Baumgart et al. measure the amount of dimeric and polymeric tubulin at mitotic centrosomes in C. elegans by light and electron microscopy. Centrosomes concentrate soluble tubulin tenfold over the cytoplasm, suggesting that centrosomal microtubule nucleation may be driven in part by concentrating tubulin.
Abstract
During mitosis, the centrosome expands its capacity to nucleate microtubules. Understanding the mechanisms of centrosomal microtubule nucleation is, however, constrained by a lack of knowledge of the amount of soluble and polymeric tubulin at mitotic centrosomes. Here we combined light microscopy and serial-section electron tomography to measure the amount of dimeric and polymeric tubulin at mitotic centrosomes in early C. elegans embryos. We show that a C. elegans one-cell stage centrosome at metaphase contains >10,000 microtubules with a total polymer concentration of 230 µM. Centrosomes concentrate soluble α/β tubulin by about 10-fold over the cytoplasm, reaching peak values of 470 µM, giving a combined total monomer and polymer tubulin concentration at centrosomes of up to 660 µM. These findings support in vitro data suggesting that microtubule nucleation in C. elegans centrosomes is driven in part by concentrating soluble tubulin.
Introduction
During mitosis, the pericentriolar material (PCM) of centrosomes grows and increases its nucleation capacity many-fold. This process, called centrosome maturation, is thought to be essential for forming a mitotic spindle in many animal systems (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2014; Sonnen et al., 2012). Despite the importance of centrosome maturation in spindle formation, the mechanisms by which centrosomes increase their nucleation capacity are unknown. In Caenorhabditis elegans, it is thought that increase of centrosome size is driven by accumulation of a coiled-coil protein called SPD-5, which forms the centrosome scaffold (Hamill et al., 2002). Growth of the scaffold is stimulated by the protein SPD-2 (Pelletier et al., 2004) and Polo kinase (Decker et al., 2011; Wueseke et al., 2016). SPD2/PLK1 dependent formation of the SPD-5 scaffold has been reconstituted in vitro (Woodruff et al., 2017). Similar pathways are thought to operate in Drosophila (Conduit et al., 2014). Once formed, it has been proposed that client proteins such as γ-tubulin, microtubules polymerases, and microtubule depolymerases partition into the scaffold, where they favor microtubule growth and nucleation (Woodruff et al., 2017). However, genetic evidence for the role of these microtubule-associated proteins in driving microtubule nucleation in mitosis is limited. In C. elegans, for instance, mutations or depletion of γ-tubulin only have marginal effects on microtubule nucleation (Hannak et al., 2002; O’Toole et al., 2012; Strome et al., 2001).
Recently, it has been suggested that nucleation could be driven in part by the partitioning of tubulin dimers into the PCM (Woodruff et al., 2017). This would raise the tubulin concentration above the critical concentration for nucleation, thus driving microtubule formation. Supporting evidence for this idea comes from biochemical reconstitutions, which have shown that tubulin and other proteins can partition into reconstituted PCM and liquid drops of the microtubule-associated protein tau (Hernández-Vega et al., 2017; Woodruff et al., 2017), as well as experiments on the role of BugZ in assembling Xenopus laevis spindles (Huang et al., 2018). Evidence for such ideas in vivo, however, is currently lacking. In particular, we lack measurements for tubulin concentration of polymerized and unpolymerized tubulin at centrosomes, and the extent of local enrichment of tubulin within the living cell.
In this study, we quantitatively measured in vivo how α/β tubulin, in the form of soluble monomers as well as microtubule polymers, is distributed across the centrosome in C. elegans one-cell embryos. We show that centrosomes concentrate soluble α/β tubulin by about 10-fold over the cytoplasm, reaching up to 470 µM. Based on our observations, we propose that microtubule nucleation in mitotic C. elegans centrosomes is mediated in part by enriching tubulin locally.
Results and discussion
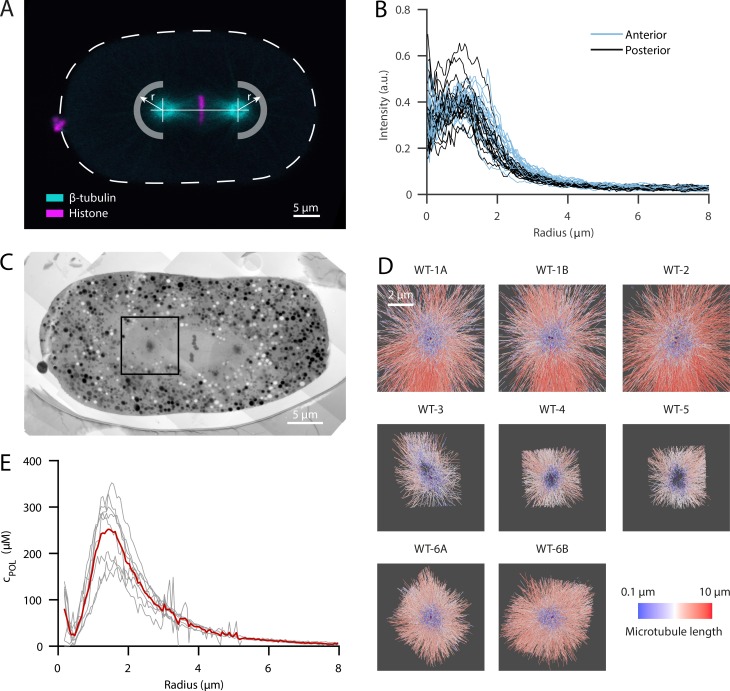
To measure the concentration of tubulin dimers at centrosomes, independent of their monomeric or polymeric state, we performed live-cell imaging using laser scanning microscopy (see Materials and methods) of one-cell C. elegans embryos expressing GFP-tagged β-tubulin (GFP::TBB-2). The fluorescence intensity was roughly symmetric with respect to the centrosome center. To obtain the intensity profile along the radial direction, we used the centrosome center as the origin of our analysis and averaged the signal along the circumferential direction in the region opposite to the spindle toward the cell cortex (Fig. 1 A). The profiles of 19 one-cell embryos in metaphase showed a peak intensity of tubulin at a radial distance of around 1.0 µm, suggesting that tubulin is locally enriched at the outer shell of the centrosome (Fig. 1 B). With further increasing distances, we observed a monotonic decay to a plateau that extended away from the centrosome. This constant intensity outside the centrosome suggests a homogenous distribution of the total tubulin in the cytoplasm. At the very center of the centrosome, we detected a decrease of tubulin signal. However, the signal at the centrosome center was still higher compared with the plateau measured at the cortex. There was no significant difference in tubulin distribution between anterior and posterior centrosomes (Fig. 1 B).
Figure 1.
LM reveals a local enrichment of tubulin at the outer shell of the centrosome which colocalizes with a high density of microtubules recorded by electron tomography. (A) Confocal live-cell imaging of metaphase spindles in one-cell C. elegans embryos labeled with β-tubulin::GFP (cyan) and histone::mCherry (magenta). For the analysis, the centrosome centers were localized manually and fluorescent intensities of β-tubulin::GFP were extracted in radial distances as indicated by arrows for half planes away from the spindle and toward the cell cortex. The anterior side is orientated to the left. Scale bar, 5 µm. (B) Radial profiles of β-tubulin::GFP after subtraction of autofluorescence outside the cell (n = 19). There is no significant difference between anterior (light blue) and posterior centrosomes (dark blue). (C) Overview of a high-pressure frozen and serial-sectioned one-cell embryo in metaphase. Scale bar, 5 µm. The black box indicates a representative area used for electron tomography. (D) Overview of all segmented centrosome models (n = 8). Six different embryos were used. A/B represent centrosomes of the same embryo. Microtubules are color coded by length in a logarithmical scale from short (blue) to long (red) and centrioles (purple). (E) The segmented microtubules of the individual centrosome regions (gray lines, n = 8) are analyzed in radial profiles as shown in A with respect to the local density as length per volume and are converted into the concentration of polymerized tubulin (cPOL). The orange curve represents the local mean value. For registration purposes, the center of the mother centriole is used to align the radial profiles.
To distinguish the soluble from the polymerized state, we performed serial-section electron tomography of eight centrosomes of six different cells in one-cell C. elegans embryos as previously reported (Fig. 1, C and D; Redemann et al., 2017) and extracted microtubule polymer concentration profiles (Fig. 1 E). For these measurements, we combined datasets from the same strain used for light microscopy (LM) and an unlabeled strain (see Materials and methods). The number of microtubules at the centrosome of a one-cell stage embryo was >10,000 (Redemann et al., 2017), with an average length of 1.1 µm. At the very center, a small peak was seen of ∼100 µM due to the presence of the centriole microtubules, which we defined as the origin of our analysis. This peak was surrounded by a void region with a dip at 430 nm, comparable to the size of the dense interphase PCM layer seen by super-resolution LM in Drosophila S2 cells and in human tissue culture cells (Lawo et al., 2012; Mennella et al., 2014). The microtubule density then increased up to a radial distance of ∼1.4 µm, with the highest polymerized tubulin concentration in the range from 170 to 350 µM. This region coincided roughly with the outer edge of the PCM. The amount of polymeric tubulin then decayed monotonically with increasing distance from the centrosome.
To calibrate the measurement from LM, where both soluble and polymerized tubulin are labeled, we combined those measurements with the spatial and quantitative information from EM. Combining the quantitative EM data with the LM data required two compensatory procedures: first, correction for shrinkage during sample preparation for EM; and second, for the blurring of intensity profiles from LM due to the point-spread function (PSF) of the microscope. After accounting for these effects, we were able to compare the radial concentration profiles from the two approaches (see Materials and methods). Briefly, the radial concentration profile of soluble tubulin (cSOL) is the difference between the calibrated concentration profile from LM and the concentration profile of polymeric tubulin (cPOL) from EM,
| (1) |
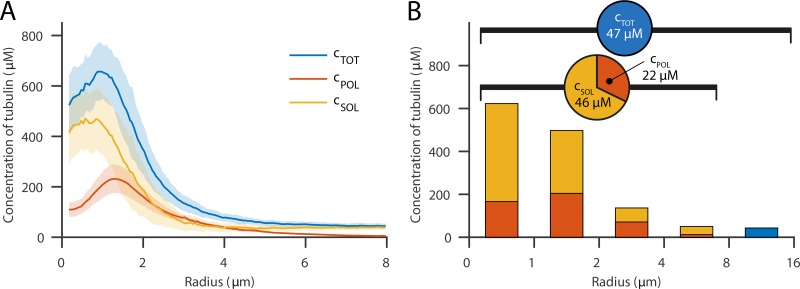
where is the calibration constant for the light intensity of the LM data (ILM). To obtain the calibration constant we used the assumption that soluble tubulin is homogenously distributed outside of the centrosome (r > 4 µm). This assumption implies that, outside the centrosome, the spatial variation of total tubulin concentration is solely due to the spatial changes of the concentration of polymerized tubulin. We implemented this condition by finding the calibration constant that minimizes the variance of the soluble tubulin outside the centrosome. By calculating the difference between the calibrated intensity profile from LM (cTOT) and the profile of the EM data (cPOL), we obtained the concentration profile of soluble tubulin cSOL = cTOT – cPOL (Fig. 2 A). This analysis revealed that the soluble tubulin concentration profile (cSOL) is significantly higher in the region of the centrosome. Furthermore, the total tubulin concentration profile peaks at a maximum value of ∼660 µM at a radial distance of 1.0 µm away from the centriole. Of this, 200 µM is polymerized and 460 µM soluble tubulin.
Figure 2.
Calibrated intensity profiles from LM by electron tomography data show the distribution of soluble and polymerized tubulin. (A) Concentrations of soluble (yellow), polymerized (red), and total (blue) tubulin concentration along the spindle axis after calibration. The soluble tubulin concentration is enriched at the centrosome and shows a peak concentration of ∼405 µM at r = 0.8 µm. Error bars are SD. (B) Fractions of polymerized and soluble tubulin analyzed in intervals along the radial distance up to the data range of the EM reconstructions and the total concentration in the remaining part. The embryo is loaded with a total tubulin concentration of ∼47 µM. Up to 8 µm, the overall tubulin is on average 68 µM, of which 22 µM is polymerized.
We used these data to measure the total polymer and monomer concentration in the embryo. For the entire cell, we obtained a concentration of 47 µM, which is comparable to data from mass spectrometry (Saha et al., 2016) and fluorescence lifetime imaging measurements (Kaye et al., 2018). By integrating from the spindle axis up to a distance of 8 µm, the maximum distance at which we obtained EM data, we measured that ∼32% of tubulin was polymerized (22 µM), and the rest remained in the soluble state (46 µM; Fig. 2 B). Because the larger fraction of this recruited tubulin is freely available in the soluble state and only a smaller fraction is assembled into microtubules, this suggests that tubulin is not a limiting component in the process of microtubule nucleation at the centrosome. It also confirms that there must be mechanisms to concentrate tubulin at centrosomes, or the tubulin would simply diffuse out of the centrosome down the concentration gradient back into the cytoplasm.
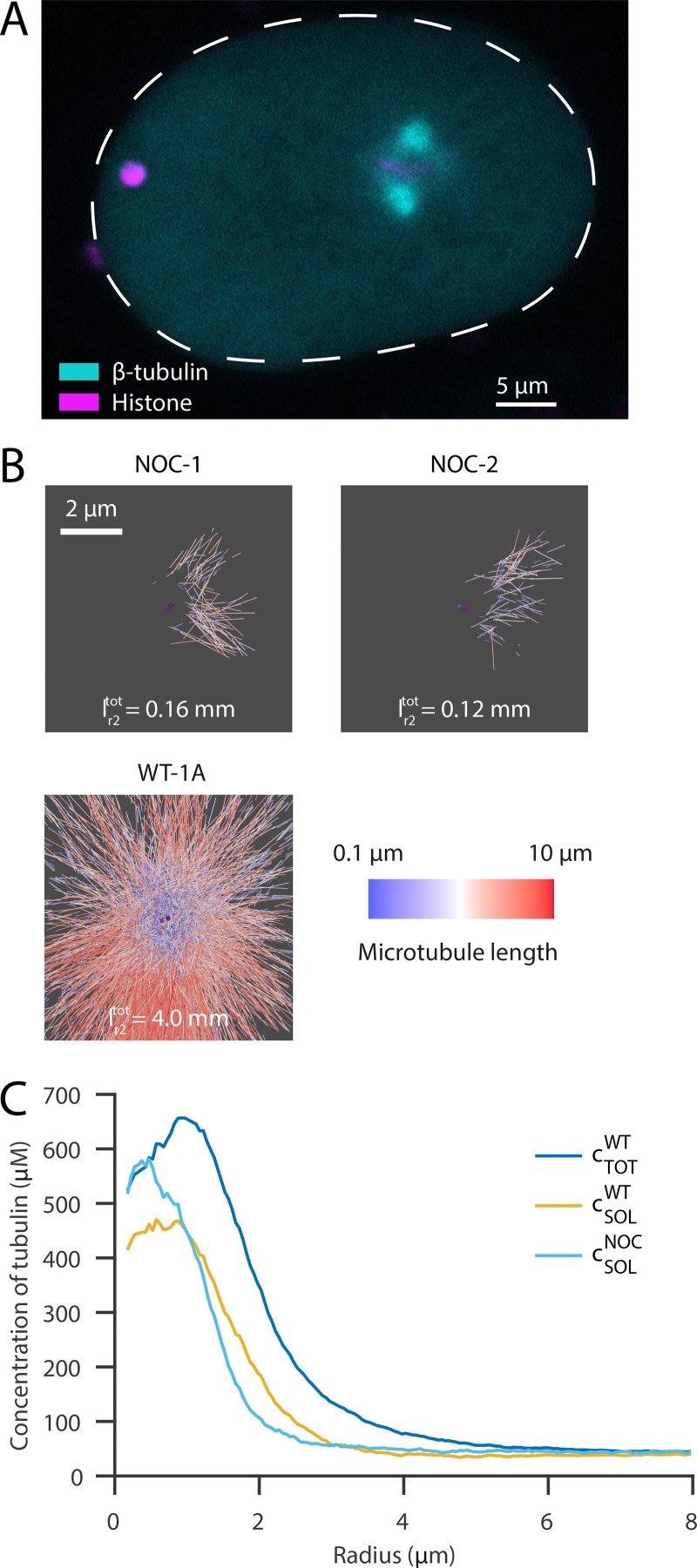
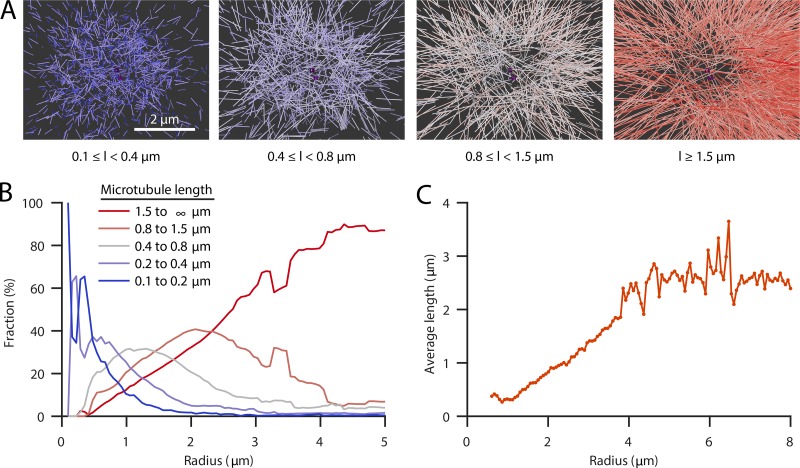
To further explore the amount of soluble versus polymerized tubulin at mitotic centrosomes, we treated embryos with nocodazole to depolymerize microtubules (Fig. 3 A; Carvalho et al., 2011; Hannak et al., 2002; Strome et al., 2001). Electron tomography confirmed that there were only a few polymerized microtubules remaining at centrosomes (the total length of microtubules within a radius of 2 µm, covering the centrosome size, measured only ∼4% compared with the WT data; Fig. 3 B). The concentration profile of soluble tubulin after nocodazole treatment showed a similar shape as that seen in WT embryos, with a peak of tubulin concentration close to the centrioles and a rapid decay outside of the centrosome (Fig. 3 C). When compared with the total concentration of tubulin in WT, the concentration profile showed no dip at the center, and peaked very close to the centrioles, suggesting that this dip is driven by microtubule polymerization. Supporting this idea, our tomographic data revealed that short microtubules are predominantly found at the centrosome center, whereas long microtubules tend to be located further outside (Figs. 1 D; and Fig. 4, A and B). In addition, we found that microtubule polymerization velocity as measured with EB2 is lower immediately outside of the centrosomes, suggesting that tubulin may be limiting once microtubules leave the centrosome (Fig. S3 B). We assume that the location of very short microtubules marks the region where nucleation predominantly takes place, and longer microtubules are in some way transported out to the periphery or stabilized (Fig. 4 C).
Figure 3.
Effect of microtubule depolymerization on the centrosomal concentration of free tubulin. (A) Confocal live-cell imaging of metaphase spindles in one-cell C. elegans embryo labeled with β-tubulin::GFP (cyan) and histone::mCherry (magenta) 100 s after nocodazole treatment. The anterior side is orientated to the left. Scale bar, 5 µm. (B) Segmentations of microtubules at the centrosomes of nocodazole-treated embryos (n = 2, top, NOC) show a significantly reduced number of microtubules compared with WT embryos (bottom). Microtubules are color-coded according to their length (short, blue; long, red; centrioles, purple). represents the total length of polymerized microtubules within a radius of 2 µm. Scale bar, 2 µm. (C) Radial mean profiles of soluble tubulin after nocodazole treatment (light blue, n = 2) compared with WT (soluble tubulin, yellow and total tubulin, blue; see Fig. 2 A).
Figure 4.
Localization of short microtubules at the centrosome. (A) Spatial graphs of a representative centrosome (WT-1A) showing groups of microtubules filtered by length (l) from short (0.1–0.4 µm, blue) to long (>1.5 µm, red) and centrioles (purple). Very short microtubules cover predominantly the core of centrosomes, and longer microtubules are found on the outer edge. Scale bar, 2 µm. (B) Plot of the fraction of microtubules of different length groups (short, blue, to long, red) at specific radii from the centrosome center. Short microtubules are predominantly found at the centrosome, whereas longer microtubules tend to be located outside the centrosome. The radial coordinate is corrected for shrinkage, based on the pooled dataset of all individual centrosome regions (n = 8; see Materials and methods). (C) Average microtubule length as a function of the radial position of the pole-proximal microtubule end from the centrosome (based on the pooled dataset of all individual centrosome regions [n = 8]).
The mechanisms driving microtubule nucleation at mitotic centrosomes have long been a mystery. Although nucleation at interphase centrosomes is prevented by lowering the concentration of γ-tubulin by RNA interference (Hannak et al., 2002; O’Toole et al., 2012), similar treatments in mitosis have little effect on microtubule nucleation. The data presented in this paper show that centrosomes contain a considerable quantity of unpolymerized tubulin. The typical critical concentration for spontaneous microtubule nucleation for bovine tubulin is thought to be 21 µM (Wieczorek et al., 2015). Therefore, the presence of such high concentrations of soluble tubulin supports the idea that an increased concentration of tubulin could be a driving force for nucleation of microtubules at centrosomes. To test whether this unpolymerized tubulin is available for polymerizing microtubules or sequestered in scaffold proteins forming the centrosome, we performed FRAP experiments in nocodazole treated embryos (see Fig. S3 A). We found that 81% of the soluble tubulin turns over in the centrosome (92% in the cytoplasm), suggesting that monomeric tubulin partitions in the centrosome, but it is not immobilized within the scaffold and available for polymerizing microtubules. The recovery time in the centrosome was measured to be 2.8 ± 0.2 s (95% CI), which is slower than in the cytoplasm (0.45 ± 0.06 s). The slower recovery timescale may arise from an increased viscosity within the centrosome or from transient binding and unbinding events with the macromolecules that make up the centrosome. A lower effective diffusion may, in principle, lead to slower microtubule growth dynamics and affect the activity of microtubule nucleation. However, it has been shown for actin that if the increase of viscosity is due to the presence of macromolecules, the polymerization growth is enhanced due to excluded volume effects (Drenckhahn and Pollard, 1986). Since the centrosome is made from scaffold proteins resembling a gel, the situation in the centrosome probably corresponds to the effect created by the presence of macromolecules and may contribute to increasing microtubule polymerization there. Moreover, recent work has shown that increased stoichiometry in phase-separated condensates increases nucleation activity in actin despite a slower turnover in the condensate compared with outside (Case et al., 2019), similar to what we find here.
We do not understand the mechanisms by which tubulin concentrates at centrosomes. However, our FRAP measurements show it does so in a form in which it can freely diffuse so that it can drive microtubule growth. One formalism would be to think of tubulin partitioning into the PCM and that the tubulin concentration at the centrosome is defined by a partition coefficient (Woodruff et al., 2017). This increased partitioning could lead to increased nucleation activity similar to what has been observed in actin (Case et al., 2019). In vitro reconstitution experiments show that tubulin is not concentrated by the main components of the PCM, SPD-5, and SPD-2, but rather by two microtubule-associated proteins, ZYG-9, which is the C. elegans homologue of XMAP215, and TPXL-1, which is the C. elegans homologue of TPX2 (Ozlü et al., 2005). Interestingly, in vitro mutants in ZYG-9 that prevent tubulin binding reduce tubulin association with centrosomes. This suggests that ZYG-9 could in part bind to and concentrate tubulin at centrosomes. However, in vivo, mutants in ZYG-9 do not prevent microtubule nucleation, indicating that in embryos, the process is more complicated and uses a combination of different tubulin-binding proteins. Subsequent work combining electron tomography, LM, and genetics will be required to elucidate these mechanisms.
Materials and methods
Worm cultivation
The following strains were used in this study. WT N2 Bristol was used for electron tomography of WT-1A, WT-1B, and WT-2. MAS91 {unc-119(ed3) III; ItIs37[pAA64; pie-1::mCherry::HIS58]; ruIs57[pie-1::GFP::tbb + unc-119(+)]} was used for the other sets of electron tomography, light microscopic analysis, and nocodazole treatment. TH315 {unc-119(ed3) III; ddEx23 [pie-1::GFP::SAS-4 (genomic introns, CAI 0.3); unc-119(+)]} was used for obtaining the point spread function at the imaging system for calibration. EU2942 ([uIs57[pie-1p::GFP::tubulin + unc-119(+)]; itIs37[pie-1p::mCherry::H2B::pie-13′UTR + unc-119(+)] IV]) was used for FRAP measurements. All strains were cultured on OP50-seeded nematode growth medium (NGM) agar plates at 16–23°C (Brenner, 1974). To enhance expression of fluorescent markers, worms were shifted to 25°C 24 h before light microscopic imaging.
LM
Young hermaphrodites of the C. elegans MAS91 strain were dissected with syringe needles in M9 buffer on a coverslip (24 mm × 60 mm) to release the embryos. We took one-cell C. elegans embryos in metaphase, a stage where spindle growth is mainly completed and the microtubules are not exposed to high pulling forces. Imaging was performed using a Zeiss LSM 710 NLO multiphoton laser scanning microscope in single-photon mode equipped with a Zeiss LCI Plan Neofluar 63× 1.3 NA water-immersion objective. Detection of the emitted fluorescent signal was performed with a QUASAR detector with 32 channels and a dichromatic 488/594 nm beam splitter for excitation/emission splitting and subsequent linear unmixing of both acquired channels. A spectral prism slider was used for selecting the range of emission detection. Confocal stacks were acquired every 20 s covering a z-range of around 15 µm with a spacing of 0.388 µm. In total, we recorded stacks of 19 embryos in metaphase for WT analysis. The time point of metaphase was defined as the stack before anaphase onset in which clear segregation of chromosomes could be observed. Point spread function was measured using the fluorescent signal of SAS-4::GFP as reference beads inside the C. elegans embryo. For this, the strain TH315 was imaged under similar conditions. Individual SAS-4 spots (n = 21) from 2- to 16-cell embryos were isolated and analyzed in cylindrical coordinates using axis-symmetry, resulting in an averaged Gaussian shape intensity profile with a full-width half maximum of 0.423 ± 0.048 µm (mean ± SD) in the plane normal to the optical axis and 1.684 ± 0.336 µm (mean ± SD) along the axis (Fig. S1).
Sample preparation for EM
Isolated early embryos were transferred into cellulose capillary tubes with a diameter of 200 µm (Leica Microsystems) for high-pressure freezing. Embryos were observed with a stereomicroscope until metaphase and then high-pressure frozen using an EMPACT2 with a rapid transfer system (Leica Microsystems; Pelletier et al., 2006; Redemann et al., 2017). The following freeze substitution was performed for 3 d at −90°C using 1% OsO4 and 0.1% uranyl acetate using an automatic freeze substitution machine (EM AFS; Leica Microsystems). Samples were embedded in a thin layer of Epon/Araldite and polymerized at 60°C for 3 d. Serial semi-thick sections (250–300 nm) were cut with an Ultracut UCT Microtome (Leica Microsystems) and collected on Formvar-coated copper slot grids. Post-staining was performed with 2% uranyl acetate in 70% methanol and 0.4% Reynolds lead citrate (Müller-Reichert et al., 2007).
Electron tomography, 3D reconstruction, and automatic segmentation of microtubules
Stained serial semi-thick sections were coated with colloidal gold (15 nm; Sigma-Aldrich) serving as fiducial markers for subsequent tomographic reconstruction. Dual-axis electron tomography was performed using a TECNAI F30 TEM (FEI) operated at 300 kV. Tilt series were acquired at every 1° in a range of ±60° with a pixel size of 2.3 nm using a Gatan US1000 CCD camera (2k × 2k). For tomographic reconstruction, the IMOD software package (http://bio3d.colorado.edu/imod) was used (Kremer et al., 1996). With this software, the computation is based on an R-weighted back-projection algorithm for each tilt axis (Gilbert, 1972). After reconstruction, tomograms were flattened and trimmed to remove unsubstantial outer edges of the volume with no information. For segmentation and automatic tracing of microtubules, we used the AMIRA software with an extension to the filament editor (Weber et al., 2012). After automatic segmentation of microtubules, 3D models were visually inspected and manually corrected. This correction included manual tracing of undetected microtubules, extending and combining individual traces as well as removing incorrectly traced structures, such as membranes and other cellular components. Using the AMIRA software, corrected 3D models were then stitched in z to obtain complete volumes of the recorded centrosomes (Weber et al., 2014). In total, we recorded eight WT and two nocodazole-treated centrosomes covering an average range of 5 µm in the slice plane. Merged datasets consist of at least seven subsequent sections for the WT preparations and four for the nocodazole-treated embryos.
Calibration of LM and EM
LM provides a measure of both soluble and polymeric local tubulin concentration. There is no distinction in signal intensity between tubulin polymerized into microtubules and in solution. We calibrated the LM data to distinguish polymeric and soluble fractions as follows. We corrected for the background signal by subtracting the median value outside the cell. Also, we controlled for the auto-fluorescence signal by measuring intensity values in unlabeled N2 WT embryos (n = 26). This signal was always below 10% of the measured intensity and decayed toward the centrosome. The coordinates of the centrosome centers, serving as the origin of our radial profiles, were manually defined by observing the datasets in FIJI in orthogonal views. Radial profiles were extracted in the image plane of the stack, which contained the manually defined center of the centrosome. For each radial position, all values in the circumferential direction were averaged for the half plane away from the spindle. Each radial profile was normalized with the integrand of the respective intensity profile from the center to r = 8 µm (the maximum distance at which we obtained EM data).
Electron tomography provides a quantitative measure of polymerized tubulin as only assembled microtubules are detectable. In total, we segmented the microtubules of eight WT one-cell C. elegans metaphase centrosomes. For registration purposes, we used the center of the mother centriole as the origin of analysis. The spindle axis was estimated either by the coordinates of the opposite mother centriole or, if not available, by manual inspection of the dataset. The segmented microtubules were analyzed with respect to the microtubule density by computing locally the microtubule length per volume. Assuming all microtubules would have the same direction and cover the entire volume, this corresponds to the number of microtubules per area. Microtubules in the mitotic spindle have predominantly 11 protofilaments (Chaaban et al., 2018), which corresponds to ∼1,354 tubulin dimers per micrometer; the corresponding length per volume relates to the molar concentration of polymerized tubulin into microtubules as c/(µM) = 2.248 ρ/(µm/µm3). We estimated the density on a 3D grid with an equidistant spacing of 100 nm. We took the boundary of the datasets into account and averaged the density values in a spherical coordinate system in the circumferential direction with respect to the spindle axis.
To compare the data from electron with those from LM, we had to correct artifacts of the respective methods. To consistently calibrate the light with the EM signal and combine both radial profiles, we compensated the sample shrinkage during the workflow of electron tomography. For this purpose, we manually measured the dimensions of embryo length and width in the maximum projection of confocal stacks before preparation for electron tomography and in serial sections after preparation (n = 9) and estimated a shrinkage factor of ∼27.5%. One possibility to correct for the PSF is to deconvolve the raw LM data. We discarded this method, however, as the PSF is not precisely known and the data are rather noisy. Instead, we convolved the shrinkage-corrected EM dataset with the PSF to compare profiles with the same recording conditions. For the convolution, we used the 3D EM data and convolved them with the Gaussian PSF on a slice that encloses the spindle axis.
To calibrate the LM data, we used the assumption that soluble tubulin is homogenously distributed outside of the centrosome (r > 4 µm) and that the spatial variation there is solely due to the spatial changes of the concentration of polymerized tubulin. To implement this, we calibrated the signal of the LM by minimizing the variance of the computed soluble tubulin in the region from 4 to 8 µm in three reconstructions. The profiles of polymerized tubulin in five additional datasets have a similar profile up to ∼4 µm.
Averaged concentrations
For the averages of total and polymerized tubulin, we used the calibrated profiles and integrated them numerically for the radial ranges as specified. We used spherical coordinates for the two half spheres away from the spindle and cylindrical coordinates for the region between the centrioles. In both cases, we used averaged profiles depending only on the distance to the center of the centrosome, either the radius or the position on the spindle axis with respect to the nearby centrosome. For radii >8 µm, we had only data from LM. Consequently, we report only total concentrations for this region and the entire cell. These averaged concentrations neglect the fraction of organelles and granules present in the cytoplasm that may exclude tubulin.
Drug treatment
For the nocodazole imaging experiment, worms were treated with perm-1 (RNAi) feeding (T01H3.4) for 17–20 h at 20°C to permeabilize the eggshell (Carvalho et al., 2011; Timmons and Fire, 1998). Worms were dissected in osmotically balanced buffer (62% ESF-921 medium; Expression Systems). Embryos were selected and transferred into a microdevice as described in Carvalho et al. (2011). For microtubule depolymerization, the medium within the microdevice was replaced at the desired stage of nuclear envelope breakdown with fresh medium containing a final concentration of 50 µg/ml nocodazole (M1404; Sigma-Aldrich). Imaging could be performed continuously, and the effect of depolymerization was visible 1 min after drug addition. The light-microscopic acquisition was performed with the same settings as for the WT experiment. This allowed us to use the same calibration coefficient as for the WT experiment.
FRAP experiments
FRAP experiments were performed with a Nikon Ti2-E microscope equipped with a Yokogawa spinning disk unit (CSU-W1; 50 µM pinhole size), 100× silicone objective (Nikon CFI SR HP Plan Apochromat Lambda S 100XC Sil, 1.35 NA), 100 mW lasers for λ 488 nm and 561 nm (for imaging of DNA and β-tubulin), an EMCCD (Andor iXon Ultra 888 EMCCD, 1024 × 1024 array, 13 µM pixel size), and a 50 mW stimulation laser used for FRAP (LUN-F λ405nm laser launch). The parameters for imaging were 2 × 2 binning, 30 MHz camera acquisition speed, 20% laser intensity for λ488 nm with 100 ms exposure time, 2% laser intensity for the FRAP 405 nm laser, and an EM gain multiplier of 300. Images were analyzed using Nikon Elements software.
Worms were grown on perm-1 RNAi feeding plates (NGM + 0.1 mM IPTG) for 18 h at 20°C, then dissected in osmotically balanced buffer containing 5 µg/ml nocodazole and 15 µm polystyrene microparticles (74964; Sigma-Aldrich) to prevent squishing when placed between a coverslip and the glass slide.
Data analysis
Data analysis was performed using either the AMIRA software (Zuse Institute Berlin) or MATLAB (R2017b; MathWorks). To reduce a bias resulting from errors in the tracing algorithm, microtubules shorter than 100 nm were excluded from all analyses. Microtubules are very stiff and consequently close to straight lines. For simplicity, we treated them as straight lines to compute the local density. For the fractions of microtubules based on length, microtubules were grouped by the end-to-end length. At a given radial position, the number of microtubules in each group was counted and compared with the total number crossing this radius. The length distribution is well captured with an exponential with an average length of 1.1 µm (Redemann et al., 2017). Extrapolating the length distribution to shorter lengths leads to a possible underestimation of the polymer mass of 0.4%, which is negligible compared with other uncertainties in this study. To provide for an estimate of the confidence in our measurements, we calculated the SD from all EM density profiles and LM intensity profiles, which we propagated to the soluble concentration. Additionally, a lower-bound calibration (which imposes the minimum of the soluble tubulin to zero; Fig. S2) shows that even in this extreme situation, soluble tubulin is enriched 10-fold at the poles, similar to our calibration based on the variance. With this alternative calibration, the overall density is 59% smaller than the estimate based on the variance approach, but the qualitative shape of the profiles is maintained, except a minimum for the soluble tubulin in the region just outside of the centrosome.
Online supplemental material
In Fig. S1, we show measurements of the size of the point spread function of our microscope. In Fig. S2, we provide a lower bound for the concentration profiles of tubulin. In Fig. S3, we show measurements of FRAP in the cell and local polymerization velocity.
Supplementary Material
Acknowledgments
The authors would like to thank Norbert Lindow, Steffen Prohaska, Martin Weigert, Oliver Wüseke, Patrick McCall, and the members of the LM and EM facility at Max Planck Institute of Molecular Cell Biology and Genetics for discussions and technical assistance.
S. Redemann received funding from the Faculty of Medicine Carl Gustav Carus of Technische Universität Dresden (Frauenhabilitationsstipendium). Research in the A.A. Hyman and F. Jülicher groups is supported by the European Commission’s seventh framework program grant Systems Biology of Mitosis (FP7_HEALTH-2009-241548/MitoSys). Research in the T. Müller-Reichert group is supported by funds from the Deutsche Forschungsgemeinschaft (MU 1423/8-1 and 8-2). Research in the J. Brugués group is funded by the Human Frontiers Science Program (CDA00074/2014). Research in the F. Jülicher, A.A. Hyman, and J. Brugués groups is funded by the Deutsche Forschungsgemeinschaft under Germany's Excellence Strategy – EXC-2068 – 390729961 – Cluster of Excellence Physics of Life of Technische Universität Dresden.
The authors declare no competing financial interests.
Author contributions: This work represents a truly collaborative effort. J. Baumgart, M. Kirchner, S. Redemann, A. Bond, J. Woodruff, J.M. Verbavatz, F. Jülicher, T. Müller-Reichert, A.A. Hyman, and J. Brugués contributed significantly to the findings, and regular group discussions guided the development of the ideas presented here. M. Kirchner, S. Redemann, A. Bond, J.M. Verbavatz, and T. Müller-Reichert did the experiments, and J. Baumgart analyzed the data. J. Baumgart, M. Kirchner, S. Redemann, J. Woodruff, T. Müller-Reichert, A.A. Hyman, and J. Brugués wrote the paper with input from the others.
References
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Olson S.K., Gutierrez E., Zhang K., Noble L.B., Zanin E., Desai A., Groisman A., and Oegema K.. 2011. Acute drug treatment in the early C. elegans embryo. PLoS One. 6:e24656 10.1371/journal.pone.0024656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L.B., Zhang X., Ditlev J.A., and Rosen M.K.. 2019. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 363:1093–1097. 10.1126/science.aau6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaaban S., Jariwala S., Hsu C.T., Redemann S., Kollman J.M., Muller-Reichert T., Sept D., Bui K.H., and Brouhard G.J.. 2018. The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth. Dev. Cell. 47:191–204.e8. [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Feng Z., Richens J.H., Baumbach J., Wainman A., Bakshi S.D., Dobbelaere J., Johnson S., Lea S.M., and Raff J.W.. 2014. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell. 28:659–669. 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker M., Jaensch S., Pozniakovsky A., Zinke A., O’Connell K.F., Zachariae W., Myers E., and Hyman A.A.. 2011. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 21:1259–1267. 10.1016/j.cub.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Drenckhahn D., and Pollard T.D.. 1986. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J. Biol. Chem. 261:12754–12758. [PubMed] [Google Scholar]
- Fu J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.F. 1972. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. II. Direct methods. Proc. R. Soc. Lond. B Biol. Sci. 182:89–102. 10.1098/rspb.1972.0068 [DOI] [PubMed] [Google Scholar]
- Hamill D.R., Severson A.F., Carter J.C., and Bowerman B.. 2002. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell. 3:673–684. 10.1016/S1534-5807(02)00327-1 [DOI] [PubMed] [Google Scholar]
- Hannak E., Oegema K., Kirkham M., Gönczy P., Habermann B., and Hyman A.A.. 2002. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J. Cell Biol. 157:591–602. 10.1083/jcb.200202047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T., Alberti S., Diez S., and Hyman A.A.. 2017. Local Nucleation of Microtubule Bundles through Tubulin Concentration into a Condensed Tau Phase. Cell Reports. 20:2304–2312. 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li T., Ems-McClung S.C., Walczak C.E., Prigent C., Zhu X., Zhang X., and Zheng Y.. 2018. Aurora A activation in mitosis promoted by BuGZ. J. Cell Biol. 217:107–116. 10.1083/jcb.201706103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye B., Stiehl O., Foster P.J., Shelley M.J., Needleman D.J., and Furthauer S.. 2018. Measuring and modeling polymer concentration profiles near spindle boundaries argues that spindle microtubules regulate their own nucleation. New J. Phys. 20:055012 10.1088/1367-2630/aac2a5 [DOI] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Mennella V., Agard D.A., Huang B., and Pelletier L.. 2014. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 24:188–197. 10.1016/j.tcb.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Reichert T., Srayko M., Hyman A., O’Toole E.T., and McDonald K.. 2007. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 79:101–119. 10.1016/S0091-679X(06)79004-5 [DOI] [PubMed] [Google Scholar]
- O’Toole E., Greenan G., Lange K.I., Srayko M., and Müller-Reichert T.. 2012. The role of γ-tubulin in centrosomal microtubule organization. PLoS One. 7:e29795 10.1371/journal.pone.0029795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlü N., Srayko M., Kinoshita K., Habermann B., O’toole E.T., Müller-Reichert T., Schmalz N., Desai A., and Hyman A.A.. 2005. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev. Cell. 9:237–248. 10.1016/j.devcel.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Pelletier L., Ozlü N., Hannak E., Cowan C., Habermann B., Ruer M., Müller-Reichert T., and Hyman A.A.. 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14:863–873. 10.1016/j.cub.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A.A., and Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Redemann S., Baumgart J., Lindow N., Shelley M., Nazockdast E., Kratz A., Prohaska S., Brugués J., Fürthauer S., and Müller-Reichert T.. 2017. C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nat. Commun. 8:15288 10.1038/ncomms15288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Weber C.A., Nousch M., Adame-Arana O., Hoege C., Hein M.Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., et al. 2016. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell. 166:1572–1584.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Powers J., Dunn M., Reese K., Malone C.J., White J., Seydoux G., and Saxton W.. 2001. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell. 12:1751–1764. 10.1091/mbc.12.6.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Weber B., Greenan G., Prohaska S., Baum D., Hege H.-C., Müller-Reichert T., Hyman A.A., and Verbavatz J.-M.. 2012. Automated tracing of microtubules in electron tomograms of plastic embedded samples of Caenorhabditis elegans embryos. J. Struct. Biol. 178:129–138. 10.1016/j.jsb.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Weber B., Tranfield E.M., Höög J.L., Baum D., Antony C., Hyman T., Verbavatz J.M., and Prohaska S.. 2014. Automated stitching of microtubule centerlines across serial electron tomograms. PLoS One. 9:e113222 10.1371/journal.pone.0113222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M., Bechstedt S., Chaaban S., and Brouhard G.J.. 2015. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 17:907–916. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., and Hyman A.A.. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 169:1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- Wueseke O., Zwicker D., Schwager A., Wong Y.L., Oegema K., Jülicher F., Hyman A.A., and Woodruff J.B.. 2016. Polo-like kinase phosphorylation determines Caenorhabditis elegans centrosome size and density by biasing SPD-5 toward an assembly-competent conformation. Biol. Open. 5:1431–1440. 10.1242/bio.020990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.