Abstract
The bromodomain family member proteins (BRDs; BET proteins) are key coregulators for estrogen receptor alpha (ERα)-mediated transcriptional enhancers. The use of BRD selective inhibitors has gained much attention as a potential treatment for various solid tumors, including ER-positive breast cancers. However, the roles of individual BET family members have largely remained unexplored. Here, we describe the role of BRDs in estrogen (E2)-dependent gene expression in ERα-positive breast cancer cells. We observed that chemical inhibition of BET family proteins with JQ1 impairs E2-regulated gene expression and growth in breast cancer cells. In addition, RNAi-mediated depletion of each BET family member (BRDs 2, 3, and 4) revealed partially redundant roles at ERα enhancers and for target gene transcription. Furthermore, we found a unique role of BRD3 as a molecular sensor of total BET family protein levels and activity through compensatory control of its own protein levels. Finally, we observed that BRD3 is recruited to a subset of ERα binding sites (ERBSs) that are enriched for active enhancer features, located in clusters of ERBSs likely functioning as ‘super enhancers’, and associated with highly E2-responsive genes. Collectively, our results illustrate a critical and specific role for BET family members in ERα-dependent gene transcription.
Introduction
Estrogen signaling plays a wide array of physiological roles in various reproductive and non-reproductive organs. When estrogens, such as the predominant naturally-occurring estrogen 17β-estradiol (E2), bind to estrogen receptor alpha (ERα), the liganded receptor dimerizes and binds to chromatin to activate target gene transcription (1,2). Sites of ERα binding, which may contain a DNA sequence motif called the estrogen response element (ERE), are often pre-bound by other chromatin-binding proteins, such as FoxA1, which serves as a ‘pioneer’ transcription factor (TF) (3,4). Upon binding to chromatin, ERα recruits additional transcription coregulators (e.g., the steroid receptor coactivator proteins, SRCs) and chromatin remodelers to establish fully active transcription enhancer complexes (2,5,6). Active ERα enhancer complexes, in turn, loop to target gene promoters to drive the recruitment the RNA polymerase II (Pol II) machinery and subsequent transcriptional output (5–8).
Estrogen signaling has been implicated in a range of pathological conditions, including ERα-positive breast and uterine cancers, obesity, and osteoporosis (1,9). Estrogen signaling through ERα exerts a potent mitogenic effect in ER-positive breast cancers (1,9). Indeed, approximately 60–70 percent of breast cancers are ER-positive at the time of diagnosis (9). Selective estrogen receptor modulator (SERM) drugs, which include a variety of ER ligands with a range agonistic and antagonistic activities (e.g., tamoxifen, raloxifene, and fulvestrant), are widely used as a first line of treatment in ER-positive breast cancer patients (10). Although SERMs have been effective as a first line of treatment for patients with ER-positive breast cancers, many patients ultimately develop resistance after prolonged usage (11). Thus, there is a great need to develop alternative therapeutic strategies for SERM-resistant breast cancers.
The BET family of proteins comprise a set of transcription coregulators (BRDs) that cooperate with a wide variety of TFs, including ERα (12,13). The BET family includes four members, BRD2, BRD3, BRD4, and BRDt, each of which contains two bromodomains that bind to acetylated lysine residues, particularly in core histones (14). BRD2, BRD3, and BRD4 are expressed in a wide variety of tissues, while BRDt expression is limited to the testes (14,15). Upon activation of TFs, histones surrounding TF binding sites are hyperacetylated by histone acetyltransferases recruited in the enhancer complex; hyperacetylated histones in turn recruit the BET family proteins through their bromodomains to regulate transcription through the promoter-bound Pol II machinery (14,16).
Recently, selective and competitive inhibitors for the bromodomains of BET family proteins have been developed, including JQ1, iBET, and their derivatives (17–19). A growing body of literature supports the efficacy of these inhibitors in various diseases, including a range of solid tumors. Inhibition of BRDs by bromodomain inhibitors modulates the recruitment of the BRDs to transcriptional enhancers and, thus, impairs the gene expression programs that are crucial for the growth of cancer cells (17–20). Previous studies on the role of BET proteins in ERα-dependent gene transcription found that JQ1 inhibits E2-dependent gene transcription in ER-positive breast cancer cells (13,21). However, the precise roles and mechanisms of individual BET family members in ERα-dependent transcription have not yet been elucidated.
In this study, we investigated the role of the BET family proteins in E2 signaling, ERα-mediated gene transcription, and the growth of ER-positive breast cancer cells. Our results point to partially redundant roles for BRDs 2, 3, and 4 in E2-dependent gene expression, with a unique role for BRD3 in modulating total BRD levels and activity. Collectively, our study demonstrates an important function of the BET family proteins in E2-dependent gene regulation.
Materials and Methods
Additional details regarding the materials and methods are provided in the Supplementary Information, included information on assays with T-47D and MDA-MB-231 cells presented in the Supplementary Figures.
Antibodies
Details for the following antibodies used are provided in the Supplementary Information.: ERα, BRD2, BRD3, BRD4, pan-acetyl H4, Myc, SRC2, SRC3, SNRP70, and β-tubulin.
Cell Culture and Treatments
MCF-7 cells were maintained in Minimum Essential Medium (MEM) Eagle with Hank’s salts (Sigma, M1018) supplemented with 5% HyClone calf serum (GE Healthcare, SH30072) and 20mM HEPES (ThermoFisher Scientific, BP310). Prior to cell proliferation, knockdown, gene expression, and ChIP experiments, the cells were grown for three days in phenol red-free MEM Eagle medium supplemented with 5% charcoal-dextran-treated calf serum. The cells were validated by genotypic and phenotypic (e.g., ERα expression assays), and were routinely verified as mycoplasma-free using a PCR-based test.
Treatment conditions for cells were as follows: 17β-estradiol (E2), 100 nM (Sigma, E8875); (+)JQ1 (the active enantiomer of JQ1, referred to herein as JQ1), 500 nM unless otherwise indicated (Cayman Chemical, 11187); and (–)JQ1 (the inactive enantiomer of JQ1), at the same concentrations as (+)JQ1 (Cayman Chemical, 11232). The cells were treated with (+)JQ1 or (–)JQ1 for 3 hours before treatment with E2. For gene expression analyses, the cells were collected after 3 hours of E2 treatment. For ChIP analyses, the cells were collected after 45 minutes of E2 treatment.
Cell Proliferation Assays
MCF-7 cells grown under estrogen-free conditions were treated with ethanol vehicle, E2, or JQ1, with fresh treatments added every 2 days. At selected time points, the cells were fixed with 10% formaldehyde and stained with 0.1% crystal violet. The crystal violet was extracted using 10% glacial acetic acid and the absorbance was read at 595 nm.
siRNA-mediated Knockdown of BRD2, BRD3, and BRD4
MCF-7 cells grown under estrogen-free conditions were transfected with commercially available siRNA oligos directed against BRD2, BRD3 or BRD4 (Sigma) using Lipofectamine RNAiMAX reagent (Invitrogen) per the manufacturer’s instructions. Treatments with E2 were performed 48 hours after siRNA transfection. The siRNA sequences are listed in the Supplementary Information.
Inducible Expression of BRD3
The lentiviral system for inducible expression of BRD3 is based on pINDUCER20. The human BRD3 cDNA was cloned by reverse transcription-PCR from MCF-7 cell total RNA and then transferred into a modified pINDUCER20 vector with the addition of a sequence encoding an HA tag at the 3’ end of the cDNA.
The pINDUCER20-BRD3-HA plasmid was co-transfected with pCMV-VSVG, pCMV-GAG-pol-Rev, and pAdVantage (Promega) into 293T cells using GeneJuice (Millipore) for recombinant lentivirus production. The supernatant containing the lentiviruses was collected 48 hours after transfection and used to infect MCF-7 cells. The infected MCF-7 cells were selected and maintained in 1 mg/ml Geneticin (Life Technologies). For induction of BRD3 expression, doxycycline hyclate (Sigma) was added to the medium at a final concentration 50 ng/mL. Twenty-four hours later, the cells were collected for Western blotting or RT-qPCR.
Kaplan-Meier Analyses
Kaplan-Meier estimators were generated using the Gene Expression-Based Outcome for Breast Cancer Online (GOBO) tool (http://co.bmc.lu.se/gobo/) (22). In our analyses, we profiled gene expression in 560 ER-positive breast cancer samples. Patients were stratified into two groups based on the expression levels of BRD2, BRD3, and BRD4 (top half = high expression; bottom half = low expression). The survival outcomes were then determined by Kaplan-Meier estimators within the GOBO tool.
Immunohistochemistry (IHC)
Immunohistochemical staining of patient samples were adapted from the Cancer Atlas of the Human Protein Atlas database, version 15 (www.proteinatlas.org).
Western Blotting
Protein lysates from MCF-7 cells were prepared using lysis buffer [20 mM HEPES (pH 7.5), 420 mM NaCl, 1.5 mM MgCl2, 0.2mM EDTA, 25% Glycerol, 0.5% NP-40, 1 mM DTT, 1x complete protease inhibitor cocktail (Roche)]. Protein expression was assessed by Western blotting with the antibodies noted above. The signals were developed using a chemilumenescent detection system. (ThermoFisher Scientific).
mRNA Expression Analysis by Quantitative Real-time PCR (RT-qPCR)
Changes in the steady-state levels of target gene mRNAs were analyzed by RT-qPCR, as described previously (5). The resulting cDNA was analyzed by qPCR using primer sets listed in the Supplementary Information. The expression levels were normalized to 18S ribosomal RNA as an internal standard. All experiments were conducted a minimum of three times with independent RNA isolations to ensure reproducibility.
Preparation of polyA+ RNA-seq Libraries
RNA-seq libraries were prepared as described previously (23). Total RNA was isolated from cells using the RNeasy Plus kit (Qiagen). PolyA+ RNA was purified from the total RNA using Dynabeads Oligo(dT)25 (Life Technologies). Strand-specific libraries were prepared according to the “deoxyuridine triphosphate (dUTP)” method, as described previously (23). After quality control analyses, the libraries were sequenced on an Illumina HiSeq 2000 (single-end sequencing, 50 nt).
Analysis of RNA-seq Data
Quality of RNA-seq reads was assessed using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The RNA-seq reads were aligned using TopHat v2.0.10 (24) on the hg19 reference genome. We used Cufflinks v.2.1.1 (25) and Cuffdiff v.2.1.1 (26) to assemble the reads into transcripts using RefSeq annotations and to call differentially regulated transcripts, respectively. Uniquely mapped reads were visualized on the UCSC genome browser as bigWig files generated using BEDtools (27). Expression of differentially expressed genes was visualized as heatmaps using Java Tree View (28). Box plots were generated using the box plot function in R.
Analysis of GRO-seq Data
GRO-seq data was analyzed as previously described (29). The GRO-seq reads surrounding ± 2.5 kb of center of the ERα peaks or surrounding the 5’ end of regulated genes nearest to the ERα peaks were visualized in box plots as Reads Per Kilobase of gene per Million mapped reads (RPKM) using box plot function in R.
Gene ontology (GO) Analyses
Gene ontology analyses were performed using DAVID (Database for Annotation, Visualization, and Integrated Discovery) (30). As input, the list of genes expressed in MCF-7 at least in one condition tested was used as a background.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described (5). The ChIPed DNA was dissolved in water and analyzed by qPCR using the primer sets listed in primer sets listed in the Supplementary Information. All experiments were conducted a minimum of three times with independent RNA isolations to ensure reproducibility.
Preparation of ChIP-seq Libraries
Fifty ng of ChIPed DNA for each condition was used to generate libraries for deep sequencing, as previously described (31). After quality control analyses, the libraries were sequenced on an Illumina HiSeq 2000 (single-end sequencing, 50 nt).
Analysis of ChIP-seq Data
Quality Control and Alignment
The quality of ChIP-seq reads was analyzed by FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). ChIP-seq reads were aligned to the hg19 reference genome using Bowtie 2 v2.2.2 using the default parameters (32), and visualized on UCSC genome browser using BbigWig files generated by BEDTools (27) and custom R scripts.
Peak Calling and Data Representation
We used an ERα binding site dataset from E2-treated MCF-7 cells from our lab in which peaks were called using the input as a control (31). This study also included FoxA1 ChIP-seq data prepared under the same conditions and in the same MCF-7 cells in which we performed the BRD3 ChIP-seq. To understand the role of BRD3 in ERα enhancer activation, we compared our ERα and BRD3 ChIP-seq datasets with our FoxA1 ChIP-seq data, as well as published BRD4 ChIP-seq data (13) and DNase-seq data (33) from MCF-7 cells treated with E2. Sequencing read densities ± 5 kb around the ERα peaks for BRD3, FoxA1, and DNase1, and ± 10 kb around the ERα peaks for acetyl H4 were calculated using annotatePeaks.pl function in HOMER software (34) and visualized as heatmaps using Java Tree View (28).
Motif Analysis
Directed motif search was performed on ± 5 kb window around the center of all the ERα peaks with and without BRD3 enrichment using the command-line version of FIMO software (35). The position weight matrix (PWM) for ESR1 was obtained from JASPAR database (36).
Genomic Data Set Availability
The new genomic data sets reported herein (BRD3 ChIP-seq ±E2 and RNA-seq ± E2 from MCF-cells) are available from the NCBI’s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) using the following accession numbers: GSE109570 and GSE109571 (see Supplementary Information). The previously reported data sets used herein include: ERα and FoxA1 ChIP-seq (31), BRD4 ChIP-seq (13), and DNase-seq data (33) from MCF-7 cells treated with or without E2. The publicly available ChIP-seq (ERα ±E2, FoxA1 ±E2) and GRO-seq (±E2) data sets from MCF-7 cells can be accessed from NCBI GEO using the following accession numbers: GSE59532 and GSE27463, respectively.
Results
Expression and activity of BET family members correlate with clinical outcomes in ER-positive breast cancer patients and ER-positive breast cancer cell growth
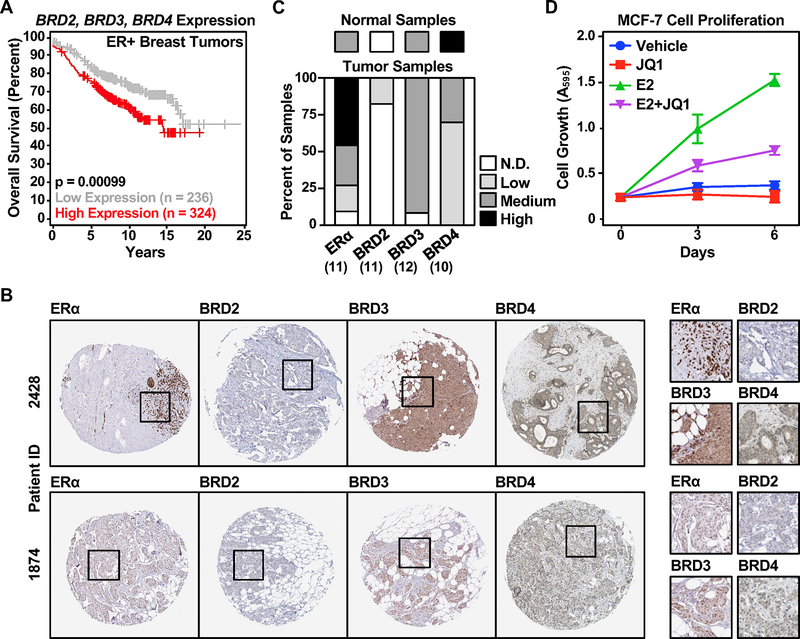
The BET family protein members have been implicated as therapeutic targets for the treatment of several solid tumors, including breast and prostate cancers (17,20). We examined the potential roles of BET family members in ER-positive breast cancers. We found that high expression of BRDs 2, 3, and 4 collectively correlates with poor overall survival for ER-positive breast cancer patients (Figure 1A). In particular, BRD3 and BRD4 expression were retained in >90% and 100% of breast tumor samples, respectively, while <20% of tumor samples had detectable levels of BRD2 (Figure 1B and 1C) (37). We examined whether the BET family members play a role in ER-positive breast cancer cell growth. Consistent with previous studies (13,21), a potent BET family protein inhibitor, JQ1, attenuated the E2-dependent growth of MCF-7 cells, an ER-positive human breast cancer cell line (Figure 1D). We observed similar effects of JQ1 on the E2-dependent growth of T-47D cells (another ER-positive human breast cancer cell line), but not MDA-MB-231 cells (an ER-negative human breast cancer cell line) (Supplementary Figure S1).
Figure 1. The expression of BET family members correlates with clinical outcomes in ER-positive breast cancers and their activity is required for the growth of ER-positive breast cancer cells.
(A) Elevated expression of the BET family proteins correlates with unfavorable clinical outcomes in ER-positive breast cancers. Kaplan-Meier plots for overall survival of ER-positive breast tumor patients with relatively high (red) or low (gray) BRD2, BRD3, and BRD4 expression.
(B) A majority of breast tumor samples express BRD3 or BRD4. Quantification of immunohistochemical standing for ERα, BRD2, BRD3, and BRD4 in breast tumor specimens in the Human Protein Atlas. Normal tissues represent samples of non-neoplastic and morphologically normal tissues that were surgically removed from three individuals with breast cancers.
(C) Representative immunohistochemical staining for ERα, BRD2, BRD3, and BRD4 in ER-positive ductal carcinoma patient samples from the Human Protein Atlas.
(D) JQ1 attenuates E2-dependent growth of MCF-7 cells. Proliferation assays of ER-positive MCF-7 breast cancer cells ± E2 treatment in the presence or absence of 250 nM JQ1.
BRDs 2, 3, and 4 are recruited to ERα enhancers through their bromodomains
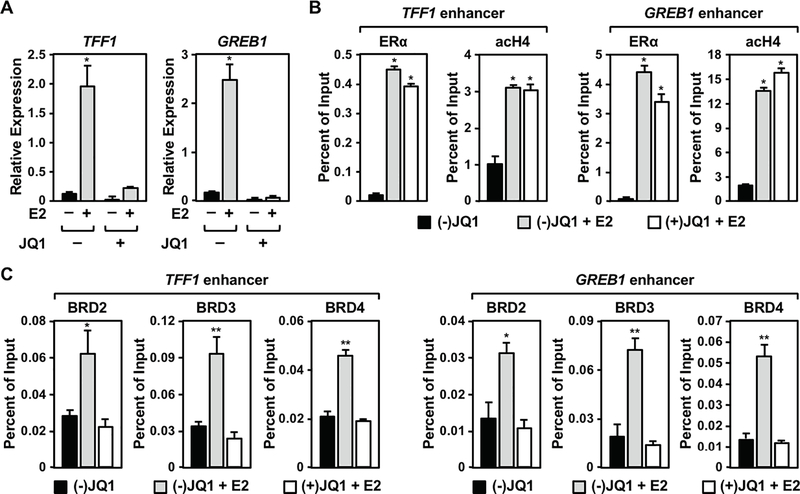
BET family members are recruited to TF binding sites at enhancers through acetylated histones during the process of enhancer assembly and transcription activation (14,16). We hypothesized that the effects of BET family member inhibition on E2-stimulated growth of MCF-7 cell is mediated by impaired E2-dependent gene expression as direct result of impaired enhancer formation. Indeed, treatment with JQ1 reduced the E2-dependent expression of well-characterized E2 target genes (i.e., TFF1, GREB1) in MCF-7 cells (Figure 2A) and T-47D cells (Supplementary Figure S2A), but not MDA-MB-231 cells (Supplementary Figure S2B). Importantly, the levels of ERα and SRCs in MCF-7 cells were unaffected by treatment with JQ1 (Supplementary Figure S3). Furthermore, JQ1 did not alter ERα recruitment or histone acetylation at the TFF1 and GREB1 enhancers (Figure 2B). However, the E2-dependent recruitment of BRDs 2, 3, and 4 (Figure 2C) and the expression of enhancer RNAs (eRNAs), a marker of active enhancers (7) (Supplementary Figure S4), was significantly reduced by JQ1 at the ERα enhancers. Collectively, our data support a model in which the recruitment of BRDs to hyperacetylated ERBSs is inhibited by JQ1 to reduce enhancer activity and target gene transcription.
Figure 2. BET proteins recruited to ERα binding sites upon E2 treatment are required for E2-dependent gene expression in MCF-7 cells.
(A) JQ1 impairs E2-dependent gene induction in MCF-7 cells. The expression of E2-induced genes in the presence of the active (+) or inactive (−) form of JQ1 determined by RT-qPCR. Asterisks indicate significant differences (p < 0.001; two-way ANOVA).
(B) JQ1 does not affect ERα binding or histone acetylation levels at ERα enhancers. The enrichment of ERα and acetyl H4 at enhancers for E2-responsitve genes in the presence of the active (+) or inactive (−) form of JQ1 determined by ChIP-qPCR. Asterisks indicate significant differences compared to the (−) JQ1, vehicle control (p < 0.001; one-way ANOVA).
(C) JQ1 inhibits the E2-dependent recruitment of the BET family proteins to ERα binding sites. The effects of JQ1 on BRD2, BRD3, and BRD4 recruitment at ERα binding sites upon E2 treatment determined by ChIP-qPCR. Asterisks indicate significant differences compared to the (−) JQ1, vehicle control (*, p < 0.05; **, p < 0.005; one-way ANOVA).
Previous studies have shown that JQ1 inhibits tumor growth by repressing transcription of the MYC gene in some Myc-dependent cancers (38). Since MYC expression is elevated by E2 treatment, we tested whether the inhibition of E2-dependent growth of MCF-7 cells by JQ1 may be due, in part, to reduced E2-dependent induction of MYC expression. Interestingly, JQ1 did not affect E2-induced MYC expression or Myc protein levels (Supplementary Figure S5), suggesting a Myc-independent mechanism of JQ1 action on the E2-induced growth of MCF-7 cells.
Inhibition of the BET family proteins promotes misregulation of the E2-dependent gene expression program
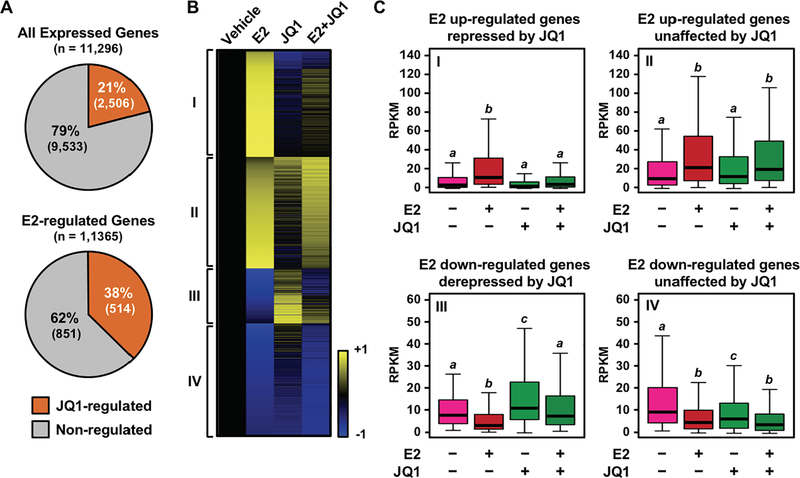
Based on the significant growth inhibitory effect of JQ1, we hypothesized that the BET family proteins may have a genome-wide effect on E2-dependent gene expression. To explore this, we performed RNA-seq assays using MCF-7 cells treated with E2 in the presence or absence of JQ1. We found that 21% of genes expressed in at least one of the conditions examined in MCF-7 cells were affected by treatment with JQ1. However, the proportion of genes affected by JQ1 increased to 38% when specifically examining E2-regulated genes, suggesting an important role of the BET family members in E2-responsive gene expression (Figure 3A). The expression of 345 out of 706 (~49%) E2-upregulated genes and 178 out of 659 (~27%) E2-downregulated genes was altered greater than 1.5 fold by JQ1 (Figure 3B and 3C), indicating that JQ1 is disproportionally affecting E2-dependent gene activation. Furthermore, gene ontology analysis identified ‘response to endogenous stimulus’ and ‘response hormone’ as the top ontology terms among gene groups whose E2-induced activation was impaired by JQ1 (Supplementary Table S1). These results support the hypothesis that the inhibitory effect of JQ1 on the E2-stimulated growth of MCF-7 cells was mediated through impaired regulation of E2-dependent gene expression.
Figure 3. BET proteins are required for the E2-dependent gene expression program in MCF-7 cells.
(A) E2-regulated genes are disproportionally affected by JQ1. Pie charts showing the percentage of genes whose expression levels were affected by JQ1 among the universes of expressed or E2-regulated genes in MCF-7 cells. The expression levels were determined by RNA-seq.
(B) A heatmap representation of E2-regulated gene expression in the presence of JQ1. Genes were called significantly regulated when the fold change of expression levels were > 2 or < 0.25 compared to the vehicle without JQ1. FDR < 0.01. Roman numerals designate four gene clusters: I, E2 up-regulated and JQ1-repressed; II, E2 up-regulated and JQ1-unaffected; III, E2 down-regulated and JQ1-derepressed genes; IV, E2 down-regulated and JQ1-unaffected genes.
(C) Box plots showing the expression levels for the top 50% of highly regulated genes within each cluster indicated in panel (B). Bars marked with different letters (a, b, c) are significantly different from each other (p < 5 × 10−10; Wilcoxon rank-sum test).
BRD3 is a critical coregulator for ERα-dependent gene expression in MCF-7 cells
The BET family bromodomain inhibitor JQ1 exhibits a range of affinities across the BET family of proteins (18). Moreover, each family member contains two bromodomains, each with different affinities toward JQ1. The N-terminal bromodomains in BRD3 and BRD4 have the highest affinities for JQ1 (Kd = ~50–60 nM), followed by their C-terminal bromodomains (Kd = ~80–90 nM). Although the affinity is lower (Kd = >120 nM), JQ1 also binds to the bromodomains of BRD2 (18). Elucidating the differential functions among BET family members is crucial to understanding their precise roles in E2-dependent gene expression.
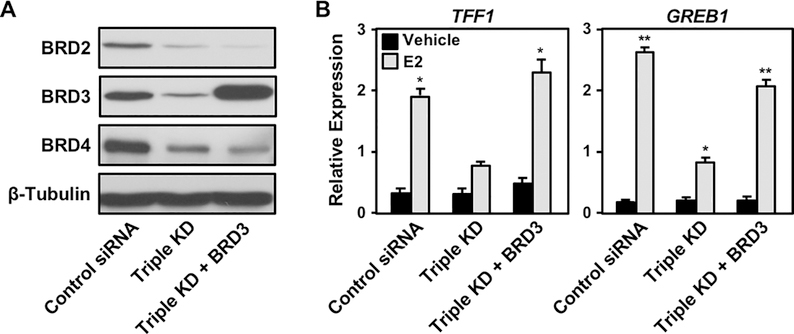
To dissect the differential functions of the BET family members, we performed siRNA-mediated knockdown of BRD2, BRD3, and BRD4 in MCF-7 cells and evaluated their individual and collective contributions to E2-dependent gene activation (Figure 4A). Depletion of a single family member did not affect E2-induced gene expression. However, depletion of BRD3 in combination with BRD2, BRD4, or both led to reduced E2-dependent gene expression (Figure. 4B). Interestingly, BRD3 protein levels were elevated compared to the control when BRD2, BRD4, or both were depleted (Figure 4A). This result suggests that elevated BRD3 protein levels upon BRD2 or BRD4 depletion compensates for the decreased levels of BRD2 and BRD4. This may explain why we did not observe impaired E2-dependent gene expression without BRD3 depletion (Figure 4B). In contrast, we did not observe elevation of BRD2 or BRD4 protein levels under any condition tested (Figure 4A), highlighting a BRD3-specific role in fine-tuning the overall levels and activity of the BET family members in MCF-7 cells.
Figure 4. BRD3 is a critical regulator of E2-dependent gene expression in MCF-7 cells.
(A) Western blot showing siRNA-mediated knockdown of BRD2, BRD3, and BRD4 individually or in combination.
(B) E2-induced gene expression is attenuated when BRD3 is depleted in combination with BRD2, BRD4, or both. E2-dependent gene expression determined by RT-qPCR after depletion of BET family proteins by siRNA individually or in combination. Asterisks indicate significant differences compared to the control siRNA under each treatment condition (*, p < 0.05; **, p < 0.005; two-way ANOVA).
We observed an analogous situation in T-47D cells, although the specific BET protein affected differed compared to MCF-cells (Supplementary Figure S6). As with the MCF-7 cells, depletion of a single family member had little to no effect on E2-induced gene expression in T-47D cells (Supplementary Figure S6, A and B). However, depletion of BRD2 in combination with BRD3, or both BRD3 and BRD4, led to reduced E2-dependent expression of TFF1 in T-47D cells (similar effects were not observed with GREB1) (Supplementary Figure S6B). Interestingly, BRD2 protein levels were elevated compared to the control when BRD3, BRD4, or both were depleted (Supplementary Figure S6A). Thus, similar compensatory mechanisms among BET family members may be active in other ER-positive cell types.
To determine if BRD3 is sufficient for E2-dependent gene expression, we ectopically expressed siRNA-resistant BRD3 in MCF-7 cells with simultaneous knockdown of BRD2, BRD3, and BRD4 (Figure 5A). Re-expression of BRD3 restored E2-dependent gene expression to a level comparable to the siRNA control. This result demonstrates that BRD3 has a role redundant with BRD2 and BRD4, and is sufficient for E2-dependent gene activation (Figure 5B). Together, these results suggest a partial functional redundancy among BET family members, and a key role of BRD3 as a molecular sensor that regulates the total levels and activity of BET proteins in breast cancer cells.
Figure 5. BRD3 is sufficient to restore E2-dependent gene expression suppressed by depletion of BET proteins in MCF-7 cells.
(A) Western blot showing restored expression of BRD3 from siRNA-resistant transgene in MCF-7 cells under siRNA-mediated simultaneous knockdown of BRD2, BRD3, and BRD4.
(B) Re-expression of BRD3 rescues E2-induced gene expression impaired by the depletion of BET proteins. E2-induced gene expression in MCF-7 cells determined by RT-qPCR upon expression of siRNA-resistant BRD3 after siRNA-mediated triple knockdown of BRD2, BRD3, and BRD4. Asterisks indicate significant differences compared to the control siRNA without treatment (*, p < 0.01; **, p < 0.0001; two-way ANOVA).
BRD3 occupies a subset of ERα binding sites
To further understand the role of BRD3 at ERα enhancers, we performed ChIP-seq assays for BRD3 and acetylated histone H4 in MCF-7 cells treated with E2. We analyzed the data in combination with ERα ChIP-seq data. Interestingly, we found that BRD3 occupancy segregates ERBSs into two groups: (1) ERBSs with BRD3 enrichment and (2) ERBSs without BRD3 enrichment (Figure. 6A). The group of ERBSs associated with BRD3 recruitment was also significantly enriched for acetylated histone H4 upon E2 treatment (Figures 6A, 6B, and 6C, Supplementary Figure S7), as might be expected given that acetylated histone H4 serves as a platform for BRD3 binding (15). Interestingly, BRD3, BRD4, and acetylated histone H4 were all enriched in broad peaks around the sharply defined ERα enhancers, perhaps illustrating the role of acetylated histone H4 in recruiting BET proteins through their acetyl-lysine-binding bromodomains (Supplementary Figure S7).
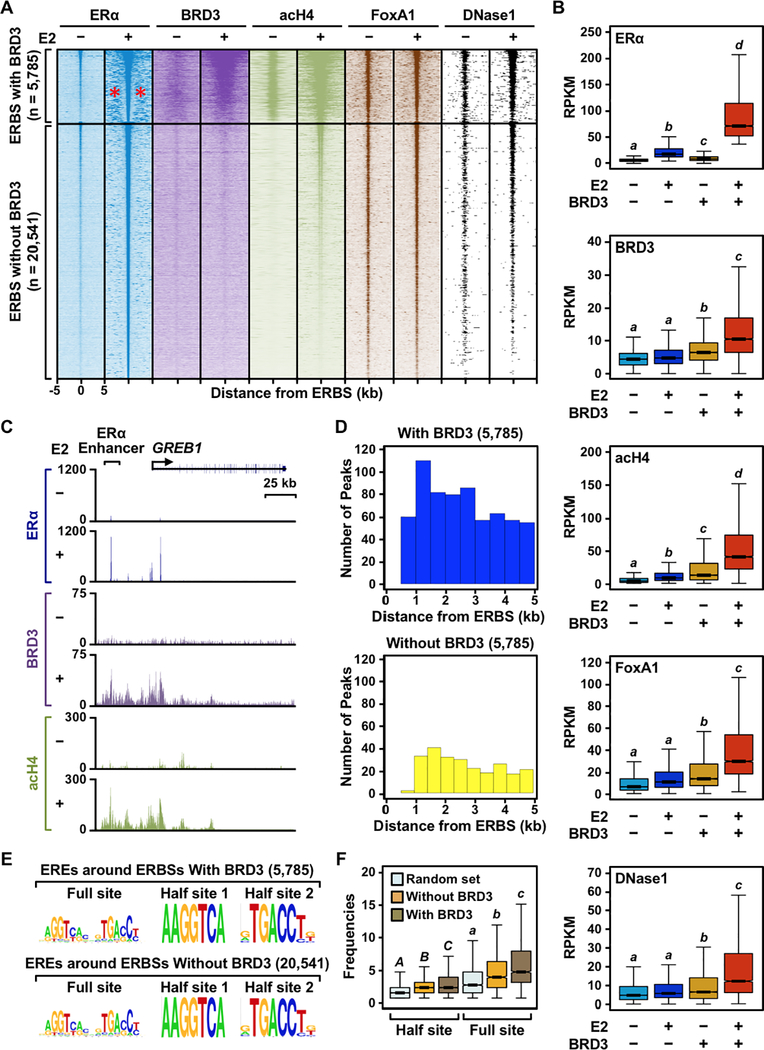
Figure 6. BRD3 is enriched at a subset of ERα binding sites that are associated with active enhancer marks.
(A) Heatmap representations of ERα, BRD3, acetyl H4, and FoxA1 ChIP-seq and DNase-seq reads surrounding ERα peaks. ERα binding sites were divided into two groups, with or without BRD3 enrichment, based on the presence of significant BRD3 ChIP-seq peaks within ± 500 bp of an ERα binding site. The genomic locations relative to the center of the ERBSs are indicated on the bottom of ERα −E2 column; although not shown, the same scales were used for the remaining datasets. The red asterisks in the ERα −E2 condition for the ERBSs with BRD3 enrichment indicate the presence of ‘satellite’ ERα binding sites.
(B) Box plots of ERα, BRD3, acetyl H4, and FoxA1 ChIP-seq and DNase-seq read counts 1 kb surrounding ERα peaks with or without BRD3 enrichment. Box plots marked with different letters (a, b, c, d) are significantly different from each other (p < 0.05; Wilcoxon rank-sum test).
(C) Genome browser tracks of ERα, BRD3, and acH4 ChIP-seq data at the GREB1 locus which has ERα binding sites enriched with BRD3.
(D) The number of ERα peaks surrounding ERα binding sites with or without BRD3 enrichment (satellite ERBSs) binned based on the distance from the BRD3-enriched or -unenriched ERα binding sites.
(E) The quality of ERE sequences does not affect the clustering of ERBSs enriched with BRD3. Motifs enriched under satellite ERBSs surrounding the central ERα binding sites with or without BRD3 enrichment.
(F) The quantity of ERα binding motifs (EREs) governs clustering of ERBSs enriched with BRD3. Box plots showing the frequencies of full or half ERα binding motifs within 10 kb of ERα binding sites with or without BRD3 enrichment. Box plots marked with different letters [(A,B,C) and (a, b, c)] are significantly different from each other within the same motif (p < 2.2 × 10−16; Wilcoxon rank-sum test).
Enhancers are enriched for specific genomic features, including binding of lineage-determining TFs (e.g. FoxA1), DNaseI accessibility, acetylated histones, additional histone modifications (e.g. H3K4me1/2 and H3K27ac), transcription coregulators, and the production of enhancer transcripts (eRNAs) (5)Shlyueva, 2014 #65;Vasquez, 2019 #122}. To understand the role of BRD3 in ERα enhancer activation, we compared our ChIP-seq datasets with publicly available ChIP-seq and DNase-seq datasets from MCF-7 cells treated with E2 (31,33). We observed that ERα binding and acetylated H4 levels were significantly induced after E2 treatment at ERBSs regardless of their association with BRD3 enrichment (Figure 6B). These results suggest that BRD3 recruitment occurs downstream of ERα binding and histone acetylation. In addition, they suggest that, although acetylated histones might be required for BRD3 recruitment, they are not sufficient.
While FoxA1 recruitment and DNaseI accessibility were not significantly induced after E2 treatment at ERBSs without BRD3 enrichment, we observed a significant E2-dependent increase in these features uniquely at ERBSs enriched with BRD3 (Figure 6B). The hormone dependence of FoxA1 recruitment at ERBSs has been debated in the literature (39,40), with one study suggesting that the apparent hormone dependence of FoxA1 binding may be influenced by sequencing read depth (39). Our results remain to be confirmed, but taken at face value, our results suggest functional interplay among BRD3, FoxA1, and chromatin remodeling at ERα enhancers, including a role for BRD3 in the enrichment of these features. Alternatively, it is also possible that FoxA1 and/or DNA accessibility affect BRD3 recruitment to ERα enhancers.
DNA sequences dictate ERα binding profiles unique to BRD3-associated ERBSs
From our ChIP-seq experiments, we found that ERα peaks associated with BRD3 enrichment are located near ‘satellite’ ERBSs (i.e., non-overlapping ERBSs located near the reference ERBS), compared to ERα peaks that are not enriched with BRD3 (Figure 6A; see the red asterisks). To characterize these satellite ERBSs, we first analyzed their genomic localization with respect to the reference ERBSs with or without BRD3 enrichment. We quantified the number of ERα peaks within a 10 kb window surrounding each reference ERBS with or without BRD3 recruitment and binned them into 500 bp intervals. As expected, we found more satellite ERBSs near reference ERBSs associated with significant peaks of BRD3 compared to reference ERBSs without significant peaks of BRD3. We observed a 30-fold greater incidence of satellite ERBSs within 1 kb if a given ERBS is associated with BRD3 compared to an ERBS not associated with BRD3 (Figure 6D).
We also noticed that the intensity of ERα binding (as determined by the area under ERα peak) in satellite ERBSs is greater when they are located near reference ERBSs associated with BRD3 compared to reference ERBSs not associated BRD3. We asked if DNA sequences under the satellite ERBSs might affect their binding. The sequence of both full and half ERE motifs was qualitatively similar for both groups of satellite ERBSs (i.e., those located near reference ERBSs associated with BRD3 and those located near reference ERBSs not associated BRD3; Figure 6E). But, the frequency of ERE occurrence was significantly higher within a 10 kb window surrounding reference ERBSs associated with BRD3 compared to reference ERBSs not associated BRD3 enrichment (Figure 6F). Taken together, our results suggest a genomic relationship between clustered ERBSs, BRD3 recruitment, and the underlying DNA sequences. These clustered, BRD3-enriched ERBSs are reminiscent of ‘super enhancers’ (41,42) (see Discussion).
BRD3 is uniquely recruited at active ERα enhancers
In a previous study, we demonstrated that approximately half of distal ERBSs produce enhancer transcripts (eRNAs) (7). Furthermore, we showed that ERBSs associated with eRNAs are enriched with various features of active enhancers, including coregulators, H3K27ac, and H3K4me1 (5,7). These results indicate that (1) only a subset of ERBSs serve as active enhancers and (2) eRNAs are a sensitive indicator enhancer activity (5,7).
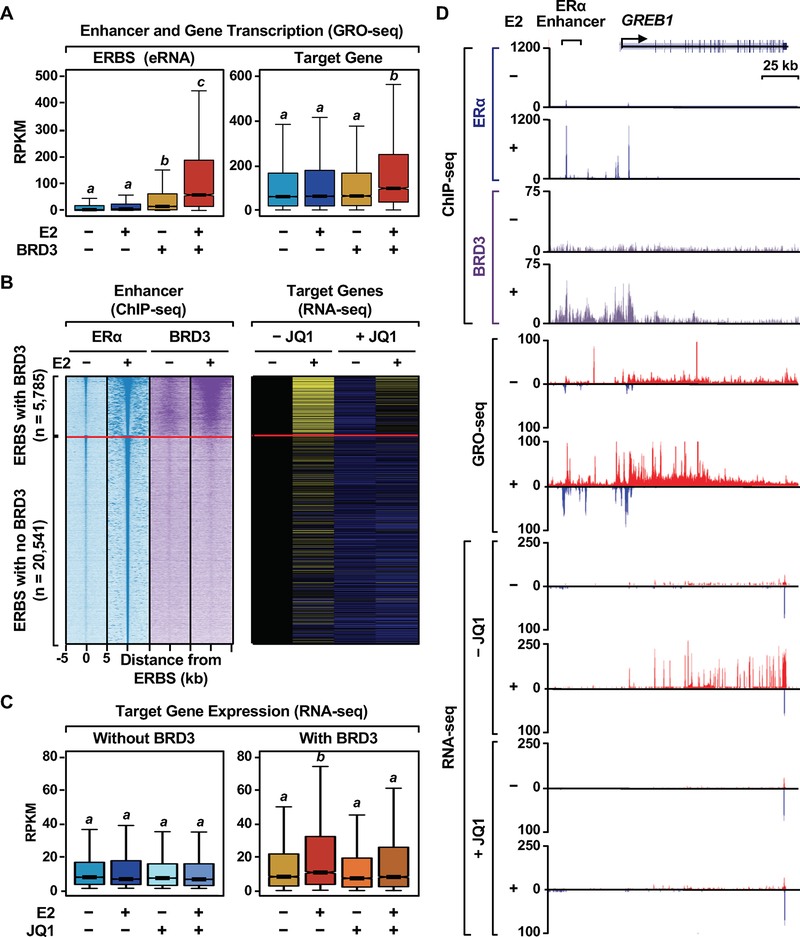
To examine the role of BRD3 in ERα enhancer activity, we analyzed enhancer transcription levels using GRO-seq from existing data sets (29). We observed significantly higher levels of enhancer transcription upon E2 treatment at ERBSs with BRD3 enrichment compared to ERBSs without BRD3 enrichment (Figure 7A). In addition, we found that E2-regulated genes located near ERBSs associated with BRD3 enrichment have significantly higher levels of E2 responsiveness (both transcription and steady-state RNA) than when they are located near ERBSs without BRD3 enrichment (Figure 7A–D, and Supplementary Figure S8), indicating that BRD3 is enriched at active ERα enhancers. Importantly, E2-dependent expression of the same set of genes was significantly impaired in the presence of JQ1 (Figure 7B–D, and Supplementary Figure S8), indicating an important role of BRD3 in determining the activity of ERα enhancers
Figure 7. BRD3-enriched ERα binding sites are associated with E2-induced genes that are affected by JQ1.
(A) Box plots showing the levels of transcription determined by GRO-seq at ERα binding sites with or without BRD3 enrichment and the target E2-regulated genes. Box plots marked with different letters (a, b, c, d) are significantly different from each other (p < 0.05; Wilcoxon rank-sum test,).
(B) JQ1-affected, E2 up-regulated genes are enriched for the target genes of BRD3-enriched ERBSs. Heatmap representations of ERα and BRD3 ChIP-seq reads, as shown in Fig. 6A, and RNA-seq of the target genes upon E2-treatment in the presence of JQ1.
(C) Box plots showing RNA-seq reads of the target genes of ERα binding sites with or without BRD3 enrichment upon E2-treatment in the presence of JQ1. Box plots marked with different letters (a, b, c, d) are significantly different from each other (p < 2.2 × 10−16; Wilcoxon rank-sum test,).
(D) Genome browser tracks of ChIP-seq data for ERα and BRD3, GRO-seq, and RNA-seq at the GREB1 locus which has ERα binding sites enriched with BRD3.
Discussion
BET family members play an important functional role at ERα enhancers in breast cancer cells
In this study, we uncovered a critical role of BET family members BRDs 2, 3, and 4 in ERα enhancer function, E2-dependent gene expression, E2-dependent breast cancer cell growth, and breast cancer outcomes. In this regard, we observed that elevated expression levels of BRDs 2, 3, and 4 correlate with unfavorable clinical outcomes in ER-positive breast cancers (Figure 1A). Inhibition of BET family members with the bromodomain inhibitor JQ1 attenuated the E2-dependent growth of ER-positive breast cancer cells (Figure 1D). The growth attenuation by JQ1 was due, in part, to a global impairment of E2-dependent gene expression (Figures 2A and 3B). Interestingly, BRDs 2, 3, and 4 exhibit functional redundancy, with BRD3 playing a unique role as a molecular sensor that coordinates the overall levels and activities of the BRDs (Figures 4 and 5). Across the genome, BRD3 is recruited to a subset of ERBSs that are (1) actively transcribed, (2) enriched for other features of active enhancers, (3) located in clusters of ERBSs, and (4) associated with highly E2-responsive genes (Figures 6 and 7). Importantly, E2-responsive genes nearest to ERBSs enriched for BRD3 recruitment were highly responsive to E2 treatment and disproportionally impaired by JQ1 (Figure 7). Our results are consistent with, but expand previous studies showing that BET proteins in general, and BRD4 in particular, are required for ERα enhancer activation and E2-dependent gene transcription (13,21).
BRD3 determines the overall levels and activities of BET family members in MCF-7 cells
BRD2, BRD3, and BRD4 are all expressed at detectable levels in MCF-7 cells (Figure 4A). Despite the low selectivity of JQ1 among BET family members (18), previous studies using this inhibitor have tended to focus on BRD4 without assessing the roles of other family members (13,43–45). This is perhaps because one of the BRD4 bromodomains was used to design and select for potent BET family bromodomain inhibitors, such as JQ1 (18). Indeed, thorough studies exploring the functional differences among the BET family members are limited.
In the current study, we sought to understand the functional differences among the BET family members. We found that the depletion of BRD 2, 3, or 4 alone was not sufficient to impair E2-dependent gene activation. In contrast, treatment with JQ1 or the simultaneous depletion of BRDs 2, 3, and 4 significantly impaired E2-dependent gene regulation (Figures 3 and 4A). Interestingly, co-depletion of BRD3 with either BRD2 or BRD4 impaired E2-dependent gene regulation (Figure 4B). In addition, BRD3 protein levels were increased when BRD2 or BRD4 were depleted, presumably to compensate for the total levels and activity of the BET proteins (Figure 4A). The increase in BRD3 protein levels presumably result in enhanced binding of BRD3 binding to chromatin at the enhancers, a possibility that will require further investigation. Studies in T-47D cells suggest that similar compensatory mechanisms among BET family members may be active in other ER-positive cell types, but may differ in the BET family members involved (Supplementary Figure S6). Collectively, our results demonstrate partial functional redundancy among BRDs 2, 3, and 4 in E2-dependent gene regulation, with a unique role for BRD3 as a molecular sensor in MCF-7 cells that fine tunes the overall levels and activity of the family members to suit the needs of the cell. In this regard, the low selectivity of JQ1 among BET family members could be an asset from a therapeutic standpoint.
Our findings on the redundant, but functionally distinct, roles of BET family members are likely to be context-specific. For instance, a previous study demonstrated a critical role of BRD2, but not BRD3 or BRD4, for insulin secretion by pancreatic β-cells (46). In contrast, other studies have demonstrated that each BET family member is required for proper inflammatory response in macrophages, enhanced cell survival in multiple myeloma models, and AR-dependent growth of prostate cancers (17,47l,48). Thus, further investigation is required on the extent of functional redundancy as well as member-specific roles of the BET family proteins to fully understand the biological contexts in which the BET family members play critical roles. Our studies in T-47D cells are a step in this direction.
BRD3 marks active ERα enhancers
Given the central role of BRD3 in E2-responsive gene expression in MCF-7 cells, we examined the genome-wide localization of BRD3 in response to E2 treatment. Interestingly, BRD3 was enriched only at a subset of ERBSs upon E2 stimulation (Figure 6A and 6B). This subset of ERBSs is associated with elevated E2-dependent transcription at the ERBSs, as well as at their target genes (Figure 7A). These observations were consistent with our previous finding that not all TF binding sites result in transcription activation and TF binding sites that are enriched for coregulators correlate with active transcription (7,49–51). In addition, treatment with JQ1 disproportionally impaired E2-dependent gene expression for genes near ERBSs with BRD3 enrichment without significantly affecting the expression levels of genes near ERBSs without BRD3 enrichment (Figure 7B and 7C).
Based on our ChIP-seq experiments, we find that BRD3-enriched ERBSs are surrounded by additional ERBSs at higher frequencies and intensities. Recent studies by the Young group and others have reported clusters of TF binding sites, termed super enhancers, at or near context-specific genes, supporting context-specific gene expression networks (41,42). Other studies have demonstrated that the genes controlled by SEs are disproportionally sensitive to perturbation of transcription coregulators, including BET family members and the basal transcription factor TFIIH (52–54). In a concordance with these studies, the clusters of ERBSs enriched with BRD3 identified in our present study may be categorized as SEs that collectively activate key ERα regulated genes (Figure 6 and 7). Taken together, our results support an integral role of BRD3 in ERα-mediated enhancer activation and E2-responsive gene expression.
Mechanisms of BET family member recruitment to ERα enhancers
Data from our ChIP-qPCR assays conducted in the presence of JQ1 indicate that the bromodomains of BRD2, BRD3, and BRD4 are required for their recruitment to ERBSs (Figure 2C). In agreement with this, previous studies have shown that BET proteins are recruited to TF binding sites through bromodomains that bind to histones that are acetylated upon TF binding (14). Interestingly, our ChIP-seq experiments revealed that a subset of ERBSs that recruit BRD3 upon E2 treatment are enriched for acetylated histone H4 at well above detectable levels compared to the surrounding regions even prior to E2 exposure. In this condition, BRD3 recruitment at these sites is at a basal level (Figure 6A). In addition, BRD3 shows a narrower distribution surrounding this subset of ERBSs, while histone acetylation is more broadly distributed (Figure 6A; note that the genomic window is different in the heatmap representation of BRD3 and acH4 ChIP-seq). Moreover, ERBSs exhibit an E2-dependent increase of histone acetylation regardless of their association with BRD3 (Figure 6A and 6B). Collectively, these observations suggest that (1) H4 acetylation that occurs prior to E2 treatment at future ERBSs segregates ERBSs into two groups: with or without BRD3 recruitment upon E2 treatment, (2) BRD3 is recruited at an intermediate step of ERα enhancer formation, downstream of histone acetylation, (3) histone acetylation is not sufficient for BRD3 recruitment, and (4) therefore, there might be another layer of regulation for BET family member recruitment during ERα enhancer complex formation in addition to histone acetylation.
Supplementary Material
Implications.
BRD3 is recruited to and controls the activity of a subset estrogen receptor alpha (ERα) transcriptional enhancers, providing a therapeutic opportunity to target BRD3 with BET inhibitors in ERα-positive breast cancers.
Acknowledgements
We would like to thank members of the Kraus lab for helpful feedback during the course of this study and Dr. Samie Jaffrey for supporting the revision of this manuscript. This work was supported by a grant from the NIH/NIDDK (DK058110), grants from the Cancer Prevention and Research Institute of Texas (CPRIT; RP160319 and RP190236), and funds from the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowment to W.L.K.
Footnotes
Disclosure Declaration: The authors have no conflicts of interest to disclose.
References
- 1.Burns KA, Korach KS. Estrogen receptors and human disease: an update. Arch Toxicol 2012;86(10):1491–504 doi 10.1007/s00204-012-0868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. Mechanisms of estrogen action. Physiol Rev 2001;81(4):1535–65. [DOI] [PubMed] [Google Scholar]
- 3.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer 2012;12(6):381–5 doi 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 4.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43(1):27–33 doi 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami S, Nagari A, Kraus WL. Dynamic assembly and activation of estrogen receptor alpha enhancers through coregulator switching. Genes Dev 2017;31(15):1535–48 doi 10.1101/gad.302182.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasquez YM, Kraus WL. The estrogen-regulated transcriptome: rapid, robust, extensive, and transient. In: X. Z, editor. Estrogen Receptor and Breast Cancer Cancer Drug Discovery and Development: Humana Press; 2019. p 95–127. [Google Scholar]
- 7.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 2013;23(8):1210–23 doi 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009;462(7269):58–64 doi 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Qiao JX, Shetty A, Wu G, Huang Y, Davidson NE, et al. Regulation of estrogen receptor signaling in breast carcinogenesis and breast cancer therapy. Cellular and molecular life sciences : CMLS 2014;71(8):1549 doi 10.1007/s00018-013-1376-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Patel HK, Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol Ther 2018;186:1–24 doi 10.1016/j.pharmthera.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 2013;45(12):1446–51 doi 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez R, Meslamani J, Zhou MM. The bromodomain: from epigenome reader to druggable target. Biochim Biophys Acta 2014;1839(8):676–85 doi 10.1016/j.bbagrm.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep 2014;8(2):460–9 doi 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012;149(1):214–31 doi 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012;12(7):465–77 doi 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi Y The bromodomain and extra-terminal domain (BET) family: functional anatomy of BET paralogous proteins. Int J Mol Sci 2016;17(11) doi 10.3390/ijms17111849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014;510(7504):278–82 doi 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468(7327):1067–73 doi 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011;478(7370):529–33 doi 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017;12(5):323–39 doi 10.1080/15592294.2016.1265710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sengupta S, Biarnes MC, Clarke R, Jordan VC. Inhibition of BET proteins impairs estrogen-mediated growth and transcription in breast cancers by pausing RNA polymerase advancement. Breast Cancer Res Treat 2015;150(2):265–78 doi 10.1007/s10549-015-3319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PloS one 2011;6(3):e17911 doi 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harbor protocols 2011;2011(8):940–9 doi 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 24.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25(9):1105–11 doi 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28(5):511–5 doi 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31(1):46–53 doi 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26(6):841–2 doi 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004;20(17):3246–8 doi 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 29.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 2011;145(4):622–34 doi 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology 2003;4(5):P3. [PubMed] [Google Scholar]
- 31.Franco HL, Nagari A, Kraus WL. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell 2015;58(1):21–34 doi 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 2009;10(3):R25 doi 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res 2012;22(6):1015–25 doi 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38(4):576–89 doi 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011;27(7):1017–8 doi 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 2014;42(Database issue):D142–7 doi 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347(6220):1260419 doi 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 38.Sun L, Gao P. Small molecules remain on target for c-Myc. Elife 2017;6 doi 10.7554/eLife.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glont SE, Chernukhin I, Carroll JS. Comprehensive genomic analysis reveals that the pioneering function of FOXA1 is independent of hormonal signaling. Cell Rep 2019;26(10):2558–65 e3 doi 10.1016/j.celrep.2019.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 2016;165(3):593–605 doi 10.1016/j.cell.2016.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153(2):307–19 doi 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155(4):934–47 doi 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell 2014;56(2):219–31 doi 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong SH, Eun JW, Choi SK, Shen Q, Choi WS, Han JW, et al. Epigenetic reader BRD4 inhibition as a therapeutic strategy to suppress E2F2-cell cycle regulation circuit in liver cancer. Oncotarget 2016;7(22):32628–40 doi 10.18632/oncotarget.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gopalakrishnan R, Matta H, Tolani B, Triche T, Jr., Chaudhary PM. Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene 2016;35(14):1797–810 doi 10.1038/onc.2015.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deeney JT, Belkina AC, Shirihai OS, Corkey BE, Denis GV. BET Bromodomain Proteins Brd2, Brd3 and Brd4 Selectively Regulate Metabolic Pathways in the Pancreatic beta-Cell. PloS one 2016;11(3):e0151329 doi 10.1371/journal.pone.0151329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol 2013;190(7):3670–8 doi 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146(6):904–17 doi 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savic D, Roberts BS, Carleton JB, Partridge EC, White MA, Cohen BA, et al. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/ enhancer-binding protein beta binding sites. Genome Res 2015;25(12):1791–800 doi 10.1101/gr.191593.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014;15(4):272–86 doi 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 51.Kouwenhoven EN, Oti M, Niehues H, van Heeringen SJ, Schalkwijk J, Stunnenberg HG, et al. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep 2015;16(7):863–78 doi 10.15252/embr.201439941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153(2):320–34 doi 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014;159(5):1126–39 doi 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 2013;24(6):777–90 doi 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.