Stress granules and paraspeckles are prototypical RNP granules localized in the cytoplasm and nucleus, respectively. An et al. show that despite spatial separation, these two granules are interconnected, and stress granules act as key regulators of stress-induced paraspeckle hyperassembly.
Abstract
Eukaryotic cells contain a variety of RNA-protein macrocomplexes termed RNP granules. Different types of granules share multiple protein components; however, the crosstalk between spatially separated granules remains unaddressed. Paraspeckles and stress granules (SGs) are prototypical RNP granules localized exclusively in the nucleus and cytoplasm, respectively. Both granules are implicated in human diseases, such as amyotrophic lateral sclerosis. We characterized the composition of affinity-purified paraspeckle-like structures and found a significant overlap between the proteomes of paraspeckles and SGs. We further show that paraspeckle hyperassembly is typical for cells subjected to SG-inducing stresses. Using chemical and genetic disruption of SGs, we demonstrate that formation of microscopically visible SGs is required to trigger and maintain stress-induced paraspeckle assembly. Mechanistically, SGs may sequester negative regulators of paraspeckle formation, such as UBAP2L, alleviating their inhibitory effect on paraspeckles. Our study reveals a novel function for SGs as positive regulators of nuclear RNP granule assembly and suggests a role for disturbed SG-paraspeckle crosstalk in human disease.
Introduction
A whole repertoire of large, microscopically visible RNA-protein complexes termed RNP granules are present in mammalian cells (Anderson and Kedersha, 2006). RNP granules are membraneless structures assembled and maintained through a combination of protein–protein, protein–RNA, and RNA–RNA interactions (Protter and Parker, 2016). The presence of multiple RNA-binding proteins featuring intrinsically disordered regions, also called low-complexity domains, in their structure allows liquid-liquid phase separation and formation of a distinct condensate surrounded by nucleoplasm or cytoplasm (Boeynaems et al., 2018; Gomes and Shorter, 2019). RNP granules regulate key processes related to mRNA localization, translation, and stability.
RNP granules are especially abundant in the nucleus, where they are often referred to as nuclear bodies (Mao et al., 2011); here belong paraspeckles (PSs), gems, and promyelocytic leukemia bodies, among others. The most well-known constitutive cytoplasmic RNP granules are processing bodies (P-bodies) and neuronal RNA transport granules (Kiebler and Bassell, 2006; Standart and Weil, 2018). RNP granules can be assembled de novo under various physiological and pathological conditions, the most prominent examples being nuclear stress bodies and stress granules (SGs) formed in stressed cells (Biamonti and Vourc’h, 2010; Buchan and Parker, 2009). SGs are large cytoplasmic RNP granules that form as a normal cellular response to medium to severe stresses (Kedersha and Anderson, 2002). Their assembly is usually caused by phosphorylation of the eukaryotic translation initiation factor 2α (eIF2α), leading to stalled translation and release of mRNA from polysomes (Kedersha et al., 2002). Several constitutive RNP granules, such as PSs and promyelocytic leukemia bodies, also respond to stress, by increasing their size and/or number (Fox et al., 2018; Lallemand-Breitenbach and de Thé, 2010). PSs are nuclear RNP granules found in the interchromatin space, adjacent to splicing speckles (Fox and Lamond, 2010). PSs contain several core and multiple additional proteins that are assembled around the longer isoform of a nuclear-retained long noncoding RNA (lncRNA) NEAT1 (Fox et al., 2018). Although PSs are believed to be stress-responsive nuclear bodies, the unifying molecular signature of PS-inducing stresses is still unclear.
Intense interest in the biology of RNP granules in the past decade has followed the discovery of their tight connection to neurodegenerative diseases, primarily amyotrophic lateral sclerosis (ALS). There is substantial genetic and experimental evidence on the involvement of disturbed SG metabolism in ALS (Li et al., 2013), and the link between PSs and ALS pathogenesis is also emerging (An et al., 2019; Nishimoto et al., 2013; Shelkovnikova et al., 2014, 2018).
Recent development of approaches to determine the composition of membraneless assemblies, including biochemical affinity purification, sorting, and proximity labeling, have allowed characterization of the proteome of cytoplasmic RNP granules such as SGs and P-bodies (Hubstenberger et al., 2017; Jain et al., 2016; Markmiller et al., 2018). In the current study, we applied an affinity purification approach to expand our knowledge of the protein composition of PSs. Analysis of the proteome of PS-like structures and reanalysis of literature data revealed that proteomes of PSs and SGs significantly overlap, which prompted us to investigate the crosstalk between these two types of RNP granules. Cell culture experiments demonstrated an unexpected role for SGs in controlling PS formation and dynamics during stress. Our data suggest that despite spatial separation, SGs and PSs are interconnected, and SGs act as key regulators of PS assembly in response to diverse stress signals.
Results
Identification of novel PS proteins (PSPs)
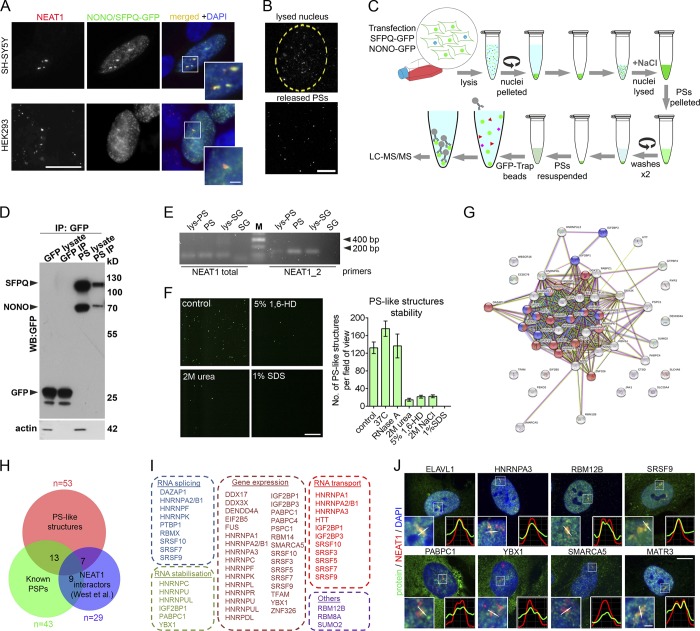
Protein components of PSs were first cataloged through a Venus-tagged human protein library screen, which allowed identification of 40 proteins (Naganuma et al., 2012). Subsequently ELAVL1 and RBFOX2 were added to this list (Mannen et al., 2016). One additional protein, MATR3, could be counted as a PSP based on its previous identification in the NONO-SFPQ complex (Zhang and Carmichael, 2001). To expand the known PS proteome (Table S1, All datasets tab), we employed purification of PS-like structures followed by proteomic analysis. Co-overexpressed GFP-tagged SFPQ and NONO formed NEAT1-positive granules in the nucleus in SH-SY5Y and HEK293 cells (Fig. 1 A), which were stable after cell lysis and their release from the nucleus (Fig. 1 B). These structures remained intact 4 h after lysis and were still detectable 16 h after lysis (74.2 ± 12, 73.8 ± 3.5, and 26.6 ± 4.8 granules per field of view 30 min, 4 h, and 16 h after lysis, respectively). We developed a protocol to purify these PS-like structures from transiently transfected cells, which involves their enrichment from the nuclear fraction, followed by affinity purification using GFP-Trap beads (Fig. 1 C). To ensure that PS-like structures but not soluble nucleoplasmic SFPQ and NONO were captured, pellets of PS-like structures enriched by centrifugation were washed to remove soluble proteins. Efficient pulldown of PS-like structures by GFP-Trap beads was confirmed by Western blot (Fig. 1 D), and RT-PCR analysis showed that these granules contain lncRNA NEAT1 (Fig. 1 E). PS-like structures were phase-separated assemblies since they were sensitive to 1,6-hexanediol, which disrupts weak hydrophobic interactions (Fig. 1 F). Similar to SG cores (Jain et al., 2016), they were not affected by RNase A treatment, were stable at 37°C for at least 1 h, and completely dissipated in the presence of 1% SDS. Unlike SG cores, they were sensitive to 2 M NaCl and 2 M urea (Fig. 1 F).
Figure 1.
Purification and proteomic analysis of PS-like structures. (A) PS-like structures formed by coexpressed SFPQ-GFP and NONO-GFP in SH-SY5Y and HEK293 cells contain NEAT1 as revealed by RNA-FISH. Cells were analyzed 24 h after transfection. Scale bars: main image, 10 µm; inset, 2 µm. (B) PS-like structures during the lysis of purified nuclei. Nuclei of SFPQ/NONO-GFP-expressing HEK293 cells were purified and monitored under fluorescent microscope during lysis. Partially lysed nucleus (top, beginning of lysis) and released PS-like structures (bottom, 30 min of lysis) are shown. Scale bar, 20 µm. (C) Workflow used for affinity purification of PS-like structures. (D) Western blot analysis of GFP-Trap bead purified PS-like structures from cells expressing SFPQ/NONO-GFP using an anti-GFP antibody. Samples from cells expressing GFP alone and processed in parallel were used as a control. For lysate, 1/20 of immunoprecipitation (IP) fraction was loaded on gel. (E) Purified PS-like structures but not SG cores contain NEAT1 as revealed by RT-PCR. Lys-PS and lys-SG, total cell lysates of cells used for PS and SG isolation, respectively. (F) Stability of PS-like structures. PS-like structures in the nuclear lysate were treated as indicated for 1 h and imaged under fluorescent microscope (40× magnification). Representative images and quantification of the numbers of GFP-positive dots per field are shown (four to eight fields analyzed). 1,6-HD, 1,6 hexanediol. Error bars represent SEM. Scale bar, 20 µm. (G) STRING functional protein association for the proteome of PS-like structures. In the network, proteins in categories RNA splicing and RNA transport are shown in blue and red, respectively. (H) Overlap between the proteome of PS-like structures, known pool of PSPs (Naganuma et al., 2012; Mannen et al., 2016; Zhang and Carmichael, 2001) and the pool of NEAT1-interacting proteins previously identified by CHART-seq (West et al., 2014). Numbers within circles indicate the number of proteins common between the datasets. Also see Table S1. (I) Biological function enrichment analysis for proteins identified within PS-like structures (also see Table S1, PS-like structures). (J) Validation of proteins identified in the proteomic analysis. Colocalization of proteins and NEAT1 in human fibroblasts was studied using a combination of NEAT1 RNA-FISH and immunocytochemistry. Fibroblasts were treated with MG132 for 4 h to induce PS clusters. MATR3 was included as a positive control. Scale bars: main image, 10 µm; inset, 1 µm.
Mass spectrometry analysis of preparations of PS-like structures from HEK293 cells identified 53 proteins (including SFPQ and NONO) enriched in these granules, 13 of which are known PSPs (Fig. 1, G and H; and Table S1, PS-like structures and All datasets tabs). Seven of these proteins were also present among 29 NEAT1 binding partners identified by capture hybridization analysis of RNA targets sequencing (CHART-seq; West et al., 2014). PS-like structures were enriched in the components of RNP complexes (19 of 53, false discovery rate [FDR] = 1.75 × 10−13). Functional enrichment analysis showed significant overrepresentation for proteins involved in the regulation of RNA splicing (FDR = 3.78 × 10−10), RNA transport (FDR = 5.32 × 10−11), and RNA stability (FDR = 5.32 × 10−11); 58% proteins (31 of 53) were also involved in the regulation of gene expression (FDR = 1.51 × 10−9; Fig. 1 I and Table S1, PS-like structures tab). Almost half of all proteins (25 of 53) contained at least one RRM domain in their structure (FDR = 6.9 × 10−39). We confirmed the enrichment of several proteins from this analysis in PSs using a combination of RNA-FISH and immunocytochemistry in human fibroblasts (Fig. 1 J). However, it should be noted that PSPs identified in the fluorescent protein screen (Naganuma et al., 2012) are likely most strongly associated with PSs, whereas some of the new proteins from the current study may be present in PSs at a level below microscopic detection limit and/or associate with PSs transiently, including via interaction with PS-localized SFPQ or NONO.
Overlap between the proteomes of PSs and SGs
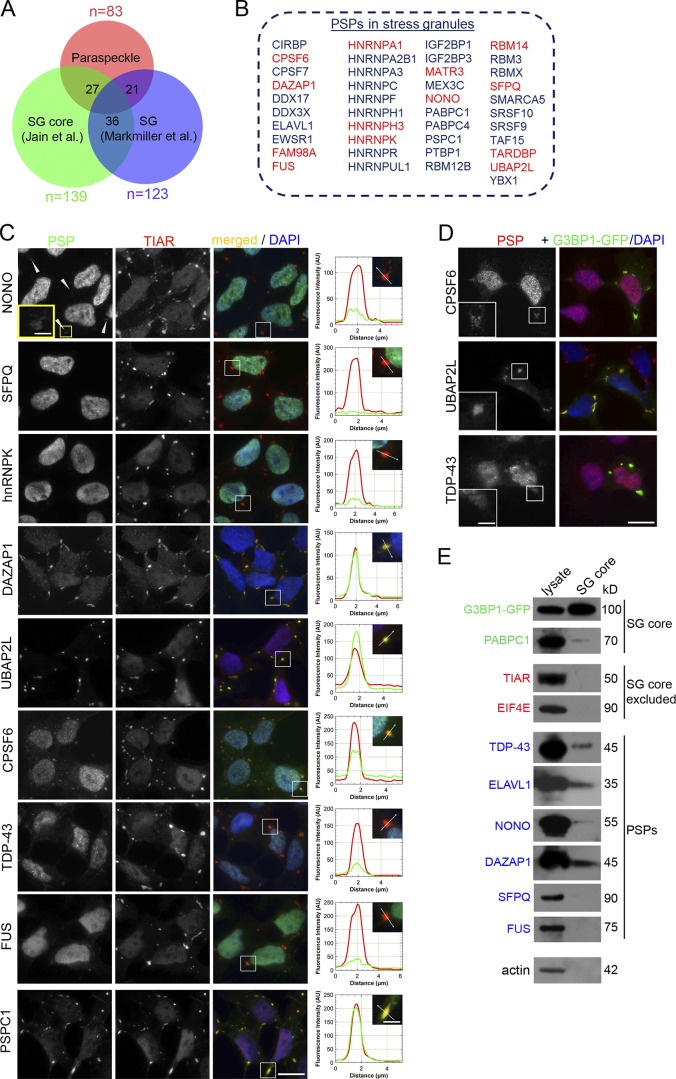
We noticed that several components of PS-like structures are predominantly cytoplasmic proteins known to contribute to SG assembly, e.g., PABPC1, YBX1, and DDX3X. We compared the list of PSPs (83 proteins, comprising known PSPs and components of PS-like structures from this study) with recently published SG proteomes (Jain et al., 2016; Markmiller et al., 2018). The uncrosslinked proteome of SG cores, which comprises 139 proteins most strongly associated with SGs (Jain et al., 2016), was found to contain 27 PSPs (19.4%, P value of overlap < 1.133 × 10−27), and the full SG proteome (411 proteins) from the same study contains 11 additional PSPs. Furthermore, 21 PSPs are present in the SG proteome obtained using APEX-based proximity labeling (Markmiller et al., 2018; P value of overlap <3.853 × 10−16). Therefore, nearly half of all PSPs (41 of 83) are associated with SGs (Fig. 2, A and B; and Table S1, All datasets and PSPs in SGs tabs). Recent analysis of public eCLIP (enhanced crosslinking and immunoprecipitation) data for RNA-binding proteins allowed identification of proteins directly binding NEAT1 (Lin et al., 2018; Nakagawa et al., 2018; Table S1, All datasets tab), and 37% of these proteins (27 of 73) are also present in the SG proteome. This overlap between PS and SG proteomes was a somewhat surprising finding given that the majority of PSPs are predominantly nuclear proteins. Among SG-recruited PSPs, 13 are either essential or important for PS assembly and 8 of them regulate levels of NEAT1 isoforms (Banerjee et al., 2017; Naganuma et al., 2012; Shelkovnikova et al., 2018; Fig. 2 B). We examined the presence of nine core PSPs in SGs induced by various stresses in SH-SY5Y cells (Figs. 2 C and S1). The only predominantly cytoplasmic PSP in this set, UBAP2L, was dramatically enriched in SGs, consistent with the previous report (Markmiller et al., 2018). PSPs abundant in both the nucleus and cytoplasm, DAZAP1 and PSPC1, were also robustly recruited into SGs. Small amounts of predominantly nuclear NONO, TDP-43, CPSF6, FUS, and hnRNPK were also present in SGs. We also verified SG recruitment for three PSPs using overexpressed G3BP1 protein (Fig. 2 D). Finally, we purified SG cores from G3BP1-GFP–expressing cells (Jain et al., 2016; Wheeler et al., 2017) and confirmed that they contain PSPs TDP-43, ELAVL1, DAZAP1, and NONO (Fig. 2 E). Therefore, multiple PSPs are recruited into SGs during stress.
Figure 2.
Multiple PSPs are recruited into SGs. (A) Overlap between PS and SG proteomes. The proteome of PSs (n = 83) comprises known PSPs and proteins recruited to PS-like structures identified in this study. SG proteomes were taken from Jain et al. (2016) and Markmiller et al. (2018). Also see Table S1. (B) List of PSPs recruited into SGs (appearing in SG proteome datasets in Jain et al. [2016] and Markmiller et al. [2018]). Core PSPs, i.e., those regulating NEAT1 levels and/or PS integrity, are given in red. (C) PSP recruitment into SGs. SH-SY5Y cells were treated with NaAsO2 for 1 h, and SGs were visualized using an anti-TIAR antibody. Intensity profiles for representative SGs are also shown. Arrowheads point to SGs weakly positive for NONO. Scale bars: main image, 10 µm; inset, 2 µm. (D) PSP recruitment into SGs formed by overexpressed G3BP1-GFP. SH-SY5Y cells expressing G3BP1-GFP were treated with NaAsO2 for 1 h. Scale bars: main image, 10 µm; inset, 2 µm. (E) Presence of PSPs in SG cores as analyzed by Western blot. SG cores were affinity purified from SH-SY5Y cells treated with NaAsO2 for 1 h.
PS hyperassembly is triggered by SG-inducing stresses
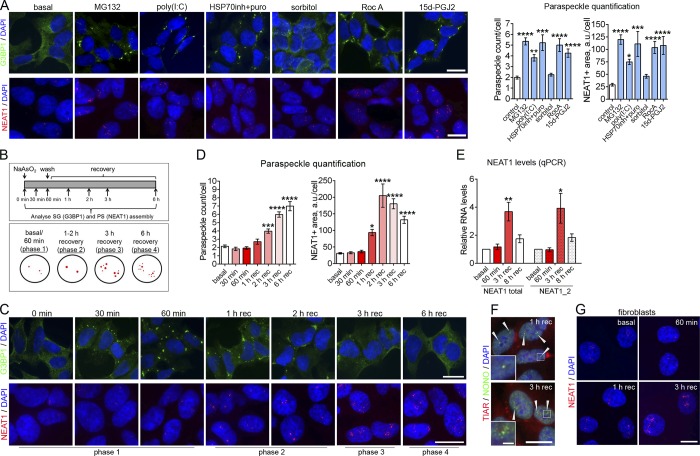
Significant overlap between PS and SG proteomes prompted us to examine possible crosstalk between these two types of RNP granules. Three SG-inducing stressors, namely, proteasome inhibition (Hirose et al., 2014), heat shock (Lellahi et al., 2018), and foreign double-strand RNA (dsRNA; Imamura et al., 2014), have been reported to enhance PS formation. We examined the effect of four additional SG-inducing stressors: osmotic shock with sorbitol (Dewey et al., 2011), exposure to a lipid molecule 15d-PGJ2 (Kim et al., 2007), eIF4A inhibitor rocaglamide A (Kedersha et al., 2016), or a combination of an HSP70 inhibitor and puromycin, which causes accumulation of misfolded proteins and release of mRNA from polysomes (Bounedjah et al., 2014). MG132 and dsRNA poly(I:C) were included as positive controls. We found that the peak of SG assembly for MG132, sorbitol, 15d-PGJ2, rocaglamide A, and HSP70 inhibitor plus puromycin takes place at 2 h, and for poly(I:C), at 6 h after addition of a stressor (Fig. 3 A, upper panel). PS assembly is critically dependent on NEAT1; therefore, we used NEAT1 RNA-FISH as the most reliable method to detect PSs. All SG-inducing treatments tested caused a significant increase in PS assembly, which occurred 2 h after the peak of SG assembly (Fig. 3 A, bottom panel). Of note, sorbitol induced smaller SGs and only a slight increase in the NEAT1-positive area (Fig. 3 A).
Figure 3.
PS hyperassembly is typical for cells subjected to a SG-inducing stress. (A) Effect of SG-inducing stressors on PS assembly. Cells were treated with indicated chemicals and analyzed for SG assembly (after 2 h) and for PS assembly (after 4 h). Cells were also transfected with poly(I:C) and analyzed for SG assembly after 6 h and for PS assembly after 8 h. SGs were visualized with anti-G3BP1 staining and PSs using NEAT1 RNA-FISH. 112–299 cells were included in analysis per condition. ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with Holm–Sidak’s test). (B) Timeline for SG and PS analysis during NaAsO2 treatment (top panel) and phases of PS assembly identified at these time points (bottom panel; NEAT1 signal is depicted in red). (C and D) Dynamics of SG and PS assembly in NaAsO2-treated cells. Representative images (C) and quantitation (D) are shown. SGs were visualized with anti-G3BP1 staining and PSs using NEAT1 RNA-FISH. In D, 87–235 cells were included into analysis per condition. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with Dunnett’s test). (E) NEAT1 levels during NaAsO2 stress as measured by qRT-PCR (n = 4). *, P < 0.05; **, P < 0.01 (Mann–Whitney U test). (F) Recruitment of a core PSP NONO into NaAsO2-induced PSs after 1 and 3 h of recovery (phases 2 and 3, respectively). Arrowheads indicate PS clusters. (G) PS assembly in human fibroblasts during the recovery from NaAsO2 stress. PSs were visualized using NEAT1 RNA-FISH. In A, C, F, and G, representative images are shown. Error bars represent SEM. Scale bars, 10 µm; inset in F, 2 µm.
What puzzled us, however, is that although sodium arsenite (NaAsO2) is the most widely used, robust trigger of SG assembly, in our previous studies we failed to detect PS hyperassembly in NaAsO2-treated cells. Since for all other stresses examined, PS hyperassembly was delayed by 2 h relative to the peak of SG formation, we hypothesized that a 60-min NaAsO2 exposure, typically used to induce SGs, is too short to have an impact on PSs. We thus examined PS formation during the recovery from NaAsO2 stress (Fig. 3 B, upper panel). SGs formed in nearly all NaAsO2-treated SH-SY5Y cells within 60 min and completely disappeared 6 h after washing off NaAsO2 (Fig. 3 C, upper panel). In line with our prediction, significant PS accumulation was detectable 2 h into the recovery from NaAsO2 (i.e., ∼2 h after the peak of SG formation, similar to other stressors; Fig. 3, C and D). In agreement with RNA-FISH data, quantitative RT-PCR (qRT-PCR) detected an approximately fourfold increase in NEAT1 levels after 3 h of recovery (Fig. 3 E). After this point, NEAT1 levels started declining and dropped approximately twofold by the 8-h recovery time point, consistent with the NEAT1 half-life of ∼8 h (Clark et al., 2012).
NEAT1 accumulation and PS formation had four distinct phases during the recovery from NaAsO2 stress: (1) basal, few small foci (individual PSs); (2) 1–2 h of recovery, two bigger foci, likely marking NEAT1 transcription sites; (3) 3 h of recovery, multiple PSs/PS clusters around the two bigger foci; and (4) 6 h of recovery, multiple individual PSs scattered in the nucleoplasm (schematically shown in Fig. 3 B, lower panel). An essential PSP NONO showed a distribution pattern similar to that of NEAT1 signal (Fig. 3 F). The same phases of NaAsO2-induced PS assembly were detected in other human cell lines such as fibroblasts (Fig. 3 G). Consistently, another study recently reported that prolonged, 24-h NaAsO2 treatment also increases NEAT1 and PS assembly (Wang et al., 2018). Therefore, an SG-inducing stress (or recovery therefrom) is usually accompanied by PS hyperassembly delayed by ∼2 h after the formation of microscopically visible SGs.
SGs regulate stress-induced PS assembly
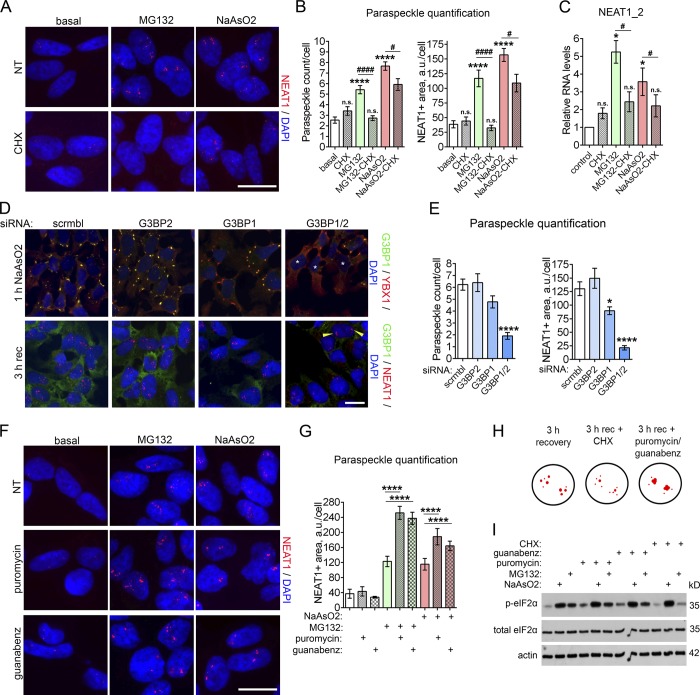
Having demonstrated that SG-inducing stresses commonly trigger PS response, we went on to examine whether SG formation has an impact on stress-induced PS assembly. SG formation can be blocked by translational inhibitors cycloheximide (CHX) and emetine, which stabilize polysomes and deplete the free RNA pool (Kedersha et al., 2005). We confirmed that cotreatment of NaAsO2- or MG132-stressed cells with CHX almost entirely prevents SG assembly (Fig. S2 A). CHX addition 1 h before stress significantly decreased NEAT1 accumulation and PS assembly in NaAsO2-treated cells and completely abolished MG132-induced PS hyperassembly, as revealed by RNA-FISH and qRT-PCR (Fig. 4, A–C). Similar results were obtained using emetine (Fig. S2 B).
Figure 4.
SGs regulate PS assembly and dynamics. (A–C) Translational inhibitor CHX, which dissipates SGs (also see Fig. S2 A), impedes NEAT1 up-regulation and PS hyperassembly during stress in NaAsO2- and MG132-treated cells. Representative images (A), quantification of PS numbers/area (B), and NEAT1_2 levels (C) are shown. PSs were visualized using NEAT1 RNA-FISH, and NEAT1_2 levels were measured with qRT-PCR. Cells were pretreated with CHX for 1 h before NaAsO2 or MG132 addition to the medium. In B, 107–293 cells were included into analysis. #, P < 0.05; **** and ####, P < 0.0001 (one-way ANOVA with Tukey’s test). In C, n = 3; * and #, P < 0.05 (Mann–Whitney U test). n.s., nonsignificant. (D and E) Depletion of both G3BP proteins disrupts stress-induced PS hyperassembly. Simultaneous siRNA knockdown of G3BP1 and G3BP2 disrupts SG assembly (D, top panel) and reduces NaAsO2-induced PS hyperassembly (D, bottom panel; and E). Representative images (D) and quantification of PS numbers/area (E) are shown. Cells were transfected with respective siRNAs, and 72 h after transfection, treated with NaAsO2. PSs were visualized using NEAT1 RNA-FISH, and SGs using anti-YBX1 staining. Asterisks indicate cells devoid of SGs, and arrowheads indicate intact PS clusters. In E, 178–258 cells were included in the analysis. *, P < 0.05; ****, P < 0.0001 (one-way ANOVA with Tukey’s test). (F and G) Puromycin and guanabenz, which prolong SG presence during stress (also see Fig. S2 D), promote NEAT1 accumulation and impair PS dynamics in stressed cells. Representative images (F) and quantification of NEAT1-positive area (G) are shown. Puromycin or guanabenz was added to the cells simultaneously with MG132 or before recovery from NaAsO2. Cells were analyzed after 4 h of MG132 treatment or after 3 h of recovery from NaAsO2. In G, 67–204 cells were included in analysis per condition. ****, P < 0.0001 (one-way ANOVA with Tukey’s test). (H) Schematic representation of the effect of CHX and puromycin/guanabenz on PSs during recovery from NaAsO2 stress. (I) Levels of p-eIF2α in cells treated with NaAsO2 or MG132 and compounds affecting SG assembly/clearance. Cells were pretreated with CHX for 1 h before NaAsO2 or MG132 addition to the medium. Puromycin or guanabenz was added to the cells simultaneously with MG132 or before recovery from NaAsO2. Cells were analyzed after 4 h of MG132 treatment or after 3 h of recovery from NaAsO2. Note that CHX addition decreases p-eIF2α levels in MG132-treated cells. The experiment was repeated three times, and a representative Western blot is shown. Error bars represent SEM. Scale bars, 10 µm.
Previously, it has been reported that simultaneous depletion of both G3BP proteins can disrupt SG assembly (Kedersha et al., 2016; Matsuki et al., 2013). We confirmed that simultaneous knockdown of G3BP1 and G3BP2, but not each protein individually, was sufficient to significantly perturb NaAsO2-induced SG formation in neuroblastoma cells (Fig. 4 D). Cells depleted of both G3BP proteins failed to efficiently assemble PSs in response to NaAsO2 or MG132 (Fig. 4, D and E; and Fig. S2 C). Interestingly, we found that G3BP1 knockdown alone slightly affects PS assembly during stress, which is probably explained by disturbed SG composition in the absence of G3BP1. Taken together, these data indicate that intact SGs are required to trigger and maintain stress-induced PS hyperassembly.
We reasoned that potentiated SG formation as well as their inhibited disassembly should promote PS assembly. Puromycin, which facilitates accumulation of stalled preinitiation complexes, the “building material” for SGs, delays SG dissolution (Kedersha and Anderson, 2002). Another compound, guanabenz, extends SG presence by inhibiting p-eIF2α dephosphorylation during stress (Ruggieri et al., 2012). Indeed, cotreatment of NaAsO2- or MG132-stressed neuroblastoma cells with puromycin or guanabenz, which leads to postponed SG clearance (Fig. S2 D), was sufficient to increase NEAT1 accumulation and significantly perturb PS dynamics during the recovery phase (Fig. 4, F–H).
We found that CHX, puromycin, or guanabenz did not affect p-eIF2α levels during NaAsO2 stress, and puromycin or guanabenz also had no effect on p-eIF2α levels in MG132-treated cells (Fig. 4 I). This indicates that SG-mediated signaling, rather than cellular stress signaling per se, regulates PS assembly. In support of this, a short, 10-min NaAsO2 exposure, which is sufficient to trigger stress signaling and increase p-eIF2α levels (Shelkovnikova et al., 2017) but does not induce SGs, failed to increase PS numbers (Fig. S2 E). Overall, our data indicate that microscopically visible SGs are critical regulators of stress-induced PS assembly.
SGs may regulate PS assembly via sequestration of specific proteins
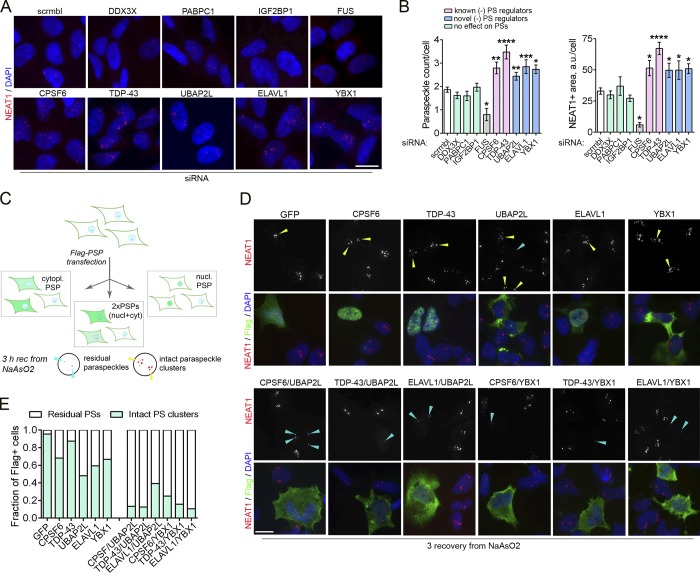
What is the molecular mechanism underlying the role of SGs in stress-induced PS assembly? Given the overlap between PS and SG proteomes and the large size of SGs, it is plausible that SGs sequester negative regulators of PS formation, which alleviates their inhibitory effect on PSs. The majority of known PSPs positively regulate PS formation, in that their down-regulation results in decreased PS numbers (Naganuma et al., 2012). Only two negative regulators of PS assembly are known, namely, CPSF6 (as part of CFIm complex; Naganuma et al., 2012) and TDP-43 (Modic et al., 2019; Shelkovnikova et al., 2018). CPSF6 and TDP-43 are mainly nuclear, and their recruitment into SGs is modest (Figs. 2 C and S1). To identify additional PS/SG proteins, negative regulators of PS assembly, we used siRNA-mediated knockdown. In our analysis, we included five protein components of PS-like structures (Fig. 2 B) known to be highly enriched in SGs: UBAP2L, YBX1, IGF2BP1, DDX3X, PABPC1 (all mainly cytoplasmic), and ELAVL1 (nuclear and cytoplasmic). As positive controls, we knocked down TDP-43 and CPSF6. Depletion UBAP2L, ELAVL1, and YBX1, as well as two positive controls, led to increased PS numbers, as predicted (Fig. 5, A and B). We verified this result using an independent set of siRNAs, which showed even more dramatic increase in PS assembly in UBAP2L-depleted cells (Fig. S3 A). Thus, UBAP2L, ELAVL1, and YBX1 are novel negative regulators of PS assembly.
Figure 5.
Overabundance of proteins acting as negative regulators of PS formation disrupts PS hyperassembly during stress. (A and B) Depletion of certain PSPs leads to increased PS numbers under basal conditions. Cells were transfected with a respective siRNA or scrambled (scrmbl) control RNA and analyzed 48 h after transfection. Representative images (A) and quantification of PS numbers/area (B) are shown. Also see Fig. S3 A. 68–264 cells were included in the analysis per condition. Cells depleted of FUS are known to lose PSs and were included as a negative control. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (one-way ANOVA with Dunnett’s test). Error bars represent SEM. Scale bar, 10 µm. (C–E) Increasing cellular levels of two negative regulators of PS formation simultaneously, by expression of Flag-tagged proteins disrupts PS hyperassembly during NaAsO2-induced stress. Experimental design is schematically shown in C. nucl, nuclear; cyt/cytopl, cytoplasmic. Cells were transfected with respective plasmids, left to express the proteins for 24 h, treated with NaAsO2, and allowed to recover (rec) for 3 h, followed by PS analysis using NEAT1 RNA-FISH. A predominantly cytoplasmic protein was coexpressed with a predominantly nuclear protein to be able to identify the coexpressing cells. GFP tagged with Flag was used as a control. Representative images (D) and quantification (E) are shown. In D, yellow arrowheads point to intact stress-induced PS clusters, and blue arrowheads, to residual PS clusters. Scale bar, 10 µm.
We next examined whether increasing cellular levels of each of these negative regulators would be sufficient to inhibit stress-induced PS assembly. We overexpressed Flag-tagged versions of these proteins and examined PSs during the recovery from NaAsO2 stress. In addition, we studied the effect of coexpression of these proteins, where a predominantly nuclear protein was coexpressed with a predominantly cytoplasmic protein, to allow identification of the coexpressing cells (Fig. 5 C). Overexpression of each protein alone was not sufficient to significantly perturb NaAsO2-induced PS hyperassembly. However, coexpression of any two PSPs led to almost undetectable PS clusters (Fig. 5, D and E). In contrast, coexpression of two RNA-binding proteins whose knockdown does not affect PSs, TAF15 and ATXN2 (Naganuma et al., 2012; Fig. S3 B), did not inhibit stress-induced PS hyperassembly (Fig. S3 C). Novel negative regulators of PS assembly UBAP2L and YBX1 are almost exclusively cytoplasmic, and ELAVL1 is both nuclear and cytoplasmic. We examined subcellular localization of UBAP2L during stress and recovery in the presence of CHX, guanabenz, or puromycin and found that this protein remained cytoplasmic under all conditions studied (Fig. S3 D). This suggests that UBAP2L sequestration into SGs inhibits its cytoplasmic functions and that the protein regulates PSs indirectly, via signaling events in the cytoplasm.
SG dysfunction has been heavily implicated in ALS (Li et al., 2013). Recently, we and others have shown that ALS is characterized by PS hyperassembly in spinal neurons, likely downstream of TDP-43 loss of function (Nishimoto et al., 2013; Shelkovnikova et al., 2018). We examined the distribution of UBAP2L in ALS motor neurons by immunohistochemistry. UBAP2L is predominantly expressed in the ventral motor neurons in the human spinal cord (Fig. 6). We found that UBAP2L is significantly depleted and sometimes undetectable in this population of neurons in ALS patients. In some cases, we also detected cytoplasmic aggregates of UBAP2L. Therefore, loss of UBAP2L function in motor neurons may contribute to PS hyperassembly in ALS.
Figure 6.
A negative regulator of PS assembly UBAP2L is depleted or aggregated in the spinal motor neurons of ALS patients. UBAP2L distribution in the spinal cord sections of control individuals and sALS and ALS-C9 patients was analyzed by immunohistochemistry. Representative images of motor neurons in the ventral horn are shown. Scale bar, 50 µm.
Discussion
Due to their localization, SGs and PSs have been traditionally viewed as RNP granules completely isolated from each other. In the current study, we show that despite spatial separation, SGs and PSs are intimately linked. Although several stresses have been reported to induce PS hyperassembly, it was not clear what all these stresses have in common, and here we identify the presence of SGs as a universal feature of PS-inducing stresses. Our study also suggests that cells can regulate nuclear body assembly by engaging certain proteins into cytoplasmic RNP granules. Conceivably, differences in the protein composition of SGs induced by various stresses (Aulas et al., 2017) should contribute to the differences in PS assembly. NEAT1 expression is restricted to mammals, whereas formation of SGs is a response conserved from yeast to humans. Therefore, in nonmammals, SGs may act to regulate other nuclear RNA granules in a similar way.
We show that SG assembly and PS hyperassembly are early and late events during stress, respectively. We propose that SGs serve as the robust, first-line defense during stress, whereas PSs orchestrate fine adjustments of cellular processes required for the recovery from stress or during prolonged stress. SGs are believed to play a general prosurvival role (Arimoto et al., 2008; Takahashi et al., 2013). Similarly, PSs have been shown to promote viability in cells subjected to proteasome inhibition, heat shock, and foreign dsRNA (Hirose et al., 2014; Lellahi et al., 2018; Shelkovnikova et al., 2018). It is tempting to speculate that the protective role of SGs may be at least partially mediated by PSs. Physiological and pathological significance of increased abundance of PSs late during stress should be addressed in future studies.
Our data suggest that sequestration of proteins that negatively regulate PS assembly may serve as a mechanism underlying PS hyperassembly during stress. On the other hand, sequestration of essential PSPs, positive PS regulators (NONO, SFPQ, hnRNPK, and FUS), into SGs is very weak; therefore, their levels in the nucleus should remain sufficient to support PS assembly. In future studies, it will be crucial to carry out quantitative analysis of protein shuttling between the two granules to establish to what extent sequestration of each negative PS regulator into SGs would affect PS formation. Of note, some negative PS regulators are present in the nucleus at very low levels; thus these proteins likely associate with PSs only transiently and regulate their formation via other proteins and/or signaling pathways, without acting as structural components of these RNP granules. Simultaneous sequestration of multiple negative PS regulators into SGs during stress likely triggers signaling cascades, leading to enhanced NEAT1 accumulation and PS hyperassembly. Since these signaling cascades would require some time to develop, PS assembly becomes delayed relative to SG formation.
A variety of protein components of SGs and PSs are affected by ALS-causative mutations (An et al., 2018; Li et al., 2013). Proteinaceous inclusions found in the neurons of ALS patients often contain SG proteins suggestive of SG-related pathological changes (Dormann et al., 2010; Liu-Yesucevitz et al., 2010), and de novo PS assembly in spinal neurons and glia is a pathological hallmark of ALS (Nishimoto et al., 2013; Shelkovnikova et al., 2018). These changes have been considered as unrelated events taking place in two separate cellular compartments. Our study provides the first evidence for the existence of a link between assembly of pathological inclusions and PS hyperassembly in neurons and glia of ALS patients. Sequestration of negative regulators of PS, such as UBAP2L, into cytoplasmic inclusions or their down-regulation via other mechanisms could contribute to PS hyperassembly in ALS. UBAP2L is not an RNA-binding protein, but it regulates the integrity of the two RNP granules central to ALS pathogenesis. Even though its genetic link to ALS has not been reported, UBAP2L dysfunction might be a common phenotype for etiologically different ALS cases.
Finally, both SGs and PSs have been implicated in multiple types of cancer (Anderson et al., 2015; Fox et al., 2018). SGs can increase tumor cell fitness and chemotherapy resistance (El-Naggar and Sorensen, 2018). Similarly, PSs can promote survival of oncogene-targeted cells (Adriaens et al., 2016). Studies into the crosstalk between SGs and PSs in neurodegenerative diseases and cancer should reveal novel molecular mechanisms underlying these diseases as well as inform therapeutic developments going forward.
Materials and methods
Cell lines, transfection, and treatments
SH-SY5Y and HEK293 cells and human fibroblasts were maintained in a 1:1 mixture of DMEM/F12 medium supplemented with 10% FBS, penicillin-streptomycin, and GlutaMAX (all Invitrogen). G3BP1 and NONO expression plasmids were constructed by inserting human G3BP1 and NONO ORFs into pEGFP-N1 and pEGFP-C1 vectors, respectively. Plasmids for the expression of SFPQ-GFP and TDP-43-Flag were kind gifts from Archa Fox (University of Western Australia, Crawley, Australia) and Francisco Baralle (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy). pEGFP-N1-Flag plasmid was a gift from Patrick Calsou (Addgene plasmid #60360). Plasmids for the expression of Flag-tagged CPSF6, UBAP2L, YBX1, ELAVL1, and ATXN2 were purchased from Sino Biological (HG11458-CF-SIB, HG13903-CF-SIB, HG17046-CF-SIB, MG57819-CF-SIB, and HG15977-CF-SIB, respectively). The following gene-specific siRNAs were used for transient knockdown: G3BP1 (Sigma-Aldrich, EHU113241); G3BP2 (Sigma-Aldrich, EHU078891); TDP-43 (Invitrogen, Silencer Select, s23829); CPSF6 (Invitrogen, Silencer, 136160); UBAP2L (Sigma-Aldrich, EHU072151; and Invitrogen, Silencer, 122721); YBX1 (Sigma-Aldrich, EHU148801; and Invitrogen, Silencer, 115541); ELAVL1 (Sigma-Aldrich, EHU063491; and Invitrogen, Silencer, 145882); PABPC1 (Sigma-Aldrich, EHU113851); IGF2BP1 (Sigma-Aldrich, EHU020781); DDX3X (Invitrogen, Silencer, 145803); and ATXN2 (Sigma-Aldrich, EHU104101). Scrambled negative control siRNA was AllStars from Qiagen. Transfections were performed using 200 ng of siRNA, 200 ng of plasmid DNA, or 250 ng of poly(I:C) (Sigma-Aldrich) in 24-well plates using Lipofectamine 2000 (Invitrogen). Cells were treated with compounds (Sigma-Aldrich, unless stated otherwise) at the following concentrations: 0.5 mM NaAsO2 (sodium arsenite), 10 µM MG132, 20 µg/ml puromycin, 10 µg/ml CHX, 10 µg/ml emetine, 50 µM guanabenz, 50 µM 15d-PGJ2 (Cayman), 5 µM pifithrin-µ (HSP70 inhibitor, Enzo Life Sciences), 0.6 M sorbitol, and 500 nM rocaglamide A (Santa Cruz Biotechnology). In NaAsO2 recovery experiments, cells were washed twice with fresh medium. For heat shock, cells were subjected to 43°C for 1 h.
Immunocytochemistry and RNA-FISH
Immunocytochemistry, NEAT1 RNA-FISH, and microscopic analysis were performed as described earlier (Kukharsky et al., 2015). Briefly, cells were fixed on coverslips with 4% PFA for 15 min and permeabilized in cold methanol. For RNA-FISH, coverslips were soaked in 70% ethanol overnight at 4°C. Commercially available NEAT1 probe (Stellaris FISH probe for human NEAT1, 5′ segment; Biosearch Technologies) was used per the manufacturer’s protocol. For immunocytochemistry, primary antibodies diluted in blocking solution (5% goat serum in 0.1% Triton X-100/1× PBS) were applied for 1–4 h at RT or overnight at 4°C. Secondary Alexa Fluor 488– or 546–conjugated antibody was added for 1 h at RT. Nuclei were visualized with DAPI (Sigma-Aldrich). Fluorescent images were captured with a 100× objective (UPlanFI 100×/1.30) on an Olympus BX61 microscope equipped with F-View II camera and processed using CellF software (all Olympus). Quantification of PS numbers and measurement of NEAT1-positive area were performed using the “analyze particles” tool of ImageJ software (National Institutes of Health). Plot profiles were made using the respective tool of ImageJ. Images were prepared using Photoshop CS3 or PowerPoint 2003.
RNA analysis
Total RNA was purified with GenElute total RNA kit (Sigma-Aldrich). RNA from PS-like structures and SG cores was purified using TRI-reagent (Sigma-Aldrich). First-strand cDNA synthesis was performed using random primers (Promega) and Superscript IV (Invitrogen) according to the manufacturers’ instructions. PCR was performed using New England BioLabs Taq DNA polymerase (M0273). Quantitative real-time PCR was run in triplicate on a StepOne real-time PCR instrument, and data were analyzed using StepOne software v2.0 (Applied Biosystems). GAPDH was used for normalization. Primer sequences were as follows: NEAT1 total, 5′-CTCACAGGCAGGGGAAATGT-3′ and 5′-AACACCCACACCCCAAACAA-3′; NEAT1_2, 5′-TGTGTGTGTAAAAGAGAGAAGTTGTGG-3′ and 5′-AGAGGCTCAGAGAGGACTGTAACCTG-3′; GAPDH, 5′-TCGCCAGCCGAGCCA-3′ and 5′-GAGTTAAAAGCAGCCCTGGTG-3′.
Purification of PS-like structures
HEK293 cells were transfected with equal amounts of plasmids to express NONO-GFP and SFPQ-GFP or GFP alone in 6-cm dishes (1 µg plasmid/dish) using Lipofectamine 2000. The following day, after medium aspiration, cells were snap-frozen on dishes. Cells were defrosted on ice for 5 min and scraped in 1 ml of lysis buffer (50 mM Tris HCl, pH 7.4, 100 mM KOAc, 2 mM MgOAc, 0.5 mM DTT, 50 µg/ml heparin, and 0.5% NP-40) supplemented with protease inhibitors cocktail (cOmplete Mini, EDTA-free; Roche) and RNase inhibitor (murine; New England Biolabs). Cells were lysed by passing through a 25G needle seven times while on ice. Nuclei were pelleted by spinning at 1,000 g for 5 min at 4°C and washed in the lysis buffer. After resuspension in the lysis buffer supplemented with 400 mM NaCl, nuclei were left to lyse on ice for 30 min with periodic vortexing, and nuclear lysis was monitored under fluorescent microscope. Nuclear lysates were centrifuged at 1,000 g for 5 min at 4°C, and supernatant was transferred to a new tube. Supernatant was centrifuged at 17,000 g for 20 min at 4°C to obtain PS-enriched fraction. After washes, the pellet was resuspended in the lysis buffer and centrifuged again at 1000 g for 5 min at 4°C. GFP-Trap agarose beads (Chromotek) were added to the supernatant and rotated on nutator for 4 h. Beads were washed three times in the lysis buffer and three times in the lysis buffer with 400 mM NaCl and snap-frozen for subsequent liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis. Cells transfected with a plasmid to express GFP alone were processed in parallel to control samples. Samples from three independent experiments were combined for LC-MS/MS analysis. For analysis of the stability of PS-like structures, nuclear lysates (supernatant before 17,000 g centrifugation) were treated with 1% SDS, 5% 1,6-hexanediol (Sigma-Aldrich), 2 M NaCl, or 2 M urea in RNase-free water or 100 µg/ml RNase A (Qiagen) for 1 h at RT, or left at 37°C for 1 h. Images of GFP-positive dots were taken with a 40× objective (UPLFLN 40×Ph/0.75) of a BX53 microscope equipped with DP73 camera and processed using cellSens Standard 1.9 software (all Olympus). The number of GFP-positive dots was counted using the “analyze particles” tool of ImageJ.
Purification of SG cores
SG cores were purified from SH-SY5Y cells transiently transfected with a plasmid for G3BP1-GFP expression (generated in-house) as described previously (Jain et al., 2016; Wheeler et al., 2017). Briefly, SH-SY5Y cells expressing G3BP1-GFP were subjected to 0.5 mM NaAsO2 for 1 h and snap-frozen on dishes. Cells were scraped in the lysis buffer (50 mM Tris HCl, pH 7.4, 100 mM KOAc, 2 mM MgOAc, 0.5 mM DTT, 50 µg/ml heparin, 0.5% NP-40, RNase inhibitor, and protease inhibitors cocktail) and passed through a 25G needle. The lysate was centrifuged at 1000 g for 5 min, and the supernatant from this spin was centrifuged at 17,000 g for 20 min. The pellet containing SG cores was resuspended in the lysis buffer and incubated with GFP-Trap agarose beads for 4 h. Beads were washed three times with washing buffer 1 (20 mM Tris HCl and 200 mM NaCl, pH 8.0), once with washing buffer 2 (20 mM Tris HCl and 500 mM NaCl, pH 8.0), and once with washing buffer 3 (lysis buffer containing 2 M urea).
LC-MS/MS and analysis
Each sample was separated using SDS-PAGE. Gel lanes were excised and subjected to in-gel tryptic digestion using a DigestPro automated digestion unit (Intavis). The resulting peptides were fractionated using an Ultimate 3000 nano-LC system in line with an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Peptides were ionized by nano-electrospray ionization at 2.1 kV using a stainless-steel emitter with an internal diameter of 30 µm (Thermo Fisher Scientific) and a capillary temperature of 250°C. Tandem mass spectra were acquired using an LTQ-Orbitrap Velos mass spectrometer controlled by Xcalibur 2.1 software (Thermo Fisher Scientific) and operated in data-dependent acquisition mode. The raw data files were processed and quantified using Proteome Discoverer software v1.4 (Thermo Fisher Scientific) and searched against the UniProt Human database (downloaded September 14, 2017; 140,000 sequences) using the SEQUEST algorithm. Peptide precursor mass tolerance was set at 10 ppm, and MS/MS tolerance was set at 0.8 Da. Search criteria included carbamidomethylation of cysteine (+57.0214) as a fixed modification and oxidation of methionine (+15.9949) as a variable modification. Searches were performed with full tryptic digestion, and a maximum of one missed cleavage was allowed. The reverse database search option was enabled, and all peptide data were filtered to satisfy FDR of 5%. The list of proteins identified in the samples from cells expressing GFP alone was used to calculate the ratio PS/control (Table S1, PS-like structures tab). A network diagram was prepared using BioVenn online tool (http://www.biovenn.nl/index.php). Enrichment analysis was performed using String Database (https://string-db.org/).
Western blotting
Total cell lysates were prepared for Western blot by adding 2× Laemmli buffer to the samples or directly to cells on the dish followed by denaturation at 100°C for 5 min. Proteins were resolved in Mini-Protean TGX precast gels (Bio-Rad) and transferred to PVDF membrane (GE Healthcare) by semidry blotting. After blocking in 4% milk in TBST, membranes were incubated with primary and subsequently HRP-conjugated secondary (GE Healthcare) antibodies. For signal detection, WesternBright Sirius kit (Advansta) and ChemiDoc Gel Imaging System (Bio-Rad) were used. Equal loading was confirmed by reprobing membranes for β-actin.
Primary antibodies
The following commercial primary antibodies were used: G3BP1 (mouse monoclonal; BD Biosciences); TIAR (mouse monoclonal; BD Biosciences); NONO (rabbit polyclonal C-terminal; Sigma-Aldrich); SFPQ (rabbit monoclonal; ab177149, Abcam); CPSF6 (rabbit polyclonal; A301-356A, Bethyl); FUS (mouse monoclonal; 4H11, Santa Cruz); ELAVL1 (rabbit polyclonal, 11910-1-AP, Proteintech); PABPC1 (rabbit polyclonal, Cell Signaling, 4992); EIF4E (mouse monoclonal; BD Biosciences); UBAP2L (rabbit polyclonal; A300-533A, Bethyl); DAZAP1 (rabbit polyclonal; A303-984A, Bethyl); hnRNP K (rabbit polyclonal; A300-674A, Bethyl); PSPC1 (rabbit polyclonal N-terminal; Sigma-Aldrich); TDP-43 (rabbit polyclonal C-terminal; Sigma-Aldrich); HNRNPA3 (rabbit polyclonal; 25142-1-AP, Proteintech); RBM12B (rabbit polyclonal; 17137-1-AP, Proteintech); SRSF9 (rabbit polyclonal; 17926-1-AP, Proteintech); SMARCA5 (rabbit polyclonal; 13066-1-AP, Proteintech); MATR3 (rabbit polyclonal; 12202-2-AP, Proteintech); YBX1 (rabbit polyclonal; 20339-1-AP, Proteintech); GFP (mouse monoclonal; sc-9996, Santa Cruz); Flag (DYKDDDDK tag, mouse monoclonal; 9A3, Cell Signaling); eIF2α phosphorylated at Ser51 (rabbit monoclonal; ab32157, Abcam); total eIF2α (rabbit monoclonal; D7D3, Cell Signaling); and β-actin (mouse monoclonal; A5441, Sigma-Aldrich). Antibodies were used at 1:1,000 dilution for all applications unless stated otherwise.
Analysis of human tissue samples
Human spinal cord paraffin sections from clinically and histopathologically characterized ALS patients and neurologically healthy individuals were obtained from the Sheffield Brain Tissue Bank. Consent was obtained from all subjects for autopsy, histopathological assessment, and research in accordance with local and national Ethics Committee–approved donation. Three control cases, three sALS, and three ALS-C9 cases were included in the study. Human spinal cord sections were 7 µm thick. Rehydrated sections were subjected to microwave antigen retrieval in sodium citrate buffer. After incubation with the primary anti-UBAP2L antibody overnight at 4°C, secondary HRP-conjugated anti-rabbit IgG antibody (Vector Laboratories) was applied for 1.5 h at RT. Vectastain Elite ABC Universal Plus Kit (Vector Laboratories) and DAB (Sigma-Aldrich) were used for detection. Images were taken with a 20× objective (UPLFLN 20×Ph/0.5) of a BX53 microscope equipped with DP73 camera and processed using cellSens Standard 1.9 software (all Olympus).
Statistical analysis
In all cases, error bars represent SEM and n indicates the number of biological replicates. Mean values of biological replicates were compared using an appropriate test. P values >0.05 were considered not significant. All sample sizes and P values are indicated in the figure legends. For comparing more than two groups, one-way ANOVA with a post hoc test was used; data distribution was assumed to be normal due to large numbers of observations. Post hoc tests used in each case are stated in the figure legends. For comparing two groups, due to small numbers of observations, nonparametric Mann–Whitney U test was used. Statistical analysis was performed using GraphPad Prism 6 software.
Online supplemental material
Fig. S1 shows the analysis of PSP recruitment into SGs induced by heat shock, proteasome inhibition, and dsRNA stimulation. Fig. S2 contains additional data on the regulation of PS assembly by SGs. Fig. S3 includes additional data on the analysis of proteins, negative regulators of PS assembly. Table S1 provides details of all protein datasets used in the study.
Supplementary Material
Acknowledgments
We acknowledge the Sheffield Brain Tissue Bank for providing human materials and the University of Bristol Proteomics Facility for their help with proteomic analysis. We also thank Vladimir Buchman for critical reading of the manuscript.
This study was supported by fellowships from the Medical Research Foundation and the Motor Neurone Disease Association (Shelkovnikova/Oct17/968-799) to T.A. Shelkovnikova. H. An and J.T. Tan are recipients of a Cardiff University/China Scholarship Council PhD studentship and an International Association for the Exchange of Students for Technical Experience training studentship, respectively.
The authors declare no competing financial interests.
Author contributions: T.A. Shelkovnikova conceived the study; H. An, J.T. Tan, and T.A. Shelkovnikova designed and performed experiments and analyzed data; T.A. Shelkovnikova wrote the manuscript with input from all authors.
References
- Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W., Radaelli E., Vermi W., et al. . 2016. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 22:861–868. 10.1038/nm.4135 [DOI] [PubMed] [Google Scholar]
- An H., Skelt L., Notaro A., Highley J.R., Fox A.H., La Bella V., Buchman V.L., and Shelkovnikova T.A.. 2019. ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol. Commun. 7:7 10.1186/s40478-019-0658-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Williams N.G., and Shelkovnikova T.A.. 2018. NEAT1 and paraspeckles in neurodegenerative diseases: A missing lnc found? Noncoding RNA Res. 3:243–252. 10.1016/j.ncrna.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2006. RNA granules. J. Cell Biol. 172:803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N., and Ivanov P.. 2015. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta. 1849:861–870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., and Takekawa M.. 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10:1324–1332. 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- Aulas A., Fay M.M., Lyons S.M., Achorn C.A., Kedersha N., Anderson P., and Ivanov P.. 2017. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130:927–937. 10.1242/jcs.199240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Vest K.E., Pavlath G.K., and Corbett A.H.. 2017. Nuclear poly(A) binding protein 1 (PABPN1) and Matrin3 interact in muscle cells and regulate RNA processing. Nucleic Acids Res. 45:10706–10725. 10.1093/nar/gkx786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G., and Vourc’h C.. 2010. Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2:a000695 10.1101/cshperspect.a000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., et al. . 2018. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 28:420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O., Desforges B., Wu T.D., Pioche-Durieu C., Marco S., Hamon L., Curmi P.A., Guerquin-Kern J.L., Piétrement O., and Pastré D.. 2014. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42:8678–8691. 10.1093/nar/gku582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.B., Johnston R.L., Inostroza-Ponta M., Fox A.H., Fortini E., Moscato P., Dinger M.E., and Mattick J.S.. 2012. Genome-wide analysis of long noncoding RNA stability. Genome Res. 22:885–898. 10.1101/gr.131037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C.M., Cenik B., Sephton C.F., Dries D.R., Mayer P. III, Good S.K., Johnson B.A., Herz J., and Yu G.. 2011. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31:1098–1108. 10.1128/MCB.01279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B., et al. . 2010. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29:2841–2857. 10.1038/emboj.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naggar A.M., and Sorensen P.H.. 2018. Translational control of aberrant stress responses as a hallmark of cancer. J. Pathol. 244:650–666. 10.1002/path.5030 [DOI] [PubMed] [Google Scholar]
- Fox A.H., and Lamond A.I.. 2010. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2:a000687 10.1101/cshperspect.a000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.H., Nakagawa S., Hirose T., and Bond C.S.. 2018. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem. Sci. 43:124–135. 10.1016/j.tibs.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Gomes E., and Shorter J.. 2019. The molecular language of membraneless organelles. J. Biol. Chem. 294:7115–7127. 10.1074/jbc.TM118.001192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Bénard M., Fox A.H., and Pierron G.. 2014. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 25:169–183. 10.1091/mbc.e13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A., Courel M., Benard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M., et al. . 2017. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell. 68:144–157.e5. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. . 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 53:393–406. 10.1016/j.molcel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., and Parker R.. 2016. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 164:487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969. 10.1042/bst0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., and Anderson P.. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., and Anderson P.. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212:845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., and Anderson P.. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M.A., and Bassell G.J.. 2006. Neuronal RNA granules: movers and makers. Neuron. 51:685–690. 10.1016/j.neuron.2006.08.021 [DOI] [PubMed] [Google Scholar]
- Kim W.J., Kim J.H., and Jang S.K.. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 26:5020–5032. 10.1038/sj.emboj.7601920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukharsky M.S., Quintiero A., Matsumoto T., Matsukawa K., An H., Hashimoto T., Iwatsubo T., Buchman V.L., and Shelkovnikova T.A.. 2015. Calcium-responsive transactivator (CREST) protein shares a set of structural and functional traits with other proteins associated with amyotrophic lateral sclerosis. Mol. Neurodegener. 10:20 10.1186/s13024-015-0014-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., and de Thé H.. 2010. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2:a000661 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellahi S.M., Rosenlund I.A., Hedberg A., Kiær L.T., Mikkola I., Knutsen E., and Perander M.. 2018. The long noncoding RNA NEAT1 and nuclear paraspeckles are up-regulated by the transcription factor HSF1 in the heat shock response. J. Biol. Chem. 293:18965–18976. 10.1074/jbc.RA118.004473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.R., King O.D., Shorter J., and Gitler A.D.. 2013. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201:361–372. 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Schmidt B.F., Bruchez M.P., and McManus C.J.. 2018. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 46:3742–3752. 10.1093/nar/gky046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., et al. . 2010. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 5:e13250 10.1371/journal.pone.0013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannen T., Yamashita S., Tomita K., Goshima N., and Hirose T.. 2016. The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J. Cell Biol. 214:45–59. 10.1083/jcb.201601024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Zhang B., and Spector D.L.. 2011. Biogenesis and function of nuclear bodies. Trends Genet. 27:295–306. 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.C., Krach F., Yang D., Sen A., et al. . 2018. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell. 172:590–604.e13. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., and Fujii M.. 2013. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 18:135–146. 10.1111/gtc.12023 [DOI] [PubMed] [Google Scholar]
- Modic M., Grosch M., Rot G., Schirge S., Lepko T., Yamazaki T., Lee F.C.Y., Rusha E., Shaposhnikov D., Palo M., et al. . 2019. Cross-Regulation between TDP-43 and Paraspeckles Promotes Pluripotency-Differentiation Transition. Cell. 74:951–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., and Hirose T.. 2012. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31:4020–4034. 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Yamazaki T., and Hirose T.. 2018. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol. 8:180150 10.1098/rsob.180150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto Y., Nakagawa S., Hirose T., Okano H.J., Takao M., Shibata S., Suyama S., Kuwako K., Imai T., Murayama S., et al. . 2013. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 6:31 10.1186/1756-6606-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D.S.W., and Parker R.. 2016. Principles and Properties of Stress Granules. Trends Cell Biol. 26:668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.P., Mazur J., Bankhead P., Hiet M.S., Kallis S., Alvisi G., et al. . 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 12:71–85. 10.1016/j.chom.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T.A., Dimasi P., Kukharsky M.S., An H., Quintiero A., Schirmer C., Buée L., Galas M.C., and Buchman V.L.. 2017. Chronically stressed or stress-preconditioned neurons fail to maintain stress granule assembly. Cell Death Dis. 8:e2788 10.1038/cddis.2017.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T.A., Kukharsky M.S., An H., Dimasi P., Alexeeva S., Shabir O., Heath P.R., and Buchman V.L.. 2018. Protective paraspeckle hyperassembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 13:30 10.1186/s13024-018-0263-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T.A., Robinson H.K., Troakes C., Ninkina N., and Buchman V.L.. 2014. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 23:2298–2312. 10.1093/hmg/ddt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standart N., and Weil D.. 2018. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 34:612–626. 10.1016/j.tig.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., and Fujii M.. 2013. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol. 33:815–829. 10.1128/MCB.00763-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hu S.B., Wang M.R., Yao R.W., Wu D., Yang L., and Chen L.L.. 2018. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 20:1145–1158. 10.1038/s41556-018-0204-2 [DOI] [PubMed] [Google Scholar]
- West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., and Kingston R.E.. 2014. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 55:791–802. 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J.R., Jain S., Khong A., and Parker R.. 2017. Isolation of yeast and mammalian stress granule cores. Methods. 126:12–17. 10.1016/j.ymeth.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., and Carmichael G.G.. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 106:465–475. 10.1016/S0092-8674(01)00466-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.