The retromer complex is responsible for the endosomal sorting of numerous nutrient transporters. In this study, Curnock et al. show that in response to nutrient deprivation, retromer is transcriptionally regulated by TFEB and acts to support adaptive nutrient acquisition through endosomal recycling of the glutamine transporter, SNAT2 (SLC38A2).
Abstract
Endosomal recycling maintains the cell surface abundance of nutrient transporters for nutrient uptake, but how the cell integrates nutrient availability with recycling is less well understood. Here, in studying the recycling of human glutamine transporters ASCT2 (SLC1A5), LAT1 (SLC7A5), SNAT1 (SLC38A1), and SNAT2 (SLC38A2), we establish that following amino acid restriction, the adaptive delivery of SNAT2 to the cell surface relies on retromer, a master conductor of endosomal recycling. Upon complete amino acid starvation or selective glutamine depletion, we establish that retromer expression is upregulated by transcription factor EB (TFEB) and other members of the MiTF/TFE family of transcription factors through association with CLEAR elements in the promoters of the retromer genes VPS35 and VPS26A. TFEB regulation of retromer expression therefore supports adaptive nutrient acquisition through endosomal recycling.
Introduction
By deciding the fate of signaling receptors, nutrient and solute transporters, and cell–cell and cell–matrix adhesion molecules, the endosomal network plays an essential role in cell-, tissue-, and organ-level physiology (Grant and Donaldson, 2009; Hsu et al., 2012; Cullen and Steinberg, 2018). Key to the function of the network is its ability to sort integral proteins (termed cargoes) between two fates: degradation within the lysosome or retrieval from this fate for subsequent recycling and reuse at the cell surface or the TGN. To achieve this, cargoes are sorted and segregated between two functionally distinct subdomains that reside on the cytosolic face of the endosomal vacuole. While ESCRTs (endosomal sorting complexes required for transport) define the functional organization of the degradative subdomain (Frankel and Audhya, 2018), the opposing retrieval subdomain is composed of a number of distinct multi-protein complexes that include retromer (McNally and Cullen, 2018).
Initially identified in yeast as a pentameric complex (Seaman et al., 1998), the mammalian retromer is a stable heterotrimer of VPS35, VPS29, and VPS26, of which two isoforms, VPS26A and VPS26B, are expressed in humans (Haft et al., 2000; Seaman, 2004; Arighi et al., 2004; Kerr et al., 2005). Retromer functions as a molecular scaffold that promotes the formation and organization of a retrieval subdomain by engaging a range of accessory proteins that include cargo adaptors such as sorting nexin 3 (SNX3) and sorting nexin 27 (SNX27) and the actin polymerizing Wiscott-Aldrich syndrome protein and SCAR homologue (WASH) complex (Strochlic et al., 2007; Harterink et al., 2011; Gomez and Billadeau, 2009; Temkin et al., 2011; Harbour et al., 2012; Steinberg et al., 2013; Gallon et al., 2014; Lucas et al., 2016). While it is clear that hundreds of diverse cargo proteins transit through these dynamic retrieval subdomains en route to the cell surface and the TGN (Burd and Cullen, 2014), how retromer function is regulated and how this impacts the function of the retrieval subdomain remains unclear. What is evident is that disruption of retrieval subdomain function, either through mutations and/or reduced expression of retromer and retromer accessory proteins, is associated with the pathoetiology of a number of neurological disorders that include Alzheimer’s and Parkinson’s diseases (Small and Petsko, 2015; McMillan et al., 2017).
Here, by exploring the role of retromer in the retrieval and recycling of glutamine transporters as part of a cellular adaptive response to amino acid withdrawal, we identify that the expression of retromer is controlled by the MiTF/TFE family of transcription factors, master regulators of cellular nutrient sensing and energy metabolism (Sardiello et al., 2009; Settembre et al., 2011). This establishes a point of transcriptional regulation through which the sensing of nutrient availability can be synchronized with the regulation of cargo retrieval and recycling by the endosomal network.
Results and discussion
Retromer regulates endosomal recycling of glutamine transporters
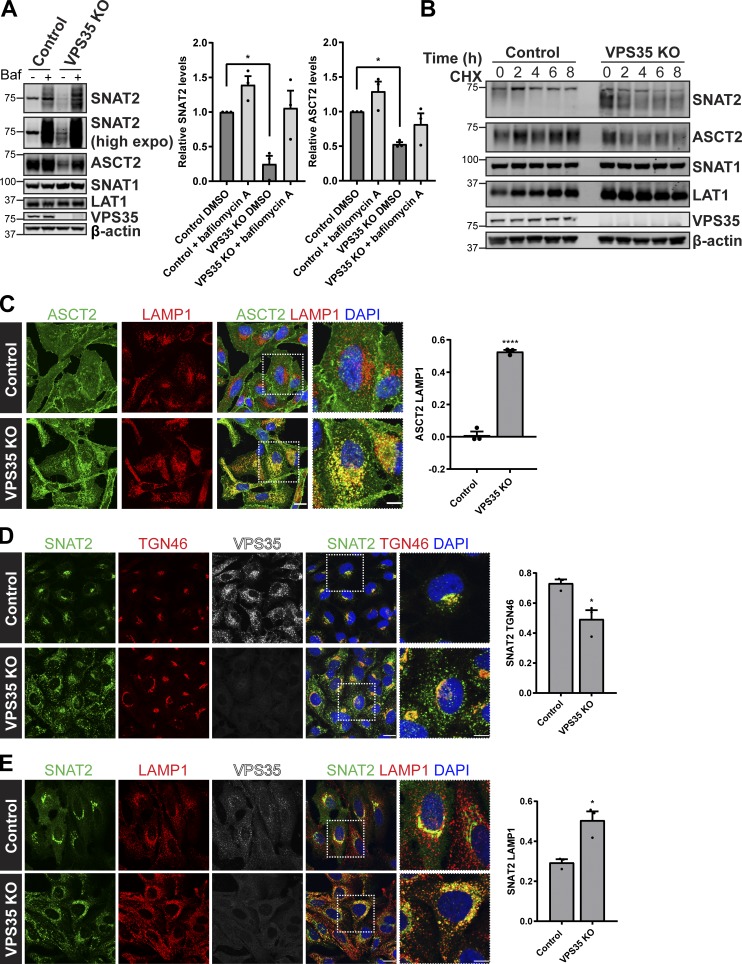
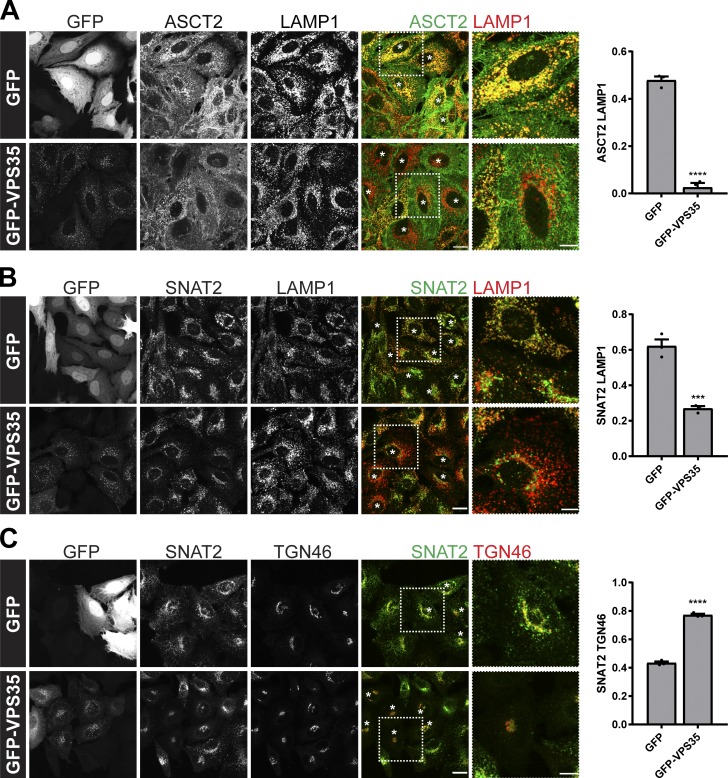
In nutrient replete conditions, glutamine uptake and sustained mTORC1 activation requires the cell surface transporter ASCT2 (SLC1A5) and a heterodimeric bidirectional antiporter, LAT1 (SLC7A5)/CD98hc (SLC3A2), that coordinates the efflux of cytosolic glutamine with the uptake of essential amino acids, such as leucine (Nicklin et al., 2009). Two additional transporters, SNAT1 (SLC38A1) and SNAT2 (SLC38A2), can functionally replace glutamine uptake mediated by ASCT2 (Bröer et al., 2016). In examining the whole-cell levels of these transporters in a previously characterized CRISPR-Cas9–generated retromer-null HeLa cell line (targeting the VPS35 subunit; Simonetti et al., 2017), we noted that while the expression levels of SNAT1 and LAT1 were unaffected, as were their degradation kinetics upon initiation of a cycloheximide block, the expression levels of SNAT2 and ASCT2 were both significantly reduced (Fig. 1, A and B). Their reduced expression was rescued upon addition of bafilomycin A and was therefore consistent with an enhanced rate of lysosome-mediated degradation upon retromer knockout (Fig. 1 A). To further validate this phenotype, we examined the endogenous localization of ASCT2 and SNAT2 in cells grown in nutrient-rich media. For ASCT2, this was defined by a steady-state enrichment at the cell surface, while for SNAT2, its steady-state localization was defined by enrichment at the TGN (Fig. 1, C and D). In the retromer knockout cells, however, the steady-state distribution of both transporters shifted to an enrichment defined by colocalization with LAMP1-labeled late endosomes and lysosomes (Fig. 1, C and E). This observation was entirely consistent with the reduced expression of these transporters and was indicative of their missorting into the degradative fate. Retromer therefore serves to promote the retrieval and recycling of ASCT2 and SNAT2 (but not LAT1 and SNAT1) away from the lysosomal degradative fate, a conclusion supported by the rescue of the steady-state enrichment of ASCT2 to the cell surface and SNAT2 to the TGN upon expression of GFP-tagged VPS35 in the retromer knockout cells (Fig. 2, A–C). Together, these data are consistent with evidence that ASCT2 requires binding to the retromer-associated cargo adaptor SNX27 for its retrieval and recycling (Kvainickas et al., 2017; Yang et al., 2018) and, moreover, establishes that SNAT2 may constitute a previously unrecognized cargo for retromer-mediated endosome to TGN retrograde transport.
Figure 1.
Retromer-dependent cargo recycling of the glutamine transporters ASCT2 and SNAT2. (A) Representative immunoblotting analysis for the indicated proteins in control and VPS35 knockout cells treated with DMSO or bafilomycin A1 (100 nM for 16 h). The quantification represents means ± SEM; n = three independent experiments; one-way ANOVA followed by Dunnett’s multiple comparison. (B) Immunoblot analysis of the total levels of the indicated proteins in control and VPS35 knockout HeLa cells treated with 10 µg/ml cycloheximide (CHX) over the specified time course. Data representative of three independent experiments. (C) Colocalization analysis of endogenous ASCT2 (green) and LAMP1 (red) in control and VPS35 knockout HeLa cells. The graph represents means ± SEM. Pearson’s correlation; n = three independent experiments with >100 cells per condition; Student’s t test (unpaired). (D) IF analysis of endogenous SNAT2 (green), TGN46 (red), and VPS35 (white) in control and VPS35 knockout HeLa cells. (E) Representative IF images of endogenous SNAT2 (green), LAMP1 (red), and VPS35 (white) in control and VPS35 knockout HeLa cells. Graphs in D and E represent means ± SEM. Pearson’s correlation of three independent experiments with >150 cells per condition; Student’s t test (unpaired). Scale bars, 20 µm and 10 µm in magnified merge panels in C–E. *P < 0.05; ****P < 0.0001. KO, knockout.
Figure 2.
Retromer maintains the cell surface abundance and TGN localization of the glutamine transporters ASCT2 and SNAT2, respectively. (A and B) VPS35 knockout HeLa cells were transiently transfected with GFP-VPS35 or GFP alone. After 48 h of incubation, cells were fixed and immunostained with antibodies to endogenous ASCT2 (green) and LAMP1 (red; A) or SNAT2 (green) and LAMP1 (red; B). (C) Colocalization analysis of endogenous SNAT2 (green) and TGN46 (red) in VPS35 knockout HeLa cells transiently transfected with GFP-VPS35 or GFP alone. White asterisks indicate transfected cells. Graphs represent means ± SEM. Pearson’s correlation of three independent experiments with >150 cells per condition; Student’s t test (unpaired). Scale bars, 20 µm and 10 µm in magnified merge panels. ***P < 0.001; ****P < 0.0001.
Retromer-mediated sorting is required as part of an adaptive response to glutamine deprivation
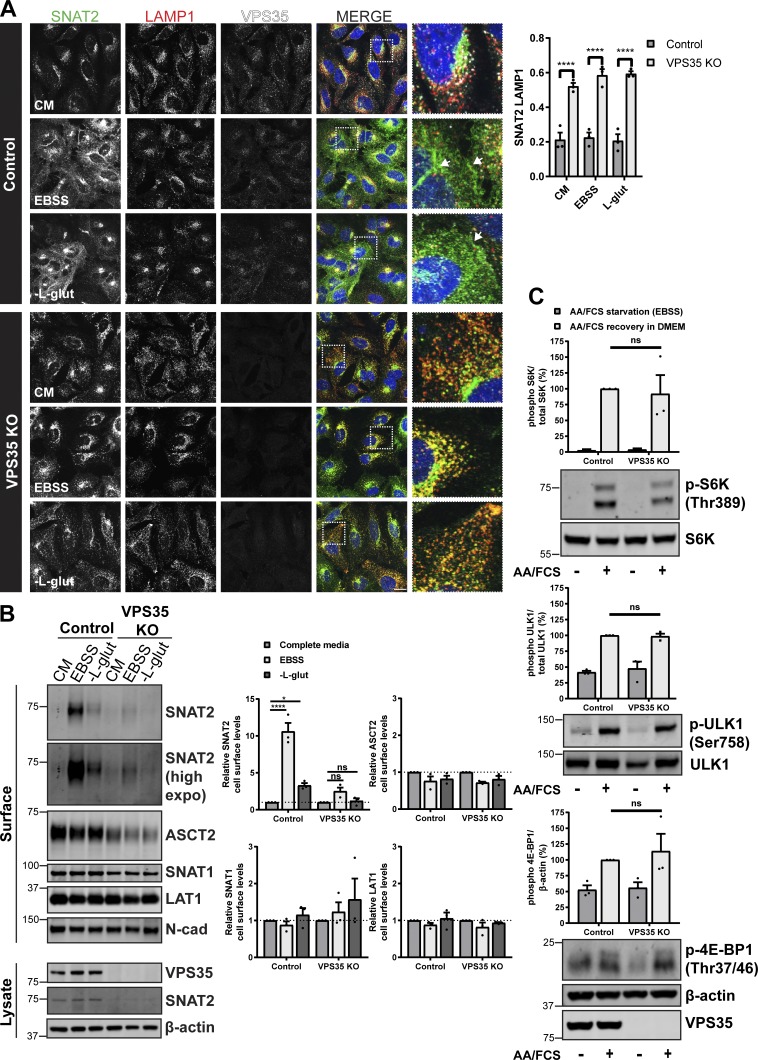
To date, mammalian studies of the role of retromer in the sorting of nutrient transporters have primarily focused on cells grown in nutrient-rich conditions (Steinberg et al., 2013; Kvainickas et al., 2017; Yang et al., 2018). For glutamine transporters, nutrient withdrawal induces an adaptive response in which transporters reconfigure their cell surface expression to scavenge extracellular amino acids in an attempt to rebalance nutrient supply with cellular demand (Hyde et al., 2001; Nardi et al., 2015; Bröer et al., 2018; Hoffmann et al., 2018). We therefore subjected wild type and retromer knockout cells to either complete amino acid removal (Earle’s balanced salt solution [EBSS]) or the selective removal of glutamine and examined the resulting effects on glutamine transporter trafficking. Immunofluorescence (IF) analysis revealed that, following complete amino acid starvation, endogenous SNAT2 shifted from its steady-state TGN localization to a vesicular dispersion throughout the cell with some enrichment at the cell surface (Fig. 3 A and Fig. S1). To quantify the cell surface level of SNAT2, we turned to restricted biotinylation of cell surface proteins coupled with their identification by streptavidin affinity capture and Western analysis. This established that under conditions of complete amino acid withdrawal there was an ~10-fold increase in the cell surface expression of SNAT2 (Fig. 3 B). Similarly, selective withdrawal of glutamine also induced the appearance of SNAT2 at the cell surface, but to a lesser extent (an approximate threefold elevation) than observed upon complete amino acid removal (Fig. 3, A and B; and Fig. S1). There was no significant change to the cell surface levels of ASCT2, SNAT1, or LAT1, either upon complete or selective amino acid withdrawal (Fig. 3 B). In the retromer knockout cell line, however, we failed to observe a SNAT2 adaptive response following complete amino acid or selective glutamine starvation (Fig. 3, A and B; and Fig. S1). SNAT2 remained mislocalized and sequestered in LAMP1-positive late endosomes and lysosomes under both nutrient-deprived conditions (Fig. 3 A). Cell surface biotinylation further confirmed that, in the absence of retromer, SNAT2 was unable to relocate to the cell surface in response to nutrient insufficiency (Fig. 3 B). These data establish that by promoting the retrieval and recycling of key glutamine transporters away from the lysosomal degradative fate, retromer both maintains and allows adaptive remodeling of their steady-state subcellular localizations in response to amino acid availability. As retromer controls the cell surface expression of various other nutrient transporters, including the glucose transporter GLUT1 and the arginine transporter SLC7A1 (Steinberg et al., 2013), the role of this complex in adaptive responses is likely to play a broader role in aiding the balance between nutrient supply and cellular demand. Indeed, the observed effect of retromer silencing on the proliferative rate of melanoma cells (Zhou et al., 2016) may, in part, be due to a restriction in nutrient supply to these rapidly growing cells (Zhang et al., 2017). However, to exclude the alternative possibility that, in the absence of retromer, cells are no longer able to sense and respond to changes in amino acid availability, we examined mTORC1 activity defined by phosphorylation of S6K (Thr389), ULK1 (Ser758), and 4E-BP1 (Thr37/46) in response to nutrient addition following starvation. At a single time point after amino acid stimulation, we observed a similar level of phosphorylation recovery between control and VPS35 knockout cells (Fig. 3 C). This indicates that cells can still respond to changes in nutrient availability in the absence of retromer, despite the perturbed trafficking of various nutrient transporters (Steinberg et al., 2013; Cui et al., 2019).
Figure 3.
Retromer is required for cellular adaptation to nutrient insufficiency. (A) IF analysis of SNAT2 (green), LAMP1 (red), and VPS35 (white) in control and VPS35 knockout HeLa cells following overnight (16 h) amino acid starvation (EBSS), L-glutamine starvation, or incubation in complete media (CM). Arrows indicate the redistribution of SNAT2 to the cell periphery. Bars represent mean ± SEM. Pearson’s correlation; n = three independent experiments with >100 cells per condition; two-way ANOVA followed by Sidak’s multiple comparison. Scale bar, 20 µm. (B) Control and VPS35 knockout HeLa cells were subjected to overnight (16 h) amino acid starvation (EBSS), L-glutamine starvation, or maintained in complete media (CM). Cells were surface biotinylated, and streptavidin agarose was used to capture biotinylated membrane proteins. Surface abundance of the indicated proteins was assessed by quantitative immunoblotting. The quantification shows the mean ± SEM; n = three independent experiments; two-way ANOVA followed by Dunnett’s multiple comparison. (C) Representative immunoblots showing the levels of phosphorylated S6K, ULK1, and 4E-BP1 in control and VPS35 KO cells starved with EBSS for 50 min and then stimulated with complete 1× amino acids and FCS containing DMEM (+) or maintained in EBSS for 10 min (−). Graphs represent the quantification of three independent experiments. Error bars denote ±SEM. (A and B) *P < 0.05; ****P < 0.0001. KO, knockout.
Promoters of the retromer VPS35 and VPS26A genes contain TFEB-responsive CLEAR elements
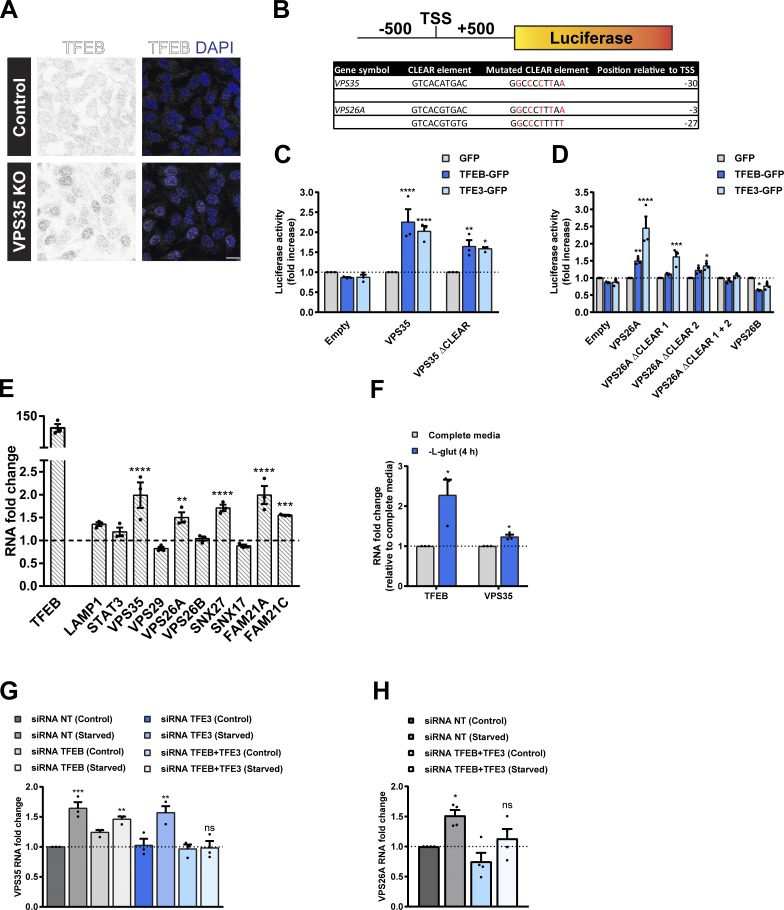
A major controller of the cellular adaptive response to nutrient availability is the transcription factor EB (TFEB; Napolitano and Ballabio, 2016). By transcriptionally regulating cellular processes that favor catabolic metabolism, TFEB and other members of the MiTF/TFE family of transcription factors serve to elevate the supply of nutrients to meet the demands of the cell during nutrient restriction as well as an array of other cellular stresses (Sardiello et al., 2009; Settembre et al., 2011). When nutrients are abundant, mTORC1 phosphorylates TFEB, coupling it to the cytosolic chaperone 14-3-3, thereby retaining it in the cytosol and rendering it inactive (Settembre et al., 2012; Martina et al., 2012; Roczniak-Ferguson et al., 2012; Martina et al., 2014). Upon amino acid deprivation, TFEB is dephosphorylated and translocates to the nucleus (Settembre et al., 2011; Roczniak-Ferguson et al., 2012). Here, it regulates gene expression by recognizing and binding to CLEAR (coordinated lysosomal expression and regulation) elements, palindromic 10-bp motifs enriched in the promoters of genes required for the biogenesis of lysosomes and the process of autophagy (Sardiello et al., 2009; Settembre et al., 2011). Our focused analysis of glutamine transporters has established that retromer controls the cell surface trafficking of key transporters under homeostatic conditions and as part of the adaptive response to amino acid removal. Given this role and the ability of retromer to control the cell surface expression of other nutrient transporters (Steinberg et al., 2013), we examined whether retromer knockout cells display an activation of TFEB that would be indicative of a reduced ability to control cellular nutrient homeostasis. Indeed, IF analysis revealed an increased nuclear localization of endogenous TFEB in retromer knockout cells (Fig. 4 A). This raised the intriguing possibility of a potential link between TFEB-mediated transcriptional activation and the control of retromer expression as part of a cellular adaptive response to nutrient availability.
Figure 4.
TFEB and TFE3 promote transcription of the retromer complex in a CLEAR element-dependent manner. (A) Representative IF images of endogenous TFEB in control and VPS35 knockout (KO) HeLa cells. Scale bar, 20 µm. (B) Schematic of the promoter-luciferase reporter constructs used in C and D indicating the location (relative to TSS) and sequence of CLEAR elements in the retromer genes VPS35 and VPS26A and the sequence of the mutated CLEAR elements that were generated. (C and D) Luciferase activity measured in HEK293T cells lentivirally transduced with GFP, TFEB-GFP, or TFE3-GFP and transiently expressing the indicated wild type or mutated CLEAR element VPS35 (C) or VPS26A or VPS26B (D) promoter-luciferase reporter construct. Bars represent a fold increase in luciferase activity relative to cells infected with GFP from three independent experiments; two-way ANOVA followed by Dunnett’s multiple comparison. (E) Quantitative RT-PCR analysis of the retromer complex gene expression in HeLa cells lentivirally transduced with GFP or TFEB-GFP. Data were normalized to ACTB. Bars represent a fold change relative to GFP-transduced cells; n = three independent experiments; two-way ANOVA followed by Sidak’s multiple comparison. (F) mRNA levels of TFEB and VPS35 in HeLa cells treated with L-glutamine starvation for 4 h. Values normalized to ACTB and expressed as a fold change relative to untreated cells (complete media); n = three independent experiments; Student’s t test (unpaired). (G and H) HeLa cells were treated with nontarget siRNA or siRNA to TFEB and/or TFE3 for 72 h. Cells were then maintained in complete media (control) or in EBSS (starved) for 4 h. Total RNAs were extracted, and mRNA transcript abundance was assessed using primers specific for VPS35 (G) or VPS26A (H). Data were normalized to ACTB. Bars represent a fold change relative to nontarget siRNA control cells from at least three experiments; one-way ANOVA followed by Dunnett’s multiple comparison. (C–H) Data represent mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. NT, nontarget.
In silico promoter analysis identified putative CLEAR elements in the promoter regions of the retromer genes VPS35, VPS29, and VPS26A (but not the promoter region of VPS26B) and retromer interactors SNX27 (Temkin et al., 2011; Steinberg et al., 2013) and the FAM21A and FAM21C components of the actin-polymerizing WASH complex (Table S1; Gomez and Billadeau, 2009). We identified a single putative CLEAR site in the promoter region of VPS35 and two in the promoter region of VPS26A (Table S1). To assess whether TFEB upregulates expression of retromer via binding to these putative CLEAR sites, we transfected luciferase reporter constructs containing the promoter regions (500 bp upstream and downstream of the transcription start site [TSS]) of human VPS35, VPS26A, or VPS26B into HEK293T cells lentivirally transduced with TFEB-GFP or with TFE3-GFP, another member of the MiTF/TFE family of transcription factors (Fig. 4 B). Overexpression of TFEB or TFE3 increased the luciferase activity of the VPS35 and VPS26A reporters, but did not elevate the activity of the VPS26B reporter (Fig. 4, C and D). Compared with the wild type promoter activity, mutagenesis of the single VPS35 or either of the two VPS26A CLEAR sites reduced activity in TFEB- or TFE3-expressing cells (Fig. 4, C and D). Depletion of both CLEAR sites in VPS26A completely abolished TFEB- and TFE3-dependent luciferase activity (Fig. 4 D). This functional validation of CLEAR elements in the predicted promoters of VPS35 and VPS26A supports a global ChIP-seq data analysis that has previously suggested these genes, among 471 other genes, as putative targets of TFEB transcriptional control (Palmieri et al., 2011).
To further establish the link between TFEB and retromer, we next examined the effect of TFEB overexpression on retromer gene expression. In HeLa cells lentivirally transduced to overexpress TFEB, the expression of the retromer genes VPS35 and VPS26A, and genes encoding for retromer accessory protein SNX27, and the WASH complex components FAM21A and FAM21C were all increased (Fig. 4 E). Reflecting the lack of a CLEAR element in the predicted promoter of VPS26B, we did not observe an increase in the level of VPS26B (Fig. 4 E). TFEB-mediated transcriptional control of retromer expression therefore preferentially regulates the level of VPS26A-containing retromer over the VPS26B retromer, which may reflect their suggested functionally distinct roles in endosomal cargo sorting (Bugarcic et al., 2011).
TFEB and TFE3 regulate retromer expression upon nutrient deprivation
Glutamine starvation leads to a reduction in mTOR activity and translocation of endogenous TFEB into the nucleus (Fig. S2, A and B). This prompted us to test whether transcriptional regulation of retromer, or TFEB itself, was affected upon withdrawal of glutamine alone. Assessing VPS35 and TFEB mRNA transcript abundance by quantitative RT-PCR revealed an increase in VPS35 and TFEB mRNA levels in cells cultured for 4 h in the absence of glutamine (Fig. 4 F). TFEB shares partial redundancy with TFE3 (Martina et al., 2014), and complete amino acid starvation is recognized to activate both transcription factors (Raben and Puertollano, 2016). For this reason, we determined that the increase in VPS35 mRNA abundance in response to cellular stress was dependent on TFEB and TFE3. Starvation of HeLa cells for 4 h increased VPS35 mRNA levels, but suppression of either TFEB or TFE3 alone was not sufficient to prevent the increase (Fig. 4 G and Fig. S3 A). Combined suppression of TFEB and TFE3 abolished the starvation-induced increase in VPS35 mRNA (Fig. 4 G) and VPS26A mRNA (Fig. 4 H). This establishes that, in response to nutrient deprivation, the retromer genes VPS35 and VPS26A are among the repertoire of genes upregulated by TFEB and TFE3.
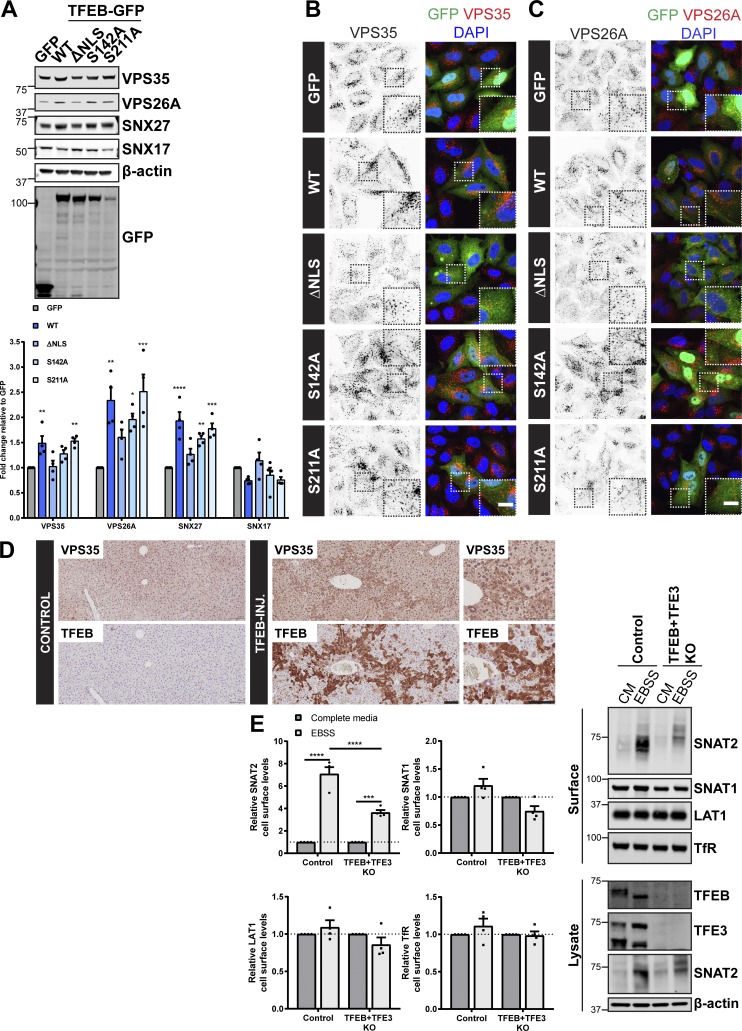
In agreement with quantitative RT-PCR data (Fig. 4 E), in HeLa cells lentivirally transduced to overexpress wild type or constitutively active forms of TFEB (S142A and S211A), but not an inactive form in which the nuclear localization sequence was removed (ΔNLS), we observed an increase in the total protein abundance of endogenous VPS35, VPS26A, and SNX27, but not the non–retromer-associated protein SNX17 (Fig. 5 A; Steinberg et al., 2012; Böttcher et al., 2012; McNally et al., 2017). A similar increase in the total protein abundance of endogenous VPS35, VPS26A, and SNX27 was observed in HEK293T cells overexpressing TFEB and the MiTF/TFE family members MITF and TFE3 (Fig. S3 B). IF analysis revealed that transient overexpression of wild type or constitutively active forms of TFEB (S142A and S211A), but not the inactive form (ΔNLS), increased the immunofluorescent intensity of endogenously labeled VPS35- and VPS26A-positive endosomes (Fig. 5, B and C). Moreover, in liver samples taken from TFEB-injected mice, histological analysis revealed elevated levels of VPS35 expression (Fig. 5 D). Together, thesein cellulo and in vivo data establish retromer as a target for TFEB-mediated transcriptional control.
Figure 5.
TFEB regulates expression of the retromer complex in cellulo and in vivo. (A) HeLa cells were infected with lentiviruses expressing the indicated GFP-tagged TFEB or GFP alone. Cells were lysed, and total cell lysates were examined for the indicated proteins by immunoblotting. Data were normalized to β-actin. Quantification represents mean ± SEM fold change relative to GFP-expressing cells; n = four independent experiments; one-way ANOVA followed by Dunnett’s multiple comparison. (B and C) IF analysis of endogenous retromer component VPS35 (B) or VPS26A (C) in HeLa cells transiently transfected with the indicated GFP-tagged TFEB construct or GFP alone. (D) Immunohistochemistry of liver sections from mice starved for 24 h and injected with an adenoviral control vector (control) or human TFEB under control of a liver-specific promoter (TFEB-INJ). Tissues were stained for VPS35 or TFEB. Scale bars, 10 µm (B and C) and 100 µm (D). (E) Control and TFEB + TFE3 knockout HeLa cells were subjected to overnight (16 h) amino acid starvation (EBSS) or maintained in complete media (CM). Cells were surface biotinylated, and streptavidin agarose was used to capture biotinylated membrane proteins. Surface abundance of the indicated proteins was assessed by quantitative immunoblotting. The quantification shows the mean ± SEM; n = four independent experiments; two-way ANOVA followed by Sidak’s multiple comparison. (A and E) *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. KO, knockout.
Finally, we have established that the SNAT2 adaptive response to nutrient starvation requires retromer-mediated endosomal sorting (Fig. 3, A and B). We therefore considered whether TFEB and TFE3 are necessary to evoke the starvation-induced and retromer-dependent increase in the cell surface levels of SNAT2. Cell surface biotinylation revealed that, in the absence of TFEB and TFE3, the relocation of SNAT2 to the cell surface in response to nutrient insufficiency was perturbed (Fig. 5 E; note that SNAT2 gene expression is not regulated by TFEB overexpression [Fig. S3 C]). Together, these data imply that TFEB/TFE3 regulation of retromer gene expression is required for SNAT2 redistribution to the cell surface upon prolonged nutrient withdrawal.
In sum, as far as we are aware, our study provides the first functional evidence of the transcriptional control of retromer expression. In identifying the central role of the TFEB-containing MiTF/TFE family in this process, our data lead to the proposal of a regulatory pathway whereby TFEB-mediated expression of retromer and retromer accessory proteins serves to modulate retromer-mediated cargo retrieval and recycling through the endosomal network. We have provided initial evidence for a functional role of this regulated pathway in the endosomal sorting of key glutamine transporters as part of the adaptive response to amino acid restriction. With TFEB playing a central role in adaptive responses to a variety of other cellular stresses (Napolitano and Ballabio, 2016), the synchronized transcriptional control of retromer expression, and hence cargo retrieval and recycling through the endosomal network, is likely to be an important process in the long-term adaptation of cells to these stresses. Finally, with clinical studies having revealed perturbed retromer function in age-related Alzheimer’s and Parkinson’s diseases (Small and Petsko, 2015; McMillan et al., 2017), the identification of a TFEB–retromer pathway may offer an unexplored strategy through which to enhance retromer expression and thereby increase its neuroprotective role in these and other neurological diseases.
Materials and methods
Antibodies
Antibodies used in this study were mouse monoclonal antibodies raised against SNAT2 (clone C-6, Santa Cruz, sc-514037; IF), SNX27 (clone 1C6, Abcam, Ab77799; Western blot [WB]), GFP (clones 7.1 and 13.1, Roche, 11814460001; WB), LAMP1 (clone H4A3, Developmental Studies Hybridoma Bank; IF), N-cadherin (clone 32, BD Biosciences, 610920; WB), p70 S6 kinase (clone H-9, Santa Cruz, sc-8418; WB), CD71/transferrin receptor (clone H68.4, Santa Cruz, sc-65882; WB), and β-actin (Sigma-Aldrich, A1978; WB); rabbit monoclonal antibodies raised against VPS35 (Abcam, ab157220; WB), ASCT2 (clone D7C12, Cell Signaling Technology, 8057; WB), phospho-p70S6 kinase (Thr389, clone 108D2, Cell Signaling Technology, 9234; WB), phospho-4E-BP1 (Thr37/46, clone 236B4, Cell Signaling Technology, 2855; WB), and ULK1 (clone D8H5, Cell Signaling Technology, 8054; WB); rabbit polyclonal antibodies raised against VPS35 (Abcam, 97545; IF), SNX17 (Proteintech, 10275–1-AP; WB), SLC1A5/ASCT2 (Sigma-Aldrich, HPA035240; IF), SNAT2/SLC38A2 (MBL, BMP081; WB), SNAT1/SLC38A1 (Proteintech, 12039-1-AP; WB), TFEB (Cell Signaling Technology, 4240; IF), TFE3 (Sigma-Aldrich, HPA023881; WB), VPS26 (Abcam, ab23892; IF), VPS26 (Abcam, ab137447; WB), VPS29 (Abcam, ab98929; WB), phospho-ULK1 (Ser757, Cell Signaling Technology, 6888; WB), and LAT1 (Cell Signaling Technology, 5347; WB); and a sheep polyclonal antibody raised against human TGN46 (Bio-Rad, AHP500G; IF). For Odyssey detection of WB, the following secondary antibodies were used: goat anti-mouse 680 (Invitrogen) and goat anti-rabbit 800 (Invitrogen).
Drug treatments
Cells were incubated for the indicated time at 37°C in medium containing DMSO (Sigma-Aldrich), 100 nM bafilomycin A1 (Tocris Bioscience), or 10 µg/ml cycloheximide (Sigma-Aldrich).
Cell culture conditions and transfections
All cell lines were maintained at 37°C and in a 5% CO2 atmosphere, cultured in DMEM containing 4.5 g/liter glucose (Sigma-Aldrich, D5796), and supplemented with 10% (vol/vol) FBS. The clonal VPS35 knockout HeLa cell line was previously described (Simonetti et al., 2017). The clonal TFEB/TFE3 knockout cell line was a kind gift from Dr. Richard Youle (National Institute of Neurological Disorders and Stroke, Porter Neuroscience Research Center, Bethesda, MD). DNA was transiently transfected into cells using FuGENE 6 transfection reagent (Promega) according to the manufacturer’s instructions. For siRNA-based knockdown, cells were reverse transfected using DharmaFECT 1 (GE Healthcare) and then transfected again 24 h later according to the manufacturer’s instructions. 48 h after the second transfection, cells were processed for RNA extraction. TFEB was suppressed using Dharmacon siGENOME Human TFEB (cat. #D-009798-03). For TFE3 suppression, Dharmacon ON-TARGETplus TFE3 siRNA SMARTpool (cat. #L-009363-00) was used. The nontarget control siRNA used was Dharmacon ON-TARGETplus nontargeting siRNA no. 2 (cat. #D-001810-02).
Starvation and glutamine withdrawal
For complete starvation, cells were incubated in EBSS (Thermo Fisher Scientific) for the indicated time. For glutamine deprivation experiments, normal culture media were replaced with modified DMEM without L-glutamine (Thermo Fisher Scientific, 11960) containing 10% dialyzed FBS (Sigma-Aldrich, F0392) for the indicated time.
Generation of lentiviral cell lines
For lentiviral production, constructs were cloned into the pXLG3 viral vector and cotransfected into HEK293T cells with Pax2 and pMDG2 plasmids. The virus was harvested 72 h after transfection. The virus was filtered before HeLa or HEK293T cells were transduced with a viral titration to produce stably transduced cells.
Quantitative WB analysis
For Western blotting, cells were lysed in PBS with 2% Triton X-100 containing Pierce protease and phosphatase inhibitor tablets (Thermo Fisher Scientific). The protein concentration was determined with a bicinchoninic acid assay kit (Thermo Fisher Scientific), and equal amounts were resolved on NuPAGE 4–12% precast gels (Invitrogen). Blotting was performed on polyvinylidene fluoride membranes (Immobilon-FL, EMD Millipore) followed by detection using the Odyssey infrared scanning system (LI-COR Biosciences).
Cell surface biotinylation
Cells were surface biotinylated with membrane-impermeable biotin (Thermo Fisher Scientific) at 4°C to prevent endocytosis. Following biotinylation, cells were lysed (2% Triton X-100, PBS, and Pierce protease and phosphatase inhibitor EDTA-free tablets, pH 7.5), and lysates were cleared by centrifugation. Equal amounts of protein from the control and indicated knockout were then added to streptavidin Sepharose to capture biotinylated proteins. Streptavidin beads and lysates were incubated for 30 min at 4°C before washing in PBS containing 1.2 M NaCl and 1% Triton X-100. Proteins were eluted in 2× NuPAGE LDS Sample Buffer (Life Technologies) by boiling at 95°C for 10 min and then were separated by SDS-PAGE and subjected to quantitative WB analysis.
Luciferase assay
HEK293T cells stably expressing GFP, TFEB-GFP, or TFE3-GFP seeded at 50% confluency in DMEM without phenol red into 96-well plates were transfected with 50 ng of pLightSwitch promoter vector or pLightSwitch empty vector as a control (SwitchGear Genomics) using FuGENE 6 (Promega) transfection reagent. 48 h after transfection, luciferase activity was assayed using LightSwitch assay reagent according to the manufacturer’s instructions. The luciferase activity was expressed as a fold increase versus GFP-expressing cells.
RNA extraction, reverse transcription, and quantitative PCR
Total RNA was extracted from cells using an RNeasy Mini Kit (Qiagen). cDNA was synthesized and real-time quantitative RT-PCR was performed using the SuperScript III Platinum SYBR Green One-Step quantitative RT-PCR Kit (Thermo Fisher Scientific). The quantification of gene expression was performed in triplicate. Amplification of the sequence of interest was normalized with a housekeeping gene, ACTB. Quantification was performed using comparative threshold cycle (Ct) method of analysis. The value was expressed as a fold change relative to RNA from GFP-expressing or untreated control cells. Intron-spanning quantitative PCR primers were designed using the assay design center of the Universal ProbeLibrary (Roche).
IF staining
Cells were washed in PBS and fixed in 4% PFA and then washed in PBS and permeabilized with 0.1% Triton X-100 or 0.1% saponin. Fixed cells were then blocked in 1% BSA and incubated in primary antibody and appropriate secondary antibody (Alexa Fluor; Thermo Fisher Scientific) in 0.1% BSA. PBS-washed coverslips were mounted onto glass slides with Mowiol (Sigma-Aldrich).
Mouse models/histology
All mice used were males and maintained in a C57BL/6 strain background. Hepatic transduction was achieved by intravenous administration (retro-orbital) of 2 × 1013 viral particles/kilogram. The HDAdTFEB virus was described previously (Settembre et al., 2013). Mice were analyzed 1 mo after infection. At least five animals per group were analyzed. Livers were dissected, post-fixed with buffered 4% paraformaldehyde overnight at 4°C and then dehydrated in a graded series of ethanol, cleared with xylene, and infiltrated with paraffin. Formalin-fixed, paraffin-embedded liver sections (4 µm) were analyzed using immunohistochemistry against TFEB (1:200; Cell Signaling Technology, 4240) and VPS35 (1:100; Abcam, ab157220). Sections were subjected to a heat-mediated antigen retrieval procedure (10 mM citrate buffer, pH 6.0), followed by a 1-h preincubation with normal goat serum. After blocking of endogenous peroxidase activity for 15 min in 0.1% H2O2 in water, sections were incubated with primary antibodies diluted in 1% BSA in PBS. Incubation with primary antibodies was performed for 2 h at room temperature. Following incubation with a secondary antibody (MACH 4 Universal HRP-Polymer; Biocare Medical, M4U534), immune reactions were revealed using NovaRed and counterstained with hematoxylin, dehydrated, and mounted. All the experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee of Cardarelli Hospital in Naples and authorized by the Italian Ministry of Health.
Image acquisition and image analysis
Microscopy images were collected with a confocal laser-scanning microscope (SP5 AOBS; Leica Microsystems) attached to an inverted epifluorescence microscope (DMI6000; Thermo Fisher Scientific). A 63× 1.4 NA oil immersion objective (Plan Apochromat BL; Leica Biosystems) and the standard SP5 system acquisition software and detector were used. Images were captured using Application Suite AF software (version 2.7.3.9723; Leica Microsystems) and then analyzed with the Volocity 6.3 software (PerkinElmer). For colocalization studies, Pearson’s correlation (measuring the correlation in the variation between two channels) and Manders’s colocalization coefficient (measuring the degree to which the corresponding channel overlaps with the other channel) were measured using the Costes method to set automatic thresholds. Immunohistochemistry sections were visualized at 20× magnification with the Leica DM5000 microscope and acquired with the Leica LAS V4.4 software. Sections were digitized using a Scan-Scope slide scanner (Leica scn400), and images were generated using the Leica digital image hub.
DNA plasmids
TFEB-GFP was sub-cloned into the pXLG3 lentiviral vector. TFE3 and MITF were amplified from HeLa cDNA and cloned into the pEGFPN1 vector (Takara Bio) and pXLG3 lentiviral vector. The GFP VPS35 construct has previously been described (McGough et al., 2014). The gRNA for CRISPR genome editing was cloned into the CRISPR-Cas9 plasmid px330. The VPS35 gRNA used in this study was 5′-GTGGTGTGCAACATCCCTTG-3′. The VPS35, VPS26A, and VPS26B promoter regions (±500 TSS) were amplified from genomic DNA isolated from HEK293T cells and cloned into the pLightSwitch promoter reporter vector (SwitchGear Genomics).
Site-directed mutagenesis
Point mutations in the TFEB constructs and CLEAR mutant promoters were generated using site-directed mutagenesis. PCR primers that contain desired mutations were designed according to the online tool QuikChange Primer Design (Agilent Genomics). CLEAR mutant promoters for VPS35 and VPS26A SDM CLEAR motif 1 could not be generated by site-directed mutagenesis and were synthesized by Eurofins Genomics.
Statistical analysis
Statistical analyses were performed using Prism 7 (GraphPad Software). For comparing two groups, statistical analyses were performed using Student’s t test (unpaired and two-tailed). For multiple comparisons the following statistical tests were used: one-way ANOVA followed by Dunnett’s multiple comparison, or two-way ANOVA followed by Sidak’s multiple comparison or Dunnett’s multiple comparison. Data distribution was assumed to be normal, but this was not formally tested. All quantified WB and confocal colocalization data are the mean of at least three independent experiments. Graphs represent means and SEM.
Online supplemental material
Fig. S1 illustrates that retromer is required for SNAT2 cell surface redistribution following nutrient deprivation. Fig. S2 shows that L-glutamine withdrawal results in a reduction in mTOR activity and translocation of endogenous TFEB into the nucleus. Fig. S3 shows that retromer expression is regulated by the MITF family of transcription factors. Table S1 presents the distribution of CLEAR elements in relation to the TSS in the promoters of the retromer complex genes and accessory binding partners.
Supplementary Material
Acknowledgments
We thank the Wolfson Bioimaging Facility at the University of Bristol for their support.
This work was supported by grants from the Medical Research Council (MR/L007363/1 and MR/P018807/1) and the Wellcome Trust (104568/Z/14/2) to P.J. Cullen. Italian Telethon Foundation (TGM16CB6); European Research Council Advanced Investigator no. 694282 (LYSOSOMICS) to A. Ballabio); US National Institutes of Health (R01-NS078072) to A. Ballabio); and the Associazione Italiana per la Ricerca sul Cancro (IG2018 22103) to A. Ballabio.
The authors declare no competing financial interests.
Author contributions: Initial conceptualization, R. Curnock and P.J. Cullen; evolution of conceptualization, all authors; investigation, R. Curnock and A. Calcagni; writing – original draft, R. Curnock and P.J. Cullen; writing – review and editing, all authors; funding acquisition, A. Ballabio and P.J. Cullen; supervision, A. Ballabio and P.J. Cullen.
References
- Arighi C.N., Hartnell L.M., Aguilar R.C., Haft C.R., and Bonifacino J.S.. 2004. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165:123–133. 10.1083/jcb.200312055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher R.T., Stremmel C., Meves A., Meyer H., Widmaier M., Tseng H.Y., and Fässler R.. 2012. Sorting nexin 17 prevents lysosomal degradation of β1 integrins by binding to the β1-integrin tail. Nat. Cell Biol. 14:584–592. 10.1038/ncb2501 [DOI] [PubMed] [Google Scholar]
- Bröer A., Rahimi F., and Bröer S.. 2016. Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 291:13194–13205. 10.1074/jbc.M115.700534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröer A., Fairweather S., and Bröer S.. 2018. Disruption of amino acid homeostasis by novel ASCT2 inhibitors involves multiple targets. Front. Pharmacol. 9:785 10.3389/fphar.2018.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugarcic A., Zhe Y., Kerr M.C., Griffin J., Collins B.M., and Teasdale R.D.. 2011. Vps26A and Vps26B subunits define distinct retromer complexes. Traffic. 12:1759–1773. 10.1111/j.1600-0854.2011.01284.x [DOI] [PubMed] [Google Scholar]
- Burd C., and Cullen P.J.. 2014. Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6:a016774 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Carosi J.M., Yang Z., Ariotti N., Kerr M.C., Parton R.G., Sargeant T.J., and Teasdale R.D.. 2019. Retromer has a selective function in cargo sorting via endosome transport carriers. J. Cell Biol. 218:615–631. 10.1083/jcb.201806153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., and Steinberg F.. 2018. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19:679–696. 10.1038/s41580-018-0053-7 [DOI] [PubMed] [Google Scholar]
- Frankel E.B., and Audhya A.. 2018. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 74:4–10. 10.1016/j.semcdb.2017.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallon M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R.B., Teasdale R.D., Collins B.M., and Cullen P.J.. 2014. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. USA. 111:E3604–E3613. 10.1073/pnas.1410552111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T.S., and Billadeau D.D.. 2009. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 17:699–711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., and Donaldson J.G.. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608. 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft C.R., de la Luz Sierra M., Bafford R., Lesniak M.A., Barr V.A., and Taylor S.I.. 2000. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Mol. Biol. Cell. 11:4105–4116. 10.1091/mbc.11.12.4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M.E., Breusegem S.Y., and Seaman M.N.. 2012. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem. J. 442:209–220. 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Harterink M., Port F., Lorenowicz M.J., McGough I.J., Silhankova M., Betist M.C., van Weering J.R.T., van Heesbeen R.G.H.P., Middelkoop T.C., Basler K., et al. . 2011. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13:914–923. 10.1038/ncb2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T.M., Cwiklinski E., Shah D.S., Stretton C., Hyde R., Taylor P.M., and Hundal H.S.. 2018. Effects of sodium and amino acid substrate availability upon the expression and stability of the SNAT2 (SLC38A2) amino acid transporter. Front. Pharmacol. 9:63 10.3389/fphar.2018.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V.W., Bai M., and Li J.. 2012. Getting active: protein sorting in endocytic recycling. Nat. Rev. Mol. Cell Biol. 13:323–328. 10.1038/nrm3332 [DOI] [PubMed] [Google Scholar]
- Hyde R., Christie G.R., Litherland G.J., Hajduch E., Taylor P.M., and Hundal H.S.. 2001. Subcellular localization and adaptive up-regulation of the System A (SAT2) amino acid transporter in skeletal-muscle cells and adipocytes. Biochem. J. 355:563–568. 10.1042/bj3550563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr M.C., Bennetts J.S., Simpson F., Thomas E.C., Flegg C., Gleeson P.A., Wicking C., and Teasdale R.D.. 2005. A novel mammalian retromer component, Vps26B. Traffic. 6:991–1001. 10.1111/j.1600-0854.2005.00328.x [DOI] [PubMed] [Google Scholar]
- Kvainickas A., Orgaz A.J., Nägele H., Diedrich B., Heesom K.J., Dengjel J., Cullen P.J., and Steinberg F.. 2017. Retromer- and WASH-dependent sorting of nutrient transporters requires a multivalent interaction network with ANKRD50. J. Cell Sci. 130:382–395. 10.1242/jcs.196758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Gershlick D.C., Vidaurrazaga A., Rojas A.L., Bonifacino J.S., and Hierro A.. 2016. Structural mechanism for cargo recognition by the retromer complex. Cell. 167:1623–1635.e14. 10.1016/j.cell.2016.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Chen Y., Gucek M., and Puertollano R.. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8:903–914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Lishu L., Jeong-A L., Patange S., Raben N., and Puertollano R.. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7:ra9 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I.J., Steinberg F., Jia D., Barbuti P.A., McMillan K.J., Heesom K.J., Whone A.L., Caldwell M.A., Billadeau D.D., Rosen M.K., and Cullen P.J.. 2014. Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24:1670–1676. (erratum appears in Curr. Biol. 2014. 24:1678). 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan K.J., Korswagen H.C., and Cullen P.J.. 2017. The emerging role of retromer in neuroprotection. Curr. Opin. Cell Biol. 47:72–82. 10.1016/j.ceb.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.E., and Cullen P.J.. 2018. Endosomal retrieval of cargo: Retromer is not alone. Trends Cell Biol. 28:807–822. 10.1016/j.tcb.2018.06.005 [DOI] [PubMed] [Google Scholar]
- McNally K.E., Faulkner R., Steinberg F., Gallon M., Ghai R., Pim D., Langton P., Pearson N., Danson C.M., Nägele H., et al. . 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19:1214–1225. 10.1038/ncb3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano G., and Ballabio A.. 2016. TFEB at a glance. J. Cell Sci. 129:2475–2481. 10.1242/jcs.146365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi F., Hoffmann T.M., Stretton C., Cwiklinski E., Taylor P.M., and Hundal H.S.. 2015. Proteasomal modulation of cellular SNAT2 (SLC38A2) abundance and function by unsaturated fatty acid availability. J. Biol. Chem. 290:8173–8184. 10.1074/jbc.M114.625137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., et al. . 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 136:521–534. 10.1016/j.cell.2008.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20:3852–3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Raben N., and Puertollano R.. 2016. TFEB and TFE3: Linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol. 32:255–278. 10.1146/annurev-cellbio-111315-125407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. . 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Seaman M.N. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165:111–122. 10.1083/jcb.200312034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J., McCaffery J.M., and Emr S.D.. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. . 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. . 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., et al. . 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15:647–658. 10.1038/ncb2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti B., Danson C.M., Heesom K.J., and Cullen P.J.. 2017. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J. Cell Biol. 216:3695–3712. 10.1083/jcb.201703015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S.A., and Petsko G.A.. 2015. Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat. Rev. Neurosci. 16:126–132. 10.1038/nrn3896 [DOI] [PubMed] [Google Scholar]
- Steinberg F., Heesom K.J., Bass M.D., and Cullen P.J.. 2012. SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J. Cell Biol. 197:219–230. 10.1083/jcb.201111121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F., Gallon M., Winfield M., Thomas E.C., Bell A.J., Heesom K.J., Tavaré J.M., and Cullen P.J.. 2013. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15:461–471. 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic T.I., Setty T.G., Sitaram A., and Burd C.G.. 2007. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 177:115–125. 10.1083/jcb.200609161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P., Lauffer B., Jäger S., Cimermancic P., Krogan N.J., and von Zastrow M.. 2011. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13:715–721. 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Follett J., Kerr M.C., Clairfeuille T., Chandra M., Collins B.M., and Teasdale R.D.. 2018. Sorting nexin 27 (SNX27) regulates the trafficking and activity of the glutamine transporter ASCT2. J. Biol. Chem. 293:6802–6811. 10.1074/jbc.RA117.000735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pavlova N.N., and Thompson C.B.. 2017. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 36:1302–1315. 10.15252/embj.201696151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Wiener H., Su W., Zhou Y., Liot C., Ahearn I., Hancock J.F., and Philips M.R.. 2016. VPS35 binds farnesylated N-Ras in the cytosol to regulate N-Ras trafficking. J. Cell Biol. 214:445–458. 10.1083/jcb.201604061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.