Kinetochore localization of CENP-C, which is a key and conserved kinetochore component, is regulated during cell cycle progression. Watanabe et al. demonstrate that CDK1-mediated CENP-C phosphorylation regulates mitotic kinetochore localization via binding of CENP-C to the CENP-A nucleosome.
Abstract
The kinetochore is essential for faithful chromosome segregation during mitosis. To form a functional kinetochore, constitutive centromere-associated network (CCAN) proteins are assembled on the centromere chromatin that contains the centromere-specific histone CENP-A. CENP-C, a CCAN protein, directly interacts with the CENP-A nucleosome to nucleate the kinetochore structure. As CENP-C is a hub protein for kinetochore assembly, it is critical to address how the CENP-A–CENP-C interaction is regulated during cell cycle progression. To address this question, we investigated the CENP-C C-terminal region, including a conserved CENP-A–binding motif, in both chicken and human cells and found that CDK1-mediated phosphorylation of CENP-C facilitates its binding to CENP-A in vitro and in vivo. We observed that CENP-A binding is involved in CENP-C kinetochore localization during mitosis. We also demonstrate that the CENP-A–CENP-C interaction is critical for long-term viability in human RPE-1 cells. These results provide deeper insights into protein-interaction network plasticity in centromere proteins during cell cycle progression.
Introduction
In eukaryotes, genetic material is equally distributed to daughter cells during mitosis. This process is achieved by the attachment of sister chromatids to the bipolar mitotic spindle followed by their segregation into daughter cells. The kinetochore, a large protein complex that is formed on the centromere of each sister chromatid, ensures faithful chromosome segregation by directly associating with the spindle microtubules (Fukagawa and Earnshaw, 2014; Hara and Fukagawa, 2017, 2018; McKinley and Cheeseman, 2016).
The location of the centromere is specified by the histone H3 variant CENP-A (Palmer et al., 1987), which is incorporated into chromatin as an octameric nucleosome along with canonical histones (H4, H2A, and H2B; Black and Cleveland, 2011; Palmer et al., 1987; Westhorpe and Straight, 2013). Various kinetochore proteins are assembled on centromeric chromatin containing CENP-A nucleosomes. Among these kinetochore proteins, the constitutive centromere-associated network (CCAN), which consists of 16 components (CENP-C, CENP-H, CENP-I, CENP-K, CENP-L, CENP-M, CENP-N, CENP-O, CENP-P, CENP-Q, CENP-R, CENP-S, CENP-T, CENP-U, CENP-W, and CENP-X), localizes to the centromere throughout the cell cycle (Amano et al., 2009; Foltz et al., 2006; Hori et al., 2008a; Izuta et al., 2006; Nishino et al., 2012; Okada et al., 2006), forming a base for functional kinetochore architecture via recruitment of the KMN (KNL1, Mis12, and Ndc80 complexes) network that binds to the microtubules during mitosis (Alushin et al., 2010; Cheeseman et al., 2006; DeLuca et al., 2006; Hara and Fukagawa, 2017; McKinley and Cheeseman, 2016; Nagpal and Fukagawa, 2016; Pesenti et al., 2016).
CENP-C, a CCAN component, is a key hub protein for kinetochore assembly (Fukagawa and Brown, 1997; Fukagawa et al., 1999; Klare et al., 2015; Kwon et al., 2007; Saitoh et al., 1992; Weir et al., 2016). CENP-C has multifunctional domains that bind to various proteins, including the Mis12 complex (Dimitrova et al., 2016; Petrovic et al., 2010, 2016; Przewloka et al., 2011), the CENP-L–CENP-N complex (Chittori et al., 2018; McKinley et al., 2015; Nagpal et al., 2015; Pentakota et al., 2017; Tian et al., 2018), the CENP-H–CENP-I–CENP-K–CENP-M complex (CENP-H complex; Basilico et al., 2014; Klare et al., 2015), CENP-B (Fachinetti et al., 2015), and the CENP-A nucleosome (Fachinetti et al., 2013; Falk et al., 2015; Guo et al., 2017; Kato et al., 2013). Previous studies using chicken (gCENP-C) and human CENP-C (hCENP-C) demonstrated that the middle region associates with the CENP-L–CENP-N and CENP-H complexes, and the C-terminal region binds to the CENP-A nucleosome (Klare et al., 2015; McKinley et al., 2015; Nagpal et al., 2015). We have also found that the gCENP-C C-terminal region interacts with kinetochores during mitosis, but not during interphase (Nagpal et al., 2015), suggesting that CENP-C alters kinetochore binding of its C-terminal region during cell cycle progression. These findings lead to critical questions: how is the cell cycle–dependent CENP-A–CENP-C interaction regulated, and what is its biological significance?
To address these questions, we focused on the conserved CENP-A nucleosome interaction motif in the CENP-C C-terminal region and found that this motif is required for mitotic kinetochore localization of the CENP-C C-terminal fragment in both chicken and human cells. We identified a conserved threonine residue (threonine 651 [T651] in gCENP-C and T734 in hCENP-C) in CENP-C as a key CDK1-phosphorylation site, which regulates mitotic kinetochore localization of CENP-C in both chicken and human cells. We also showed that the CDK1 phosphorylation facilitates the binding of CENP-C to the CENP-A nucleosome. These results demonstrate that the CENP-A–CENP-C interaction mode changes between interphase and mitosis via CDK1-mediated phosphorylation, suggesting that such change is important for proper kinetochore function.
Results
CDK1-mediated phosphorylation of CENP-C is required for localization of its C-terminal fragment to kinetochores
hCENP-C has two CENP-A–binding regions: a central domain and a CENP-C motif (Kato et al., 2013). Sequences around the central domain are conserved in human, mouse, and frog CENP-C, but not in gCENP-C (Fig. S1 A) or other model organisms (Kato et al., 2013). In contrast, the CENP-C motif is conserved among species from yeast to human (Kato et al., 2013). Therefore, we first focused on the C-terminal region of CENP-C containing the CENP-C motif to investigate the CENP-A–CENP-C interaction (Fig. 1, A and B).
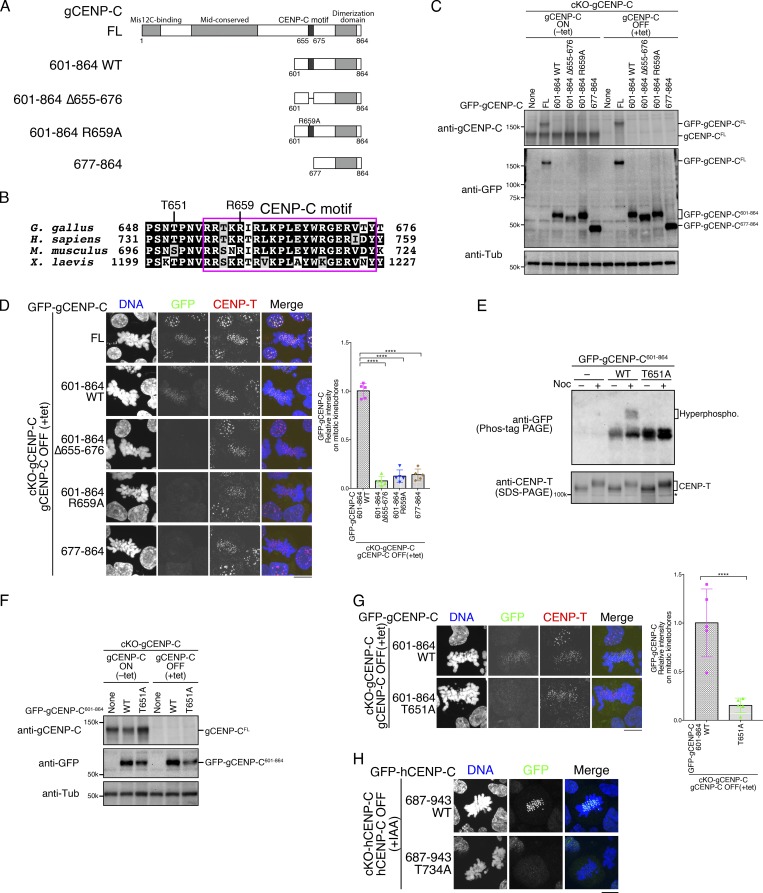
Figure 1.
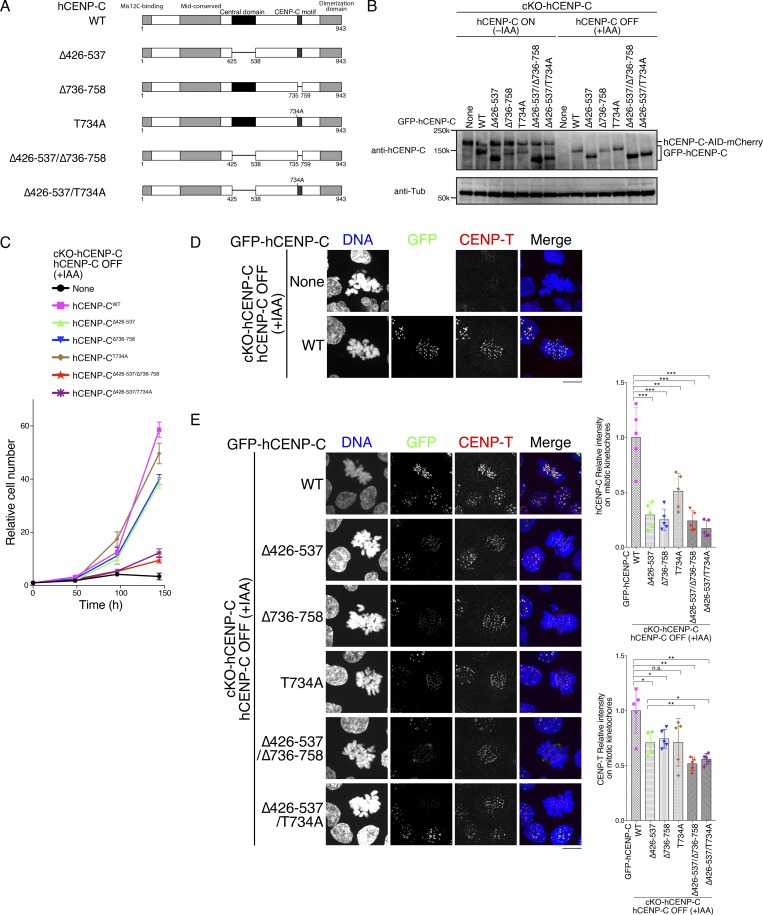
CENP-C phosphorylation near the CENP-C motif is required for mitotic kinetochore localization of the CENP-C C-terminal fragment. (A) Schematic representation of gCENP-C. gCENP-C contains 864 aa (gCENP-CFL), the gCENP-C C-terminal fragment (gCENP-C601–864 WT), and its mutants (gCENP-C601–864 Δ655–676, gCENP-C601–864 R659A, and gCENP-C677–864). (B) Amino acid sequence alignment of the CENP-C C-terminal region. The magenta box indicates the CENP-C motif. (C) Expression of full-length WT gCENP-C (gCENP-CFL) was conditionally turned off by tet addition in gCENP-C cKO (cKO-gCENP-C) cells (None). GFP-fused gCENP-C full-length, gCENP-C601–864 WT, and its mutants (full length [FL] 601–864 WT, 601–864 Δ655-676, 601–864 R659A, and 677–864) were stably expressed in cKO-gCENP-C cells. These cells were cultured in the presence or absence of tet (+tet: CENP-C OFF or −tet: CENP-C ON) for 48 h. α-Tubulin (Tub) was probed as a loading control. (D) Localization of GFP-fused gCENP-CFL, gCENP-C 601–864, gCENP-C 601–864 Δ655–676, gCENP-C 601-864 R659A, and gCENP-C 677-864 in mitotic and interphase cKO-gCENP-C cells at 48 h after addition of tet (green). CENP-T was stained as a kinetochore marker (red). DNA was stained by DAPI (blue). Scale bar indicates 10 µm. GFP signals on kinetochores in mitotic cells were quantified. Bar graph indicates mean with SD (n = 5; ****, P < 0.0001; NS, P > 0.05; unpaired t test, two tailed). See also Fig. S3 A. (E) Phos-tag PAGE analysis of GFP-fused gCENP-C601–864 WT or T651A in cKO-gCENP-C cells enriched in mitosis by nocodazole (Noc) treatment. CENP-T was examined using SDS-PAGE as a control for phosphorylation in mitotic-arrested cells. cKO-CENP-C without GFP-gCENP-C601–864 expression was also analyzed (None). Asterisk shows a nonspecific signal. See also Fig. S1 B. (F) Expression of GFP-fused gCENP-C 601–864 T651A in cKO-gCENP-C cells. The cells shown were cultured in the presence or absence of tet (+tet: gCENP-C OFF or −tet: gCENP-C ON) for 48 h. α-Tubulin was probed as a loading control. (G) Localization of GFP-fused gCENP-C601–864 WT and gCENP-C601–864 T651A in mitotic and interphase cKO-gCENP-C cells at 48 h after addition of tet (green). CENP-T was stained as a kinetochore marker (red), and DNA was stained by DAPI (blue). Scale bar indicates 10 µm. GFP signals on kinetochores in mitotic cells were quantified. Bar graph indicates mean with SD (n = 5; ****, P < 0.0001; NS, P > 0.05, unpaired t test, two tailed). See also Fig. S3 B. (H) Localization of GFP-fused hCENP-C687-943 WT and hCENP-C687-943 T734A in mitotic and interphase cKO-hCENP-C RPE-1 cells (green). Full-length WT hCENP-C (hCENP-C-AID-mCherry) was conditionally degraded by IAA. The cells were cultured in the presence of IAA (+IAA: hCENP-C OFF) for 48 h. DNA was stained with DAPI (blue). Scale bar indicates 10 µm. See also Fig. S1, C–E.
Since the C-terminal fragment of gCENP-C (aa 601–864) localizes to the kinetochore during mitosis, but not during interphase (Nagpal et al., 2015), we examined whether the CENP-C motif is required for kinetochore localization of the C-terminal fragment. GFP-fused full-length gCENP-C (gCENP-CFL), C-terminal fragment (aa 601–864: gCENP-C601–864 WT), or its mutants were stably expressed in CENP-C conditional knockout (cKO; cKO-gCENP-C) DT40 cells (Kwon et al., 2007) in which WT full-length gCENP-C expression was turned off in the presence of tetracycline (tet; Fig. 1, A and C). Because CENP-C appears to form a dimer through a dimerization domain in its C-terminal region (Cohen et al., 2008), it is necessary to remove WT full-length CENP-C to precisely test the localization of the CENP-C C-terminal fragment. In contrast to GFP-gCENP-CFL, which localized to kinetochores in both interphase and mitotic cells, GFP-gCENP-C601–864 WT localized to kinetochores in mitotic, but not interphase, cells (Fig. 1 D), as seen in our previous study (Nagpal et al., 2015). Interestingly, deletion of the CENP-C motif (aa 655–676 in gCENP-C) from gCENP-C601–864 prevented its localization to the kinetochore in mitosis (Figs. 1 D and S3 A; gCENP-C601–864 Δ655–676 and gCENP-C677–864). In addition, when we substituted a critical arginine residue (R659 in gCENP-C) for binding to the CENP-A nucleosome in vitro (Fig. 1, A and D; Kato et al., 2013) with alanine (R659A: gCENP-C601–864 R659A), this GFP-gCENP-C601–864 R659A did not localize to interphase or mitotic kinetochores (Figs. 1 D and S3 A), indicating that the CENP-C motif is required for the mitotic kinetochore localization of the CENP-C C-terminal fragment in DT40 cells.
To examine how the mitosis-specific kinetochore localization of the C-terminal fragment via the CENP-C motif is regulated, we focused on the phosphorylation of CENP-C, because CENP-C is phosphorylated during mitosis (Fig. S1 B). Indeed, gCENP-C601–864 was phosphorylated in cells treated with nocodazole, which is a microtubule-depolymerizing drug that causes mitotic arrest by activating the mitotic checkpoint (Fig. 1 E). There are three minimal CDK1 consensus sites in gCENP-C601–864; among them is T651, which is adjacent to the CENP-C motif and is conserved among human, mouse, chicken, and frog, but not in fish (Kral, 2015; Fig. 1 B). Alanine substitution of T651 (T651A) prevented the hyperphosphorylation of gCENP-C601–864 in nocodazole-treated cells (Fig. 1 E), suggesting that T651 is phosphorylated during mitosis. Strikingly, when we expressed gCENP-C601–864 with the T651A mutation in cKO-gCENP-C DT40 cells in the presence of tet (Fig. 1 F), GFP-CENP-C601–864 T651A did not localize to the kinetochores during mitosis (Figs. 1 G and S3 B), just as in mutants lacking the CENP-C motif (gCENP-C601–864 Δ655–676, gCENP-C601–864 R659A, or gCENP-C677–864). These results indicated that T651 phosphorylation is required for kinetochore localization of the CENP-C C-terminal fragment during mitosis, implying that this phosphorylation could regulate interaction of the CENP-C motif with centromere chromatin.
We then tested whether this identified phosphoregulation is conserved in hCENP-C. GFP-fused hCENP-C C-terminal fragment (hCENP-C687–943) or a mutant in which the conserved minimal CDK1 consensus site (T734) was substituted with alanine (T734A: hCENP-C687–943 T734A), was stably expressed in cKO-hCENP-C RPE-1 cells whereby two endogenous hCENP-C gene alleles were replaced with hCENP-C-AID-mCherry (Fig. S1 C), and hCENP-C-AID-mCherry protein was degraded using the auxin-inducible degron (AID) system (Fig. S1, D and E; Holland et al., 2012; Nishimura et al., 2009). As observed in the gCENP-C C-terminal fragment, stably expressed hCENP-C687–943 localized to kinetochores during mitosis, but not during interphase, in indole-3-acetic acid (IAA)–treated cKO-hCENP-C RPE-1 cells (Fig. 1 H; hCENP-C687–943 WT). Strikingly, hCENP-C687–943 T734A did not localize to mitotic kinetochores in cKO-hCENP-C RPE-1 cells (Fig. 1 H; hCENP-C687–943 T734A), as was observed for gCENP-C601–864 T651A in cKO-gCENP-C DT40 cells, indicating that phosphoregulation is conserved in hCENP-C.
T651 phosphorylation of gCENP-C C-terminal region by CDK1 enhances its binding to the CENP-A nucleosome in vitro
Localization analyses (Fig. 1) suggested that T651 phosphorylation of gCENP-C regulates the CENP-A nucleosome binding of the CENP-C motif. To test this idea, we investigated binding of phosphorylated gCENP-C C-terminal fragment to the CENP-A nucleosome in vitro. First, we purified and phosphorylated maltose-binding protein (MBP)–fused gCENP-C601–864 using active cyclin B–CDK1 purified from starfish oocytes (Figs. 2 A and S1 F). The T651A mutation prevented the CDK1-dependent mobility shift on the Phos-tag gel (Fig. 2 A), indicating that T651 is directly phosphorylated by CDK1 in vitro. We noted that a band for gCENP-C601–864 T651A was slightly shifted by CDK1, possibly due to existence of other phosphorylation sites in this region. Using phosphorylated and unphosphorylated MBP-gCENP-C601–864, we pulled down reconstituted chicken CENP-A nucleosomes whereby the CENP-A N-terminal region was replaced with the H3 N-terminal region to stabilize the nucleosome (Figs. 2 B and S1 G). We confirmed that hybrid CENP-A nucleosomes, but not H3 nucleosomes, specifically bound to gCENP-C (Fig. S1 H). Phosphorylation by CDK1 increased the levels of CENP-A nucleosome pulled down by MBP-gCENP-C601–864 (Fig. 2 B). However, incubation with CDK1 did not affect CENP-A nucleosome binding of gCENP-C601–864 T651A (Fig. 2 B), suggesting that T651 phosphorylation of gCENP-C601–864 by CDK1 facilitates its CENP-A nucleosome binding.
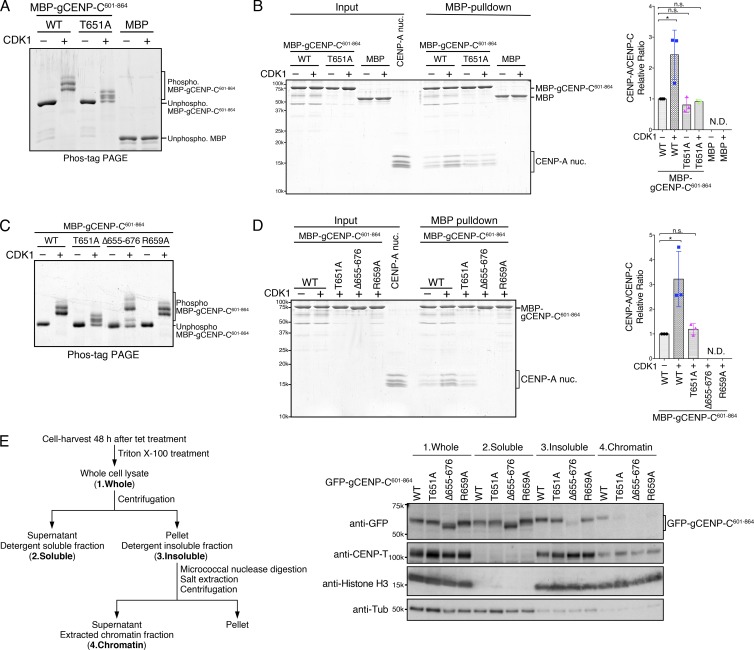
Figure 2.
T651 phosphorylation of the gCENP-C C-terminal fragment facilitates its binding to the CENP-A nucleosome in vitro. (A) Phos-tag PAGE analysis of purified MBP fused to gCENP-C601–864 WT (WT) and gCENP-C601–864 T651A (T651A) phosphorylated by CDK1. MBP was used as a negative control. Unphosphorylated proteins were also examined. See also Fig. S1 F. (B) MBP pull-down of CENP-A nucleosomes with MBP-gCENP-C601–864 WT or T651A. The phosphorylated or unphosphorylated MBP-gCENP-C601–864 and the chicken CENP-A nucleosome (CENP-A nuc.; input) were incubated and pulled down by MBP (MBP-pulldown). The signal intensities of all histones in CENP-A nucleosomes, which were precipitated with MBP proteins, were quantified. The signal intensities were normalized to MBP-gCENP-C601–864 (CDK1−) signals. Bar graph indicates mean with SD (n = 3; *, P < 0.05, unpaired t test, two tailed). See also Fig. S1, G and H. (C) Phos-tag PAGE analysis of purified MBP-gCENP-C601–864 WT, T651A, Δ655–676, and R659A phosphorylated by CDK1. Unphosphorylated proteins were also examined. See also Fig. S1 I. (D) MBP pull-down of the CENP-A nucleosome with MBP-gCENP-C601–864 WT and its mutants phosphorylated by CDK1. Each protein was phosphorylated by CDK1, as in C. MBP pull-down and signal quantification performed as in B. Bar graph indicates mean with SD (n = 3; *, P < 0.05, unpaired t test, two tailed). (E) cKO-gCENP-C cells expressing the indicated GFP-gCENP-C601–864 proteins were fractionated (left scheme) and examined by immunoblotting (right panels). CENP-T and histone H3 were used as controls for the insoluble and chromatin fractions. α-Tubulin (Tub) was used as a control for the soluble fraction.
Previous structural analysis showed that a short peptide of hCENP-C containing the CENP-C motif, but not the CDK1-phosphorylation site, bound to the H3 nucleosome containing a CENP-A C-terminal tail (Kato et al., 2013). We also purified and phosphorylated MBP-gCENP-C601–864 mutants in which the CENP-C motif was deleted (Δ656–676) or mutated (R659A; Figs. 2 C and S1 I). CENP-A nucleosomes were not detected in complexes pulled down with gCENP-C601–864 mutants, even in mutants phosphorylated by CDK1 (Fig. 2 D), suggesting that T651 phosphorylation facilitates CENP-A nucleosome binding of gCENP-C601–864 via the CENP-C motif.
Consistent with the in vitro assays, gCENP-C601–864 T651A, gCENP-C601–864 R659A, and gCENP-C601–864 Δ655–676 did not bind to chromatin in DT40 cells, whereas gCENP-C601–864 WT did (Fig. 2 E), suggesting that gCENP-C T651 phosphorylation facilitates CENP-A nucleosome binding of the CENP-C motif both in vitro and in vivo.
T651 phosphorylation of gCENP-C regulates its precise kinetochore localization during mitosis
Next, we investigated the significance of the CENP-C motif and T651 phosphorylation in the context of full-length gCENP-C. We introduced a deletion of the CENP-C motif or T651A mutation into the endogenous gCENP-C gene locus using CRISPR/Cas9 in chicken DT40 cells (Fig. S2, A and B; gCENP-CΔ648–676 or gCENP-CT651A). These cell lines expressed comparable levels of gCENP-C-GFP protein, based on immunoblotting analysis (Fig. 3 A). However, CENP-C levels on mitotic kinetochores were lower (∼60%) in cells expressing gCENP-CΔ648–676 and gCENP-CT651A than in cells expressing gCENP-CWT (Fig. 3, B and C; and Fig. S3, C and D). In contrast, CENP-T and outer kinetochore protein Ndc80 (also known as Hec1) levels at the mitotic kinetochores in cells expressing either gCENP-C-GFPΔ648–676 or gCENP-C-GFPT651A were similar to levels in cells expressing gCENP-C-GFPWT (Fig. 3, B and C; and Fig. S3, C and D). These results indicate that the CENP-C motif and T651 phosphorylation contribute to kinetochore localization of CENP-C during mitosis in its full-length context.
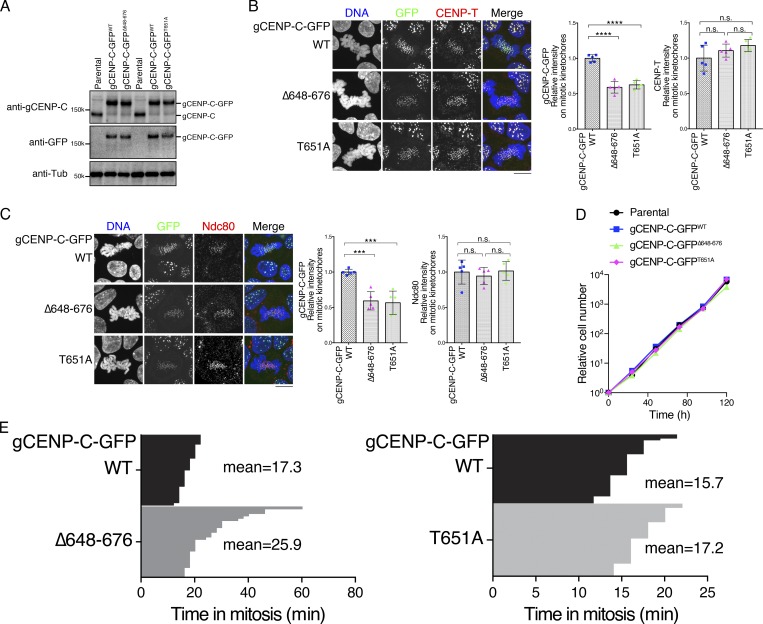
Figure 3.
CENP-A binding and T651 phosphorylation of gCENP-C are involved in kinetochore localization of gCENP-C during mitosis. (A) Expression of the gCENP-C-GFP proteins (WT, Δ648–676, and T651A) introduced by CRISPR/Cas9 using immunoblotting with anti-gCENP-C and anti-GFP antibodies. Parental WT DT40 cells were also examined. α-Tubulin (Tub) was probed as a loading control. See also Fig. S2, A and B. (B) Localization of GFP-fused gCENP-C WT, Δ648–676, or T651A (green). CENP-T was stained as a kinetochore marker (red), and DNA was stained by DAPI (blue). Scale bar indicates 10 µm. GFP and CENP-T signals on kinetochores in mitotic cells were quantified in each cell line. Bar graph indicates mean with SD (n = 5; ****, P < 0.0001; NS, P > 0.05, unpaired t test, two tailed). See also Fig. S3 C. (C) Localization of Ndc80 in DT40 cells expressing GFP-fused gCENP-C WT, Δ648–676, or T651A. Scale bar indicates 10 µm. GFP (green) and Ndc80 (red) signals on kinetochores in mitotic cells were quantified in each cell line. Bar graph indicates mean with SD (n = 5; ***, P < 0.001; NS, P > 0.05, unpaired t test, two tailed). DNA was stained by DAPI (blue). See also Fig. S3 D. (D) Growth of DT40 cells expressing GFP-fused gCENP-C WT, Δ648–676, or T651A and parental WT DT40 cells. The presented graphs are representative results of three independent experiments. (E) Duration of mitosis in cells expressing gCENP-C-GFP WT, Δ648–676, or T651A, based on live-cell imaging. Average times for mitosis were calculated with gCENP-C-GFPΔ648–676/H2B-RFP cells (n = 30) and gCENP-C-GFPT651A/H2B-RFP cells (n = 40).
However, despite the reduction of CENP-C levels on mitotic kinetochores in DT40 cells expressing gCENP-C-GFPΔ648–676 or gCENP-C-GFPT651A, they grew similarly to cells expressing gCENP-C-GFPWT (Fig. 3 D), indicating that CENP-A nucleosome binding of the CENP-C C-terminal region is dispensable for cell viability in DT40 cells (see below). We also examined mitotic progression and chromosome segregation with live-cell imaging. Although cells expressing gCENP-C-GFPΔ648–676 or gCENP-C-GFPT651A exhibited slightly delayed mitotic progression, obvious defects in chromosome segregation were not observed (Fig. 3 E).
Mitotic kinetochore localization of gCENP-C depends on both CENP-A and the CENP-H complex in DT40 cells
Although the Δ648–676 and T651A mutations significantly reduced full-length gCENP-C-GFP levels on mitotic kinetochores, their GFP signals were still detectable. In contrast, signals of GFP-gCENP-C601–864 Δ655–676 and GFP-gCENP-C601–864 T651A were undetectable on mitotic kinetochores (Fig. 1, D and G), suggesting that other regions of gCENP-C could also be involved in the mitotic kinetochore localization of gCENP-C in DT40 cells. Since the CENP-H and CENP-L–CENP-N complexes are required for kinetochore localization of CENP-C during interphase (Fukagawa et al., 2001; Kwon et al., 2007; McKinley et al., 2015; Nagpal et al., 2015), the CENP-H complex might also contribute to mitotic kinetochore localization of CENP-C. To test this possibility, we introduced Δ648–676 or T651A mutation into the endogenous chicken gCENP-C gene locus using CRISPR/Cas9 in CENP-H cKO (cKO-CENP-H) DT40 cells (Fukagawa et al., 2001), in which CENP-H expression is turned off by the addition of tet (Fig. 4 A and Fig. S2, A and C). Consistent with previous studies (Fukagawa et al., 2001; Kwon et al., 2007), the kinetochore localization of gCENP-CWT-GFP was abolished in interphase cKO-CENP-H cells after tet addition, but ∼60% of gCENP-CWT-GFP still remained in mitotic cells (Figs. 4 B and S3 E). However, both gCENP-CΔ648–676-GFP and gCENP-CT651A-GFP were undetectable on the kinetochores in cKO-CENP-H interphase and mitotic cells after tet addition (+tet Δ648–676 or T651A in Figs. 4 B and S3 E). Compared with parental cKO-CENP-H cells (Fukagawa et al., 2001), cKO-CENP-H cells expressing various CENP-C mutants showed more severe growth defects by 48 h after tet addition (Fig. 4 C). These results indicated that mitotic kinetochore localization of CENP-C depends on both the CENP-A nucleosome and CENP-H complex in DT40 cells.
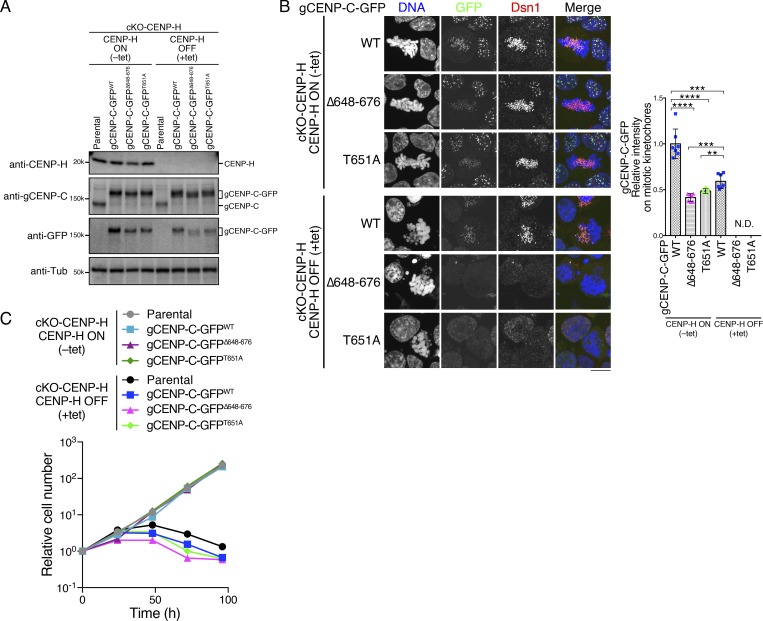
Figure 4.
Mitotic kinetochore localization of gCENP-C depends on both the CENP-A and CENP-H complex in DT40 cells. (A) Expression of each GFP-fused gCENP-C and CENP-H in CENP-H cKO (cKO-CENP-H) cells cultured in the presence or absence of tet (+tet: CENP-H OFF or −tet: CENP-H ON) for 48 h by immunoblotting. Parental cKO-CENP-H cells were also examined. α-Tubulin (Tub) was probed as a loading control. See also Fig. S2, A and C. (B) Localization of gCENP-C-GFP WT and its mutants shown in A in cKO-CENP-H cells (green). Dsn1 was stained as a kinetochore marker (red), and DNA was stained by DAPI (blue). Scale bar indicates 10 µm. The GFP signals on kinetochores in mitotic cells were quantified. Bar graph indicates mean with SD (n = 7; ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; NS, P > 0.05, unpaired t test, two tailed). See also Fig. S3 E. (C) Growth of cKO-CENP-H cells expressing GFP-fused gCENP-C WT, Δ648–676, or T651A. Cell numbers were examined at the indicated time after tet addition (+tet: CENP-H OFF) and were normalized to those at 0 h for each line. Untreated cells were also examined (−tet: CENP-H ON). Each graph shows results of one experiment.
CENP-A binding of hCENP-C is essential for long-term cell viability, and hCENP-C phosphorylation by CDK1 controls the CENP-A–CENP-C interaction in human RPE-1 cells
The CENP-A–CENP-C interaction is dispensable for cell viability in chicken DT40 cells (Fig. 3). However, previous studies have suggested the importance of the CENP-A–CENP-C interaction in human kinetochore assembly (Falk et al., 2015; Guo et al., 2017; Kato et al., 2013). Therefore, we evaluated the importance of the CENP-A–CENP-C interaction and CDK1-mediated phosphorylation using a genetic complementation assay with cKO-hCENP-C human RPE-1 cells (Fig. S1 C). As hCENP-C has two CENP-A–binding regions (the central domain and the CENP-C motif; Fig. 5 A), we tested various CENP-A–binding region mutants of hCENP-C as GFP fusions (Fig. 5, A and B; and Figs. S1 C and S4 A). GFP-hCENP-C lacking the central domain (hCENP-CΔ426–537) or the CENP-C motif (hCENP-CΔ736–758) suppressed viability defects of IAA-treated cells in the same way as hCENP-CWT (Fig. 5 C). However, a mutant hCENP-C lacking both the central domain and the CENP-C motif (hCENP-CΔ426–537/Δ736–758) did not suppress growth defects in IAA-treated cKO-hCENP-C human RPE-1 cells (Fig. 5 C), indicating that the CENP-A–CENP-C interaction is essential for long-term viability in human RPE-1 cells and that either one of the CENP-A–binding motifs is sufficient for cell viability. This is different from chicken DT40 cells, in which the CENP-A–CENP-C interaction is dispensable for cell growth (Fig. 3 D).
Figure 5.
CENP-A-binding of hCENP-C is essential for long-term viability, and CDK1-mediated phosphorylation of hCENP-C controls the CENP-A–CENP-C interaction in human RPE-1 cells. (A) Schematic representation of hCENP-C (WT, 943 aa) and its mutants: hCENP-C lacking the central domain (Δ426–537), lacking the CENP-C motif (Δ736–758), with phospho-dead mutation (T734A), and their combinational mutants (Δ426–537/Δ736–758, Δ426–537/T734A). (B) Expression of GFP-fused hCENP-C WT and its mutants (shown in A) in cKO-hCENP-C human RPE-1 cells. Full-length WT hCENP-C (hCENP-C-AID-mCherry) was conditionally degraded by IAA (None). The indicated cells were cultured in the presence or absence of IAA (+IAA: hCENP-C OFF or −IAA: hCENP-C ON) for 48 h. α-Tubulin (Tub) was probed as a loading control. (C) Growth of cKO-hCENP-C human RPE-1 cells expressing GFP-fused hCENP-C WT and its mutants (shown in A) after IAA addition. Parental cKO-hCENP-C human RPE-1 cells (None) were also examined. The presented graphs are mean results of three independent experiments. See also Fig. S4. (D) GFP and CENP-T localization in cKO-hCENP-C human RPE-1 cells (None) or cells expressing GFP-hCENP-CWT at 48 h after IAA addition (green). CENP-T was stained with an anti-human CENP-T (red). DNA was stained by DAPI (blue). Scale bar indicates 10 µm. (E) Localization of GFP-fused hCENP-C WT and its mutants after IAA addition (green). CENP-T was stained with an anti-human CENP-T antibody (red). DNA was stained by DAPI (blue). Scale bar indicates 10 µm. GFP and CENP-T signals on kinetochores were quantified in each cell line. Bar graph indicates mean with SD in the right graphs (n = 5; ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, P > 0.05, unpaired t test, two tailed). See also Fig. S4 B.
In addition to the necessity of the CENP-A–CENP-C interaction, the localization hierarchy of CENP-C and other CCAN proteins in human cells is also slightly different from that of chicken cells. While CENP-H and CENP-T signals are detectable in CENP-C–deficient mitotic DT40 cells (Fukagawa et al., 2001; Hori et al., 2008a; Kwon et al., 2007), CENP-T signals were strongly reduced in CENP-C knockout RPE-1 cells (Fig. 5 D), suggesting that CENP-T localization occurs downstream of CENP-C in human RPE-1 cells, as shown in HeLa (Klare et al., 2015; McKinley et al., 2015) and DLD-1 cells (Hoffmann et al., 2016).
We found that levels of hCENP-CΔ426–537 and hCENP-CΔ736–758 were reduced on mitotic kinetochores in cKO-hCENP-C human RPE-1 cells after IAA addition (Figs. 5 E and S4 B). However, as both mutants suppress growth defects in CENP-C knockout cells, the reduced levels are sufficient for CENP-C to function. On the other hand, the deletion of both CENP-A binding motifs (hCENP-CΔ426–537/Δ736–758) further reduced the levels of kinetochore hCENP-C (Figs. 5 E and S4 B). In addition, CENP-T levels at kinetochores were also reduced in cells expressing hCENP-CΔ426–537/Δ736–758 (Figs. 5 E and S4), and this reduction might cause cell lethality. However, CENP-A levels at kinetochores were not altered in these mutant cells (Fig. S4, C and D), which is similar to a previous report (Cao et al., 2018). While Guo et al. (2017) demonstrated that CENP-A levels are decreased in CENP-C mutants, their quantification method is different from our analyses. These data indicate that the CENP-A–CENP-C interaction is critical for sustaining normal levels of CENP-C localization on mitotic kinetochores and its defects lead to loss of CENP-T from kinetochores in human RPE-1 cells.
Finally, we examined the significance of CDK1-mediated phosphorylation of hCENP-C. We introduced the hCENP-CT734A mutant into cKO-hCENP-C human RPE-1 cells. The mutant cells did not show growth defects after IAA addition (Fig. 5 C), which is similar to hCENP-CΔ426–537 or hCENP-CΔ736–758. Consistent with this, binding of recombinant hCENP-C426–943 T734A (phosphorylated or unphosphorylated) to the CENP-A nucleosome was comparable to that of phosphorylated hCENP-C426–943, which has both a central domain and CENP-C motif (Fig. S5, A–D), suggesting that T734 phosphorylation of hCENP-C did not appear to facilitate CENP-A binding in the presence of the central domain. However, cKO-hCENP-C human RPE-1 cells expressing an hCENP-C mutant with T734A with central domain deletion (hCENP-CΔ426–537/T734A) showed severe growth defects after IAA addition (Fig. 5 C), and also reduced hCENP-C and CENP-T levels on mitotic kinetochores (Figs. 5 E and S4 B). A recent study suggests that the central domain preferentially binds to the CENP-A nucleosome over the CENP-C motif (Ali-Ahmad et al., 2019). Our in vitro assays also suggest that the central domain preferentially binds to the CENP-A nucleosome over the CENP-C motif peptide, even if it is phosphorylated (Fig. S5, E–G). In contrast, our genetic assays indicate that the central domain and phosphorylated CENP-C motif are equivalently used in RPE-1 cells (Fig. 5 C). Given that the cells expressing hCENP-CΔ426–537/T734A exhibited severe growth defects (Fig. 5 C), we conclude that phosphorylation of hCENP-C by CDK1 at T734 controls the CENP-A–CENP-C interaction during mitosis in human RPE-1 cells.
The CENP-A–CENP-C interaction is critical for the CCP in DT40 cells
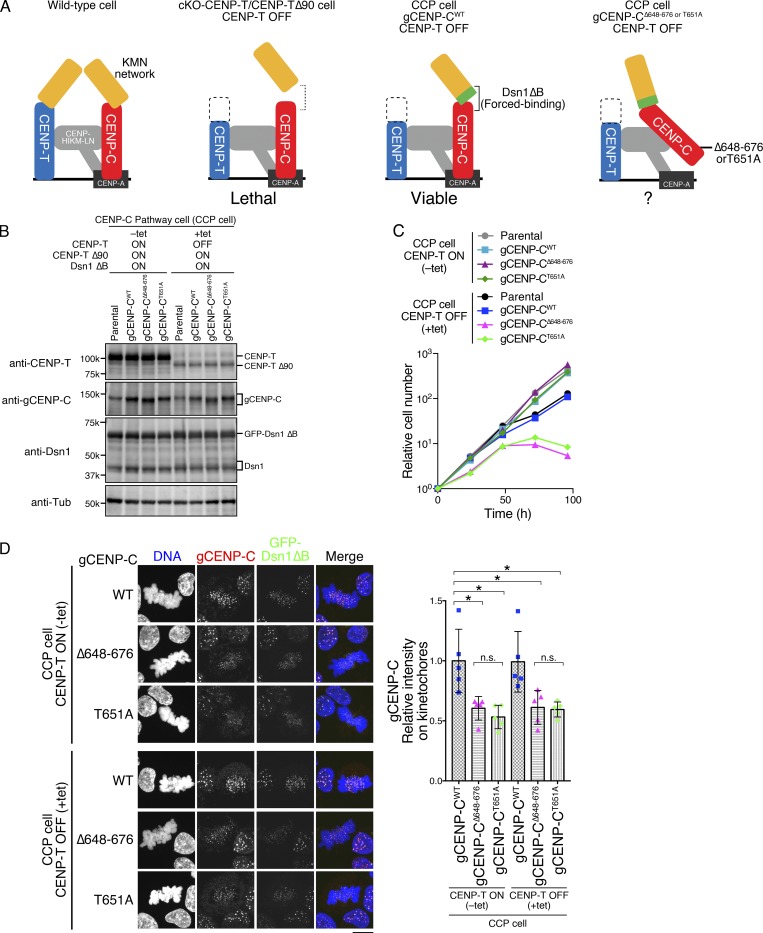
While the CENP-A–CENP-C interaction is crucial for long-term viability in human cells (Fig. 5), it is dispensable in chicken DT40 cells (Fig. 3). We have previously shown two distinct pathways that recruit the KMN network onto the kinetochores (CENP-C and CENP-T pathways; Fukagawa and Earnshaw, 2014; Hara and Fukagawa, 2017, 2018; Hori et al., 2013). As the CENP-T pathway is dominant in DT40 cells, the CENP-T pathway may suppress defects in the CENP-C pathway (CCP) without the CENP-A–CENP-C interaction. Therefore, removal of the CENP-T pathway is needed to test this hypothesis. However, the CCP by itself is insufficient for chromosome segregation in DT40 cells (Fig. 6 A). We have recently generated a DT40 cell line that relies solely on the CCP by expression of a Dsn1 mutant lacking the basic domain (aa 93–114: Dsn1 ΔB), which facilitates binding of CENP-C to the Mis12 complex in cells lacking the CENP-T pathway (cKO-CENP-T/CENP-T Δ90; Hara et al., 2018; Fig. 6 A). CCP cells are viable without the CENP-T–Ndc80 interaction, resulting in only the CCP being functional for KMN recruitment in CCP cells (Fig. 6 A).
Figure 6.
CENP-A–CENP-C binding is essential for the CCP. (A) Schematic representation of kinetochores in WT and several mutant DT40 cell lines. In WT, the KMN network is recruited to kinetochores through two pathways: the CENP-C and CENP-T pathways (left). cKO-CENP-T cells expressing CENP-T Δ90 (cKO-CENP-T/CENP-T Δ90) died after inhibition of WT CENP-T expression by tet treatment (CENP-T OFF; second from left; Hara et al., 2018). This lethality is suppressed by expressing a Dsn1 mutant lacking the basic domain (aa 93–114; Dsn1 ΔB), which increases binding affinity of CENP-C to the KMN network (third from left: CCP cell; Hara et al., 2018). The CCP cells rely solely on the CCP to recruit the KMN network and enable us to test the importance of the CENP-A–binding domains in the CCP (right). (B) Expression of each gCENP-C construct (Fig. S2, D and E) in CCP cells cultured in the presence or absence of tet (+tet: CENP-T OFF or −tet: CENP-T ON) for 48 h. Expression of CENP-T, gCENP-C, and Dsn1 proteins was examined by immunoblotting. Parental CCP cells were also examined. α-Tubulin (Tub) was probed as a loading control. (C) Growth of CCP cells expressing gCENP-C WT, Δ648–676, or T651A. Cell proliferation was examined in the presence or absence of tet (+tet: CENP-T OFF or −tet: CENP-T ON) at the indicated time after tet addition. Parental CCP cells were also examined. Each graph shows the results of one experiment. (D) Localization of gCENP-C WT, Δ648–676, or T651A in CCP cells by immunostaining with an anti-gCENP-C antibody (red). GFP-Dsn1 ΔB shows kinetochores (green). DNA was stained by DAPI (blue). Scale bar indicates 10 µm. The CENP-C signal intensities on kinetochores in mitotic cells were quantified. Bar graph indicates mean with SD (n = 5; *, P < 0.05; **, P < 0.01; NS, P > 0.05, unpaired t test, two tailed). See also Fig. S3 F.
We introduced Δ648–676 or T651A mutation into the endogenous gCENP-C gene locus in CCP cells (Fig. 6 A and Fig. S2, D and E). While CCP cells expressing WT gCENP-C grew in the presence of tet, neither gCENP-CΔ648–676 nor gCENP-CT651A supported the growth of CCP cells after tet addition (Fig. 6, B and C). We confirmed that levels of gCENP-CΔ648–676 or gCENP-CT651A at mitotic kinetochores were reduced by ∼50% compared with those of WT CENP-C (Figs. 6 D and S3 F), suggesting that the remaining CENP-C appears to be insufficient for the CCP. These results indicated that CENP-A nucleosome binding of CENP-C is critical in the CCP.
Inhibition of the CENP-A–CENP-C interaction causes mitotic defects even when Ndc80 levels on kinetochores are comparable to unperturbed cells
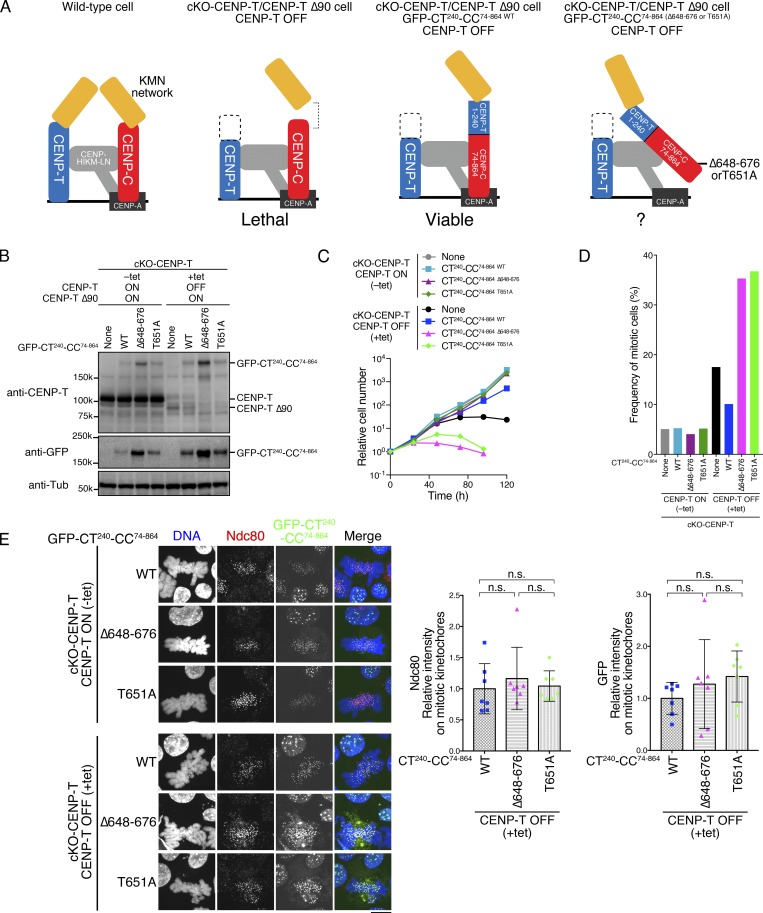
Since a reduction in CENP-C at kinetochores causes a reduction in the levels of Ndc80 (Kwon et al., 2007), it is possible that the CENP-A–CENP-C interaction defect might affect Ndc80 recruitment and lead to lethality in the CCP cells. We previously found that the CENP-T N-terminal region worked on the CENP-C scaffold (CENP-T and CENP-C chimeric constructs: aa 1–240 CENP-T fused with aa 74–864 gCENP-C [CT240-CC74–864 WT]). This chimera suppresses the deletion of the CENP-T pathway in cKO-CENP-T/CENP-T Δ90 via recruitment of Ndc80 onto kinetochores (Hara et al., 2018; Fig. 7 A). As the CENP-T C-terminal region is necessary for other CCAN functions, we need to maintain expression of CENP-T Δ90 in these cells. Importantly, CT240-CC74–864 WT recruits Ndc80 on the kinetochore at levels comparable to those of WT DT40 cells (Hara et al., 2018). We further examined the importance of the CENP-A–CENP-C interaction in cKO-CENP-T/CENP-T Δ90–expressing CT240-CC74–864 chimeras (Fig. 7 A). The expression of CT240-CC74–864 WT suppresses the growth defect in cKO-CENP-T/CENP-T Δ90 after tet addition (WT; Fig. 7, B and C; Hara et al., 2018). In contrast, cKO-CENP-T/CENP-T Δ90 cells expressing CT240 fused with CC74–864 Δ648–676 or CC74–864 T651A died with accumulation of mitotic cells after tet addition, even when they were more highly expressed than cells expressing the WT chimera (Fig. 7, B–D). Due to the higher expression of CT240-CC74–864 mutant proteins, CT240-CC74–864 and Ndc80 levels in mitotic kinetochores of cKO-CENP-T/CENP-T Δ90 cells expressing CT240-CC74–864 Δ648–676 or CT240-CC74–864 T651A were comparable with those of cells expressing CT240-CC74–864 (Figs. 7 E and S3 G). This result indicated that, even though sufficient levels of Ndc80C for cell viability in cells expressing CT240-CC74-864 WT were recruited to kinetochores, cells died without the CENP-C–CENP-A interaction. All together, the CCP kinetochore requires the CENP-A–CENP-C interaction that may be important for sustaining the tension generated by the microtubule-pulling force via the Ndc80 complex.
Figure 7.
Inhibition of the CENP-A–CENP-C interaction causes defects on kinetochores with Ndc80 levels comparable to unperturbed cells. (A) Schematic representation of kinetochores in WT and several mutant DT40 cell lines. WT (left) and cKO-CENP-T/CENP-T Δ90 (second from left) lines are described in Fig. 6 A. Expression of the GFP-fused gCENP-C-CENP-T chimera (aa 1–240 CENP-T fused with aa 74–864 gCENP-C: CT240-CC74–864 WT) suppresses lethality of cKO-CENP-T/CENP-T Δ90 cells (third from left). This chimera associates with chromatin via the CENP-C C-terminal region and binds to KMN via the CENP-T N-terminal region, which recruits Ndc80 to the kinetochore at levels comparable to WT cells (Hara et al., 2018). Therefore, the importance of the CENP-A–CENP-C interaction is testable on kinetochores with WT levels of Ndc80 (right). (B) Expression of GFP-fused CT240-CC74–864 WT, CT240-CC74–864 Δ648–676, or CT240-CC74–864 T651A in cKO-CENP-T/CENP-T Δ90 cells cultured in the presence or absence of tet (+tet: CENP-T OFF or −tet: CENP-T ON) for 48 h. Expression of CENP-T and chimeric proteins was examined by immunoblotting. cKO-CENP-T/ΔCENP-T Δ90 cells were also examined (None). α-Tubulin (Tub) was probed as a loading control. (C) Growth of cKO-CENP-T/CENP-T Δ90 cells expressing GFP-CT240-CC74–864 WT or its mutants. Cell proliferation was examined in the presence or absence of tet (+tet: CENP-T OFF or −tet: CENP-T ON). Time shown represents hours after tet addition. Each graph is a result by one experiment. (D) Mitotic index of cKO-CENP-T cells expressing GFP-CT240-CC74–864 WT or its mutants. The indicated cells were cultured in the presence or absence of tet (+tet: CENP-T OFF or −tet: CENP-T ON) for 24 h, fixed, and stained with DAPI. Mitotic index of each line (n = 474 for none, n = 458 for WT, n = 420 for Δ648–676, and n = 407 for T651A in −tet and n = 468 for none, n = 476 for WT, n = 425 for Δ648–676, and n = 422 for T651A in +tet) was measured by microscopic observation. (E) Localization of Ndc80 in cKO-CENP-T/CENP-T Δ90 cells expressing GFP-CT240-CC74–864 WT or its mutants. The indicated cells were cultured in the presence or absence of tet (+tet: CENP-T OFF or −tet: CENP-T ON) for 48 h, fixed, and stained with an anti-Ndc80 antibody (red). GFP-CT240-CC74–864 is in green. DNA was stained by DAPI (blue). Scale bar indicates 10 µm. GFP and Ndc80 signals on the mitotic kinetochores in each cell line treated with tet were quantified. Bar graph indicates mean with SD (n = 7; NS, P > 0.05, unpaired t test, two tailed). See also Fig. S3 G.
Discussion
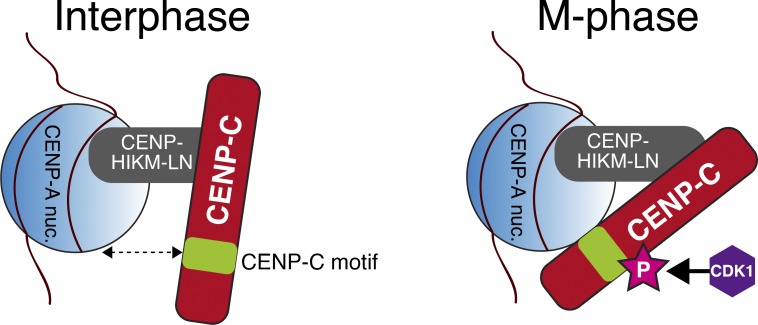
We previously proposed that the CENP-C C-terminal region binds to the CENP-A nucleosome during mitosis, but not during interphase (Nagpal et al., 2015). Here, we demonstrated that T651 of gCENP-C (T734 of hCENP-C) in the CENP-C C-terminal region is phosphorylated by CDK1 during mitosis and that this phosphorylation facilitates the CENP-A–CENP-C interaction, increasing CENP-C kinetochore localization during mitosis (Fig. 8).
Figure 8.
A model of kinetochore localization of CENP-C during interphase and mitosis. CENP-C localizes to interphase centromeres through interaction of the CENP-C middle region with the CENP–HIKM–LN complex. During mitosis, T651 or T734 in gCENP-C or hCENP-C, respectively, is phosphorylated by CDK1 (depicted as “P”), and this phosphorylation facilitates binding of the CENP-C motif to the CENP-A nucleosome (CENP-A nuc.). Kinetochore localization of CENP-C depends on both CENP–HIKM–NL and the CENP-A nucleosome during mitosis.
While phosphorylated gCENP-C601–864 WT increases affinity to the CENP-A nucleosome, unphosphorylated gCENP-C601–864 WT still binds to CENP-A nucleosomes in vitro (Fig. 2). Importantly, T651 phosphorylation of gCENP-C still requires the CENP-C motif for CENP-A nucleosome binding in vitro, because the T651-phosphorylated gCENP-C601–864 with the CENP-C motif mutation (Δ655–676 or R659A) did not bind to CENP-A nucleosomes (Fig. 2). Thus, we propose that T651 phosphorylation positively regulates the CENP-C motif binding to the CENP-A nucleosome. One possible idea of how this binding is up-regulated in vitro is that phosphorylation may induce conformational changes in the CENP-C C-terminal region to efficiently bind to the CENP-A nucleosomes. However, it is also possible that T651 phosphorylation makes another direct contact site, which is not sufficient for stable interaction, in the CENP-C C-terminal region to support the CENP-C motif.
Our cell biological analyses (Figs. 1, 3, and 5) reveal that T651 phosphorylation of gCENP-C (T734 phosphorylation of hCENP-C) is also critical for CENP-A nucleosome binding in vivo. One plausible explanation for how the CDK1-mediated phosphorylation up-regulates the CENP-A–CENP-C interaction in vivo is that the phosphorylation might efficiently expose the CENP-C motif to facilitate binding to CENP-A nucleosomes in cells, as seen in our in vitro assay. It is also possible that other proteins interact with the phosphorylated threonine and facilitate binding of CENP-C to CENP-A nucleosomes in vivo. Given that the threonine residues corresponding to T651 of gCENP-C are conserved in vertebrates other than fishes (Kral, 2015), this phosphoregulation appears to be conserved in those species. It will be important to clarify the molecular mechanisms of how CDK1-mediated CENP-C phosphorylation facilitates binding to the CENP-A nucleosomes in future studies.
The CENP-A–CENP-C interaction is essential for long-term viability in human cells (Δcentral domain/CENP-C motif), but not in chicken cells (ΔCENP-C motif; Figs. 3 and 5). One explanation for this difference is that the CCP might be critical in human RPE-1 cells, while it is dispensable in chicken DT40 cells (Hara et al., 2018). However, this explanation might not be the case, because the CENP-T pathway appears to form a major linkage between CCAN and spindle microtubules even in human cells, based on measurement of CENP-C and CENP-T copy numbers in human kinetochores (Suzuki et al., 2015). Another possible explanation is the difference between human and chicken cells in the kinetochore localization hierarchy of CENP-C and CENP-T. CENP-T localization in the interphase centromere largely depends on CENP-C in human cells (Hoffmann et al., 2016; Klare et al., 2015), but not in chicken cells (Hori et al., 2008a). In fact, we observed a reduction in CENP-T in cKO-hCENP-C RPE-1 cells during both interphase and mitosis (Fig. 5 D), suggesting that hCENP-C is also involved in CENP-T kinetochore localization in mitotic cells. Deletion of the CENP-C motif, or T734A mutation, combined with deletion of the central domain caused reduction of both CENP-C and CENP-T levels on kinetochores (Fig. 5 E), which might not be strong enough to support chromosome segregation. Based on the results of our present study, the latter explanation seems more plausible.
Although the CENP-C motif is dispensable in chicken and human cells, it is retained in both species. In fact, the motif is highly conserved from yeast to humans. More curiously, phosphoregulation of the CENP-A–CENP-C interaction is also conserved in chicken and human cells. We have previously showed that the CENP-T pathway is dominant over the CCP, depending on CDK1 (Hara et al., 2018). However, it is possible that the strength of balance between the CENP-C and CENP-T pathways could vary in other cell types or situations by modulating CDK1 activity. For instance, kinase activity of another mitotic kinase, Aurora B, which phosphorylates various kinetochore proteins, is variable between transformed and nontransformed cells (Abe et al., 2016). Clarifying whether the CENP-C motif is essential in various cell types or biological contexts will be important in future studies. It is also possible that the CENP-C motif might have other roles distinct from CENP-A binding in such situations.
In this study, we demonstrated that the CDK1-mediated phosphorylation of CENP-C is critical for dynamic changes of CENP-C binding to the CENP-A nucleosome between interphase and mitosis. Given that CENP-C constitutively localizes to the centromere throughout the cell cycle, our findings suggest that interactions between CENP-C and other inner kinetochore proteins fluctuate during the cell cycle and that phosphoregulation determines the order and timing of these changes.
Materials and methods
Chicken DT40 cells
A chicken DT40 cell line CL18 was used as the WT cell (Buerstedde et al., 1990). DT40 cells were cultured at 38.5°C in DMEM (Nacalai Tesque) supplemented with 10% FBS (Sigma), 1% chicken serum (Thermo Fisher), and Penicillin-Streptomycin (Thermo Fisher; DT40 culture medium).
The gCENP-C cKO (cKO-gCENP-C) cell line was described previously (Kwon et al., 2007). The cKO-gCENP-C cell line expressing GFP-gCENP-C full-length (cKO-gCENP-C/GFP-gCENP-CFL) or GFP-gCENP-C601-864 WT (cKO-gCENP-C/GFP-gCENP-C601–864 WT; Nagpal et al., 2015) was reestablished in this study by transfection of a plasmid encoding GFP-fused gCENP-CFL or gCENP-C601–864 WT, and a neomycin resistance gene, into cKO-gCENP-C cells using electroporation. The transfected cells were selected in DT40 culture medium containing 2 mg/ml G418 (Santa Cruz Biotechnology). The cKO-gCENP-C cell line expressing GFP-gCENP-C601–864 Δ655–676, GFP-gCENP-C601–864 R659A, GFP-gCENP-C601–864 T651A, or GFP-gCENP-C677–864 was generated by transfection of a plasmid encoding GFP-fused gCENP-C with indicated mutations as described above (cKO-gCENP-C/GFP-gCENP-C601–864 Δ655–676, cKO-gCENP-C/GFP-gCENP-C601–864 R659A, cKO-gCENP-C/GFP-gCENP-C601–864 T651A, or cKO-gCENP-C/GFP-gCENP-C677–864).
The cKO-CENP-T cell line expressing CENP-T Δ90 (cKO-CENP-T/CENP-T Δ90) and the cKO-CENP-T cell line expressing CENP-T Δ90, GFP-CENP-T240-gCENP-C74–864 (cKO-CENP-T/CENP-T Δ90/GFP-CT240-CC74–864 WT) were described before (Hara et al., 2018; Nishino et al., 2013). The cKO-CENP-T/CENP-TΔ90 cell line expressing GFP-CT240-CC74–864 Δ648–676 or GFP-CT240-CC74–864 T651A was generated by transfection of a plasmid encoding GFP-CT240-CC74–864 with the corresponding mutant sequence and neomycin resistance gene into cKO-CENP-T/CENP-TΔ90 cells with another plasmid expressing EcoGPT using electroporation. The transfected cells were selected in DT40 culture medium containing 2 mg/ml G418, 25 µg/ml mycophenolic acid, and 125 µg/ml xanthine (cKO-CENP-T/CENP-T Δ90/GFP-CT240-CC74–864 Δ648–676, cKO-CENP-T/CENP-T Δ90/GFP-CT240-CC74–864 T651A).
To express GFP-fused gCENP-C under control of the endogenous gCENP-C promoter in CL18 cell or CENP-H cKO (cKO-CENP-H; Fukagawa et al., 2001) cell lines, gCENP-C ORF cDNA corresponding to aa 647–864 coded on exons 12–18 fused with GFP was targeted into exon 12 of gCENP-C gene using CRISPR/Cas9 system–mediated homologous recombination (see Plasmid constructions and Southern blotting sections). Since the gCENP-C cDNA in the donor cassette was followed by puromycin- or histidinol-resistance gene (puroR, HisD), the targeted cells were selected in DT40 culture medium containing 0.5 µg/ml puromycin or 1 mg/ml L-Histidinol dihydrochloride (CL18 or cKO-CENP-H/gCENP-C-GFPWT). The mutant gCENP-C cDNAs (Δ648–676, T651A) were also targeted as described above (CL18 or cKO-CENP-H/gCENP-C-GFPΔ648–676, CL18 or cKO-CENP-H/gCENP-C-GFPT651A).
The cell line in which KMN network recruitment relies solely on the CCP cells was described previously (Hara et al., 2018; cKO-CENP-T cell line expressing CENP-T Δ90 and GFP-Dsn1 Δ93-114 [GFP-Dsn1 ΔB]). To express gCENP-CWT or its mutants (Δ648–676, T651A) under control of the endogenous gCENP-C promoter in the CCP cell line, gCENP-C ORF cDNA corresponding to aa 647–864 coded on exons 12–18 was targeted into exon 12 of the gCENP-C gene as above (CCP/gCENP-CWT, CCP/gCENP-CΔ648–676, CCP/gCENP-CT651A).
RFP-fused histone H2B was expressed in cells expressing gCENP-C-GFPWT or its mutants for live-cell imaging as described previously (Hori et al., 2008b; gCENP-C-GFPWT/H2B-RFP, gCENP-C-GFPΔ648–676/H2B-RFP, gCENP-C-GFPT651A/H2B-RFP).
Human RPE-1 cells
RPE-1 cells were cultured at 37°C in DMEM supplemented with 10% FBS and penicillin-streptomycin (RPE-1 culture medium).
hCENP-C targeting in RPE-1 cells was done as previously described (Fachinetti et al., 2015). Briefly, TALENs were designed to the C-terminal region of hCENP-C gene (5′-GAGGAAAGTGTTCTTC-3′ and 5′-GGTTGATCTTTCATC-3′). RPE-1 cells were cotransfected with the TALEN expression vectors and a donor cassette containing the two homology arms for hCENP-C C-terminal region and an AID and mCherry cassettes. Single mCherry-positive cells were then isolated by FACS and tested for efficiency of hCENP-C targeting and AID-fused hCENP-C depletion by PCR, immunoblotting, and immunofluorescence (cKO-hCENP-C). To establish the cKO-hCENP-C RPE-1 cell line expressing GFP-fused hCENP-C687–943 WT, hCENP-C687–943 T734A, hCENP-CWT, hCENP-CΔ426–537, hCENP-CΔ736–758, hCENP-CT734A, hCENP-CΔ426–537/Δ736–758, or hCENP-CΔ426–537/T734A, these sequences were integrated into the genome using the Sleeping Beauty transposon system (Mátés et al., 2009; cKO-hCENP-C/GFP-hCENP-C687–943 WT, cKO-hCENP-C/GFP-hCENP-C687–943 T734A, cKO-hCENP-C/GFP-hCENP-CWT, cKO-hCENP-C/GFP-hCENP-CΔ426–537, cKO-hCENP-C/GFP-hCENP-CΔ736–758, cKO-hCENP-C/GFP-hCENP-CΔT734A, cKO-hCENP-C/GFP-hCENP-CΔ426–537/Δ736–758, cKO-hCENP-C/GFP-hCENP-CΔ426–537/T734A). Since the transgene cassette has a neomycin resistance gene, the cell lines were selected in RPE-1 culture medium containing 500 µg/ml G418.
Cell culture
DT40 cells were cultured at 38.5°C in DT40 culture medium. cKO cell lines for gCENP-C, CENP-T, CENP-H, and CCP cells were described previously (Fukagawa et al., 2001; Hara et al., 2018; Hori et al., 2008a; Kwon et al., 2007), and gene inactivation in these cKO cell lines was performed by the addition of tet (Sigma) at 2 µg/ml (final concentration) to the culture medium. To establish the cell lines, which stably express the transgenes, plasmid constructs were linearized by appropriate restriction enzymes and transfected into DT40 cells using Gene Pulser II electroporator (Bio-Rad). For CRISPR/Cas9-mediated homologous recombination, plasmid constructs were transfected into DT40 cells using the Neon Transfection System (Thermo Fisher).
RPE-1 cells were cultured at 37°C in RPE-1 culture medium. Plasmid constructs were transfected into RPE-1 cells with the Neon Transfection System. To degrade AID-fused hCENP-C, cells were treated with 200 µM IAA (Sigma).
Cell counting
To count the number of DT40 cells, 50 µl cultured medium was mixed with 0.4 wt/vol% Trypan Blue Solution (Wako), and the mixture was counted using Countess II (Thermo Fisher).
To count the number of RPE-1 cells, the culture medium was removed by aspirator. Then, 2.5 g/liter of Trypsin 1 mmol/liter EDTA solution (Nacalai Tesque) was added and incubated for 3 min at 37°C. To stop trypsinization, RPE-1 culture medium was added. The cell solution was mixed with same volume of 0.4 wt/vol% Trypan Blue Solution, and cell numbers were counted using Countess II.
Plasmid constructions
gCENP-C (described before) was used as a full-length sequence (Fukagawa and Brown, 1997). To generate GFP-fused gCENP-C full-length, 601–864, and 677–864, the sequences were amplified using PCR and cloned into pEGFP-C3 (Clontech; pEGFP-C3-gCENP-CFL, pEGFP-C3-gCENP-C601–864, and pEGFP-C3-gCENP-C677–864). The deletion of 655–676 (Δ655–676) or alanine substitution of T651 or arginine 659 (T651A, R659A) was introduced into pEGFP-C3-gCENP-C601–864 using PCR (pEGFP-C3-gCENP-C601–864 Δ655–676, pEGFP-C3-gCENP-C601–864 T651A, and pEGFP-C3-gCENP-C601–864 R659A). To generate GFP-CENP-T240-gCENP-C74–864 Δ648–676 and GFP-CENP-T240-gCENP-C74–864 T651A, these sequences were deleted or mutated from pEGFP-C3 CENP-T240-gCENP-C74–864 WT (Hara et al., 2018) using PCR (pEGFP-C3-CT240-CC74–864 Δ648–676, pEGFP-C3-CT240-CC74–864 T651A).
To express GFP-fused gCENP-C under control of gCENP-C endogenous promoter in DT40 cells, a gCENP-C-GFP sequence was integrated into an endogenous gCENP-C locus using CRISPR/Cas9 genome editing. Three single guide RNAs (sgRNAs 1, 2, and 3) against genomic sequence around the start of gCENP-C exon12 were designed (Optimized CRISPR Design; Hsu et al., 2013) and cloned into pX335 plasmid, which has SpCas9 nickase (D10A; Addgene, 54233 [Cong et al., 2013]; pX335-gCENP-C 1, 2, and 3). sgRNA 3 was also cloned into pX330 plasmid (Addgene, 42330; pX330-gCENP-C 3). gCENP-C cDNA corresponding to aa 647–864 coded on exons 12–18 fused with GFP and puroR of HisD expressed by β-actin promoter were inserted between ∼1 kb 5′ and 3′ homology arms flanking the start of gCENP-C exon12 and cloned into pBlueScriptII KS (pBSKS-gCENP-C-GFPWT-puroR or HisD). Note that in some experiments, we used pBSKS-gCENP-C-GFPWT-puroR or pBSKS-gCENP-C-GFPT651A-puroR, which has a longer 3′ homology arm (pBSKS-gCENP-C-GFPWT-puroR [long 3′], pBSKS-gCENP-C-GFPT651A-puroR [long 3′]). gCENP-C mutants were also cloned into the gCENP-C homology arms (pBSKS-gCENP-C-GFPT651A or Δ648–676 -puroR or -HisD). To knock in the gCENP-C WT or mutants cDNA without GFP, the gCENP-C cDNA coding aa 647–864 was cloned into the homology arms as done above (pBSKS-gCENP-CWT, T651A or Δ648–676 -puroR or -HisD).
cKO-hCENP-C RPE-1 cell line expressing GFP-fused hCENP-C687–943 WT, hCENP-C687–943 T734A, hCENP-CWT, hCENP-CΔ426–537, hCENP-CΔ736–758, hCENP-CT734A, hCENP-CΔ426–537/Δ736–758, or hCENP-CΔ426–537/T734A were established using Sleeping Beauty transposon system (Mátés et al., 2009). hCENP-C687–943 WT, hCENP-C687–943 T734A, hCENP-CWT, hCENP-CΔ426–537, hCENP-CΔ736–758, hCENP-CT734A, hCENP-CΔ426–537/Δ736–758, or hCENP-CΔ426–537/T734A was cloned into pEGFP-C3. These GFP-fused hCENP-C expression cassette with the neomycin resistance gene cassette was cloned into pT2/HB (Addgene, 26557; pT2/HB-hCENP-C687–943 WT, pT2/HB-hCENP-C687–943 T734A, pT2/HB-hCENP-CWT, pT2/HB-hCENP-CΔ426–537, pT2/HB-hCENP-CΔ736–758, pT2/HB-hCENP-CT734A, pT2/HB-hCENP-CΔ426–537/Δ736–758, pT2/HB-hCENP-CΔ426–537/T734A). The plasmid was transfected to RPE-1 cells together with pCMV (CAT) T7-SB100 (Addgene, 34879) using the Neon Transfection System.
MBP-fused gCENP-C601–864 was cloned into pMAL-c5x (New England Biolabs; pMAL-c5x-gCENP-C601–864 WT). The deletion of aa 655–676 (Δ655–676) or alanine substitution of T651 or arginine 659 (T651A, R659A) was introduced into pMAL-c5x-gCENP-C601–864 using PCR (pMAL-c5x-gCENP-C601–864 Δ655–676, pMAL-c5x-gCENP-C601–864 T651A, and pMAL-c5x-gCENP-C601–864 R659A).
MBP-fused hCENP-C426–943 WT was cloned into the pMAL-c5x (pMAL-c5x-hCENP-C426–943 WT). The alanine substitution of T734 was introduced into pMAL-c5x-hCENP-C426–943 using PCR (pMAL-c5x-hCENP-C426–943 T734A). MBP-fused hCENP-C426–537 or hCENP-C501–537 were cloned into the pMal-TEV-6His plasmid (Nishino et al., 2013; pMAL-TEV-hCENP-C501–537 and pMAL-TEV-hCENP-C426–537). These plasmids have a stop codon before the 6xHis sequence to express the proteins without 6xHis-tag.
Chicken histone H3.2 was cloned into pHCE-AMPFREE (Takara; pHCE-AMPFREE-gH3.2). His-tagged chicken H4 was cloned into pET15b (Novagen; pET15b-gH4). N-terminally 6xHis-SUMO-tagged chicken H2A and untagged chicken H2B was cloned into an in-house–modified version of pETDuet-1 (pETDuet-1-H2A/H2B).
Southern blotting
Genomic DNA was extracted from DT40 cells. Briefly, ∼2 × 106 cells were resuspended in 200 µl lysis buffer (100 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 5 mM EDTA-NaOH, pH 8.0). Following the addition of 100 µl lysis buffer containing Proteinase K (60 µg) and SDS (0.6% final), the lysate was incubated for 1 h at 55°C. Genomic DNA was precipitated by adding 300 µl isopropanol, washed with 70% ethanol, and resuspended with 50 µl TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA-NaOH, pH 8.0). 10 μl extracted genomic DNA was digested with PstI. The digested genomic DNAs were separated on a 0.7% agarose gel and transferred to Biodyne B membrane 0.45 µm (Pall Corporation) using the VacuGene XL Vacuum Blotting System (GE Healthcare). The membrane was incubated in 0.4 N NaOH for 20 min and then in 5× SSC (1×SSC: 150 mM NaCl and 15 mM sodium citrate, pH 7.0). After air-drying, the transfer DNA was cross-linked by UV radiation (120 mJ/cm2). For the DNA probe, a fragment of the gCENP-C genome locus was amplified with 5′-GGGGACTCACTGGAGCTTTTG-3′ (forward) and 5′-TGTTCTTGCACTGAATGTACA-3′ (reverse) primers by PCR and cloned into pGEM-T Easy (Promega). The clone fragment was reamplified with the same primers by PCR, purified with MagExtractor-PCR and Gel Clean up (TOYOBO), and labeled with 32P using the Megaprime DNA Labeling system (GE Healthcare) with [α-32P]dCTP (10 µCi/µl; PerkinElmer). After the labeling, unincorporated nucleotides were removed by Illustra ProbeQuant G-50 Micro Columns (GE Healthcare). After prehybridization in Church Mix (0.5 M phosphate buffer, pH 7.0, 7% SDS, 0.15 mM BSA, and 1 mM EDTA-NaOH, pH 8.0), the labeled probe was denatured for 5 min at 96°C and hybridized with the transferred genomic DNA on the membrane for overnight in Church Mix at 62°C. After washing three times with 2× SSC supplemented with final 0.5% SDS, hybridized fragments were detected by Typhoon FLA 9500 (GE Healthcare).
Immunoblotting
For whole-cell samples, DT40 cells were harvested, washed with PBS, and suspended in 1× Laemmli sample buffer (LSB; final 104 cells/µl), followed by sonication and heating for 5 min at 96°C. In Fig. 1 F, DT40 cells were harvested at 11 h after 100 ng/ml nocodazole treatment, washed with PBS, and suspended in 1× LSB (final 103 cells/µl), followed by sonication and heating for 5 min at 96°C. Proteins were separated on SuperSep Ace, 5–20% (Wako), or handmade SDS-PAGE (7.5 or 10%) and transferred to Immobilon-P (Merck) using HorizeBLOT (ATTO). To examine phosphorylation of proteins, samples were separated on Phos-tag-7.5%SDS-PAGE (25 µM Phos-tag Acrylamide [Wako] and 50 µM MnCl2) and transferred after EDTA treatment as per the manufacturer’s protocol.
RPE-1 cells were trypsinized and harvested, washed with PBS, and suspended in 1× LSB (final 5 × 103 cells/µl) followed by sonication and heating for 5 min at 96°C. Proteins were separated on SuperSep Ace, 5–20%, and transferred to Immobilon-P using HorizeBLOT.
Primary antibodies used in this study were rabbit anti-gCENP-C (Fukagawa et al., 1999), rabbit anti-chicken CENP-T (Hori et al., 2008a), rabbit anti-chicken CENP-H (Fukagawa et al., 2001), rabbit anti-chicken Dsn1 (Hara et al., 2018), rabbit anti-GFP (MBL), mouse anti-α-tubulin (Sigma), rat anti-Histone H3 (a gift from Hiroshi Kimura, Tokyo Tech, Tokyo, Japan; Nozawa et al., 2010), and guinea pig anti-hCENP-C (Ando et al., 2002). Secondary antibodies were HRP-conjugated anti-rabbit IgG (Jackson ImmunoResearch), HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch), and HRP-conjugated anti-guinea pig (Sigma). To increase sensitivity and specificity, Signal Enhancer Hikari (Nacalai Tesque) was used for all antibodies. The antibodies were incubated with the blotted membranes for 1 h at room-temperature or for overnight at 4°C. Proteins reacting with antibodies were detected with ECL Prime (GE Healthcare) and visualized with ChemiDoc Touch (Bio-Rad). Acquired images were processed using Image Lab 5.2.1 (Bio-Rad) and Photoshop CC (Adobe).
Immunofluorescence
DT40 cells were cytospun onto glass slides. The cells were fixed with 3% PFA in 250 mM Hepes-NaOH, pH 7.4, for 15 min; permeabilized in 0.5% NP-40 in PBS for 10 min; blocked with 0.5% BSA in PBS for 5 min; and incubated with primary antibodies anti-chicken Ndc80 (Hori et al., 2003), anti-chicken Dsn1 (Hara et al., 2018), anti-gCENP-C (Fukagawa et al., 1999), or anti-chicken CENP-T (Hori et al., 2008a), diluted with 0.5% BSA in PBS for 1 h at 37°C. After washing three times with 0.5% BSA in PBS, the cells were incubated with Cy3-conjugated anti-rabbit IgG (Jackson ImmunoResearch) diluted with 0.5% BSA in PBS for 1 h at 37°C and then washed with 0.5% BSA in PBS three times. The cells were postfixed with 3% PFA in 250 mM Hepes-NaOH, pH 7.5, for 10 min, washed with PBS, and stained DNA with 1 µg/ml DAPI in PBS for 10 min. The stained samples were washed with PBS and mounted with VECTASHIELD mounting medium (Vector Laboratories).
RPE-1 cells were harvested and cytospun onto glass slides after trypsinization with 2.5 g/liter-Trypsin-1 mmol/l EDTA solution. The cells were fixed with 3% PFA in 250 mM Hepes-NaOH, pH 7.4, for 15 min, permeabilized in 0.5% NP-40 in PBS for 10 min, blocked with 0.5% BSA in PBS for 5 min, and incubated with primary antibodies rat anti-human CENP-T (immunogen: HNPDSDSTPRTLLRRVLDTC, a gift from Kinya Yoda, Nagoya University, Nagoya, Japan) or mouse anti-human CENP-A (Ando et al., 2002), diluted with 0.5% BSA in PBS for 1 h at 37°C. After washing three times with 0.5% BSA in PBS, the cells were incubated with Dylight649-conjugated anti-Rat IgG (Jackson ImmunoResearch) or Alexa Fluor 647–conjugated anti-mouse IgG (Jackson ImmunoResearch) diluted with 0.5% BSA in PBS for 1 h at 37°C and then washed with 0.5% BSA in PBS three times. The cells were postfixed with 3% PFA in 250 mM Hepes-NaOH, pH 7.5, for 10 min, washed with PBS, and stained DNA with 1 µg/ml DAPI in PBS for 10 min. The stained samples were washed with PBS and mounted with VECTASHIELD Mounting Medium.
Immunofluorescence images of DAPI, GFP, Cy3, Dylight649, and Alexa Fluor 647 signals were acquired at 0.2-µm intervals in the z-axis using a Zyla 4.2 sCMOS camera (Andor) mounted on a Nikon Ti inverted microscope with an objective lens (Plan Apo lambda 100×/1.45 NA; Nikon) with a spinning disk confocal unit (CSU-W1; Yokogawa) controlled with NIS-Elements (Nikon) at RT. The images in figures are the maximum intensity projection of the Z-stack generated with Fiji (Schindelin et al., 2012). Acquired images were processed using Fiji (Schindelin et al., 2012) and Photoshop CC.
Live-cell imaging
Live-cell imaging for gCENP-C-GFPWT/H2B-RFP, gCENP-C-GFPΔ648-676/H2B-RFP, or gCENP-C-GFPT651A/H2B-RFP cells was performed with a Cell Voyager (Yokogawa) with an oil-immersion objective lens (UPLSApo 60×, NA 1.35) in a temperature-controlled box at 37°C. Time-lapse images were recorded at 2-min intervals. Average times for mitosis were calculated with 30 gCENP-C-GFPΔ648-676/H2B-RFP cells and 40 gCENP-C-GFPT651A/H2B-RFP cells.
Recombinant proteins
To express MBP-fused gCENP-C601–864 WT or mutants, an Escherichia coli strain, Rosetta2(DE3) (Merck), was transformed with pMAL-c5x-gCENP-C601–864 WT, pMAL-c5x-gCENP-C601–864 T651A, pMAL-c5x-gCENP-C601–864 Δ655–676, or pMAL-c5x-gCENP-C601–864 R659A. Cells expressing MBP-gCENP-C601–864 WT or mutants were resuspended in column buffer (20 mM Hepes-NaOH, pH 7.5, 300 mM NaCl, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, and 1 mM DTT) and lysed by sonication. The cell lysates were clarified by centrifugation (40,000 × g) at 4°C. The supernatant was incubated with Amylose resin (New England Biolabs) for 2 h at 4°C. The resins were washed with high-salt wash buffer (20 mM Hepes-NaOH, pH 7.5, 1 M NaCl, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, and 1 mM DTT). MBPs were eluted with elution buffer (20 mM Hepes-NaOH, pH 7.5, 500 mM NaCl, 5% Glycerol, 20 mM maltose, and 1 mM DTT). Purified proteins were applied to a 5-ml Hi-Trap SP column (GE Healthcare) in salt gradient between A buffer and B buffer (A buffer: 20 mM Hepes-NaOH, pH 7.5, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, and 1 mM DTT; B buffer: 20 mM Hepes-NaOH, pH 7.5, 1 M NaCl, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, and 1 mM DTT; gradient A to B: 20–100%). The proteins were further purified with Superdex 200 pg (GE Healthcare) or Superdex 75 pg (GE Healthcare) in SEC buffer (20 mM Hepes-NaOH, pH 7.5, 500 mM NaCl, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, and 1 mM DTT). Relevant fractions were pooled, concentrated with Amicon-Ultra-0.5 30k (Merck), frozen in liquid nitrogen, and stored at −80°C.
To express MBP-fused hCENP-C426–943 WT or hCENP-C426–943 T734A, an E. coli strain, Rosetta2(DE3), was transformed with pMAL-c5x-hCENP-C426–943 WT or pMAL-c5x-hCENP-C426–943 T734A. Cells expressing MBP-hCENP-C426–943 WT or T734A were resuspended in high-salt column buffer supplemented with 0.1% Triton X-100 (20 mM Hepes-NaOH, pH 7.5, 1M NaCl, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, 2 mM DTT, and 0.1% Triton X-100 [TMS-Triton]) and lysed by sonication. The cell lysates were clarified by centrifugation (30,000 ×g) at 4°C. The supernatant was incubated with Amylose resin (New England Biolabs) for 2 h at 4°C. The resins were washed with high-salt wash buffer containing 2 mM DTT. MBPs were eluted with elution buffer containing 1 mM EDTA-NaOH, pH 8.0, and 4 mM DTT. The purified proteins were applied to a 5-ml Hi-Trap SP column in salt gradient between A buffer containing 2 mM DTT and B buffer containing 2 mM DTT (gradient A to B: 15–100%). The proteins were further purified with Superdex 200 pg in SEC buffer containing 4 mM DTT. Relevant fractions were pooled, concentrated with Amicon-Ultra-0.5 30k, frozen in liquid nitrogen, and stored at −80°C.
To express MBP-fused hCENP-C501–537, an E. coli strain, Rosetta2(DE3) was transformed with pMAL-TEV-hCENP-C501–537. The protein was purified by MBP affinity using the same protocol as for MBP-fused hCENP-C426–943. The purified proteins were applied to a 5-ml Hi-Trap Q HP column (GE Healthcare) in salt gradient between A buffer containing 2 mM DTT and B buffer containing 2 mM DTT (gradient A to B: 7.5–100%). Relevant fractions were pooled and frozen in liquid nitrogen and stored at −80°C.
To express MBP-fused hCENP-C426–537, an E. coli strain, Rosetta2(DE3), was transformed with pMAL-TEV-hCENP-C426–537. The protein was purified by MBP affinity according to the same protocol for MBP-fused hCENP-C426–943. The purified protein was digested by homemade TEV protease overnight at 4°C. The digested proteins were applied to a 5-ml Hi-Trap SP column in salt gradient between A buffer containing 2 mM DTT and B buffer containing 2 mM DTT (gradient A to B: 10–100%). The proteins were further purified with Superdex 200 pg in SEC buffer containing 4 mM DTT. Relevant fractions were pooled, frozen in liquid nitrogen, and stored at −80°C.
Recombinant histones and nucleosome reconstitution
193 bp 601 DNA for nucleosome was purified as described before (Arimura et al., 2012; Tanaka et al., 2004). Briefly, the 601-containing plasmid was purified with isopropanol and treated with RNase. The plasmid was extracted with phenol twice and further purified with PEG and ethanol precipitation. The plasmid was digested with EcoRV, and the digested 601 DNA was purified with PEG and ethanol precipitation. The 601 DNA was suspended with TE buffer.
His-tagged chicken histone H3.2 was expressed using pHCE-AMPFREE-gH3 in E. coli (BL21(DE3)). His-tagged chicken H4 was expressed using pET15-b-gH4 in E. coli (BL21(DE3)). Cells expressing H3.2-His or H4-His were resuspended in column buffer1 (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% Glycerol, 5 mM DTT, and 1× complete EDTA-free proteinase inhibitor [Roche]) and lysed by sonication on ice. The cell lysates were clarified by centrifugation (35,000 ×g) at 4°C. The pellet was resuspended with buffer1 and lysed by sonication on ice. The lysates were clarified by centrifugation (35,000 ×g) at 4°C, and the supernatants were removed. The pellet was resuspended with buffer 2 (50 mM Tris-HCl, pH 8.0, 7 M Guanidine-HCl, 5% Glycerol, and 5 mM DTT) and lysed by sonication on ice. The lysate was incubated with agitation for overnight at 4°C. The lysate was clarified by centrifugation (35,000 ×g) at 4°C and the pellet removed. The supernatant was incubated with Ni-NTA agarose (QIAGEN) for 30 min at 4°C. Ni-NTA agarose was washed with buffer 3 (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 6 M Urea, 5% Glycerol, and 5 mM Imidazole, pH 8.0) four times. Proteins were eluted with buffer4 (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 6 M Urea, 5% Glycerol, 300 mM Imidazole, pH 8.0, and 1 mM DTT) and dialyzed with buffer 5 (10 mM Tris-HCl, pH 7.5, and 5 mM DTT). His-tag of H3.2 proteins were cleaved with Thrombin (Wako; 2 U thrombin/1 mg Histone) for 75 min at RT. The cleaved proteins were applied to 5 ml Hi-Trap SP column in salt gradient between buffer 6 and buffer 7 (buffer 6: 20 mM CH3COONa, pH 5.2, 200 mM NaCl, 6 M Urea, 1 mM EDTA-NaOH, pH 8.0, and 5 mM DTT; buffer 7: 20 mM CH3COONa, pH 5.2, 900 mM NaCl, 6 M Urea, 1 mM EDTA-NaOH, pH 8.0, and 5 mM DTT; gradient buffer 6 to buffer 7: 10–100%). Relevant fractions were pooled. His-Tagged H4 was also purified using 5 ml Hi-Trap SP column as above. H3.2 and His-tagged H4 were dialyzed to Mili-Q water four times, lyophilized, and stocked as powder at 4°C. H3 aa 1–64 fused chicken CENP-A aa 55–131 protein (CENP-A) was expressed using a pET15b vector in E. coli and purified (a gift from Hitoshi Kurumizaka, University of Tokyo, Tokyo, Japan). Human CENP-A (Tachiwana et al., 2011) was provided by Hitoshi Kurumizaka as a lyophilized powder.
To generate H3.2, chicken CENP-A, or human CENP-A and H4 tetramer, these protein powders were suspended in unfolding buffer (20 mM Tris-HCl, pH 7.5, 7 M Guanidine-HCl, and 20 mM DTT) and refolded by serial dialysis in folding buffer 1 (1 M NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA-NaOH, pH 8.0, and 10 mM DTT) for 4 h at 4°C, folding buffer 2 (500 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA-NaOH, pH 8.0, and 10 mM DTT) for 4 h at 4°C, folding buffer 3 (200 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, pH 8.0, and 10 mM DTT) overnight at 4°C, and folding buffer 4 (150 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, pH 8.0, and 10 mM DTT) for 2 h at 4°C. The refolded proteins were purified with Superdex 200 pg in folding buffer 4; relevant fractions were pooled and frozen in liquid nitrogen and kept at −80°C.
To generate H2A/H2B dimer, H2A and H2B were coexpressed as N-terminally 6xHis-SUMO–tagged H2A and untagged H2B using pETDuet-1-H2A/H2B in E. coli Rosseta2(DE3). Cells expressed H2A/H2B dimer were resuspended in suspension buffer (20 mM Hepes-NaOH, pH 7.5, 2 M NaCl, 5% Glycerol, and 0.5 mM TCEP) and lysed by sonication. The lysate was clarified by centrifugation and applied to a Ni-NTA agarose column. After extensive column washing, bound proteins were eluted in suspension buffer containing 300 mM Imidazole. Fractions containing the H2A/H2B were collected and the 6xHis-SUMO tag was removed by in-house-made SENP7 protease (Fukagawa Lab) treatment. The complex was further purified using Hi-Load Superdex 16/60 75 pg (GE Healthcare) in a buffer (20 mM Hepes-NaOH, pH 7.5, 2 M NaCl, and 1 mM TCEP). Relevant fractions were pooled, frozen in liquid nitrogen, and kept at −80°C.
To generate chicken or human nucleosome, H3.2 and H4 tetramer or chicken or human CENP-A and H4 tetramer were mixed with the H2A/H2B dimer and 601 DNA. KCl was added to the mixture to 2 M concentration. The mixture was dialyzed to salt gradient buffer (2 M to 200 mM KCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA-NaOH, pH 8.0, and 10 mM DTT) overnight at 4°C. The mixture was dialyzed to final buffer (100 mM KCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA-NaOH, pH 8.0, and 10 mM DTT) for 3 h at 4°C, and the pellet in the mixture was removed by centrifugation (5,000 ×g) at 4°C. To stabilize nucleosome, the supernatant was kept incubator for 1 h at 37°C. The nucleosome was checked by native PAGE and detected DNA with EtBr.
Cyclin B–CDK1 purification from starfish oocytes extracts
The cyclin B–CDK1 kinase complex was purified with p13suc1-affinity chromatography from extracts of starfish oocytes in metaphase of meiosis I as previously described (Okumura et al., 1996). p13suc1-affinity gel was prepared with CNBr-activated Sepharose 4B (GE Healthcare) according to the instruction manual. Ligand concentration was at 5 mg p13suc1 purified proteins per 1 ml gel. Purified p13suc1 proteins were prepared as described previously (Brizuela et al., 1987), with slight modification. E. coli strain BL21(DE3)pLysS cells transformed with pRK172 containing yeast suc1+ gene (pRK172 suc1+) were cultured in LB medium containing 50 µg/ml ampicillin and 30 µg/ml chloramphenicol at 37°C until the culture reached OD 600 nm = 0.6. Then, IPTG was added to a final concentration of 1 mM and further cultured for 3 h at 30°C. The cells cultured in 1 liter LB were harvested and resuspended in 30 ml of ice-cold lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 10% Glycerol, 20 µg/ml leupeptin, 0.3 mM PMSF, and 0.16% Triton X-100). The cell suspension was frozen, thawed, and then lysed by sonication on ice. The lysate was clarified by high-speed centrifugation at 10,000 ×g for 30 min at 4°C. The pellet was sonicated again. The cell lysate was prepared two more times. From total 90 ml of cell lysate, p13suc1 proteins were purified by sequential column chromatography. The lysate was loaded onto 90 ml of S-Sepharose (GE Healthcare), and its flow-through was loaded onto 90 ml of Q-Sepharose (GE Healthcare). p13suc1 was eluted with a 0–500 mM NaCl gradient in Tris-HCl, pH 7.5, at a peak of 150 mM. The fractions of p13suc1 protein were confirmed by 15% SDS-PAGE and Coomassie brilliant blue (CBB) staining, the positive fractions were pooled, and p13suc1 was pelleted with 80% ammonium sulfate. The pellet was resuspended with TBS (50 mM Tris-HCl, pH 7.5, and 150 mM NaCl) with <15 ml and loaded onto 475 ml of Sephacryl S-100HR (GE Healthcare) equilibrated with TBS. Purified p13suc1 was dialyzed against borate buffer (0.2 M Borate, pH 8.2, and 0.2 M NaCl) for the coupling reaction.
Starfish (Asterina pectinifera [renamed Patiria pectinifera]) were collected from the sea during their breeding season. They were kept in laboratory aquaria with artificial seawater (Rei-sea marine II; Iwaki Pumps) at 14°C. Ovaries filled with fully grown oocytes were dissected from female starfish and put into Ca2+-free artificial seawater, CaF-ASW (476 mM NaCl, 10 mM KCl, 36 mM MgCl2, 18 mM MgSO4, and 20 mM H3BO3, pH 8.2), in a 150-mm dish. The ovaries were washed three times with CaF-ASW by transferring ovaries into another dish every 5–10 min. 1/100 vol of 1 M CaCl2 was added to the dish and in incubate for 5 min, followed by addition of 1/25 vol of 1 M KCl to induce spawning from the ovaries. Spawned oocytes were separated from shrunk ovary fragments using a tea strainer. Isolated oocytes were washed several times with CaF-ASW. A maturation-inducing substance, 1-methyladenine, was added at a final concentration of 10−5 M. Confirming that the maturing oocytes entered into metaphase of meiosis I using microscopy, the oocytes were packed by centrifugation at 500 ×g. To remove the jelly coat of the oocytes, the oocyte pack was washed three times with pH 5.5 CaF-ASW while changing the temperature of the CaF-ASW (16°C, 8°C, and 0°C). The oocyte pack was then washed with ice-cold buffer C (160 mM Na-β-glycerophosphate, 40 mM EGTA, 30 mM MgCl2, 2 mM DTT, 200 mM KCl, 200 mM sucrose, 0.6 mM PMSF, and 40 µg/ml leupeptin, pH 7.3). Then, 10 ml of the oocyte pack was suspended in 20 ml of ×1/2 diluted buffer C and homogenized with five strokes of a glass/Teflon homogenizer. The homogenate was centrifuged at 14,000 ×g for 15 min at 4°C, followed by recentrifugation of the supernatant at 140,000 ×g for 40 min at 4°C to recover high-speed extracts, which were frozen in liquid nitrogen and kept at −80°C for further use.
The oocyte extracts (20 ml, 400 mg protein) containing active cyclin B–CDK1 complex were cleared by centrifugation at 140,000 ×g for 40 min at 4°C after thawing and then preincubated with Sepharose 4B beads (10 ml) for 1 h at 4°C with gentle stirring. The mixture was filtered with an empty chromatographic column, and flow-through extracts were batch-loaded on 3 ml of p13suc1-Sepharose 4B equilibrated with buffer D (80 mM Na-β-glycerophosphate, 20 mM EGTA, 15 mM MgCl2, and 1 mM DTT, pH 7.3) supplemented with 0.01% Brij35. After 60-min incubation at 4°C under gentle agitation, the p13suc1 beads were packed into a chromatographic column. The column was washed with 15 ml of buffer D supplemented with 500 mM NaCl, followed by wash with 15 ml of buffer D and then with 15 ml of buffer D supplemented with 30% ethylene glycol. Finally, starfish cyclin B–CDK1 was eluted with 15 ml of buffer D containing both 500 mM NaCl and 50% ethylene glycol. Eluted fractions were dialyzed against buffer D supplemented with 10% sucrose and 0.1% NP-40. The dialyzed preparation was concentrated with Vivaspin 2-50K (GE or Sartorius). Concentrated CDK1 solution was frozen in liquid nitrogen and kept at −80°C.
CENP-C phosphorylation by CDK1 in vitro
MBP-gCENP-C601–864 WT or mutants or MBP (2 mg/ml) was incubated in the presence or absence of the active cyclin B–CDK1 (relative H1 kinase activity: 1.4 pmol P/min μl) in reaction buffer (10 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 100 mM ATP, and 1× complete EDTA-free proteinase inhibitor) for 1 h at 25°C. To examine the phosphorylated proteins, they were separated on SuperSep Ace, 5–20%, or Phos-tag-5.0% SDS-PAGE (25 µM Phos-tag acrylamide and 50 µM MnCl2) and stained with CBB stain one (ready to use; Nacalai Tesque). After the reaction was terminated by adding EDTA-NaOH, pH 8.0 (final concentration, 5 mM), these reacted samples were bound to Amylose resin for 2 h at 4°C and washed with high-salt wash buffer. The samples were eluted with elution buffer and dialyzed to column buffer for overnight at 4°C using Slide-A-Lyzer Gamma Irradiated Dialysis Cassette Extra-Strength (Thermo Fisher). Samples were frozen in liquid nitrogen and kept at −80°C.
MBP-hCENP-C426–943 WT or MBP-hCENP-C426–943 T734A (1 mg/ml) was phosphorylated as above, except the reaction buffer contained 300 mM NaCl. Phosphorylated proteins were examined using SuperSep Ace, 5–20%, and Phos-tag-5.0% SDS-PAGE as above. After the reaction was terminated by adding EDTA-NaOH, pH 8.0 (final concentration, 5 mM), the samples were stored on ice ≤5 d.
Pull-down assay
CDK1-phosphorylated or unphosphorylated MBP-gCENP-C601–864 WT or mutants or MBP (30 pmol) was incubated with Amylose resin (15 µl, bed volume; New England Biolabs) for 2 h at 4°C in 250 µl MBP-binding buffer (20 mM Hepes-NaOH, pH 7.5, 300 mM NaCl, 5% Glycerol, 1 mM EDTA-NaOH pH 8.0, 1 mM DTT, and 1× complete EDTA-free proteinase inhibitor). The resin was centrifuged at 5,800 ×g at 4°C and washed with MBP-binding buffer three times, followed by three washes with pull-down buffer (20 mM Hepes-NaOH, pH 7.5, 300 mM NaCl, 10% Glycerol, 0.1% NP-40, 1 mM DTT, and 1× complete EDTA-free proteinase inhibitor). 30 pmol of reconstituted chicken nucleosome was mixed with salt control buffer (30 mM Hepes-NaOH, pH 7.5, 500 mM NaCl, 20% Glycerol, 0.2% NP-40, 1 mM DTT, and 1× complete EDTA-free proteinase inhibitor) at a 1:1 ratio. The volume of nucleosome mix was adjusted to 50 µl with pull-down buffer and incubated with the Amylose resin bound to MBP-gCENP-C601–864 WT or mutants or MBP for 90 min on ice. The resin was flash-spun down at 5,800 ×g at 4°C and washed with pull-down buffer three times. Subsequently, the resin was flash-spun down at 13,000 ×g at 4°C, and the remaining buffer was completely removed. The proteins were eluted with 2× LSB, separated on 15% SDS-PAGE (except Fig. S1 E; SuperSep Ace, 10–20%), and stained with CBB stain one. CDK1-phosphorylated or unphosphorylated MBP-hCENP-C426-943 WT or MBP-hCENP-C426-943 T734A (30 pmol) was pulled down with human CENP-A nucleosome as above. The proteins were eluted with 2× LSB, separated on SuperSep Ace, 5–20%, and stained with CBB stain one).
Electrophoretic mobility shift assay
The human CENP-A nucleosome (0.1 µM) was mixed with hCENP-C426–537 or MBP-hCENP-C501–537 at the indicated molar ratio to human CENP-A nucleosome in EMSA buffer (20 mM Hepes-NaOH, pH 7.5, 100 mM NaCl, 5% Glycerol, 1 mM EDTA-NaOH, pH 8.0, 2 mM DTT, and 1× complete EDTA-free proteinase inhibitor; Roche) and incubated for 30 min on ice. These samples were separated on SuperSep Ace, 5–20%, with native PAGE, and the CENP-A–CENP-C complex was visualized with ethidium bromide.
Competition assay for the CENP-A–CENP-C interaction
Phosphorylated and unphosphorylated hCENP-C727–767 were synthesized (Biologica). Each peptide or MBP-hCENP-C501–537 was added at the indicated molar ratio to 0.1 µM CENP-A nucleosome, which was preincubated with hCENP-C426–537 (0.5 µM), as above, for 30 min on ice in EMSA buffer. These samples were separated on SuperSep Ace, 5–20%, with native PAGE, and the CENP-A–CENP-C complex was visualized with ethidium bromide.
Cell fractionation
cKO-gCENP-C cells expressing GFP-gCENP-C601–864 WT or mutants were treated with 2 µg/ml tet for 48 h and harvested and washed with PBS twice, followed by another wash with TMS (10 mM Tris-HCl, pH 7.5, 5 mM MgCl2, and 0.25 M sucrose). Cells were suspended in TMS supplemented with 0.5% Triton X-100 (TMS-Triton) and incubated for 15 min on ice (whole-cell sample). The cells were spun at 17,000 ×g for 15 min at 4°C. The supernatant was recovered as a detergent-soluble fraction. The precipitate (detergent-insoluble fraction) was washed and suspended in buffer A (15 mM Hepes-NaOH, pH 7.4, 15 mM NaCl, 60 mM KCl, 0.34 M sucrose, 0.5 mM spermidine, 0.15 mM spermine, 1 mM DTT, and 1× complete EDTA-free proteinase inhibitor; final 108 cells/ml), supplemented with final 1 mM CaCl2 and 2.5 × 102 gel units/ml Micrococcal Nuclease (New England Biolabs), and incubated for 10 min at 37°C, and then final 10 mM EDTA-NaOH, pH 8.0, was added to terminate the reaction. After centrifugation, the precipitate was resuspended with 10 mM EDTA-NaOH, pH 8.0 (9/200 vol buffer A used above), supplemented with 5 M NaCl (1/200 vol buffer A used above). The suspension was diluted with buffer B (20 mM Tris-HCl, pH 8.0, 5 mM EDTA-NaOH, pH 8.0, 0.5 M NaCl, 0.2% Tween 20, and 1× complete EDTA-free proteinase inhibitor; Roche; final 108 cells/ml) and centrifuged. The supernatant was collected as the chromatin fraction..
CRISPR/Cas9-mediated homologous recombination
CL18 cells were transfected with pX335-gCENP-C 1 and pBSKS-gCENP-C-GFPWT-puroR (long 3′). The cell lines in which one of the alleles was targeted by gCENP-C-GFPWT were selected by 0.5 µg/ml puromycin. One cell line was selected after Southern blot analysis and transfected with pX335-gCENP-C 3 and pBSKS-gCENP-C-GFPWT-HisD to target the remaining allele (CL18/gCENP-C-GFPWT) and selected by 1 mg/ml L-Histidinol dihydrochloride. To generate the CL18 cell line expressing gCENP-C-GFPT651A from the endogenous promoter, the gCENP-CT651A cassettes were targeted into the gCENP-C alleles as above, except that sgRNA (pX335-gCENP-C 2) was used for the first targeting with the puroR cassette (CL18/gCENP-C-GFPT651A). The CL18 cell line expressing gCENP-C-GFPΔ648–676 (CL18/gCENP-C-GFPΔ648–676) was generated by single transfection of pX330-gCENP-C 3, pBSKS-gCENP-C-GFPΔ648–676-puroR, and pBSKS-gCENP-C-GFPΔ648–676-HisD and selected by 0.5 µg/ml puromycin and 1 mg/ml L-Histidinol dihydrochloride.
cKO-CENP-H cell lines expressing gCENP-C-GFP WT, Δ648, or T651A were established by the single transfection as done for CL18/gCENP-C-GFPΔ648–676 (cKO-CENP-H/gCENP-C-GFPWT, Δ648–676 or T651A).
To generate the CCP cell line expressing gCENP-CWT, gCENP-CWT-puroR, and -HisD, cassettes were sequentially targeted as done for CL18/gCENP-C-GFPWT, except that the following plasmids were used in the targeting: pX335-gCENP-C 3 and pBSSK-gCENP-CWT-puroR for the first targeting and pX335-gCENP-C#3 and pBSSK-gCENP-CWT-HisD for the second targeting (CCP/gCENP-CWT). The CCP cell line expressing gCENP-CΔ648–676 or gCENP-CT651A was established as done for CL18/gCENP-C-GFPΔ648–676, except that the donor plasmids were pBSKS-gCENP-CWT, Δ648–676 or T651A-puroR or -HisD (CCP/gCENP-CΔ648–676 or T651A).
Quantification and statistical analysis
The fluorescence signal intensities of gCENP-C-GFP or Ndc80, CENP-T, and gCENP-C on kinetochores were quantified using Imaris (Bitplane). The fluorescence signals on ∼50–100 kinetochores in each of five (Fig. 1, D and G; Fig. 3, B and C; and Fig. 6 D) or seven (Figs. 4 B and 7 E) cells were quantified and subtracted with mean of background signals in nonkinetochore regions. The mean of the kinetochore signals in each cell is shown in Fig. 1, D and G; Fig. 3, B and C; and Figs. 4 B, 6 D, and 7 E. To show difference of the signals among kinetochores in single cell, the signals of all kinetochores quantified of each cell are shown in Fig. S3 (A–G). Data are shown as mean ± SD. The unpaired t test (two tailed) was used, with P < 0.05 considered significant. For RPE-1 cells, the fluorescence signal intensities of various GFP-hCENP-C or CENP-T on kinetochores were quantified using Imaris. The fluorescence signals on >50 kinetochores in each of five cells were quantified and subtracted with mean of background signals in nonkinetochore regions. The mean of the kinetochore signals in each cell is shown in left graphs of Figs. 5 E and S4 C. To show differences in signals among kinetochores in a single cell, the signals of all kinetochores quantified in each cell are shown in right graphs of Fig. S4, B and D. Data are shown as mean ± SD. The unpaired t test (two tailed) was used, with P < 0.05 considered significant.
For pull-down assays, the intensities of MBP-gCENP-C or MBP were measured with Fiji (Schindelin et al., 2012). The intensities of all histones in the CENP-A nucleosome were measured as the signals for the CENP-A nucleosome and normalized by the intensities of MBP-gCENP-C or MBP or MBP-hCENP-C. The intensity values were normalized with the value of CENP-A/gCENP-C601–864 WT (CDK1–) or CENP-A/hCENP-C426–943 WT (CDK1−) of each assay. The graphs of the intensities were made using GraphPad Prism (GraphPad Software). Data are shown as mean ± SD of three independent assays. The unpaired t test (two tailed) was used, with P < 0.05 considered significant.
For growth curve of RPE-1 cells, cell numbers were normalized with those at 0 h for each cell line to show relative cell numbers. Data are shown as mean ± SD of three independent assays.
Online supplemental material
Fig. S1 shows gCENP-C phosphorylation and purification of recombinant proteins used in experiments for Fig. 2 and demonstrates the establishment of a cKO line for hCENP-C. Fig. S2 shows the CRISPR/Cas9-mediated gene editing of the gCENP-C gene locus. Fig. S3 shows signal intensities of each kinetochore for graphs of Figs. 1, 3, 4, 6, and 7. Fig. S4 shows characterization of hCENP-C cKO cells expressing various hCENP-C mutants. Fig. S5 shows the results of in vitro binding of recombinant hCENP-C to the CENP-A nucleosome.
Supplementary Material
Acknowledgments
The authors are very grateful to T. Hori for fruitful discussion; Y. Fukagawa, R. Fukuoka, K. Oshimo for technical assistance; and K. Yoda, H. Kurumizaka, and H. Kimura for reagents.
This work was supported by the Japan Society for the Promotion of Science (KAKENHI grants 17H06167, 16H06279, and 15H05972 to T. Fukagawa; KAKENHI grant 16K18491 to M. Hara; and KAKENHI grant 18K06084 to M. Ariyoshi).
The authors declare no competing financial interests.
Author contributions: R. Watanabe and M. Hara designed and performed all experiments in this study. M. Ariyoshi purified and reconstituted the nucleosomes. E. Okumura purified cyclin B–CDK1. S. Hervé and D. Fachinetti established the hCENP-C-AID RPE-1 cell line. T. Fukagawa established various mutant RPE-1 cell lines, and R. Watanabe performed experiments using these cell lines. M. Hara and T. Fukagawa supervised all experiments and wrote the manuscript, with input from all authors.
References
- Abe Y., Sako K., Takagaki K., Hirayama Y., Uchida K.S., Herman J.A., DeLuca J.G., and Hirota T.. 2016. HP1-Assisted Aurora B Kinase Activity Prevents Chromosome Segregation Errors. Dev. Cell. 36:487–497. 10.1016/j.devcel.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Ahmad A., Bilokapić S., Schäfer I.B., Halić M., and Sekulić N.. 2019. CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail. EMBO Rep. 20:e48913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., and Nogales E.. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810. 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., and Fukagawa T.. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182. 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S., Yang H., Nozaki N., Okazaki T., and Yoda K.. 2002. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22:2229–2241. 10.1128/MCB.22.7.2229-2241.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura Y., Tachiwana H., Oda T., Sato M., and Kurumizaka H.. 2012. Structural analysis of the hexasome, lacking one histone H2A/H2B dimer from the conventional nucleosome. Biochemistry. 51:3302–3309. 10.1021/bi300129b [DOI] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., et al. . 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 3:e02978 10.7554/eLife.02978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., and Cleveland D.W.. 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 144:471–479. 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela L., Draetta G., and Beach D.. 1987. p13suc1 acts in the fission yeast cell division cycle as a component of the p34cdc2 protein kinase. EMBO J. 6:3507–3514. 10.1002/j.1460-2075.1987.tb02676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde J.M., Reynaud C.A., Humphries E.H., Olson W., Ewert D.L., and Weill J.C.. 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9:921–927. 10.1002/j.1460-2075.1990.tb08190.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Zhou K., Zhang Z., Luger K., and Straight A.F.. 2018. Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Mol. Biol. Cell. 29:751–762. 10.1091/mbc.E17-10-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A.E., Bai Y., and Subramaniam S.. 2018. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 359:339–343. 10.1126/science.aar2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., and Simons K.T.. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491. 10.1091/mbc.e08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Dimitrova Y.N., Jenni S., Valverde R., Khin Y., and Harrison S.C.. 2016. Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell. 167:1014–1027.e12. 10.1016/j.cell.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E., and Cleveland D.W.. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15:1056–1066. 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D., Han J.S., McMahon M.A., Ly P., Abdullah A., Wong A.J., and Cleveland D.W.. 2015. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev. Cell. 33:314–327. 10.1016/j.devcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S.J., Guo L.Y., Sekulic N., Smoak E.M., Mani T., Logsdon G.A., Gupta K., Jansen L.E., Van Duyne G.D., Vinogradov S.A., et al. . 2015. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 348:699–703. 10.1126/science.1259308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., and Brown W.R.. 1997. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 6:2301–2308. 10.1093/hmg/6.13.2301 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., and Earnshaw W.C.. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 30:496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Pendon C., Morris J., and Brown W.. 1999. CENP-C is necessary but not sufficient to induce formation of a functional centromere. EMBO J. 18:4196–4209. 10.1093/emboj/18.15.4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., and Ikemura T.. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603–4617. 10.1093/emboj/20.16.4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L.Y., Allu P.K., Zandarashvili L., McKinley K.L., Sekulic N., Dawicki-McKenna J.M., Fachinetti D., Logsdon G.A., Jamiolkowski R.M., Cleveland D.W., et al. . 2017. Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun. 8:15775 10.1038/ncomms15775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., and Fukagawa T.. 2017. Critical Foundation of the Kinetochore: The Constitutive Centromere-Associated Network (CCAN). Prog. Mol. Subcell. Biol. 56:29–57. 10.1007/978-3-319-58592-5_2 [DOI] [PubMed] [Google Scholar]
- Hara M., and Fukagawa T.. 2018. Kinetochore assembly and disassembly during mitotic entry and exit. Curr. Opin. Cell Biol. 52:73–81. 10.1016/j.ceb.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Hara M., Ariyoshi M., Okumura E.I., Hori T., and Fukagawa T.. 2018. Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat. Cell Biol. 20:1378–1388. 10.1038/s41556-018-0230-0 [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Dumont M., Barra V., Ly P., Nechemia-Arbely Y., McMahon M.A., Hervé S., Cleveland D.W., and Fachinetti D.. 2016. CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly. Cell Reports. 17:2394–2404. 10.1016/j.celrep.2016.10.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Fachinetti D., Han J.S., and Cleveland D.W.. 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 109:E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., and Fukagawa T.. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362. 10.1242/jcs.00645 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. . 2008a CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052. 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., and Fukagawa T.. 2008b CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 19:843–854. 10.1091/mbc.e07-06-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. . 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31:827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., and Yoda K.. 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684. 10.1111/j.1365-2443.2006.00969.x [DOI] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113. 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare K., Weir J.R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., and Musacchio A.. 2015. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210:11–22. 10.1083/jcb.201412028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral L. 2015. Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. F1000 Res. 4:474 10.12688/f1000research.6823.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., and Fukagawa T.. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168. 10.1091/mbc.e07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., et al. . 2009. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41:753–761. 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2016. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17:16–29. 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., Sekulic N., Guo L.Y., Tsinman T., Black B.E., and Cheeseman I.M.. 2015. The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol. Cell. 60:886–898. 10.1016/j.molcel.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal H., and Fukagawa T.. 2016. Kinetochore assembly and function through the cell cycle. Chromosoma. 125:645–659. 10.1007/s00412-016-0608-3 [DOI] [PubMed] [Google Scholar]
- Nagpal H., Hori T., Furukawa A., Sugase K., Osakabe A., Kurumizaka H., and Fukagawa T.. 2015. Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell. 26:3768–3776. 10.1091/mbc.E15-07-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., and Fukagawa T.. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501. 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436. 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R.S., Nagao K., Masuda H.T., Iwasaki O., Hirota T., Nozaki N., Kimura H., and Obuse C.. 2010. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12:719–727. 10.1038/ncb2075 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Okumura E., Sekiai T., Hisanaga S., Tachibana K., and Kishimoto T.. 1996. Initial triggering of M-phase in starfish oocytes: a possible novel component of maturation-promoting factor besides cdc2 kinase. J. Cell Biol. 132:125–135. 10.1083/jcb.132.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Wener M.H., Andrews B.S., and Margolis R.L.. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805–815. 10.1083/jcb.104.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G.P., Weir J.R., Vetter I.R., Musacchio A., and Luger K.. 2017. Decoding the centromeric nucleosome through CENP-N. eLife. 6:e33442 10.7554/eLife.33442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesenti M.E., Weir J.R., and Musacchio A.. 2016. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 37:152–163. 10.1016/j.sbi.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. . 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852. 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A.J., Keller Y., Liu K., Overlack J., John Y.N., Dimitrova S., Jenni S., van Gerwen P., Stege S., Wohlgemuth P., et al. . 2016. Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell. 167:1028–1040.e15. 10.1016/j.cell.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., and Glover D.M.. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405. 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H. III, Maurer M., Rothfield N.F., and Earnshaw W.C.. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 70:115–125. 10.1016/0092-8674(92)90538-N [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Badger B.L., and Salmon E.D.. 2015. A quantitative description of Ndc80 complex linkage to human kinetochores. Nat. Commun. 6:8161 10.1038/ncomms9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y., et al. . 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 476:232–235. 10.1038/nature10258 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Tawaramoto-Sasanuma M., Kawaguchi S., Ohta T., Yoda K., Kurumizaka H., and Yokoyama S.. 2004. Expression and purification of recombinant human histones. Methods. 33:3–11. 10.1016/j.ymeth.2003.10.024 [DOI] [PubMed] [Google Scholar]
- Tian T., Li X., Liu Y., Wang C., Liu X., Bi G., Zhang X., Yao X., Zhou Z.H., and Zang J.. 2018. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 28:374–378. 10.1038/cr.2018.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J.R., Faesen A.C., Klare K., Petrovic A., Basilico F., Fischböck J., Pentakota S., Keller J., Pesenti M.E., Pan D., et al. . 2016. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature. 537:249–253. 10.1038/nature19333 [DOI] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25:334–340. 10.1016/j.ceb.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.