NPC since a fusion of Mex67 to the nucleoporin Nup116 rescues a deletion of MEX67. Thus, Mex67 functions as a mobile NPC component, which receives mRNA export substrates in the central channel of the NPC to facilitate their translocation to the cytoplasm.
Introduction
Directional export of mRNAs from the nucleus to the cytoplasm is a crucial step for gene expression in every eukaryotic cell. Transport is mediated by transport factors that guide the mRNA through nuclear pore complexes (NPCs), which are highly specialized transport channels embedded in the nuclear envelope. The mRNA export machinery has been characterized in multiple species, and many of its constituents are highly conserved (reviewed in Heinrich et al., 2017; Köhler and Hurt, 2007).
Export of most mRNAs depends on the transport receptors Mex67-Mtr2 (Santos-Rosa et al., 1998; Strässer et al., 2000; Weis, 2002), which are also involved in the export of ribosomal subunits (Faza et al., 2012; Yao et al., 2007). Mex67 (TAP/NXF1 in metazoa) in complex with Mtr2 mediates transport across the NPC via interactions with several nucleoporins that contain repetitive domains enriched in phenylalanine (F) and glycine (G; Strässer et al., 2000; Terry and Wente, 2007). These FG repeats form a meshwork in the central channel of the NPC and enable selective access for shuttling nuclear transport receptors carrying different cargo between the nucleus and the cytoplasm. At steady-state, Mex67 is mainly localized at the NPC (Katahira et al., 1999; Köhler and Hurt, 2007; Rodriguez et al., 2004; Strässer et al., 2000; Terry and Wente, 2007). However, it was shown to shuttle between the nucleus and cytoplasm (Segref et al., 1997). Reminiscent of the mechanism of protein transport across the NPC, the current model for mRNA export therefore postulates that Mex67-Mtr2 binds to a mature messenger RNP (mRNP) substrate in the nucleoplasm and then chaperones the mRNP through the NPC channel via successive interactions with distinct FG-repeat nuclear pore proteins (Nups; Hautbergue et al., 2008; Stewart, 2007; Strässer et al., 2000; Strässer and Hurt, 2001).
Once the mRNA reaches the cytoplasmic side of the NPC, the final steps in mRNA export depend on two Nups localized to the cytoplasmic face of the NPC, Nup159 and Nup42, which recruit two essential mRNA export factors, the DEAD-box ATPase Dbp5 and its ATPase activator Gle1 (Adams et al., 2017; Hodge et al., 2011; Noble et al., 2011; Weirich et al., 2006). Dbp5 is proposed to impart directionality to the export process by remodeling the mRNPs emerging from the NPC in an ATP-dependent manner and displacing export factors like Mex67-Mtr2 (Cole and Scarcelli, 2006b; Folkmann et al., 2011; Liker et al., 2000; Lund and Guthrie, 2005; Montpetit et al., 2011; Snay-Hodge et al., 1998; Tran et al., 2007; Weirich et al., 2006). Disassembly of the mRNP on the cytoplasmic side of the NPC would prevent its return to the nucleus, resulting in unidirectional mRNA translocation. In addition, the release of mRNA export factors including Mex67-Mtr2 would allow them to return to the nucleus, where they could function in additional rounds of mRNA export (Stewart, 2007). However, direct evidence of such a shuttling mRNA export model is lacking, and it is unclear how Mex67 and Dbp5 function in mRNA export.
To better understand the function of Mex67 and its interplay with the NPC and the export factor Dbp5, we used advanced fluorescence microscopy techniques including FRAP, fluorescence correlation spectroscopy (FCS), and fluorescent intensity measurements. This allowed us to quantitatively characterize the interactions of Mex67 at the NPC. In line with recent observations in mammalian cells (Ben-Yishay et al., 2019), we show that the interaction of Mex67 at the NPC does not depend on cargo and is not modulated by the activity of Dbp5. Furthermore, we uncovered that Mex67 does not have to leave the NPC to mediate mRNA export. This suggests that Mex67 behaves similar to a mobile nucleoporin.
Results and discussion
The majority of Mex67 is not bound to mRNA in the nucleus
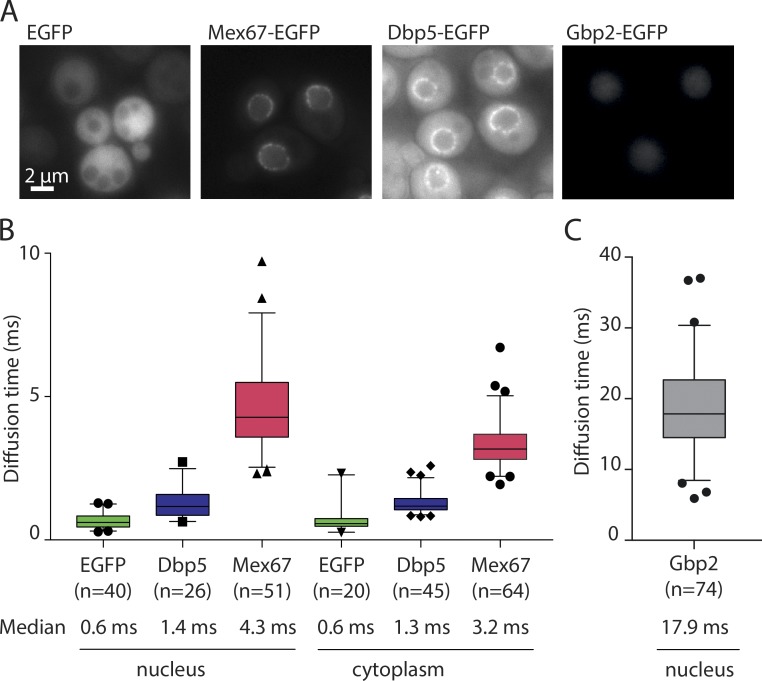
The binding of mRNA to its transport receptor Mex67, a crucial step in mRNA export, has been proposed to occur in the nucleoplasm (Dieppois et al., 2006; Gilbert and Guthrie, 2004; Strässer and Hurt, 2001). We reasoned that mRNA binding could be detected by assessing the diffusion of Mex67, as free proteins diffuse faster than large mRNP particles. GFP, for example, exhibits an apparent diffusion coefficient of 11 µm2/s in the yeast nucleus (Slaughter et al., 2007), while diffusion coefficients reported for mRNAs are in the range of 0.15 to 0.74 µm2/s (Wu et al., 2012). To measure the diffusion of Mex67 and Dbp5 in living cells, we used FCS, which allows to determine the mean duration required for a fluorescent molecule to diffuse through the confocal volume (termed diffusion time).
First, we measured the diffusion of control proteins. EGFP exhibited the expected fast diffusion behavior with diffusion times of 0.6 ms in both the nucleus and the cytoplasm (Fig. 1 B). In contrast, Gbp2, a poly(A+) RNA-binding protein that acts as surveillance factors for the selective export of spliced mRNAs, exhibited a median diffusion time of 17.9 ms in the nucleus (Fig. 1 C), corresponding to an apparent diffusion coefficient of 0.6 µm2/s, which is consistent with mRNA diffusion and indicates that the majority of Gbp2 molecules are bound to mRNPs or other slowly moving complexes. Next we assessed the diffusion of Mex67 and Dbp5 in the nucleus and the cytoplasm. Dbp5 exhibited diffusion times of 1.4 ms and 1.3 ms, respectively (Fig. 1 B), similar to EGFP. Thus, in both compartments the majority of Dbp5 is in a rapidly diffusing, unbound state. This is consistent with the model that Dbp5 binds mRNA predominantly at the NPC, where it functions to remodel it at the cytoplasmic filaments during late steps of mRNA export (Tran et al., 2007; von Moeller et al., 2009; Zhao et al., 2002).
Figure 1.
Diffusion of mRNA export factors measured by FCS. (A) Representative fluorescence wide-field microscopy images of yeast strains expressing GFP-tagged mRNA export factors. Scale bar, 2 µm. (B and C) Diffusion times measured by FCS for the indicated GFP-fusion proteins. Data are pooled from three independent biological replicates. In the boxplots, the line represents the median, the box represents the 25th to 75th percentile, whiskers extend to the 5th to 95th percentile, and black symbols represent data points outside this range.
The diffusion times of Mex67-EGFP in the nucleus and cytoplasm were 4.3 and 3.2 ms, respectively. These values, although higher than for Dpb5, are still much lower than for factors mainly associated with slowly diffusing mRNA molecules such as Gbp2. This indicates that, while a small fraction of Mex67 could be present in slowly diffusing complexes, the majority of Mex67 is not bound to large mRNPs.
Mex67 is dynamically associated with the NPC
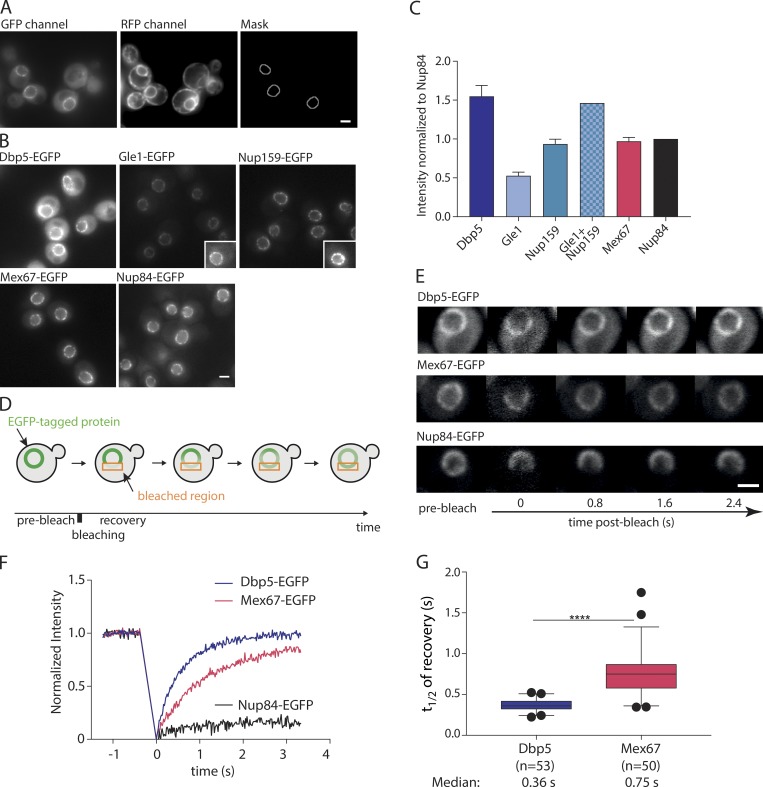
Mex67 is concentrated at NPCs under steady-state conditions (Fig. 1 A), where its main function—namely mRNA export—is performed. Therefore, we aimed to further characterize the interaction of Mex67 with the NPC. To determine the amounts of Mex67 and other mRNA export factors at the NPC, we applied a recently developed quantitative image analysis tool termed NuRIM (nuclear rim intensity measurement; Rajoo et al., 2018; Vallotton et al., 2019). In NuRIM, the nuclear envelope is segmented based on a red fluorescent marker targeted to the lumen of the endoplasmic reticulum (DsRed-HDEL), which then allows for the precise quantification of GFP signals at the nuclear envelope (Fig. 2 A). NuRIM was applied to Mex67 and Dbp5 as well as the nuclear pore components Nup84, Nup159, and Gle1 (Fig. 2 B). We found that Mex67 is present at the NPC in an amount comparable to Nup84, which is present in 16 copies per NPC (Rajoo et al., 2018). In contrast, Dbp5 has a 1.5 times higher copy number at the NPC than Nup84 (Fig. 2 C), suggesting that at steady-state, ∼24 Dbp5 molecules reside at the NPC. Interestingly, this corresponds to the summed levels of the two Dbp5-binding partners at the NPC, Gle1 (8 copies) and Nup159 (16 copies).
Figure 2.
Mex67 and Dbp5 bind dynamically to the NPC. (A) Example for the production of the NuRIM analysis mask for quantification of GFP intensity on the nuclear envelope using DsRed-HDEL signal for nuclear envelope segmentation. (B) Representative fluorescence images of mRNA export factors and related Nups used for intensity analysis. Insets: Example cells with increased contrast settings for better visualization. (C) Quantification of nuclear envelope intensities by NuRIM. Bars show the means of three biological replicates, and the error bars represent the SEM. At least 1,000 cells were analyzed per condition and replicate. (D) Schematic illustration of FRAP at the nuclear envelope. (E–G) FRAP of Dbp5, Mex67, and Nup84 tagged with GFP. (E) Representative FRAP images. (F) FRAP curves normalized to prebleach and postbleach values. Mean curves of ≥50 cells are shown. (G) Half time of recovery retrieved from fitting individual FRAP curves with a single component model are plotted in boxplots. The line represents the median, the box represents the 25th to 75th percentile, and whiskers extend to the 5th to 95th percentile. ****, P < 0.001, Mann–Whitney test. Scale bars, 2 µm.
Next, we determined the dynamics of Mex67 and Dbp5 binding at the NPC using FRAP (Fig. 2, D–G). Both Mex67 and Dbp5 quickly recovered within a few seconds after bleaching (Fig. 2, E–G). The median half time of recovery at the nuclear envelope was 0.36 s for Dbp5 and 0.75 s for Mex67 (Fig. 2 G). In contrast, the stable nucleoporin Nup84 showed no significant recovery in this timeframe (Fig. 2 F), indicating that NPC movement does not contribute to the measured dynamics. Thus, both Mex67 and Dbp5 are highly mobile at the NPC and are present in stoichiometries similar to Nups.
Mex67 binding at the NPC does not depend on, but is modulated by, mRNA and pre-ribosomes
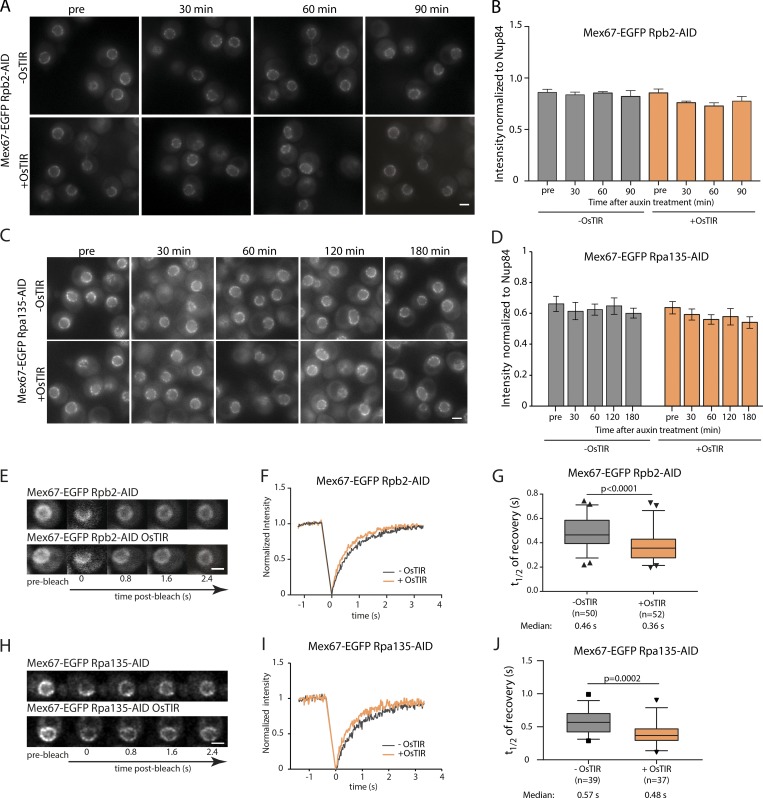
Since Mex67 is thought to facilitate the passage of both mRNAs and pre-ribosomal subunits through the FG-repeat network of the NPC, we next asked whether the localization and dynamics of Mex67 are influenced by the presence of its cargo. To lower nuclear mRNA levels, we used the auxin-inducible degron (AID; Nishimura et al., 2009) to acutely deplete the essential RNA Pol II subunit Rpb2. Auxin treatment led to a rapid and specific degradation of Rpb2, reduction in mRNA transcription, and a concomitant loss of viability (Fig. S1, A and B). Similarly, we inactivated RNA polymerase I to prevent the production of pre-ribosomes via auxin-induced degradation of the essential second largest subunit, Rpa135. Depletion of Rpa135 led to reduced viability (Fig. S1 C) as well as to reduced nucleolar size (Fig. S1 D), indicative of impaired RNA Pol I function.
Neither inhibition of mRNA transcription by auxin-mediated depletion of Pol II, nor inhibition of ribosome biogenesis by depletion of Pol I, led to a change in Mex67 abundance at the nuclear envelope (Fig. 3, A–D). This suggests that cargo is not required for Mex67 interaction with the NPC. Thus, mRNA and ribosomal cargo do not influence the number of binding sites that are occupied by Mex67 at the NPC. However, we observed a small but significant increase in the turnover dynamics of Mex67 at the nuclear envelope in both mRNA and pre-ribosome depletion conditions (Fig. 3, E–J). This could suggest that cargo binding either increases the affinity of Mex67 to FG-repeats, e.g., by generating multivalency in the binding of Mex67, or reduces the mobility of Mex67 in the FG repeat network.
Figure 3.
Mex67 dynamics, but not its amount at the NPC, is affected by the presence of cargo molecules. (A–D) NuRIM quantification of Mex67-EGFP intensity at the nuclear envelope upon depletion of RNA Pol II (A and B) or RNA Pol I (C and D) via auxin induced degradation. (A and C) Representative wide-field fluorescence microscopy images at different times after auxin treatment. (B and D) Intensity of Mex67-EGFP at the nuclear envelope. Mean of three biological replicates is shown. Error bars represent SEM. At least 300 cells were analyzed per condition and replicate. (E–J) FRAP analysis of Mex67-EGFP dynamics at the nuclear envelope upon depletion of RNA Pol II or RNA Pol I via auxin induced degradation. (E and H) Representative images of FRAP experiments. (F and I) FRAP curves normalized to prebleach and postbleach values. Mean curves of ≥50 cells are shown. (G and J) Half time of recovery retrieved from fitting FRAP. The line represents the median, the box represents the 25th to 75th percentile, whiskers extend to the 5th to 95th percentile, and black dots represent data points outside this range. Data in F and G are pooled from three biological replicates, and data in I and J from two. P values are from a Mann–Whitney test. Scale bars, 2 µm.
Mex67 binding at the NPC is not affected by Dbp5
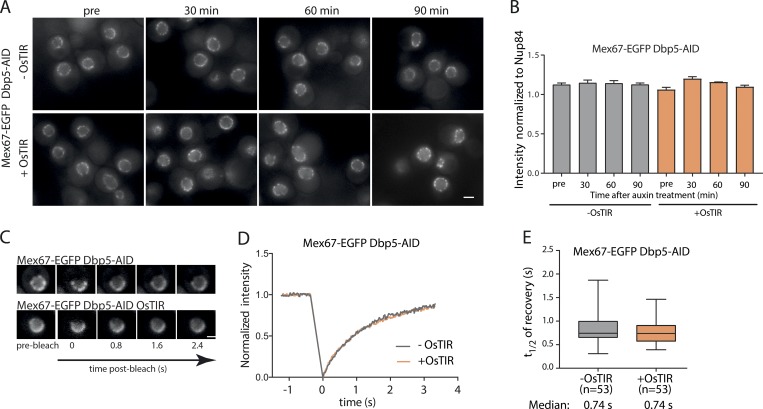
Dbp5 was proposed to remodel mRNPs at the cytoplasmic side of the NPC and to remove Mex67 and other mRNA export factors from mRNA via its ATPase activity (Cole and Scarcelli, 2006a). To test whether ATP hydrolysis by Dbp5 affects Mex67 dynamics at the NPC, we depleted either Dbp5 itself or Gle1, which greatly stimulates ATP hydrolysis by Dbp5, using the AID system. Surprisingly, depletion of Dpb5 (Fig. S2, A and B) affected neither the steady-state levels of Mex67 at the NPC (Fig. 4, A and B) nor its turnover (Fig. 4, C–E). Depletion of Gle1, which led to a reduction of Dbp5 at the NPC (Fig. S2 C), also had no effect on Mex67 intensity and dynamics at the NPC (Fig. S2, D and E). These results argue that the interaction of Mex67 with the FG-repeat network of the NPC is not regulated by the activity of Dbp5. This is incompatible with the hypothesis that the removal of mRNA export factors from the RNA by Dbp5 releases these factors into the cytoplasm (Cole and Scarcelli, 2006a). Thus, while Dbp5 may decrease the interaction of Mex67 with mRNA, it does not influence its interaction with the NPC.
Figure 4.
Mex67 binding to the NPC is independent of Dbp5. (A and B) NuRIM quantification of Mex67-EGFP intensity at the nuclear envelope upon depletion of Dbp5 via auxin induced degradation. (A) Representative wide field fluorescence microscopy images at different times after auxin treatment. (B) Intensity of Mex67-EGFP at the nuclear envelope. Mean of three biological replicates is shown. Error bars represent SEM. At least 300 cells were analyzed per condition and replicate. (C) FRAP analysis of Mex67-EGFP dynamics at the nuclear envelope upon depletion of Dbp5 via auxin-induced degradation. (C) Representative images of FRAP experiments. (D) FRAP curves normalized to prebleach and postbleach values. Mean curves of ≥50 cells are shown. (E) Half time of recovery retrieved from fitting individual FRAP curves. The line represents the median, the box represents the 25th to 75th percentile, and the whiskers extend to the 5th to 95th percentile. Data in D and E are pooled from three biological replicates. Scale bars, 2 µm.
The localization of Mex67 at the NPC is mediated by multiple FG repeats
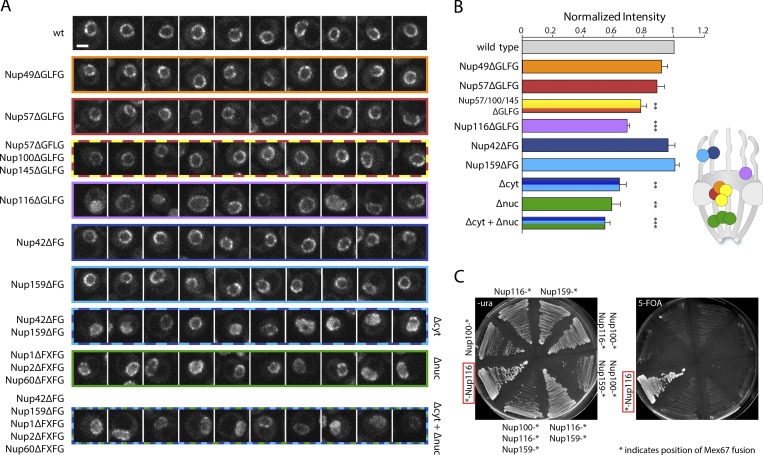
As binding of Mex67 to the NPC does not depend on the presence of mRNA and is not modulated by Dbp5, we wanted to further characterize the FG-binding sites of Mex67 at the NPC by applying NuRIM in a panel of FG repeat deletion mutants (Strawn et al., 2004; Fig. S3 A). First, we examined GLFG (glycine–leucine–phenylalanine–glycine) deletions of Nups localized in the center of the NPC. The deletion of the adhesive GLFG repeats of either Nup49 or Nup57 alone or the combination of Nup57, Nup100, and Nup145 repeats had only moderate effects on Mex67 localization, reducing its level at the nuclear envelope by ∼10% (Fig. 5, A and B). In contrast, deletion of the GLFG repeats of Nup116 led to a more severe reduction, with a loss of ∼20% of Mex67 from the NPC. In addition, an increased pool of nucleoplasmic Mex67 could be detected in a subset of nup116ΔGLFG cells. Next, we analyzed the effect of deleting FG repeat regions of asymmetrically localized Nups. While the deletion of the FG repeats of either Nup42 or Nup159 alone had no effect on Mex67 intensity at the nuclear envelope, the co-deletion of both these FG repeats on the cytoplasmic face of the NPC (Δcyt) led to a severe displacement of Mex67 from the NPC (reduction by ∼40%) and increased levels of Mex67 in the nuclear interior. Both nucleoplasmic enrichment and loss from the nuclear envelope were heterogenous phenotypes and varied in different cells (Fig. 5 A). A similar severe mislocalization of Mex67 was also observed in a triple deletion of the FxFG repeats of the three nuclear basket Nups, Nup1, Nup2, and Nup60 (Δnuc), which was exacerbated in cells where the deletion of cytoplasmic and nucleoplasmic FG repeats were combined (Δcyt+Δnuc). Note that nuclei with very low EGFP signal are excluded by the NuRIM algorithm, leading to an overestimation of the average intensity at the nuclear envelope in these severely compromised strains (Fig. 5 B). Overall, our results suggest that Mex67 does not have a single, predominant NPC interaction site, but instead multiple FG repeats contribute, directly or indirectly, to the steady-state localization of Mex67 at the NPC.
Figure 5.
Mex67 relies on multiple FG repeats for binding the NPC and fulfills its essential function only at the NPC. (A) Example nuclei are shown for different strains expressing Nups that have deleted FG/GFLG/FxFG regions as indicated. Images are single slices of z-stacks acquired on a spinning disk microscope. Slices were selected for being equatorial planes using an ER marker channel. Scale bar, 2 µm. (B) Intensity of Mex67 at the nuclear envelope using NuRIM in different strains with deletions of various FG repeats. Shown is the mean of three biological replicates normalized to wild-type cells. Error bars represent the SEM. Stars indicate significant P values of a paired t test compared with wild type (**, P < 0.01; ***, P < 0.001). Schematic indicates approximate positions of the NPC components in the NPC structure. Note that this applies to the body of the protein and not necessarily to its FG repeat region. (C) N-terminal fusion of Mex67 to Nup116 rescues deletion of MEX67 locus. Strains in which a deletion of the MEX67 locus was covered with Mex67 expressed from a plasmid containing the URA3 selection marker gene were engineered to express fusions of Mex67 to different nucleoporins. Streaks of these strains on 5-FOA–containing plate indicate rescue only by the N-terminal Nup116 fusion. Asterisk indicates position of Mex67 fusion.
The essential function of Mex67 is restricted to the NPC
Our results so far do not support a model where Mex67 functions as a shuttling transport receptor that binds to its substrate in the nucleus and is released into the cytoplasm at the termination of export. Instead, they suggest that Mex67 behaves like a dynamic nuclear pore component, where it resides in the FG-repeat network independently of mRNA. We therefore tested whether the essential function of Mex67 could be entirely restricted to the NPC, and asked whether a protein fusion of Mex67 to a nucleoporin could rescue a deletion of the endogenous MEX67 gene. To this end, we generated strains where Mex67 was fused to different components of the NPC in a background where a deletion of MEX67 was covered by a plasmid carrying the full MEX67 locus as well as the URA3 selection marker (Segref et al., 1997). Since the deletion of the GLFG repeats of Nup116 as a single component had the most drastic effect on Mex67 localization, we generated N- as well as C-terminal fusions of Mex67 to Nup116. In addition, C-terminal fusions with Nup100, Nup1, and Nup159 were tested. Nup-Mex67 fusion strains were tested for growth on 5-fluoroorotic acid (5-FOA) plates to identify strains that are viable in the absence of the wild-type Mex67 protein (Fig. 5 C). Remarkably, the N-terminal fusion of Mex67 to Nup116 supported growth on 5-FOA, suggesting that Mex67 is able to perform its essential function even when it is permanently tethered to the NPC via Nup116. After loss of the Mex67 plasmid, the Mex67-Nup116 strain exhibited only slight growth defects when compared with the parental strain at 30°C or 37°C, although it showed delayed growth at lower temperatures (Fig. S3 B). Western blotting confirmed that the only Mex67 variant detectable in this strain is a large fusion protein (Fig. S3 C). In addition, a C-terminal EGFP tag on Mex67-Nup116 was used to verify the identity of the high molecular weight band as the Mex67-Nup116 fusion protein and to confirm the exclusive and stable localization of the fusion protein to the nuclear envelope (Fig. S3, D and E). Fusion of Mex67 to the flexible FG repeat region of Nup116 thus allows it to perform its function even when stably tethered to the NPC, showing that its essential function is spatially restricted to the NPC.
Conclusion
In summary, we have shown that the RNA export factor Mex67 functions as a mobile nuclear pore component that dynamically binds to the NPC independently of cargo. Furthermore, the interaction of Mex67 with the NPC is not modulated by the DEAD-box ATPase Dbp5. These findings are consistent with a recent report in mammalian cells (Ben-Yishay et al., 2019). Since Dbp5 is thought to impart directionality to the mRNA export process by releasing export factors from cargo, this suggests that the action of Dbp5 leads to the release of mRNA from Mex67 but not of Mex67 from the NPC. By fusing Mex67 to a nucleoporin, we further demonstrate that the essential function of Mex67 occurs entirely at the NPC and propose that Mex67 can receive its export cargo within the central channel of the NPC to facilitate its translocation across the FG-repeat diffusion barrier. In this model, loading of the mature mRNP onto Mex67-Mtr2 happens at the NPC. It will now be interesting to further determine which steps of mRNP maturation occur in the nucleoplasm and which processes are performed at the NPC.
Materials and methods
Plasmid construction
Plasmids were constructed using standard molecular biology techniques. Fragments generated by PCR were verified by sequencing. pKW3124 was cloned by restriction cloning inserting a Nop1-RFP fragment from pRS416-NOP1-RFP (Rothstein laboratory, Columbia University, New York, NY) into pRS406 with NotI/SalI. In pKW3235 and pKW3570, the A206->K206 mutation was introduced to obtain monomeric GFP (Zacharias et al., 2002). pKW4216 used for mating type switching in URA+ strains was derived from YCp50-HO (Russell et al., 1986). pKW4547 was constructed by insertion of Mex67 into a pFA6a-hisMX6 plasmid with a long N-terminal linker of 110 amino acids. pKW4648 was constructed by replacing EGFP in pKW4588 (Vallotton et al., 2019) with Mex67 using RecA cloning and changing the selection marker from URA3 to LEU2 using restriction cloning. pKW4688 was derived from pKW4648 by addition of a segment of the Nup116 promoter upstream of the selection marker to allow for single integration. All plasmids used in this study are listed in Table S1. Primers are listed in Table S2.
Yeast strain construction
Saccharomyces cerevisiae strains were constructed using standard yeast genetic techniques either by transformation of a linearized plasmid or of a PCR amplification product with homology to the target site (Baudin et al., 1993). Strains used for FCS were tagged with a monomeric GFP version obtained by the A206->K206 mutation. Strains with a deletion of MEX67 covered by shuffle plasmid were based on the Mex67 shuffle strain received from the Hurt group (Segref et al., 1997). Strains with different deletions of FG repeats were based on strains received from the Wente group (Strawn et al., 2004) by integration of an ER marker (DsRed-HDEL) and tagging of Mex67 with EGFP. All yeast strains are listed in Table S3.
Yeast culture conditions
Cells were cultured in YPD (for Western blots and growth assays) or complete synthetic medium supplemented with extra adenine (for microscopy) in the presence of 2% glucose. Cells were grown at 25°C for most experiments. All experiments were performed with cells in exponential growth phase. For microscopy experiments, cells were usually inoculated from saturated cultures into fresh medium, grown overnight to OD600 0.3–0.8, and then imaged. Alternatively, cells were diluted in the morning and grown for an additional two or three cell cycles before the start of the experiment. Auxin treatment was performed with auxin (500 µM indole-3-acetic acid; 500 mM stock in ethanol) and IP6 (4 µM phytic acid dipotassium salt, 40 mM stock in water). As a solvent control, cells were treated with IP6 and ethanol.
Microscopy
Cells were pregrown and inoculated in 1 ml complete synthetic medium in 24-well plates overnight so that they reached an OD of 0.3–0.8 for the experiment. Cells were then transferred to 384-well plates (Matrical) or 8-well Ibidi dishes coated with concanavalin A (stock solution of 0.2 mg/ml, air-dried). All imaging of live cells was performed at 25°C.
Images for Fig. 5 A were acquired on a Spinning Disk microscope (Yokogawa Confocal Scanner Unit CSU-W1-T2) built on a Nikon TiE body and controlled with the VisiVIEW software and a 100× NA 1.49 CFI Apo total internal reflection fluorescence objective. The camera was an iXon Ultra EMCCD (Andor), and excitation lasers were a diode pumped solid-state (DPSS) 488 nm (200 mW) and a diode 561 nm (200 mW) laser. Z scanning was performed in streaming mode with a LUDL BioPrecision2 Piezo Stage with 100-ms exposure times per frame. Filters were Dichroic quad-band DAPI/GFP/RFP/CY5, splitting filter to camera ports, 561LP, emission filters GFP/ET525/50 and mCherry ET630/75, respectively.
Western blotting
Cells were grown overnight at 25°C and diluted to an OD600 of 0.2. Cells were grown for an additional two or three cell cycles at 25°C shaking until they reached an OD600 of 0.6–0.9. A volume of yeast culture of was spun down, supernatant was discarded, and cells were incubated in 1 ml of 1 M NaOH for 10 min. Afterward, cells were spun down and resuspended in 30–60 µl 1× SDS buffer/DTT 100 mM, boiled for 5 min at 95°C, and stored at −20°C. The samples were thawed and boiled again for 5 min at 95°C. The samples were usually loaded on 12% resolving SDS-PAGE gel or, in case of the fusions Mex67-Nup116, on precast NuPAGE 4–12% Bis-Tris Protein Gels (Invitrogen). Sample loading was adjusted based on equivalent OD600 in each lane. The SDS-PAGE was run for 50–60 min at 200 V. The proteins were blotted onto a nitrocellulose membrane (Amersham Protran 0.2 NC, GE Healthcare) for 1–2 h at 300 mA. Blots were blocked in 5% milk in TBS and 0.1% Tween 20 (TBST) for at least 2 h or overnight. The first antibody was applied overnight in 1–5% milk in TBST on a rotating wheel at 4°C. The membrane was washed three times in TBST for 20 min and stained with the secondary antibody in 1–5% milk in TBST for 40 min at RT. The blot was washed with TBST three times for 20–40 min before imaging with the CLx ODYSSEY Li-COR. Antibodies used were rabbit anti-Mex67 (Dargemont Lab, Diderot University, Paris, France; 1:2,000), mouse monoclonal anti-GFP (Roche, 11814460 001; 1:2,000), monoclonal mouse anti V5 (Invitrogen, R960-25; 1:2,000), mouse monoclonal anti-yeast PGK (22C5D8; Thermo Fisher Scientific, 4592250; 1:3,000), goat anti mouse IgG Alexa Fluor 680 (Thermo Fisher Scientific, A-21057; 1:10,000), and goat anti-rabbit IgG IRDye800CW (Li-COR Biosciences, 926-32211; 1:10,000).
NuRIM fluorescence intensity measurements
NuRIM fluorescence intensity quantification experiments were performed on a temperature-controlled Nikon Ti Eclipse equipped with a Spectra X LED lamp using a Apochromat VC 100× objective NA 1.4 (Nikon; filters: spectra emission filters 475/28 and 542/27 and DAPI/FITC/Cy3/Cy5 Quad HC Filterset with 410/504/582/669 HC Quad dichroic and a 440/521/607/700 HC quadband filter [Semrock]) with exposure times of 500 ms in the GFP and 1 s in the RFP channel. 30 image frames were imaged per sample and condition.
Images were processed using NuRIM code (Rajoo et al., 2018; Vallotton et al., 2019) in MATLAB (MathWorks). Briefly, this method uses the HDEL-DsRed signal to generate a mask that segments the nuclear envelope and then extracts intensity from this mask. Machine learning is applied to remove the contribution from variable background signals.
FRAP
FRAP experiments were performed on the Leica TCS SP8 STED/FLIM/FCS or a Leica TCS SP8-AOBS microscope using a 63× 1.4 NA Oil HC PL APO CS2 objective. Bidirectional scanner at speed of 8,000 Hz, NF488/561/633, an Airy unit of 1.5, and a FRAP booster for bleaching were applied for every FRAP experiment using the PMT3 (500–551 nm) and PMT5 (575–694 nm) detectors. Image size of 504 × 50 and a zoom of 3.5 were used together with line accumulation of three, yielding a frame rate of 60.61 frames/s with a pixel dwell time of 73 ns. 50 prebleach and 200 postbleach frames were acquired. A 488-nm argon laser line was used at 50% power in addition to a 561-nm DPSS laser line. The bleach was performed using 100% argon laser power for 200 ms. Imaging was conducted with 1.5% laser intensity with a gain of 800 to illuminate the GFP, and 0.3% of the 561 laser power to illuminate the HDEL with DsRed was used during prebleach and postbleach acquisition. The photobleaching was performed by manually defining an elliptical region comprising approximately one third of the cell nucleus. The mobility of GFP-labeled proteins in the bleached nuclear envelope region was evaluated by quantifying the signal recovery in the bleached region. Extracellular background (Ibg) was subtracted from the intensity of the bleached region (Ibl), and the values were bleach-corrected by normalizing for total cell intensity (Itotal) resulting in (Ibl − Ibg)/(Itotal − Ibg) (Bancaud et al., 2010). The datasets acquired during three independent experiments were processed using MATLAB with custom-written scripts. Briefly, the intensities were normalized for prebleach and postbleach values and the curves were fitted with an exponential function: ft = A ⋅ [1−exp(−r ⋅ t)] (ft = normalized fluorescence intensity, A = nonrecovering pool, r = time constant of recovery, t = time), with the method “NonlinearLeastSquares.” The measurements of three independent experimental days were pooled. Plots were generated in GraphPad Prism 7 (GraphPad).
FCS
FCS experiments were performed with a Leica TCS SP8 STED/FLIM/FCS microscope equipped with a 63× 1.2 NA W HC PL APO CS2 objective with the HyD 2 SMD (Single Molecule Detection; 500–550 nm) and HyD 4 SMD (604–750 nm). The Leica components were used in combination with the PicoHarp 300, TCSPC Module (Time Correlated Single Photon Counting system), 4 Channel Detector Router (PHR 800), and the Software SymPho Time 64 1.6. FCS experiments were conducted using the 488-nm argon laser line at 20% power set to 0.1–0.2% for acquisition and the pulsed white light laser at 70% with a laser pulse of 20 MHz at 594 nm set to 0.3–0.7% for acquisition. Cells were imaged for 30 s, and traces were recorded and saved by the PicoQuant software. FCS measurements were performed in diploid yeast cells in the center of the nucleus in order to place the entire confocal volume with high confidence within the nuclear boundary.
Analysis of FCS data was performed using a Fluctuation 4G analyzer (Wachsmuth et al., 2015). Raw photon traces with strong fluctuations were discarded. Autocorrelation curves of the accepted in vivo photon traces were calculated over a rolling time window of 0.5 to 5 s and averaged over the whole duration of the photon trace (Wachsmuth et al., 2015). This approach allows removing the contribution of slow fluctuations arising from photobleaching or small cell movement. Autocorrelation curves were fit with a one-component anomalous diffusion model including the fluorescence blinking contribution. This model gave access to a mean diffusion time corresponding to the average duration of the transit of the fluorescent molecule through the confocal volume. When fitting the autocorrelation curves, the lifetime of the dark state was fixed to 100 µs (Wachsmuth et al., 2015) and the structural parameter kappa was fixed to 5, based on experimentally determined values. Anomality parameters obtained from fits for EGFP, Dbp5, and Mex67 were 0.84 ± 0.04, 0.80 ± 0.02, and 0.80 ± 0.01 in the nucleus and 0.98 ± 0.03, 0.71 ± 0.01, and 0.84 ± 0.01 in the cytoplasm, respectively (mean ± SEM).
FISH
FISH was performed according to Dultz et al. (2018). Briefly, cells were grown into exponential growth phase at 25°C. Cells were then treated with auxin and IP6 and fixed after one (Dpb5-AID) or two (Rpb2-AID) hours for 25 min at 25°C and with 4% paraformaldehyde (EM grade 32% paraformaldehyde aqueous solution electron; Microscopy Sciences, 15714), washed with buffer B (1.2 M sorbitol and 100 mM KHPO4, pH 7.5, at 4°C) and stored at 4°C overnight. Cells were then spheroplasted for 20 min using 1% 20T zymolyase in 1.2 M sorbitol, 100 mM KHPO4, pH 7.5, 20 mM vanadyl ribonuclease complex, and 20 µM β-mercaptoethanol; washed with buffer B to stop the spheroplasting reaction; and then washed into 10% formamide (Merck Millipore, S4117) in 2× SSC (saline-sodium citrate buffer prepared from 20× SSC; Life Technologies, AM9624).
DNA probes targeting FBA1 coupled to CalFluore Red (Stellaris, LGC Biosearch; probes were synthesized by BioCat; Table S4) or oligo dT30 coupled to ATTO488 (Microsynth AG) were used. Per sample, 0.5 µl of FBA1 probe mix (stock 25 µM) or 0.25 µl of oligo dT30 (stock 50 µM) was mixed with 2 µl of salmon-sperm DNA (10 mg/ml; Life Technologies, 15632-011) and 2 µl yeast transfer RNA (10 mg/ml; Life Technologies, AM7119). The probe mix was denatured in 50 µl per sample of hybridization buffer F (20% formamide and 10 mM NaHPO4, pH 7.0) for 3 min at 95°C and then mixed with 50 µl per sample hybridization buffer H (4× SSC, 4 mg/ml BSA [acetylated], and 20 mM vanadyl ribonuclease complex). Cells (approximately corresponding to the pellet of 5-ml initial culture) were resuspended in the hybridization mix and incubated overnight at 37°C. After four washing steps (10% formamide/2× SSC; 0.1% Triton/2× SSC; 2× SSC/DAPI; and 2× SSC), cells were stored at 4°C. Cells were imaged in concanavalin A–coated 384 wells. Microscopy was performed with an inverted epi-fluorescence microscope (Nikon Ti) equipped with a Spectra X LED light source and a Hamamatsu Flash 4.0 scientific complementary metal–oxide–semiconductor camera using a PlanApo 100× NA 1.4 oil-immersion objective (Nikon) and the NIS Elements software. 27 z slices were acquired at 200-nm spacing.
Online supplemental material
Fig. S1 shows that RNA Pol II and RNA Pol I can be efficiently degraded via an AID (control experiments related to Fig. 3). Fig. S2 shows that Dbp5 can be efficiently degraded via an AID and that depletion of Gle1 leads to loss of Dbp5 from the nuclear envelope (control experiments related to Fig. 4). Fig. S3 characterizes the functionality of the Mex67-Nup116 fusion (control experiments related to Fig. 5 C). Tables S1, S2, S3, and S4 show plasmids, primers, yeast strains, and FISH probes, respectively.
Supplementary Material
Acknowledgments
We thank Malthe Wachsmuth for his help with Fluctuation Analyzer 4G for FCS data analyses. We are grateful to Justine Kusch from ScopeM, ETH Zürich, for microscopy support, and to Catherine Dargemont for providing the anti-Mex67 antibody. We thank members of the Weis group for discussions and critical reading of the manuscript. We are particularly grateful to Stephanie Heinrich for helpful discussions and advice.
K. Weis acknowledges support from the Swiss National Science Foundation (project number 31003A_179275).
The authors declare no competing financial interests.
Author contributions: This work was devised and planned by C.P. Derrer, E. Dultz, and K. Weis. Experiments were carried out by C.P. Derrer, E. Dultz, R. Mancini, and P. Vallotton. Data were analyzed by C.P. Derrer, E. Dultz, and P. Vallotton. Code for data analysis was produced by P. Vallotton and E. Dultz. The FCS experiments were supported by S. Huet. The manuscript was written by C.P. Derrer, E. Dultz, and K. Weis and revised by all authors.
References
- Adams R.L., Mason A.C., Glass L., Aditi, and Wente S.R.. 2017. Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells. Traffic. 18:776–790. 10.1111/tra.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancaud A., Huet S., Rabut G., and Ellenberg J.. 2010. Fluorescence perturbation techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, photoactivation, photoconversion, and FLIP. Cold Spring Harb. Protoc. 10.1101/pdb.top90 [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., and Cullin C.. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. 10.1093/nar/21.14.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yishay R., Mor A., Shraga A., Ashkenazy-Titelman A., Kinor N., Schwed-Gross A., Jacob A., Kozer N., Kumar P., Garini Y., and Shav-Tal Y.. 2019. Imaging within single NPCs reveals NXF1's role in mRNA export on the cytoplasmic side of the pore. J. Cell Biol. 218:2962–2981. 10.1083/jcb.201901127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C.N., and Scarcelli J.J.. 2006a Transport of messenger RNA from the nucleus to the cytoplasm. Curr. Opin. Cell Biol. 18:299–306. 10.1016/j.ceb.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Cole C.N., and Scarcelli J.J.. 2006b Unravelling mRNA export. Nat. Cell Biol. 8:645–647. 10.1038/ncb0706-645 [DOI] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., and Stutz F.. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26:7858–7870. 10.1128/MCB.00870-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E., Mancini R., Polles G., Vallotton P., Alber F., and Weis K.. 2018. Quantitative imaging of chromatin decompaction in living cells. Mol. Biol. Cell. 29:1763–1777. 10.1091/mbc.E17-11-0648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faza M.B., Chang Y., Occhipinti L., Kemmler S., and Panse V.G.. 2012. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet. 8:e1002915 10.1371/journal.pgen.1002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann A.W., Noble K.N., Cole C.N., and Wente S.R.. 2011. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2:540–548. 10.4161/nucl.2.6.17881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., and Guthrie C.. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell. 13:201–212. 10.1016/S1097-2765(04)00030-9 [DOI] [PubMed] [Google Scholar]
- Hautbergue G.M., Hung M.L., Golovanov A.P., Lian L.Y., and Wilson S.A.. 2008. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. USA. 105:5154–5159. 10.1073/pnas.0709167105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Derrer C.P., Lari A., Weis K., and Montpetit B.. 2017. Temporal and spatial regulation of mRNA export: Single particle RNA-imaging provides new tools and insights. BioEssays. 39:1600124 10.1002/bies.201600124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C.A., Tran E.J., Noble K.N., Alcazar-Roman A.R., Ben-Yishay R., Scarcelli J.J., Folkmann A.W., Shav-Tal Y., Wente S.R., and Cole C.N.. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 25:1052–1064. 10.1101/gad.2041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strässer K., Podtelejnikov A., Mann M., Jung J.U., and Hurt E.. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593–2609. 10.1093/emboj/18.9.2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., and Hurt E.. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773. 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Liker E., Fernandez E., Izaurralde E., and Conti E.. 2000. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 19:5587–5598. 10.1093/emboj/19.21.5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M.K., and Guthrie C.. 2005. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 20:645–651. 10.1016/j.molcel.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Montpetit B., Thomsen N.D., Helmke K.J., Seeliger M.A., Berger J.M., and Weis K.. 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 472:238–242. 10.1038/nature09862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Noble K.N., Tran E.J., Alcázar-Román A.R., Hodge C.A., Cole C.N., and Wente S.R.. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 25:1065–1077. 10.1101/gad.2040611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajoo S., Vallotton P., Onischenko E., and Weis K.. 2018. Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 115:E3969–E3977. 10.1073/pnas.1719398115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., and Stutz F.. 2004. Nuclear export of RNA. Biol. Cell. 96:639–655. 10.1016/j.biolcel.2004.04.014 [DOI] [PubMed] [Google Scholar]
- Russell D.W., Jensen R., Zoller M.J., Burke J., Errede B., Smith M., and Herskowitz I.. 1986. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory region. Mol. Cell. Biol. 6:4281–4294. 10.1128/MCB.6.12.4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., Panté N., and Hurt E.. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18:6826–6838. 10.1128/MCB.18.11.6826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., and Hurt E.. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256–3271. 10.1093/emboj/16.11.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter B.D., Schwartz J.W., and Li R.. 2007. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc. Natl. Acad. Sci. USA. 104:20320–20325. 10.1073/pnas.0710336105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge C.A., Colot H.V., Goldstein A.L., and Cole C.N.. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17:2663–2676. 10.1093/emboj/17.9.2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. 2007. Ratcheting mRNA out of the nucleus. Mol. Cell. 25:327–330. 10.1016/j.molcel.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Strässer K., and Hurt E.. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 413:648–652. 10.1038/35098113 [DOI] [PubMed] [Google Scholar]
- Strässer K., Bassler J., and Hurt E.. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695–706. 10.1083/jcb.150.4.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L.A., Shen T., Shulga N., Goldfarb D.S., and Wente S.R.. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197–206. 10.1038/ncb1097 [DOI] [PubMed] [Google Scholar]
- Terry L.J., and Wente S.R.. 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178:1121–1132. 10.1083/jcb.200704174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Zhou Y., Corbett A.H., and Wente S.R.. 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 28:850–859. 10.1016/j.molcel.2007.09.019 [DOI] [PubMed] [Google Scholar]
- Vallotton P., Rajoo S., Wojtynek M., Onischenko E., Kralt A., Derrer C.P., and Weis K.. 2019. Mapping the native organization of the yeast nuclear pore complex using nuclear radial intensity measurements. Proc. Natl. Acad. Sci. USA. 116:14606–14613. 10.1073/pnas.1903764116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moeller H., Basquin C., and Conti E.. 2009. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 16:247–254. 10.1038/nsmb.1561 [DOI] [PubMed] [Google Scholar]
- Wachsmuth M., Conrad C., Bulkescher J., Koch B., Mahen R., Isokane M., Pepperkok R., and Ellenberg J.. 2015. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 33:384–389. 10.1038/nbt.3146 [DOI] [PubMed] [Google Scholar]
- Weirich C.S., Erzberger J.P., Flick J.S., Berger J.M., Thorner J., and Weis K.. 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8:668–676. 10.1038/ncb1424 [DOI] [PubMed] [Google Scholar]
- Weis K. 2002. Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol. 14:328–335. 10.1016/S0955-0674(02)00337-X [DOI] [PubMed] [Google Scholar]
- Wu B., Chao J.A., and Singer R.H.. 2012. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J. 102:2936–2944. 10.1016/j.bpj.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., and Hurt E.. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 26:51–62. 10.1016/j.molcel.2007.02.018 [DOI] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., and Tsien R.Y.. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916. 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zhao J., Jin S.B., Björkroth B., Wieslander L., and Daneholt B.. 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 21:1177–1187. 10.1093/emboj/21.5.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.