Abstract
Innate and acquired resistance to anti-EGFR therapy (EGFRi) is a major limitation in the treatment of metastatic colorectal cancer (mCRC). Although RAS genes are the most commonly mutated innate and acquired oncogenes in cancer, there are a number of other mechanisms which limit the effectiveness of EGFRi. Patients with innate resistance have been found to contain BRAFV600E mutations, and possibly MET, MEK, PIK3CA, PTEN and HER2 alterations. Meanwhile, BRAFV600E mutations may also be involved in acquired resistance to EGFRi, in addition to EGFR ectodomain mutations, MET alterations, and possibly HER2 amplification. In addition, paracrine effects and cell fate mechanisms of resistance are being increasingly described as contributing to acquired resistance. Utilization of circulating tumor DNA has been paramount in monitoring the dynamic nature of acquired resistance, and has helped to guide treatment decisions, particularly in the EGFRi rechallenge setting. Herein, we provide an in-depth review of EGFRi resistance mechanisms and describe the current therapeutic landscape in the hopes of identifying effective rechallenge strategies.
Introduction
Colorectal cancer (CRC) represents a heterogeneous group of dynamic biologic phenomena with differing sets of genetic events, accompanying immune responses, and influences of exogenous factors, providing a challenge for personalized therapeutic approaches. These personalized treatments most often involve kinase inhibitors or monoclonal antibodies (moAbs) that target specific alterations known to drive the proliferation and survival of cancer cells.(1) In this scenario, the epidermal growth factor receptor (EGFR) family plays a key role in tumor growth and progression by promoting a variety of functions including proliferation, survival, invasion, and immune evasion.(2) These therapies have improved patient outcomes, however despite significant progress in strategies for cancer treatment, their use is limited by the presence of pre-existing innate resistance mechanisms or by the ability of cancer cells to acquire resistance to therapy.(2,3)
Innate Resistance to EGFRi
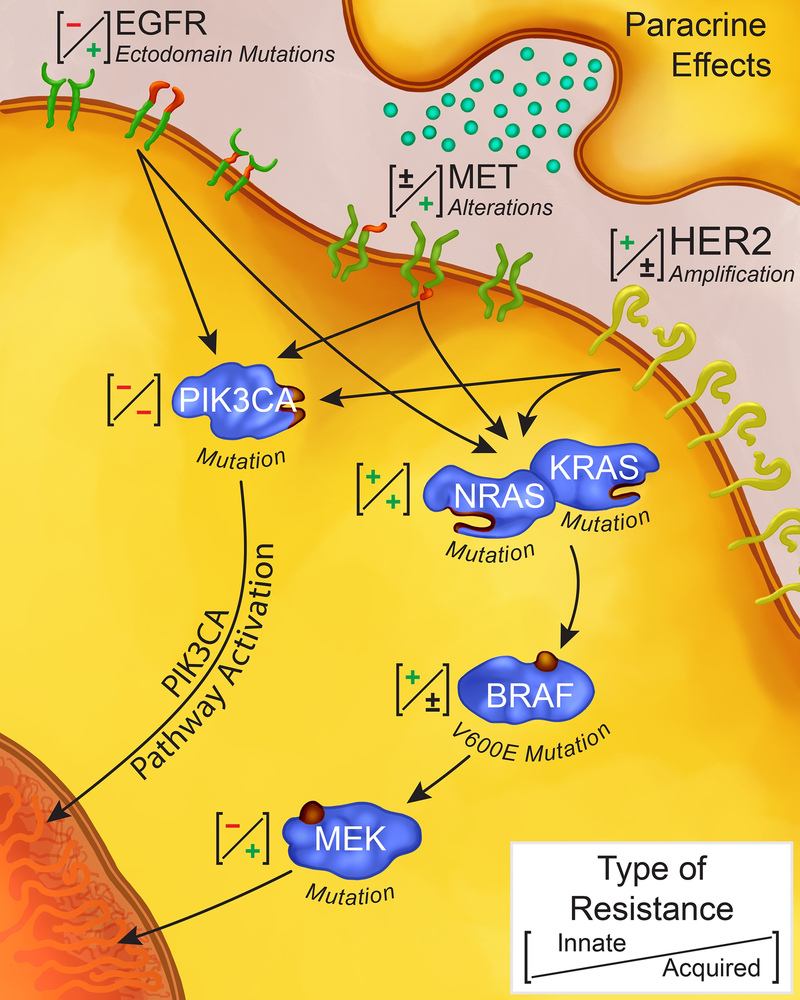
Patients with KRAS/NRAS (RAS) wildtype metastatic colorectal cancer (mCRC) have improved survival when treated with anti-EGFR (EGFRi) moAbs.(4,5) However, these agents do not benefit mCRC patients with oncogenic RAS mutations (Figure 1).(6–9) The KRAS mutation was initially identified in codons 12 and 13 of exon 2 which result in constitutive activation of the RAS-RAF-MEK-ERK (MAPK) pathway.(8–11) Activating mutations in KRAS are detected in approximately 40% of mCRC,(2,3) with good concordance between primary tumors and distant metastases.(12) Expanded RAS mutations in KRAS exon 3 or 4, or in NRAS exon 2, 3, or 4 have also been noted to predict a lack of benefit from EGFRi, increasing the prevalence of all innate RAS mutations to 50–55%.(8,13,14)
Figure 1.
Forms of innate and acquired resistance to anti-EGFR agents in metastatic colorectal cancer. (+) denotes the presence of a resistance mechanism (either innate or acquired). (–) denotes the absence of a resistance mechanism (either innate or acquired).
Though less common, alterations in BRAFV600E result in constitutive activation of the MAPK pathway and may predict lack of benefit from EGFRi.(15) In a retrospective analysis of patients with EGFRi-based chemotherapy, the progression-free survival (PFS) (median 8 vs 26 weeks, HR 3.74, 95% CI 2.44–5.75) and overall survival (OS) (median 26 vs 54 weeks, HR 3.03, 95% CI 1.98–4.63) was shorter in patients with BRAF mutated tumors than those with BRAF wildtype mCRC.(16) In a study evaluating 5-fluorouracil (5-FU) plus irinotecan (FOLFIRI) with panitumumab in the second-line setting, patients with BRAF mutated mCRC had a median PFS of 2.5 months and an OS of 4.7 months, compared with a PFS and OS of 6.9 and 18.7 months, respectively, in patients with BRAF wildtype mCRC.(17) Indeed, a large meta-analysis found limited benefit of adding EGFRi to standard regimens in RAS wildtype/BRAF mutated mCRC patients, however it was not definitively predictive.(18) Non-V600E BRAF mutations have also been identified in mCRC, however appear associated with a much better prognosis than V600E mutations, which some attribute to a lack constitutive activation of MAPK signaling.(19,20) However, in our review of over 2000 patients with mCRC, we identified only 11 non-V600E mutated BRAF tumors and none of these responded to EGFRi.(21) Others have shown mixed effect of EGFRis and interpretation of reports in these non-V600E mutated tumors is difficult given small samples sizes and differences in downstream activity of various BRAF mutations.(22)
PIK3CA mutations may also contribute to innate resistance to EGFRi. Mutations occur at a prevalence of 10%–20%, and are mainly located at hot spots in exons 9 and 20, resulting in constitutive activity of PI3K- activating downstream AKT/mTOR signaling.(23) Retrospective studies have demonstrated that PIK3CA mutations in exon 20 may be associated with lack of response to EGFRi in mCRC.(16,24) However, PIK3CA and RAS mutations co-exist and may have confounded the initial studies. Subsequent studies have failed to definitively demonstrate a role of PIK3CA in EGFRi resistance. In the same pathway, loss of function of PTEN that occurs through mutations, deletions or transcriptional silencing is present in 30 % of sporadic CRCs.(25) PTEN inactivation has been associated with non-responsiveness to anti-EGFR moAbs in mCRC patients in several studies, however other studies have failed to show that loss is anything more than prognostic(25–27) PTEN has been shown to play a role in modulating EGFR intracellular trafficking and degradation suggesting a plausible mechanism for inducing resistance.(28)
Further, innate resistance to EGFRi may be secondary to amplification of HER2 found in 3–5% of RAS wildtype mCRC patients.(29,30) Retrospective and in-vitro analyses have suggested limited activity of anti-EGFR in HER2-amplified tumors, although not universally demonstrated.(30,31) A recent review of the PFS of patients on the HERACLES trial during their EGFRi by Bregni et al. showed contrary evidence refuting the use of HER2 amplification as a biomarker and highlights the controversy and challenges in interpreting information on rare subtypes.(32) Several ongoing (NCT03384940, NCT03365882) and completed clinical studies with HER2 targeted agents after failure of EGFRi will hopefully provide patient cohorts to validate these findings.(33) HER2 and ERBB3 mutations have also been noted in CRC, however there is still limited data regarding their implications.(34) HER2 but not ERBB3 mutations appear associated with a negative prognosis, and unclear association EGFRi benefit, however unlike breast cancer where HER kinase inhibition has therapeutic benefit, the SUMMIT trial failed to show any utility of neratinib in HER2/ERBB3 mutated mCRC, suggesting a distinct biology from breast cancer.(35,36) These HER2-activating mutations produced resistance to the EGFR moAbs, cetuximab or panitumumab, when transfected into two cetuximab-sensitive CRC cell lines 57 providing pre-clinical rationale that they may be associated with EGFRi resistance.
The MET gene, which encodes the tyrosine kinase receptor for Hepatocyte Growth Factor (HGF), has been found to have a role in several tumor types. Gene amplification, overexpression, activating mutations or autocrine stimulation cause constitutive activation of MET.(37) MET(38) amplification has been proposed as an additional biomarker of innate resistance to EGFRi. However, these mechanisms may not abrogate all benefit from EGFRi as is seen with the absolute resistance noted from RAS mutations. MET amplifications have only been identified in approximately 1% of untreated mCRC.(38) Preclinical data has shown that xenograft tumors carrying MET amplification did not respond to EGFRi, and this alteration was mutually exclusive with KRAS/BRAF/NRAS, and PIK3CA mutations, as well as HER2 amplification.(38) However, the prevalence is very rare, precluding more definitive assessment of its role in innate resistance.(39)
In addition, important biologic differences based on primary tumor location have been identified. Right-sided tumors, even when controlling for stage and tumor size, have a worse prognosis.(40) Recently, retrospective reviews of multiple trials utilizing EGFRi in various lines of therapy have demonstrated that right-sided tumors appear less responsive to EGFRi.(41) This has changed clinical decision making, as planned post-hoc analyses of the CALGB 80405 and FIRE-3 studies, among others, demonstrated significant decreases in OS between patients with right-or left-sided RASWT tumors depending on which biologic was received in the first-line setting.(41–43) Patients with right-sided tumors had improved OS if bevacizumab was combined with a doublet, while patients with left-sided tumors derived more benefit from doublet plus EGFRi. The mechanism of this distinction is not well defined, but right-sided primary CRCs are known to have distinct pathobiology and characteristics, including higher rates of BRAF mutation, microsatellite instability (MSI-high), and high CpG island methylator phenotype (CIMP-high).(44) While there are multiple potential mechanisms, it is of note that expression of EGFR ligands and AREG and EREG differ between right-and left-sided tumors, in part due to their epigenetic regulation and association with CIMP-high tumors.(45) Further prospective studies are warranted with retrospective analyses ongoing to further characterize the key biological variables responsible for differences in response to EGFRi based on anatomy.
Acquired Resistance to EGFRi
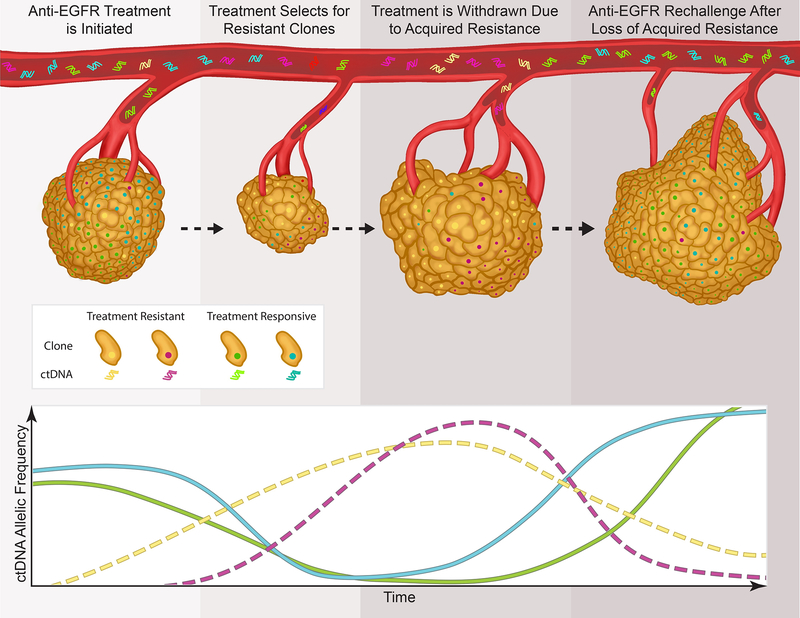
Among patients who initially respond to EGFRi, acquired mutations ultimately develop and result in secondary resistance (Figure 1). Growing utilization of plasma circulating tumor DNA (ctDNA) testing has allowed for the non-invasive detection of heterogeneous molecular abnormalities which result in the evolution of resistance to targeted therapies in mCRC(5,46–48) EGFR blockade results in evolutionary pressure and the outgrowth of subclonal populations of cells with resistance mechanisms.(49) Concurrent selection and expansion of multiple tumor subclones results in polyclonal mechanisms of resistance that drive tumor progression (Figure 2).(50,51) These data suggest that overcoming resistance will require a strategy capable of surmounting multiple heterogeneous resistance mechanisms between different tumor subclones in the same patient. The mechanisms by which tumors develop acquired resistance to therapy can typically be subcategorized into mechanisms which include activation of bypass signaling pathways, secondary alterations in the EGFR receptor, and adaptive or cell fate changes.(1,3)
Figure 2.
Selection, expansion, and loss of multiple tumor subclones at time of treatment and withdrawal of anti-EGFR highlights potential use of anti-EGFR re-challenge strategies.
MAP-Kinase Pathway Alterations as Mechanisms of Acquired Resistance to EGFRi
The most common mechanism of acquired resistance to EGFRi results from the activation of signaling pathways that “bypass” the drug target to maintain survival and proliferation.(16) Among these, RAS mutations, in addition to being a key driver of innate resistance to EGFRi antibodies in mCRC, also play a vital role in acquired resistance.(16,52–54) An estimated 50% of patients with acquired resistance will have a detectable secondary RAS mutation.(16,52,54) Acquired mutations in RAS most commonly occur in exons 3–4 (67% of NRAS and 50% of KRAS mutations versus 3.4% and 8.2%, respectively, in innate resistance).(55,56) An additional critical feature is that greater than one alteration in RAS has been frequently identified in the same patient after treatment with EGFRi.(47,48) Modeling suggests that some RAS mutations are pre-existing in the tumor but below the threshold of detection by standard assays.(47,48,57) By ctDNA, rare RAS clones detectable prior to EGFRi are present in upwards of 38% of patients, however clinical responses are still seen, reiterating the fact that these subclones may not be clinically significant in the untreated tumor.(58,59)
Another mechanism implicated in activation of EGFR downstream pathway and in consequently EGFRi resistance is promoter-specific DNA methylation. While methylation of direct effectors of this signaling pathway has not been reported, promoter methylation of RASSF1 and RASSF2 occurs in as many as 80% of CRC tumors.(60,61) RASSF1 and RASSF2 are modulators of the growth inhibitory and pro-apoptotic effects of active RAS, and consequently their inactivation is thought to promote RAS-driven tumorigenesis and could have an impact on EGFRi response. However, additional studies are still required to definitively establish its tumor-suppressor activity in CRC. Other epigenetic phenomena may also be important predictors for resistance to EGFR. Many studies have identified a subset of microRNAs (miRNAs) associated with a lack of benefit from anti-EGFR therapy, however few have robust external validation.(62,63) None of these miRNAs have been applied clinically to date, however overexpression of several, such as miR-31–5p, have been reproducibly associated with a lack of benefit from EGFRi in several studies.(64,65)
Another mechanism of acquired resistance includes mutations in the EGFR receptor(55,66) or activation of parallel receptor tyrosine kinases (RTKs).(24,38) An acquired mutation in the extracellular domain of EGFR (S492R) has been showed to cause acquired resistance to cetuximab in mCRC.(66) This mutation reduces the affinity of the ligand to the receptor and interferes with binding to cetuximab. Subsequent work defined a number of other ECD mutations, many of which appear to result in cross- resistance to both EGFRis.(46,47,67,68) The role of EGFR ECD mutations in driving resistance to EGFRi has now been documented through ctDNA analysis by several groups, and continues to be an active area of investigation in rechallenge studies.(46,47,55,66–69) In contrast to RAS mutations, ectodomain (ECD) mutations are almost never seen in untreated tumors.(47,66,68)
There is emerging evidence to suggest that alterations affecting the extracellular domain of EGFR could be also due to different mechanisms besides phosphorylation and glycosylation that need to be better explored. Liao HW discovered in their work that specific extracellular Arg methylations of EGFR render cancer cells resistant to cetuximab antibody therapy.(70) In particular, they reported a post-translational arginine methylation on the extracellular domain of EGFR by protein arginine methyltransferase (PRMT) 1 at R198 and R200 that resulted in increased ligand binding to promote EGFR receptor dimerization and activation, and alters EGFR signaling.(70)
Growth-factor signaling pathway upregulation of alternative and compensatory signaling cascades through receptors other than EGFR has also been noted to cause resistance to EGFRi. In particular, HER2 amplification has been suggested as both an intrinsic, as well as an acquired mechanism of resistance.(31,71) One explanation may be that pre-existing infrequent HER2-amplified clones may expand under the selective pressure of EGFRi, leading to disease progression. Similarly, MEK overexpression has been associated with reduced benefit from anti-EGFR therapy and in-vitro experiments demonstrate that combined EGFRi with MEKi can abrogate this resistance.(72,73) MEK mutations have been identified in colorectal cancer occurring at a low frequency (2.5% in TCGA) and may be a further mechanism of resistance, though evidence for this is weak to date.(74–76) These mutations also develop in BRAF mutant mCRC treated with targeted therapy, providing further evidence that downstream mutations are a plausible mechanism of acquired resistance.(77,78)
MET and its ligand HGF have been implicated in acquired resistance to EGFRi.(38,79) Further, in mCRC and other tumor types, MET amplification has been found to arise in tumors with pre-existing clones of MET-amplified cells which undergo positive selection.(38,80) During treatment with EGFRi, the MET-amplified cells have then been shown to become a dominant population of cells, ultimately decreasing the efficacy of further EGFRi.(38)
Activation of the PIK3CA/AKT/mTOR signaling pathway has also been implicated as a potential mechanism of acquired resistance to EGFRi.(46) However, unlike in metastatic melanoma patients who develop acquired resistance to BRAF inhibitors,(81) functionally relevant variants that cause EGFRi resistance via this pathway are relatively rare in CRC.(46) This mechanism has more definitely been implicated as a cause of acquired resistance to HER2 inhibition.
Single nucleotide polymorphisms (SNPs) in candidate genes associated with regulating the EGFR pathway communication also appeared to be of predictive value in several reports.(82) In particular, Stintzing et al have identified SNPs in genes involved in EGFR turnover that predicted clinical outcome in cetuximab-treated mCRC patients.(82) EGFR recycling could be an interesting mechanism of secondary resistance to cetuximab in mCRC and is also a putative mechanism for PTEN induced EGFRi resistance.(28,82) Further studies are needed to validate these preliminary data.
Paracrine Effects
Several groups have found that paracrine mechanisms arising from resistant clones can propagate resistance in neighboring cells without genetic drivers of resistance. In several solid tumors, cancer cells produce growth factors that sustain resistance to targeted therapies.(83) In mCRC, overexpression of a TGF-α induced EGFR–MET interaction, with subsequent MET phosphorylation and activation of downstream effectors in an EGFR-independent manner is a known acquired resistance mechanism.(84) These growth factors may have cell-specific effects or may cause effects on nearby cells. In preclinical work, tumor cells initially sensitive to cetuximab react to EGFR blockade by increasing secretion of EGFR ligands such as TGF-α and amphiregulin.(85) Consistent with this, increased levels of circulating EGFR ligands in the plasma of patients at the time of radiologic progression on cetuximab and irinotecan, suggest their potential role as a mechanism of acquired resistance to treatment.(86) This suggests that increased production of EGFR ligands by cetuximab-resistant derivatives can maintain protection of sensitive cells, thus sustaining their growth.(85) This paracrine protective mechanism may be therapeutically exploitable but demonstrates the challenges of tumor heterogeneity stemming not only from adaptive genetic events, but also microenvironmental interactions.
Cell Fate Mechanisms of Resistance
In addition to these mechanisms, adaptive changes in differentiation status and cell fate are widely associated with resistance in cancer cells. Epithelial-to-mesenchymal transition (EMT) is a complex biological process wherein epithelial cells procedurally lose their original morphology and simultaneously acquire mesenchymal characteristics.(87) It remains unclear whether EMT is a driver of EGFRi resistance.(87) This data needs additional evaluation in future studies in order to further understand the correlation between EMT and acquired resistance to EGFRi.
Dynamic Nature of Acquired Resistance
Growing utilization of ctDNA testing(48) has allowed for the non-invasive detection of heterogeneous molecular alterations which drive the evolution of resistance to targeted therapies in mCRC.(5,48,49,55,57,68,88) Moreover, because ctDNA measures fragments of DNA shed by malignant cells throughout the body, it allows detection of resistance mechanisms emerging concurrently in distinct metastatic lesions and tumor subclones.(89,90) This reassessment for resistance can be repeated easily with serial lab draws.
Khan and colleagues have demonstrated that serial ctDNA analysis can predict the time to treatment failure using mathematical modeling.(91) They noted that resistance mutations were detected in the blood up to several months prior to the identification of radiographic progression.(54,57,58,91) Based on these findings, they developed a mathematical model to identify the emergence of resistance, knowing that the tumor is composed of two separate populations of cells; those that are treatment-sensitive, and those that are treatment-resistant, both of which have determined growth and death rates. Using serial ctDNA assessments, this allowed for the prediction of time until radiographic relapse with reasonable accuracy.
Serial ctDNA analysis may also be used to predict the loss of resistance mechanisms; an important determinant of potential future therapeutic options. Our group has previously shown that in the absence of continued selective pressure from EGFRi, the relative prevalence of RAS mutated and EGFR mutated clones declines.(92) RASMT and EGFRMT alleles exponentially decay with a cumulative half-life of approximately 4 months.(5) These data are consistent with prior reports of clinical benefit with EGFRi rechallenge(93,94) and will help guide the timing of rechallenge therapies in the future. These findings provide strong support for the feasibility and validity of genomic profiling of known acquired resistance mutations to EGFRi by ctDNA to predict clonal decay, and provide strong support for rechallenge with EGFRi.
Clinical Results from EGFRi Rechallenge Studies to Date
Several prospective clinical trials examining EGFRi rechallenge are currently underway. Some results, albeit with small sample sizes, have been published to date, however results have been inconsistent. These varied results may be due to the heterogeneous prognosis of patients who are able to proceed through several lines of standard therapy and still be well enough to enroll on clinical trials. Another explanation for the discrepant results arises from the rules each trial uses to identify the population to rechallenge. Trials have used a mixture of time criteria and ctDNA to select patients for inclusion, which creates a challenge in interpreting results (see Table 1). In one trial, after an EGFRi treatment-free interval of 6 months, one group identified a response rate (RR), stable disease (SD) and disease control rate of 54%, 36%, and 90%, respectively with EGFRi rechallenge. All patients were KRASWT (codons 12 and 13), with prior response to EGFRi defined as a clinical benefit (confirmed stable disease (SD) for at least 6 months or clinical response) after a line of cetuximab plus irinotecan-based therapy followed by progressive disease. ctDNA was not tested in this study.(94) Meanwhile, in the recent CRICKET single arm phase 2 study, ctDNA was retrospectively evaluated at enrollment. Patients with tissue-based RASWT and BRAFWT with at least a partial response (PR) and progression-free survival (PFS) of at least 6 months to first-line cetuximab plus irinotecan were studied and found to have a RR, SD, and DCR rate of 21%, 32%, and 54%, respectively to EGFRi rechallenge. There was a statistically significant correlation between benefit from the first EGFRi and rechallenge.(95) All patients achieving a PR were ctDNA RASWT prior to rechallenge with EGFRi and these patients experienced a significantly longer PFS compared to ctDNA RASMT patients (4 vs 1.9 months). In other abstracts, EGFRi rechallenge strategies had RR ranging from 8–43% in these preliminary reports, and DCR ranging from 40–53% (Table 1).(96–100) Notably, a dynamic evolution of KRAS alterations between first administration and rechallenge with EGFRi was demonstrated by ctDNA in only one of the above mentioned trials.(57) Thus, there is marked heterogeneity in terms of efficacy across different trials exploring rechallenge strategies, and further large scale trials such as FIRE-4 (NCT02934529), and PULSE (NCT03765736) with serial ctDNA testing are needed to identify the best treatment strategies for these patients.
Table 1.
Summary and Clinical Results from Anti-EGFR Rechallenge Studies to Date
| Study | NCTN ID | N | Population | ctDNA | Treatment | ORR/DCR |
|---|---|---|---|---|---|---|
| Wadlow et al.(105) | NCT00842257 | 20 | Prospective KRAS wild type with prior failure of cetuximab |
Not assessed | panitumumab | 0%/45% |
| Pietrantonio et al.(106) | n/a | 30 | Retrospective KRAS wild type with prior failure of cetuximab |
Not assessed | Panitumumab | 30%/67% |
| Saif et al.(107) | n/a | 15 | Retrospective KRAS wild type with prior failure of cetuximab |
Not assessed | Panitumumab | 27%/55% |
| Santini et al.(94) | n/a | 39 | Prospective Prior EGFRi PFS ≥6 months and EGFRi free ≥6 months |
Not assessed | cetuximab + irinotecan | 54%/90% |
|
CRICKET Cremolini et al.(95) |
NCT02296203 | 28 | Prospective Prior EGFRi partial response AND PFS ≥6 months with an intervening line of therapy |
Integrated | cetuximab + irinotecan | 21%/54% |
| Tsuji et al.(96) | n/a | 36 | Prospective Prior EGFRi PFS ≥6 months with an intervening line of therapy |
Not assessed | cetuximab + irinotecan | 3%/56% |
| Nogueira et al.(96) | n/a | 15 | Retrospective Prior EGFRi with an intervening line of therapy |
Not assessed | EGFRi +/− chemotherapy | 13%/40% |
| Osawa et al.(98) | n/a | 33 | Prospective Refractory to 5-FU, irinotecan, oxaliplatin, with last EGFRi ≥ 16 weeks prior |
Integrated | cetuximab + irinotecan | 16%/56% |
|
TRECC Karani et al.(100) |
n/a | 68 | Retrospective Prior EGFRi with intervening line of therapy |
Not assessed | EGFRi +/− chemotherapy | 43%/ unknown |
| FIRE-4 Heinemann et al. | NCT02934529 | 550 | Prospective Prior FOLFIRI plus cetuximab PFS ≥6 months and intervening line of treatment |
Integrated | Randomized: cetuximab + irinotecan vs standard of care | Not yet reported |
| PULSE Strickler et al. | NCT03765736 | 500 | Prospective Umbrella protocol for patients refractory to 5-FU, irinotecan, oxaliplatin, EGFRi, and immunotherapy if MSI-H |
Integral | Multi-armed but re-challenge arm is panitumumab | Not yet reported |
| Parseghian et al. | NCT03087071 | 84 | Prospective Refractory to anti-EGFR with 3 cohorts: cfDNA detected EGFR S492R mutation OR RAS/BRAF mutation OR no cfDNA mechanism of resistance |
Integral | Panitumumab +/− trametinib if ctDNA RAS/BRAF mutation | Not yet reported |
Future Goals and Remaining Questions
ctDNA testing(48) has allowed for the detection of heterogeneous molecular alterations which underlie the evolution of resistance to targeted therapies in mCRC.(5,46–48,71) Such analyses have uncovered the role of acquired RAS and other subclonal mutations in the development of resistance to EGFRi. These findings have paved the way for ongoing clinical efforts to define the efficacy of rechallenge therapies with EGFRi. ctDNA analysis may correlate better with sensitivity of an individual patient and act as a rapid surrogate of tumor response, allowing testing to be timed with the need for an alternate treatment regimen.(101) Finally, it may allow for higher utilization and earlier access to clinical trials.
However, limitations of ctDNA testing remain. Cost is a concern, in addition to the imprecise ability to understand subclonality and trends with a single assay. Optimally, serial testing would be done. Further, the threshold for positivity of a resistance clone and treatment is not yet established and likely not an absolute, but rather an indication of gradation of benefit with reduction in subclonal burden. Premature treatment discontinuation of EGFRi based on ctDNA may sacrifice duration of tumor control, especially where available therapies are limited.
Thus, there are many remaining questions. First, is there a better way to intermittently dose EGFRi to prevent or mitigate resistance? In the frontline setting, the Phase 2 COIN-B study found no significant difference in 10 month or median failure free survival with intermittent versus continuous dosing of cetuximab.(102) However, a similar analysis has not been done in the rechallenge setting. Further, paracrine protective mechanisms could explain why, in the complex scenario of a heterogeneous disease where different subclones exist in the same patient, previously sensitive CRC cells can successfully grow despite EGFRi when they are present with their resistant derivatives.(31,38,47,55,68,92) In fact, one study observed that patients previously treated with EGFRi had an average of 5 different resistant alterations. This emphasizes the limitations of single drug therapy to overcome a broad array of resistance mechanisms, particularly as these mutations often occur in multiple functionally distinct targets in individual patients.(47) These findings support the use of ctDNA analysis to define the genomic landscape of patients with mCRC and to guide more targeted therapies in the setting of therapeutic resistance. However, much of the data to date is based on preclinical work and is yet to be examined in prospective studies.
Finally, there is a great deal of interest in methods of intercepting acquired resistance to EGFRi. The MAPK pathway is a key regulator of cellular proliferation and survival. Extracellular signal regulated kinase (ERK) is a downstream member of this pathway and plays a central role in transmitting extracellular signals from activated RTKs such as EGFR/FGFR/PDGFR/VEGFR. The feasibility of pharmacologic inhibition of the MAPK pathway in cancer has been shown through the success of BRAF and MEK inhibitors in the treatment of various cancer subtypes, especially patients who harbor BRAFMT. In mCRC, one group demonstrated that regardless of the mutation that confers resistance to EGFRi, the outcome is always sustained activation of MEK and ERK.(103) More recently, post-progression ctDNA from patients with mCRC treated with a BRAF inhibitor and EGFRi demonstrated clones that reactivated MAPK.(78,104) These data have provided a rationale for the use of combined MAPK inhibition and EGFRi to overcome resistance to EGFR antibodies. Blockade of ERK1/2 directly is postulated to overcome many of the current limitations of EGFRi in patients with acquired RAS mutations, and such studies are planned. Thus, these mechanisms may be partially analogous but further clinical data is needed. There are several ongoing phase 1/2 trials examining the role of MEK inhibition with EGFRi (NCT03087071, NCT02857270).
Several randomized clinical trials are currently ongoing to further clarify acquired resistance mechanisms to EGFRi, and will certainly assist clinicians in timing of rechallenge therapies, as well as in the discovery of therapeutic efforts to reverse resistance to EGFRi(5). Moreover, data to date support the use of ctDNA profiling to guide treatment of patients with mCRC and to track clonal evolution. Further large-scale prospective trials, in addition to education and dissemination of existing best practices is critical.
ACKNOWLEDGEMENTS
The authors acknowledge Ryan Nini for help in the design of the figures.
Financial Support: C.P was funded by a Conquer Cancer Foundation Merit Award for this work. S.K. was funded by NIH Grant [grant number R01 CA187238].
Footnotes
No conflicts of interest were disclosed by the authors.
REFERENCES
- 1.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358(11):1160–74 doi 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 2.Troiani T, Napolitano S, Della Corte CM, Martini G, Martinelli E, Morgillo F, et al. Therapeutic value of EGFR inhibition in CRC and NSCLC: 15 years of clinical evidence. ESMO Open 2016;1(5):e000088 doi 10.1136/esmoopen-2016-000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sforza V, Martinelli E, Ciardiello F, Gambardella V, Napolitano S, Martini G, et al. Mechanisms of resistance to anti-epidermal growth factor receptor inhibitors in metastatic colorectal cancer. World J Gastroenterol 2016;22(28):6345–61 doi 10.3748/wjg.v22.i28.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loree JM, Kopetz S. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol 2017;9(8):551–64 doi 10.1177/1758834017714997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, et al. Anti-EGFR Resistant Clones Decay Exponentially After Progression: Implications for Anti-EGFR Re-challenge. Ann Oncol 2018. doi 10.1093/annonc/mdy509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26(10):1626–34 doi 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 7.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359(17):1757–65 doi 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 8.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369(11):1023–34 doi 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 9.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360(14):1408–17 doi 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 2015;33(7):692–700 doi 10.1200/JCO.2014.59.4812. [DOI] [PubMed] [Google Scholar]
- 11.Tabernero J, Van Cutsem E, Diaz-Rubio E, Cervantes A, Humblet Y, Andre T, et al. Phase II trial of cetuximab in combination with fluorouracil, leucovorin, and oxaliplatin in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 2007;25(33):5225–32 doi 10.1200/JCO.2007.13.2183. [DOI] [PubMed] [Google Scholar]
- 12.Peeters M, Kafatos G, Taylor A, Gastanaga VM, Oliner KS, Hechmati G, et al. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur J Cancer 2015;51(13):1704–13 doi 10.1016/j.ejca.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Ciardiello F, Normanno N, Maiello E, Martinelli E, Troiani T, Pisconti S, et al. Clinical activity of FOLFIRI plus cetuximab according to extended gene mutation status by next-generation sequencing: findings from the CAPRI-GOIM trial. Ann Oncol 2014;25(9):1756–61 doi 10.1093/annonc/mdu230. [DOI] [PubMed] [Google Scholar]
- 14.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol 2015;26(1):13–21 doi 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 15.Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer 2015;112(12):1888–94 doi 10.1038/bjc.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11(8):753–62 doi 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 17.Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, et al. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol 2010;28(31):4706–13 doi 10.1200/JCO.2009.27.6055. [DOI] [PubMed] [Google Scholar]
- 18.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 2015;51(5):587–94 doi 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 19.Jones JC, Renfro LA, Al-Shamsi HO, Schrock AB, Rankin A, Zhang BY, et al. (Non-V600) BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J Clin Oncol 2017;35(23):2624–30 doi 10.1200/JCO.2016.71.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Jones JC, Kipp BR, Grothey A. Activity of EGFR antibody in non-V600 BRAF mutant metastatic colorectal cancer. Ann Oncol 2019;30(1):147–9 doi 10.1093/annonc/mdy477. [DOI] [PubMed] [Google Scholar]
- 21.Johnson B, Loree JM, Morris VK, Dasari A, Pant S, Raghav KPS, et al. Activity of EGFR inhibition in atypical (non-V600E) BRAF-mutated metastatic colorectal cancer. Journal of Clinical Oncology 2019;37(4_suppl):596– doi 10.1200/JCO.2019.37.4_suppl.596. [DOI] [Google Scholar]
- 22.Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548(7666):234–8 doi 10.1038/nature23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francipane MG, Lagasse E. mTOR pathway in colorectal cancer: an update. Oncotarget 2014;5(1):49–66 doi 10.18632/oncotarget.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69(5):1851–7 doi 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 25.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol 2009;20(1):84–90 doi 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 26.Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol 2009;27(16):2622–9 doi 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 27.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 2009;27(35):5924–30 doi 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 28.Shinde SR, Maddika S. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun 2016;7:10689 doi 10.1038/ncomms10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciardiello F, Normanno N. HER2 signaling and resistance to the anti-EGFR monoclonal antibody cetuximab: a further step toward personalized medicine for patients with colorectal cancer. Cancer Discov 2011;1(6):472–4 doi 10.1158/2159-8290.CD-11-0261. [DOI] [PubMed] [Google Scholar]
- 30.Raghav K, Loree JM, Morris JS, Overman MJ, Yu R, Meric-Bernstam F, et al. Validation of HER2 Amplification as a Predictive Biomarker for Anti–Epidermal Growth Factor Receptor Antibody Therapy in Metastatic Colorectal Cancer. JCO Precision Oncology 2019(3):1–13 doi 10.1200/po.18.00226. [DOI] [PubMed] [Google Scholar]
- 31.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1(6):508–23 doi 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 32.Bregni G, Sciallero S, Sobrero A. HER2 Amplification and Anti-EGFR Sensitivity in Advanced Colorectal Cancer. JAMA Oncol 2019. doi 10.1001/jamaoncol.2018.7229. [DOI] [PubMed] [Google Scholar]
- 33.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17(6):738–46 doi 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 34.Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 2015;5(8):832–41 doi 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 2018;554(7691):189–94 doi 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loree JM, Bailey AM, Johnson AM, Yu Y, Wu W, Bristow CA, et al. Molecular Landscape of ERBB2/ERBB3 Mutated Colorectal Cancer. J Natl Cancer Inst 2018;110(12):1409–17 doi 10.1093/jnci/djy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov 2008;7(6):504–16 doi 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 38.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013;3(6):658–73 doi 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghav K, Bailey AM, Loree JM, Kopetz S, Holla V, Yap TA, et al. Untying the gordion knot of targeting MET in cancer. Cancer Treat Rev 2018;66:95–103 doi 10.1016/j.ctrv.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, et al. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA 2017;317(23):2392–401 doi 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold D, Lueza B, Douillard JY, Peeters M, Lenz HJ, Venook A, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol 2017;28(8):1713–29 doi 10.1093/annonc/mdx175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brule SY, Jonker DJ, Karapetis CS, O’Callaghan CJ, Moore MJ, Wong R, et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer 2015;51(11):1405–14 doi 10.1016/j.ejca.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 43.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 2014;15(10):1065–75 doi 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 44.Loree JM, Pereira AAL, Lam M, Willauer AN, Raghav K, Dasari A, et al. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clin Cancer Res 2018;24(5):1062–72 doi 10.1158/1078-0432.CCR-17-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MS, McGuffey EJ, Morris JS, Manyam G, Baladandayuthapani V, Wei W, et al. Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br J Cancer 2016;114(12):1352–61 doi 10.1038/bjc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietrantonio F, Vernieri C, Siravegna G, Mennitto A, Berenato R, Perrone F, et al. Heterogeneity of Acquired Resistance to Anti-EGFR Monoclonal Antibodies in Patients with Metastatic Colorectal Cancer. Clin Cancer Res 2017;23(10):2414–22 doi 10.1158/1078-0432.CCR-16-1863. [DOI] [PubMed] [Google Scholar]
- 47.Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov 2018;8(2):164–73 doi 10.1158/2159-8290.CD-17-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thierry AR, Pastor B, Jiang ZQ, Katsiampoura AD, Parseghian C, Loree JM, et al. Circulating DNA Demonstrates Convergent Evolution and Common Resistance Mechanisms during Treatment of Colorectal Cancer. Clin Cancer Res 2017;23(16):4578–91 doi 10.1158/1078-0432.CCR-17-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 2014;4(11):1269–80 doi 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 50.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov 2016;6(2):147–53 doi 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366(10):883–92 doi 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol 2009;6(9):519–27 doi 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 53.Smith G, Bounds R, Wolf H, Steele RJ, Carey FA, Wolf CR. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer 2010;102(4):693–703 doi 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486(7404):532–6 doi 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arena S, Bellosillo B, Siravegna G, Martinez A, Canadas I, Lazzari L, et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clin Cancer Res 2015;21(9):2157–66 doi 10.1158/1078-0432.CCR-14-2821. [DOI] [PubMed] [Google Scholar]
- 56.Guo F, Gong H, Zhao H, Chen J, Zhang Y, Zhang L, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep 2018;8(1):6076 doi 10.1038/s41598-018-24306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21(7):827 doi 10.1038/nm0715-827b. [DOI] [PubMed] [Google Scholar]
- 58.Diaz LA Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486(7404):537–40 doi 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6(224):224ra24 doi 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandes MS, Carneiro F, Oliveira C, Seruca R. Colorectal cancer and RASSF family--a special emphasis on RASSF1A. Int J Cancer 2013;132(2):251–8 doi 10.1002/ijc.27696. [DOI] [PubMed] [Google Scholar]
- 61.van Engeland M, Roemen GM, Brink M, Pachen MM, Weijenberg MP, de Bruine AP, et al. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene 2002;21(23):3792–5 doi 10.1038/sj.onc.1205466. [DOI] [PubMed] [Google Scholar]
- 62.Cappuzzo F, Sacconi A, Landi L, Ludovini V, Biagioni F, D’Incecco A, et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin Colorectal Cancer 2014;13(1):37–45 e4 doi 10.1016/j.clcc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Manceau G, Imbeaud S, Thiebaut R, Liebaert F, Fontaine K, Rousseau F, et al. Hsa-miR-31–3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res 2014;20(12):3338–47 doi 10.1158/1078-0432.CCR-13-2750. [DOI] [PubMed] [Google Scholar]
- 64.Igarashi H, Kurihara H, Mitsuhashi K, Ito M, Okuda H, Kanno S, et al. Association of MicroRNA-31–5p with Clinical Efficacy of Anti-EGFR Therapy in Patients with Metastatic Colorectal Cancer. Ann Surg Oncol 2015;22(8):2640–8 doi 10.1245/s10434-014-4264-7. [DOI] [PubMed] [Google Scholar]
- 65.Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, Zagatti B, Radova L, Svoboda M, et al. MicroRNA expression profiling identifies miR-31–5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget 2015;6(36):38695–704 doi 10.18632/oncotarget.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med 2012;18(2):221–3 doi 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 67.Peeters M, Price T, Boedigheimer M, Kim TW, Ruff P, Gibbs P, et al. Evaluation of Emergent Mutations in Circulating Cell-Free DNA and Clinical Outcomes in Patients with Metastatic Colorectal Cancer Treated with Panitumumab in the ASPECCT Study. Clin Cancer Res 2018. doi 10.1158/1078-0432.CCR-18-2072. [DOI] [PubMed] [Google Scholar]
- 68.Van Emburgh BO, Arena S, Siravegna G, Lazzari L, Crisafulli G, Corti G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun 2016;7:13665 doi 10.1038/ncomms13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015;526(7572):263–7 doi 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest 2015;125(12):4529–43 doi 10.1172/JCI82826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med 2011;3(99):99ra86 doi 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Troiani T, Napolitano S, Vitagliano D, Morgillo F, Capasso A, Sforza V, et al. Primary and acquired resistance of colorectal cancer cells to anti-EGFR antibodies converge on MEK/ERK pathway activation and can be overcome by combined MEK/EGFR inhibition. Clin Cancer Res 2014;20(14):3775–86 doi 10.1158/1078-0432.CCR-13-2181. [DOI] [PubMed] [Google Scholar]
- 73.Bardelli A, Misale S, Arena S, Siravegna G, Lamba S, Bencardino K, et al. Concomitant blockade of EGFR and MEK overcomes acquired resistance to anti-EGFR therapy in colorectal cancer cells and patients’ avatars. Journal of Clinical Oncology 2014;32(15_suppl):2626– doi 10.1200/jco.2014.32.15_suppl.2626. [DOI] [Google Scholar]
- 74.Rankin A, Klempner SJ, Erlich R, Sun JX, Grothey A, Fakih M, et al. Broad Detection of Alterations Predicted to Confer Lack of Benefit From EGFR Antibodies or Sensitivity to Targeted Therapy in Advanced Colorectal Cancer. Oncologist 2016;21(11):1306–14 doi 10.1634/theoncologist.2016-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murugan AK, Dong J, Xie J, Xing M. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle 2009;8(13):2122–4 doi 10.4161/cc.8.13.8710. [DOI] [PubMed] [Google Scholar]
- 76.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oddo D, Sennott EM, Barault L, Valtorta E, Arena S, Cassingena A, et al. Molecular Landscape of Acquired Resistance to Targeted Therapy Combinations in BRAF-Mutant Colorectal Cancer. Cancer Res 2016;76(15):4504–15 doi 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov 2015;5(4):358–67 doi 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cappuzzo F, Janne PA, Skokan M, Finocchiaro G, Rossi E, Ligorio C, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 2009;20(2):298–304 doi 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17(1):77–88 doi 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014;4(1):80–93 doi 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stintzing S, Zhang W, Heinemann V, Neureiter D, Kemmerling R, Kirchner T, et al. Polymorphisms in Genes Involved in EGFR Turnover Are Predictive for Cetuximab Efficacy in Colorectal Cancer. Mol Cancer Ther 2015;14(10):2374–81 doi 10.1158/1535-7163.MCT-15-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74 doi 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 84.Troiani T, Martinelli E, Napolitano S, Vitagliano D, Ciuffreda LP, Costantino S, et al. Increased TGF-alpha as a mechanism of acquired resistance to the anti-EGFR inhibitor cetuximab through EGFR-MET interaction and activation of MET signaling in colon cancer cells. Clin Cancer Res 2013;19(24):6751–65 doi 10.1158/1078-0432.CCR-13-0423. [DOI] [PubMed] [Google Scholar]
- 85.Hobor S, Van Emburgh BO, Crowley E, Misale S, Di Nicolantonio F, Bardelli A. TGFalpha and amphiregulin paracrine network promotes resistance to EGFR blockade in colorectal cancer cells. Clin Cancer Res 2014;20(24):6429–38 doi 10.1158/1078-0432.CCR-14-0774. [DOI] [PubMed] [Google Scholar]
- 86.Loupakis F, Cremolini C, Fioravanti A, Orlandi P, Salvatore L, Masi G, et al. EGFR ligands as pharmacodynamic biomarkers in metastatic colorectal cancer patients treated with cetuximab and irinotecan. Target Oncol 2014;9(3):205–14 doi 10.1007/s11523-013-0284-7. [DOI] [PubMed] [Google Scholar]
- 87.Buck E, Eyzaguirre A, Barr S, Thompson S, Sennello R, Young D, et al. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther 2007;6(2):532–41 doi 10.1158/1535-7163.MCT-06-0462. [DOI] [PubMed] [Google Scholar]
- 88.Siena S, Sartore-Bianchi A, Garcia-Carbonero R, Karthaus M, Smith D, Tabernero J, et al. Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann Oncol 2018;29(1):119–26 doi 10.1093/annonc/mdx504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14(9):531–48 doi 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 90.Diaz LA Jr., Bardelli A Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32(6):579–86 doi 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, et al. Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer Discov 2018;8(10):1270–85 doi 10.1158/2159-8290.CD-17-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015;26(4):731–6 doi 10.1093/annonc/mdv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu X, George GC, Tsimberidou AM, Naing A, Wheler JJ, Kopetz S, et al. Retreatment with anti-EGFR based therapies in metastatic colorectal cancer: impact of intervening time interval and prior anti-EGFR response. BMC Cancer 2015;15:713 doi 10.1186/s12885-015-1701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santini D, Vincenzi B, Addeo R, Garufi C, Masi G, Scartozzi M, et al. Cetuximab rechallenge in metastatic colorectal cancer patients: how to come away from acquired resistance? Ann Oncol 2012;23(9):2313–8 doi 10.1093/annonc/mdr623. [DOI] [PubMed] [Google Scholar]
- 95.Cremolini C, Rossini D, Dell’Aquila E, Lonardi S, Conca E, Del Re M, et al. Rechallenge for Patients With RAS and BRAF Wild-Type Metastatic Colorectal Cancer With Acquired Resistance to First-line Cetuximab and Irinotecan: A Phase 2 Single-Arm Clinical Trial. JAMA Oncol 2018. doi 10.1001/jamaoncol.2018.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsuji TE A, Masuishi T, Satake H, Segawa Y, Tanioka H, et al. Phase II study of third-line cetuximab rechallenge in patients with metastatic wild-type K-RAS colorectal cancer who achieved a clinical benefit in response to first-line cetuximab plus chemotherapy (JACCRO CC-08). Ann Oncol 2016:27. [Google Scholar]
- 97.Nogueira JR A, Jacinto P, Ribeiro J, Bonito N, Marques M, et al. P-167Cetuximab rechallenge in metastatic colorectal cancer patients. Ann Oncol 2016;27:49–50.26487582 [Google Scholar]
- 98.Osawa ES H, Nakamura M, Ohhara Y, Shindo Y, Shiozawa M, et al. 481PPhase II study of cetuximab rechallenge in patients with ras wild-type metastatic colorectal cancer: E-rechallenge trial. Ann Oncol 2018:29. [Google Scholar]
- 99.Ohhara YSE, Osawa H et al. Liquid biopsy for optimizing the rechallenge of cetuximab in metastatic colorectal cancer: Additional study of E-Rechallenge Trial. Ann Oncol 2019. [Google Scholar]
- 100.Karani A FT, Diniz L, et al. Trecc: Re-challenge therapy with anti-EGFR in metastatic colorectal adenocarcinoma (mCRC). Ann Oncol 2019. [Google Scholar]
- 101.Husain H, Melnikova VO, Kosco K, Woodward B, More S, Pingle SC, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin Cancer Res 2017;23(16):4716–23 doi 10.1158/1078-0432.CCR-17-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wasan H, Meade AM, Adams R, Wilson R, Pugh C, Fisher D, et al. Intermittent chemotherapy plus either intermittent or continuous cetuximab for first-line treatment of patients with KRAS wild-type advanced colorectal cancer (COIN-B): a randomised phase 2 trial. Lancet Oncol 2014;15(6):631–9 doi 10.1016/S1470-2045(14)70106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med 2014;6(224):224ra26 doi 10.1126/scitranslmed.3007947. [DOI] [PubMed] [Google Scholar]
- 104.Hong DS, Morris VK, El Osta B, Sorokin AV, Janku F, Fu S, et al. Phase IB Study of Vemurafenib in Combination with Irinotecan and Cetuximab in Patients with Metastatic Colorectal Cancer with BRAFV600E Mutation. Cancer Discov 2016;6(12):1352–65 doi 10.1158/2159-8290.CD-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wadlow RC, Hezel AF, Abrams TA, Blaszkowsky LS, Fuchs CS, Kulke MH, et al. Panitumumab in patients with KRAS wild-type colorectal cancer after progression on cetuximab. Oncologist 2012;17(1):14 doi 10.1634/theoncologist.2011-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pietrantonio F, Perrone F, Biondani P, Maggi C, Lampis A, Bertan C, et al. Single agent panitumumab in KRAS wild-type metastatic colorectal cancer patients following cetuximab-based regimens: Clinical outcome and biomarkers of efficacy. Cancer Biol Ther 2013;14(12):1098–103 doi 10.4161/cbt.26343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saif MW, Kaley K, Chu E, Copur MS. Safety and efficacy of panitumumab therapy after progression with cetuximab: experience at two institutions. Clin Colorectal Cancer 2010;9(5):315–8 doi 10.3816/CCC.2010.n.046. [DOI] [PubMed] [Google Scholar]