Abstract
T cell immunotherapies are often thwarted by the limited presentation of tumor-specific antigens abetted by the downregulation of human leukocyte antigen (HLA). We showed that drugs inhibiting ALK and RET produced dose-related, increases in cell surface HLA in tumor cells bearing these mutated kinases in vitro and in vivo, as well as elevated transcript and protein expression of HLA and other antigen processing machinery. Subsequent analysis of HLA presented peptides after ALK and RET inhibitor treatment identified large changes in the immunopeptidome with the appearance of hundreds of new antigens, including T cell epitopes associated with impaired peptide processing (TEIPP) peptides. ALK inhibition additionally decreased PD-L1 levels by 75%. Therefore, these oncogenes may enhance cancer formation by allowing tumors to evade the immune system by down regulating HLA expression. Altogether, RET and ALK inhibitors could enhance T cell-based immunotherapies by upregulating HLA, decreasing checkpoint blockade ligands, and revealing new, immunogenic, cancer-associated antigens.
Keywords: HLA, Immunotherapy, Neoantigens, ALK, RET
INTRODUCTION
Emerging therapies such as checkpoint blockade, CAR T cells, TCR engineered cells, and adoptive T cell transfer have focused attention on the presentation of cancer-associated antigens that are the target of these T cell-based therapies. Though the mechanisms of action behind these therapies vary tremendously, the core component of them is inducing the ability of T cells to kill cancer cells after their T cell receptors (TCRs) recognize the appropriate peptides complexed with human leukocyte antigen (HLA) (1,2). Peptide/HLA complexes that are recognized trigger a cytolytic response by the T cell (3,4). However, the low density surface presentation of tumor-associated peptide/HLA antigens, the lack of immunogenic new antigens, and the ability of some cancers to downregulate the antigen presentation machinery can hinder the ability of T cells to recognize and destroy their target (5-7). Multiple studies, including those performed in lung, melanoma, bladder, and colorectal carcinomas, have shown that up to two-thirds of tissue samples or cell lines harbor alterations in HLA (8). These alterations include loss of the entire HLA class I locus, defective antigen presentation machinery (like beta 2-microglobulin mutations), and loss of specific HLA loci (9-12). Thus, cancer cells use downregulation of HLA as a potential mechanism of immune escape (13).
We previously hypothesized that increasing the surface levels of HLA on cancer cells utilizing small molecule drugs could increase both the number and diversity of antigens presented, thereby increasing the efficacy of T cell-based immunotherapies (14). Inhibition of the mitogen-activated protein kinase (MAPK) pathway leads to increased transcript, protein, and surface levels of HLA in a STAT1-mediated manner (14). STAT1 increases HLA by activating transcription of the interferon regulatory factor 1 (IRF1), a transcription factor that binds to a interferon-stimulated response element (ISRE) and activates transcription of HLA- molecules (15). HLA increase led to amplified cytotoxicity of TCR mimic antibodies to selected epitopes in vitro. However, some MAPK inhibitors are not selective for tumor cells and may cause T cell dysfunction, potentially limiting the effectiveness of this approach (16-19).
Aberrations in the MAPK pathway or kinases that feed into this pathway are involved in the pathogenesis of many cancers. For example, 70% of papillary thyroid cancers have non-overlapping mutations in BRAF, Ras, or RET (REarranged during Transfection) (20). Binding of RET ligands and its co-receptor leads to dimerization, autophosphorylation and activation of downstream signaling pathways like MAPK and PI3K (20-22). The most common genetic alteration, RET/PTC1, a fusion of the 3’ portion of RET with the 5’ end of CCDC6 (Coil coil domain containing 6) (20), drives transcriptional activation and constitutive phosphorylation (23). RET fusions considered capable of oncogenic transformation are seen in about 30% of papillary thyroid cancer and 1-2% of non-small cell lung cancer (NSCLC) (24).
Mutated anaplastic lymphoma kinase (ALK) is another receptor tyrosine kinase that signals through the MAPK pathway. ALK is minimally expressed in adult tissues but mutations leading to expression are seen in a variety of cancers (25). An oncogenic fusion protein product of one such fusion, nucleophosmin-anaplastic lymphoma (NPM-ALK) results from the translocation between chromosome 2 and 5 and is found in approximately 75-80% of all ALK positive anaplastic lymphomas (ALCLs) (26). Homodimers or NPM/NPM-ALK heterodimers lead to constitutive activation of ALK and subsequent activation of downstream signaling pathways like MAPK and PI3K (27).
Here, inhibition of both mutant RET and ALK in cancer cells led to downregulation of ERK output and subsequent upregulation of antigen presentation machinery. Large changes in the HLA class I presented peptide ligandome were seen following ALK and RET inhibition, leading to the appearance of thousands of new T cell epitopes, some of which were immunogenic to human T-cells. Among the new T-cell epitopes we found were “impaired peptide processing peptides” (TEIPP), which are predicted to be found only on cells with defects in antigen processing and presentation(28-30). RNA-Seq and mass spectrometry data gave us insight into the changes in gene expression and HLA upregulation that led to this dramatic repertoire shift. Overall, we demonstrated that the expanded HLA capacity after ALK and RET inhibition gave rise to specific T-cell epitopes that potentially represent new specific targets for immunotherapies.
MATERIAL AND METHODS
Cells lines, inhibitors, and antibodies
The Karpas 299 (HLA-A*03, HLA-A*11), SUDHL-1 (HLA-A*02), and SUP-M2 cells were obtained from the Anas Younes lab at MSKCC and were maintained in RPMI-1640 with 10% FBS and 2mM L-glutamine. The TPC1 cell line (HLA-A*02, HLA-A*24) was obtained from the James Fagin lab at MSKCC and maintained in DME media with 5% FBS and 2mM L-glutamine. All obtained cells were tested for Mycoplasma. TT cells and LC-2/ad cells were purchased from ATCC and Sigma-Aldrich, respectively. Cells were maintained in culture 4-12 weeks. Cells were tested for mycoplasma approximately quarterly. For cells not recently purchased, cells were authenticated by flow cytometry of relevant markers and if clonal outgrowths appeared, sorted for purity. TT cells were maintained with Ham’s F12 medium supplemented with 10% FBS and 2mM L-glutamine. LC-2/ad cells were maintained with RPMI-1640: Ham’s F12 (1:1) medium supplemented with 10% FBS and 2mM L-glutamine. ALK inhibitors crizotinib, ceritinib, alectinib, and ruxolitinib were purchased from Selleck Chemicals. The RET inhibitor, AST 487, was purchased from MedChemExpress. The RET inhibitor, BLU6864 was generously provided to us by Blueprint Medicines. Multi-kinase inhibitor Cabozantinib which also targets RET and RAF inhibitor CEP-32496 were purchased from Selleck Chemicals. Antibodies for western blots for phospho-ERK (catalog 4370S), ERK (catalog 4696S), beta-2-microglobulin (catalog 12851S), STAT3 (4904S), pSTAT3 (catalog 9131S), STAT1 (catalog 9175S), pSTAT1 (catalog 9167S) and GAPDH (catalog 3683S) were purchased from Cell Signaling. Antibodies for western blot for anti-HLA-A (catalog sc-23446), goat anti-mouse IgG-HRP, mouse anti-rabbit IgG-HRP, anti-CD30, and donkey anti-goat IgG-HRP were purchased from Santa Cruz. Antibodies for flow cytometry to HLA-A02 (BB7.2) and HLA-A,B,C (W6/32) were purchased from eBioscience; the PD-L1 (MIH1) antibody was purchased from eBioscience; ESK1 and hIgG1 (catalog ET901) were provided by Eureka Therapeutics.
Flow cytometry
5×104 cells were seeded in a 12 well plate and treated with drug for 72hrs. Adherent cells were seeded one day before treatment. At 72hrs, cells were harvested, washed with PBS and incubated on ice with appropriate fluorophore conjugated antibodies diluted in FACS buffer for 1hr. Gating strategy: Cells were then washed and incubated 30 min with a viability dye (propidium iodide at 1ug/mL) and live cells only were analyzed by on a Guava flow cytometer with FlowJo software.
Western blots
Cells were seeded in a 60mm dish or 6 well plates. After the appropriate time point, cells were harvested, lysed in RIPA buffer (Thermo Instruments) and protein concentration was quantified by a lowry assay (using the Bio-Rad DC Protein Assay; #5000116 on a Spectramax device from Molecular Devices). Protein loading levels were equalized per lane and separated on SDS gels (Bio-Rad). Protein was transferred to a nitrocellulose membrance using semi-dry transfer (BioRad) Membranes were blocked with Omniblock (American Bio) and incubated with respected antibodies. When needed, secondary antibodies with HRP were incubated for 1hr. Enhanced chemiluminescent substrate for HRP enzymes was used to image protein levels (Thermo-Fischer; #34095) on a ChemiDoc MP imager with Soft Max Pro software (Biorad).
Real time PCR
In brief, cells were treated and incubated with appropriate small molecule inhibitors for 48hrs and RNA was extracted using Qiagen RNA Easy Plus (Qiagen; #74134). cDNA was created using qScript cDNA SuperMix (Quantabio; #95048). qPCR was performed using PerfeCTa FastMix II (Quantabio; #95118) and TaqMan real time probes purchased from Life Technologies: HLA-A (Hs01058806_g1), beta-2 microgobulin (Hs00187842_m1), TAP1 (Hs00388677_m1), TAP2 (Hs00241060_m1), and TBP (Hs00427620_m1). Data were normalized to baseline expression of each analyzed gene separately. For details see (60).
Antibody-Dependent Cellular Cytotoxicity
For each experiment reported, fresh peripheral blood mononuclear cells were derived from a healthy donor by Ficoll density centrifugation after receiving informed consent on Memorial Sloan Kettering Institutional Review Board-approved protocols. TPC1 cells were treated with DMSO, 10nM AST487, or 100nM trametinib for 72 hrs to increase surface HLA, after which cells were thoroughly washed in PBS to remove the drugs. Cells were then labeled with 1uCi/well chromium-51 for 1hr at 37°C. Chromium labeled TPC1 cells were co-cultured with PBMCs and ESK1 (a human IgG1 reactive with WT1 peptide/HLA-A*02:01) or its isotype control (hIgG1). Different effector:target ratios were used and after 5hrs of incubation at 37 °C, the supernatant was harvested and chromium levels were measured through standard chromium-51 release assay on a Top Count machine (Perkin Elmer).
RNA-seq
For RNA-seq analysis total RNA was extracted using the RNeasy Mini Kit (Qiagen) after treatment of TPC-1 cells with either DMSO, AST487 or Cabozantinib for 72 hours. Purified polyA mRNA was subsequently fragmented, and first and second strand cDNA synthesis performed using standard Illumina mRNA TruSeq library preparation protocols. Double stranded cDNA was subsequently processed for TruSeq dual-index Illumina library generation. For sequencing, pooled multiplexed libraries were run on a HiSeq 2500 machine on RAPID mode. Approximately 10 million 76bp single-end reads were retrieved per replicate condition. Resulting RNA-Seq data was analyzed by removing adaptor sequences using Trimmomatic, aligning sequencing data to GRCh37.75(hg19) with STAR, and genome wide transcript counting using HTSeq to generate a RPKM matrix of transcript counts. This RPKM matrix was further log (log2) transformed and normalized per gene to obtain the Z-score. Differential gene expression was analyzed by looking at fold changes between experimental conditions.
Animal studies
All animal experiments were conducted in accordance with and the approval of the MSKCC IACUC (Institutional Animal Care and Use Committee) protocols. Female NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) and NRG (NOD.Cg-Rag1tm1MomIl2rgtm1Wjl/SzJ) mice were purchased from the Jackson Laboratory at 5-10 weeks old. For experiments in vivo with RET and ALK, 2.5-6 ×106 tumor cells in PBS were subcutaneously injected into the flank of mice. When tumors were palpable (2-3 mm), mice were treated daily with drugs or vehicles through oral gavage of drugs in 200ul of water. At day 4 or 7, tumors were harvested and flow cytometry was conducted to determine the effect of inhibitors on HLA and PD-L1 on the tumor cells. N=5 for all treatment groups in vivo (one outlier was excluded from the vehicle group in the RET experiment, but results remained significant: P value with outlier included 0.031, P value without outlier 0.016)). TPC1 cells were transduced with luciferase and GFP on an SFG vector and this allowed gating of the tumor cells in flow cytometry. A CD30 antibody was used in the ceritinib experiments.
Immunopurification of HLA class I ligands.
Immunopurification affinity columns were prepared as described previously (31). In brief, 40 mg of Cyanogen bromide-activated-Sepharose® 4B (Sigma-Aldrich, Cat# C9142) was activated with 1mM hydrochloric acid (Sigma-Aldrich, Cat# 320331) for 30 min. Subsequently, 0.5 mg of W6/32 antibody (BioXCell, BE0079; RRID: AB_1107730) was coupled to sepharose in presence of binding buffer (150mM sodium chloride, 50 mM sodium bicarbonate, pH 8.3; sodium chloride: Sigma-Aldrich, Cat# S9888, sodium bicarbonate: Sigma-Aldrich, Cat#S6014) for at least 2 hours at room temperature. Sepharose was blocked for 1 h with glycine (Sigma-Aldrich, Cat# 410225). Columns were equilibrated with PBS for 10 min. TPC1 cells were treated with DMSO, 10nM AST487, or 100nM cabozantinib. Karpas 299 cells were treated with DMSO, 100nM crizotinib or 100nM ceritinib for 72h. 20 – 30 × 106 cells were harvested and washed three times in ice-cold sterile PBS (Media preparation facility MSKCC). Afterwards, cells were lysed in 1 ml 1% CHAPS (Sigma-Aldrich, Cat# C3023) in PBS, supplemented with 1 tablet of protease inhibitors (cOmplete, Cat# 11836145001) for 1 hour at 4°C. This lysate was spun down for 1 hour at 20,000 g at 4°C. Supernatant was run over the affinity column through peristaltic pumps at 1 ml/min overnight at 4°C. Affinity columns were washed with PBS for 15 min, run dry, and HLA complexes subsequently eluted three times with 200 μl 1% trifluoracetic acid (TFA, Sigma/Aldrich, Cat# 02031). For separation of HLA ligands from their HLA complexes tC18 columns (Sep-Pak tC18 1 cc VacCartridge, 100 mg Sorbent per Cartridge, 37-55 μm Particle Size, Waters, Cat# WAT036820) were prewashed with 80% acetonitrile (ACN, Sigma-Aldrich, Cat# 34998) in 0.1% TFA and equilibrated with two washes of 0.1% TFA. Samples were loaded, washed again with 0.1% TFA and eluted in 400 μl 30% ACN in 0.1%TFA. Sample volume was reduced by vacuum centrifugation for mass spectrometry analysis.
LC-MS/MS analysis of HLA ligands
Samples were analyzed by a high resolution/high accuracy LC-MS/MS (Lumos Fusion, Thermo Fisher). Peptides were desalted using ZipTips (Sigma Millipore, Cat. # ZTC18S008) according to manufactures instructions and concentrated using vacuum centrifugation prior to being separated using direct loading onto a packedin-emitter C18 column (75um ID/12cm, 3 μm particles, Nikkyo Technos Co., Ltd. Japan). The gradient was delivered at 300nl/min increasing linear from 2% Buffer B (0.1% formic acid in 80% acetonitrile) / 98% Buffer A (0.1% formic acid) to 30% Buffer B / 70% Buffer A, over 70 minutes. MS and MS/MS were operated at resolutions of 60,000 and 30,000, respectively. Only charge states 1, 2 and 3 were allowed. 1.6 Th was chosen as the isolation window and the collision energy was set at 30%. For MS/MS, the maximum injection time was 100ms with an AGC of 50,000.
Mass spectrometry data processing
Mass spectrometry data was processed using Byonic software (version 2.7.84, Protein Metrics, PaloAlto, CA) through a custom-built computer server equipped with 4 Intel Xeon E5-4620 8-core CPUs operating at 2.2 GHz, and 512 GB physical memory (Exxact Corporation, Freemont, CA). Mass accuracy for MS1 was set to 10 ppm and to 20 ppm for MS2, respectively. Digestion specificity was defined as unspecific and only precursors with charges 1,2, and 3 and up to 2 kDa were allowed. Protein FDR was disabled to allow complete assessment of potential peptide identifications. Oxidization of methionine, N-terminal acetylation, phosphorylation of serine, threonine and tyrosine were set as variable modifications for all samples. All samples were searched against the UniProt Human Reviewed Database (20,349 entries, http://www.uniprot.org, downloaded June 2017). Peptides were selected with a minimal log prob value of 2 resulting in a 1% false discovery rate and were HLA assigned by netMHC 4.0 with a 5% rank cutoff.
Peptide Stimulation
Peripheral blood mononuclear cells were again derived from healthy donors after receiving informed consent (see above). T cells were isolated by Ficoll density centrifugation and stimulated with pools of peptides that were selected from the population of: (1) new peptides that appeared after RET inhibitor treatment, (2) peptides found on cells before RET inhibitor treatment, and (3) irrelevant peptides not found on the target cells. CD14+ cells were isolated from PBMCs by negative immunomagnetic cell separation using an isolation kit (Miltenyi Biotec). CD14 cells were used for stimulation in week one and autologous dendritic cells were generated for use thereafter. The purity of the cells was always more than 98%. Monocyte-derived dendritic cells (DCs) were generated from CD14+ cells, by culturing the cells in RPMI 1640 medium supplemented with 1% AP, 500 units/mL recombinant IL-4, and 1,000 units/mL GM-CSF. On days 2 and 4 of the incubation, fresh medium with IL-4 and GM-CSF was either added or replaced half of the culture medium. On day 6, maturation cytokine cocktail was added (IL-4, GM-CSF, 500 IU/mL IL-1, 1,000 IU/mL IL-6, 10 ng/ml TNF-α, and 1 ug/mL PGE-2). An interferon-gamma ELISpot was performed at the beginning of week three. For this HA-Multiscreen plates (Millipore) were coated with 100 uL of mouse anti-human IFN-gamma antibody (10 ug/mL; clone 1-D1K; Mabtech #3420-2A) in PBS, incubated overnight at 4°C, washed with PBS to remove unbound antibody, and blocked with RPMI 1640/10% autologous plasma (AP) for 2 h at 37°C. Purified CD3+ T cells (> 98% pure) were plated with either autologous CD14+ (10:1 E: APC ratio) or autologous DCs (30:1 E: APC ratio). Various test peptides were added to the wells at 20 ug/mL. Negative control wells contained APCs and T cells without peptides or with irrelevant peptides. Positive control wells contained T cells plus APCs plus 20 ug/mL phytohemagglutinin (PHA, Sigma). All conditions were done in triplicates. Microtiter plates were incubated for 20 h at 37°C and then extensively washed with PBS/0.05% Tween and 100 uL/well biotinylated detection antibody against human IFN-g (2 ug/mL; clone 7-B6-1; Mabtech) was added. Plates were incubated for an additional 2 h at 37°C and spot development was done as described. Spot numbers were read and determined by Zellnet Consulting In. T cell killing of target cells was measured at week five as described in the section of ADCC above.
Statistics
P values were calculated with GraphPad Prism 7 using an unpaired t test for flow cytometry experiments, RNA quantitation and in vivo experiments. Error bars indicate SEM for in vivo experiments and SD for flow cytometry and RNA quantitation experiments. All flow cytometry and RNA quantitation experiments were performed in technical triplicates and with a minimum of 2 biological replicates. Western blots were done at least two-three times. Representative demonstration blots are shown only. Values are reported in figures with “*” equal to P≤0.05, “**” equal to P≤0.01, “***” equal to P≤0.001, and “****” equal to P≤0.0001. No symbol indicates not statistically significant (P>0.05).
RESULTS
ALK inhibition increased HLA expression through MAPK pathway suppression
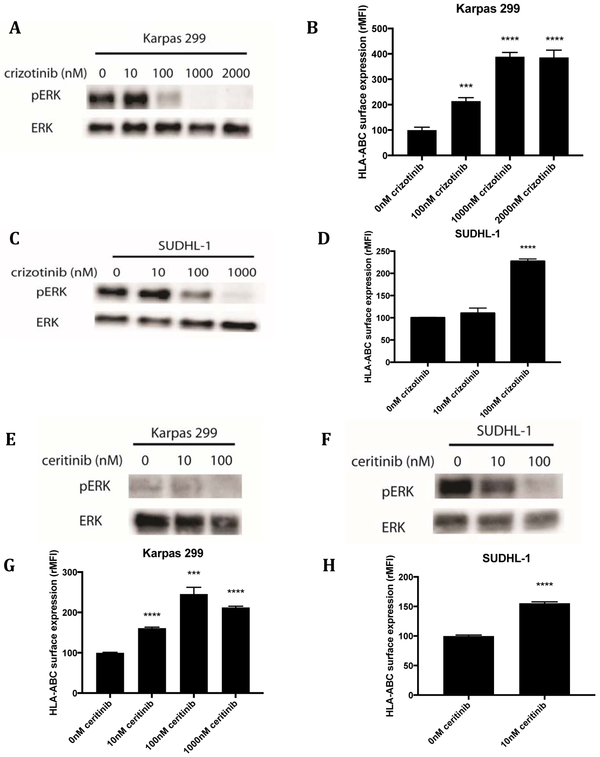
Crizotinib is a small molecule tyrosine kinase inhibitor that is Food and Drug Administration (FDA) approved for the treatment of mutated ALK positive non-small cell lung cancer (NSCLC) (32). Increasing concentrations of crizotinib on Karpas 299, a NPM-ALK fusion oncogene-positive ALCL cell line, showed a dose-related, reduction of pERK at 3 hours of treatment, indicating inhibition of ALK shuts down the MAPK pathway (Fig. 1A). Flow cytometric analysis of HLA levels after a 72 hour incubation of Karpas 299 cells with crizotinib showed an inverse dose-response. Decreasing levels of pERK were associated with increased levels of surface HLA class I complexes (Fig. 1B). HLA levels on Karpas 299 lymphoma cells treated with 1uM crizotinib increased 4-fold compared to control cells treated with DMSO. The plateau in surface HLA upregulation that was seen at higher concentrations of crizotinib correlated with complete shut-down of ERK phosphorylation at lower doses. Similar results were seen with SUDHL-1, another NPM-ALK mutated fusion protein positive ALCL line (Fig. 1C,D).
Figure 1: ALK inhibition decreased pERK levels and increased surface HLA levels in ALK mutated cell lines.
(A) Karpas 299 cells were treated with increasing concentrations of crizotinib for 3hrs and pERK and ERK (loading control) were measured by western blot. (B) After 72hrs of crizotinib treatment, flow cytometry was used to measure cell surface HLA-A, B, C on Karpas 299 cells. Similarly, SUDHL-1 cells were treated with crizotinib and (C) pERK and ERK and (D) cell surface HLA molecules were measured. Western blot for ERK and pERK on (E) Karpas 299 cells and (F) SUDHL-1 cells treated with the second-generation ALK inhibitor, ceritinib, for 3hrs. Flow cytometry analysis of HLA-A, B, C expression in (G) Karpas 299 cells and (H) SUDHL-1 cells treated with ceritinib for 72hrs. W6/32-APC antibody was used to measure HLA-A,B,C.
To confirm that ALK inhibition was the mechanistic target for HLA regulation, another small molecule ALK inhibitor, ceritinib (LDK378), which is approved for crizotinib-resistant NSCLC, was investigated (33). Treatment of Karpas 299 and SUDHL-1 with increasing concentrations of ceritinib also shut down pERK levels (Fig. 1E,F). Cells were comparatively more sensitive to ceritinib than crizotinib; however, cell surface HLA increased in both cell lines in a dose-dependent manner (Fig. 1G,H). Similar results were seen with alectinib, another second-generation ALK inhibitor (Supplemental Fig. S1A-D). We also showed that the effect on HLA is temporary, but increased HLA levels are still seen 6 days later, indicating its potential for clinical use (Supplemental Fig. S1E). A representative flow cytometry histogram of the HLA increases is also provided as an example of the typical raw data (Supplemental Fig. S1F). As these three inhibitors share ALK as a target, but have different off-target kinases, similar results with the three inhibitors in two different cell lines providee strong confidence that the increase in HLA seen was a result of ALK inhibition and not another kinase (33-35). The dependence on MAPK inhibition for HLA upregulation was further confirmed with the EML4-ALK protein positive fusion cell line H2228. In these cells, crizotinib did not change pERK levels, and consequently cell surface HLA levels did not change either. Ceritinib resulted in minimal changes in pERK levels and only a minimal increase in surface HLA levels was seen (Supplemental Fig. S1G, H). Overall, using several inhibitors of ALK in multiple cell lines, the inhibition of ERK output by the drugs correlated positively with cell surface HLA levels.
RET inhibition increased HLA expression through MAPK pathway suppression
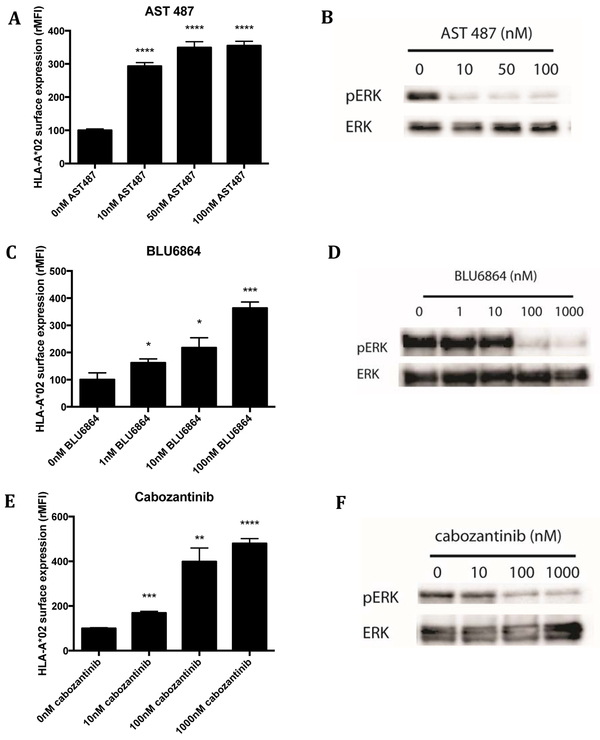
RET is a receptor tyrosine kinase that also positively feeds into the MAPK pathway. Specific targeting of RET with AST487 inhibits growth of thyroid cell lines with activating RET mutations, such as TPC1 (36). In concordance with the results for ALK inhibition, treatment of TPC1 cells with AST487 for 72h led to a 3 to 4- fold increase in cell surface HLA class I levels (Supplemental Fig. S2A). In addition to pan-HLA increases, we investigated HLA upregulation of one of the most common HLA alleles, HLA-A*02:01, which was also increased (Fig. 2A). Inhibition of pERK was seen even at concentrations as low as 10nM AST487 (Fig. 2B).
Figure 2: RET inhibition in TPC1 cells led to decreased pERK levels and increased surface expression of HLA.
TPC1 cells, a papillary thyroid carcinoma line with a RET/PTC1 rearrangement, were treated with the RET inhibitor, AST487. (A) After 72hrs, cell surface HLA-A*02 was measured through flow cytometry. (B) pERK and ERK (loading control) were measured at 24hrs by western blot. Similar results were observed with two other RET inhibitors: BLU6864 and cabozantinib. (C) Cell surface HLA expression and (D) pERK and ERK expression after BLU6864 treatment. (E) Cell surface HLA expression and (F) pERK and ERK expression after Cabozantinib treatment. BB7-APC antibody was used to measure HLA-A*02.
Two other small molecule kinase inhibitors of RET showed similar effects. BLU6864, one of the first inhibitors designed to selectively inhibit RET, and cabozantinib small molecule inhibitor of RET, MET, and VEGF2 that is also FDA approved for treatment of medullary thyroid cancer, were tested on TPC1 cells in the same manner (37), . Both inhibitors at 100nM led to a 4-fold increase in surface pan-HLA, as well as HLA-A*02 specifically. Again, a dose response relationship was seen with increasing concentrations of drug. Western blot analysis confirmed decreasing pERK levels with inhibitor treatment (Fig. 2C-F; Supplemental Fig. S2B, C). By use of these different RET inhibitors, we confirmed that inhibition of RET was most likely causing the increase in HLA (36,37). To further validate that RET inhibition increased HLA, we applied siRNAs to knockdown RET expression. Knockdown of RET by 2 different siRNAs resulted in increased HLA expression (Supplemental Fig. S2D).
To determine if these findings were reproduced with other RET mutations or in other cancer types, we tested a lung cancer cell line LC-2/ad, which harbors the same CCDC6-RET fusion as TPC1 (38). We also examined TT cells, which are a medullary thyroid cell line that is driven by a C634W mutation leading to dimerization and activation (39). HLA levels of the TT line increased in a AST487 dose-related manner (Supplemental Fig. S2E). In the other line, only small increases were seen with RET inhibition (Supplemental Fig. S2F). Use of cabozantinib and CEP-32496 also had minimal effects on HLA upregulation (Supplemental Fig. S2G,H) in these 2 lines. Due to more robust upregulation in TPC1, we used these cells for other RET inhibition studies.
ALK and RET inhibition increased HLA through alterations in transcript and protein expression
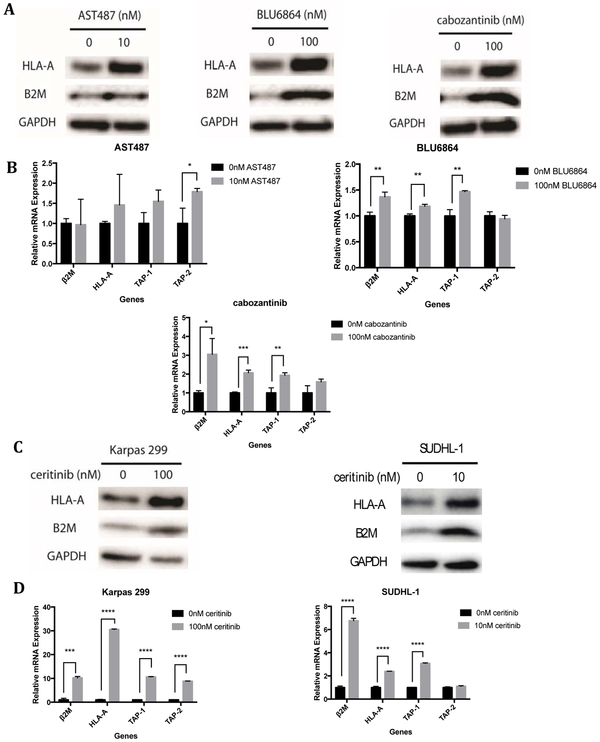
Nascent HLA molecules reside in the endoplasmic reticulum until they associate with beta-2-microglobulin, after which TAP1 and TAP2 transport the proteasome-cleaved peptides into the ER and antigenic peptides are loaded onto the complex. This complex is shuttled to the cell surface and later recycled back through endosomes(3). Therefore, the increase in cell surface HLA could have been the result of increased transcription or translation, increased stabilization by peptide loading and beta-2-microglobulin association, or reduced degradation. The increase in surface HLA seen from RTK inhibition resulted from an increase in protein levels of HLA, indicating an effect on total molecule numbers (Fig 3; Supplemental Fig. S3). The drugs also caused variable increases in transcript levels of HLA and other proteins involved in antigen processing machinery, though in general there was an increase in either HLA and/or TAP1, TAP2, or beta-2-microglobulin (Fig. 3A-D; Supplemental Fig. S3A-D). The magnitude of change seen in each of the proteins varied based on the cell line and inhibitor used and was not always coordinated across all proteins, suggesting other differences among these cell lines in the control of these processes. The increase in TAP1 and TAP2 and beta-2-microglobulin, indicated the potential for more peptide loading in the ER and more stabilization of the cell surface HLA.
Figure 3: The regulation of HLA increase was at the transcript level.
(A) Representative western blots probing for HLA-A, beta-2-microglobulin (B2M), and GAPDH (loading control) in TPC1 cells at 72hrs after RET inhibitor treatments. (B) HLA and antigen processing machinery (TAP1, TAP2 and Beta-2 microglobulin) transcript levels measured by qPCR at 48hrs after RET inhibitor treatment. (C) Western blots for HLA-A, beta-2-microglobulin (B2M), and GAPDH (loading control) and (D) RNA levels of for HLA-A, beta-2-microglobulin (B2M), and TAP-1 and TAP-2 in Karpas 299 cells and SUDHL-1 cells after ceritinib treatment. qPCR experiments were in performed in technical triplicate.
The role of the JAK/STAT pathway in HLA expression after MAPK inhibition
STAT1 is a primary regulator of HLA and other antigen presentation machinery proteins (15, 40). STAT1 increases HLA by activating transcription of IRF1, a transcription factor that binds to ISRE and activates transcription of HLA-A, HLA-B, HLA-C, and HLA-F (15,41). This STAT1-mediated increase in surface HLA is also reported with EGFR inhibitors (42,43). EGFR is a receptor tyrosine kinase that feeds into the MAPK pathway, and when inhibited, leads to decreases in pERK (44,45). STAT1 is driving the changes in HLA mRNA, protein and cell surface expression after MAPK pathway inhibition (14, 15,40). Activated MAPK associated kinases (including ERK1and ALK) directly reduce activated pSTAT1, which promotes proteasomal degradation of pSTAT1 via PIAS1 (46-49). Hence, inhibition of these pathways should directly increase pSTAT1 output.
Next, we asked if there were other activators of STAT1 that may play a role in the upregulation of HLA after MAPK pathway inhibition. JAK is a primary activator of STAT1 when stimulated with IFN gamma (15, 40); however, specific inhibition of JAK with ruxolitinib had no effect on the upregulation of HLA expression in either ALK mutated Karpas 299 cells or RET mutated TPC1 cells after specific inhibition of their respective oncogenic kinases (Supplemental Fig S4A). As a control, ruxolitinib blocked IFNγ-mediated upregulation of HLA at these doses (Supplemental Fig S4B.)
We also asked if there was evidence of cytokine secretion by the cancer cells in response to ALK or RET inhibition that might account for indirect activation of the more traditional JAK/STAT pathway (Supplemental Fig S5). Alectinib inhibition in Karpas 299 lymphoma had no effect on IFNα, IFNγ, IL4, and reduced IL6 and TNFα secretion (Supplemental Fig S5). AST487 inhibition in TPC1 thyroid cells had no effect on IFNα, IFNγ, IL4, IL6 or TNFα secretion (Supplemental Fig S5). Therefore, it was unlikely that these inhibitors acted to upregulate the JAK/STAT pathway either directly, as shown above, or indirectly, by increased cytokine release. As a positive control, IFNγ increased both IL4 and IL6 in these cells, which was reduced by ruxolitinib (Supplemental Fig S5). Therefore, the dominant mechanism for the activity seen here in response to ALK or RET inhibition appeared to be loss of the direct reduction in pSTAT1 by the MAPK associated enzymes.
Increase of HLA expression after MAPK inhibition was produced in vivo
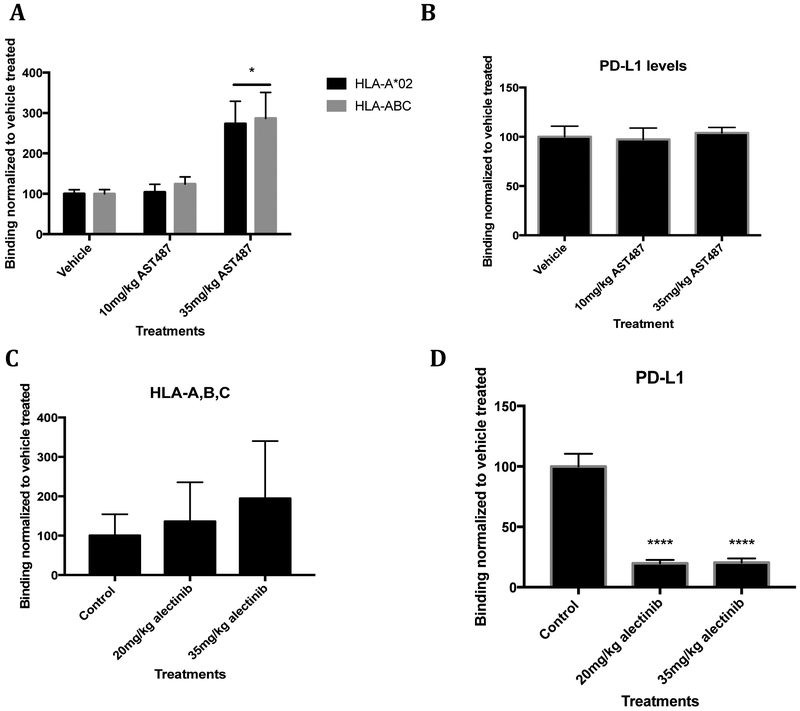
To determine if the RET and ALK inhibitors produced similar effects in live animals, we treated mice bearing TPC1 and Karpas 299 tumors with RET inhibitors. The highly immunodeficient NRG (NOD-Rag1null IL2rgnull) mice were subcutaneously injected with luciferase tagged TPC1 cells in their flank. When the tumors were palpable, mice were given vehicle or AST 487 through once daily oral gavage for 7 days. Afterwards, cells were harvested immediately and stained with antibodies against HLA-A*02 and HLA-ABC. Dose-related increases in all HLA class I levels were seen with AST487 treatment, indicating that HLA also can be upregulated in vivo by RET inhibition (Fig. 4A). Moreover, PD-L1 levels were measured and no increase in PD-L1 was seen (Fig. 4B). These data suggest a potential for RET inhibition in combination with T cell-based therapies or checkpoint blockade inhibition. NSG mice were also injected with Karpas 299 cells and treated with vehicle or ALK inhibitors, crizotinib or alectinib. Levels of HLA increased modestly in a dose-dependent manner for both drugs, though not all mice responded (Fig. 4C; Supplemental Fig. S6A). PD-L1 decreased with increasing doses of crizotinib and alectinib (Fig. 4D, Supplemental Fig. S6B). Treatment with alectinib dropped PD-L1 by approximately 75% (Fig. 4D). The dramatic decreases in PD-L1 were seen in vitro as well (Supplemental Fig. S6C,D). ALK inhibition was able to decrease levels of nectin-2, another checkpoint ligand that binds to TIGIT (Supplemental Fig. S7). However, ALK inhibition did not affect all checkpoint ligands, as levels of galectin-9, the ligand of TIM-3, stayed constant (Supplemental Fig. S7). RET inhibition did not alter either of these ligands (Supplemental Fig. S7). Altogether, these effects could have a profound impact on using RET and ALK inhibitors with therapies that rely on T cells.
Figure 4: Increased tumoral surface HLA expression and decreased tumoral PD-L1 expression in vivo during ALK inhibition.
(A) TPC1 cells were subcutaneously injected into NRG mice and harvested after 7 days of AST487 treatment or vehicle treatment (n=5). Cell surface HLA-A*02:01 and HLA-A, B, C were measured (with BB7 and W6/32 respectively). (B) PD-L1 levels were measured after RET inhibition. (C, D) Karpas 299 cells were subcutaneously injected into NSG mice, treated with alectinib, and harvested (n=5). HLA-A, B, C and PD-L1 levels were measured by flow cytometry.
RET inhibition altered the surface immunopeptidome and increased peptide presentation
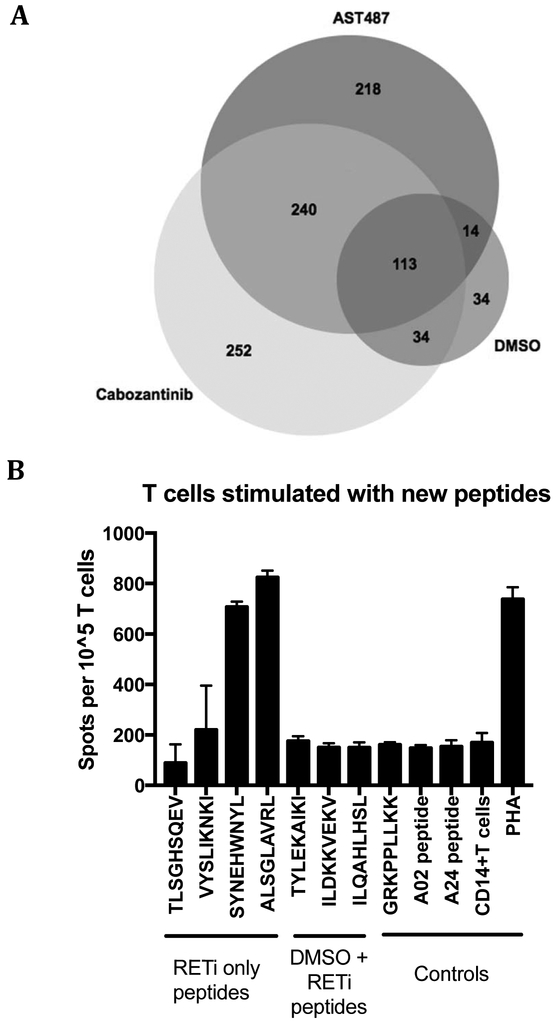
The increases in cell surface HLA and antigen presentation machinery after drug treatment suggested it was possible that the peptide repertoire would also be altered with RTK inhibition. Such changes could provide a second rationale for the use of these inhibitors as immune adjuvants or immunotherapies by enabling presentation of novel tumor associated antigens or neoantigens to be recognized by T cell based therapies (50,51). We treated cells with AST487 or cabozantinib for 3 days and then analyzed the peptides presented by HLA class I by mass spectrometry. We profiled the peptides acquired from 3 independent experiments, comparing untreated cells with drug treated cells, and determined the number and sequences of unique peptides that were complexed with cell surface HLA molecules. Peptides detected in all 3 runs were analyzed preferentially (Fig. 5A), as these HLA ligands represented the most robust group. RET inhibitor treated groups yielded 3-fold higher amounts of unique HLA ligands compared to the DMSO group: 639 for Cabozantinib, 585 for AST487 and 195 for the DMSO vehicle control group (Fig. 5A). Half of the peptides seen only in the treated subgroups (240 unique ligands) were shared between the two treatment groups (Fig. 5A). 34 HLA presented peptides from the DMSO group could not be identified anymore after drug treatment (Fig. 5A). Overall, this analysis showed a large increase in detectable HLA ligands for the TKI treated subgroups. We also analyzed the changes in unique HLA ligands when the new peptide was found in only one or two of the three biological mass spectrometry replicates performed; in this case, the number of new peptides increased 5 fold after drug treatment (Supplemental Fig. S8A). A similar shift in new antigens and peptide repertoire was seen with ALK inhibition by the three different ALK inhibitors when treating Karpas 299 cells (Supplemental Fig. S8B).
Figure 5: Mass spectrometry of eluted HLA class I presented peptides show a change in peptide number and repertoire after RET inhibition.
(A) HLA bound peptides from lysate of TPC1 cells treated with DMSO (control), 10nM AST487, or 100nM cabozantinib were analyzed by mass spectrometry (n=3). Only peptides found in all three separate runs were counted. Each circle encompasses the unique peptides identified after treatment; overlaps of circles show presence of each peptide in 2 or more groups. 458 and 492 new peptides appeared in each of the two RET inhibitor groups, respectively. (B) IFN-gamma ELISpot data for T cells stimulated with new arising peptides after RET inhibitor treatment (TLSGHSQEV, VYSLIKNKI, SYNEHWNYL, ALSGLAVRL). A representative figure is shown of two. PHA was used as a positive control. An irrelevant peptide (GRKPPLLKK) and CD14+ cells were used as negative controls.
The appearance of hundreds of new cell surface peptides complexed with HLA class I that are not present before drug treatment increased the chance of presentation of immunogenic peptides with this group. We tested the immune response of HLA-A*02:01 positive healthy donors to a small sample of the newly presented HLA ligands. Through an IFN-gamma ELISpot assay, we showed several of the peptides arising after drug treatment were immunogenic. Human T cells were stimulated against a sample pool of four of the HLA ligands (TLSGHSQEV, VYSLIKNKI, SYNEHWNYL, ALSGLAVRL). Two of the four new antigens were able to elicit T cell mediated IFN gamma response to autologous CD14+ cells presenting those corresponding peptides (Fig. 5B). No response of these cells was seen against several control peptides that were found before drug treatment on TPC1 cells (TYLEKAIKI, ILDKKVEKV, ILQAHLHSL) or to an irrelevant peptide (GRKPPLLKK) (Fig. 5B).
To further understand the possible biochemical mechanisms behind the repertoire shift, we first looked at the motifs of the peptides found in each treatment group to determine if drug treatment was altering protein cleavage and processing (Supplemental Fig. S8C). Processing of the peptides did not appear to be altered substantially, as the frequency of amino acids in each position was similar before and after treatment, thus not accounting for these large changes in the repertoire (Supplemental Fig. S8C). We next performed RNA-seq on the cells treated with RET inhibitors to determine if the drugs were altering protein expression and thus the ligandome. We also analyzed the protein derivation of the peptides in the ligandome in comparison to the upregulated proteins in each cell group (Supplemental Fig. S8D-G). Network analysis of new peptides after RET inhibition showed enrichment in the cell cycle arrest and negative regulation of the cell cycle pathways (Supplemental Fig. S8D, E). Among the new peptides in the RET inhibitor treated cells, 2 out of the 16 known TEIPPs with spontaneous immune responses in healthy donors presented on HLA-A*02:01 were detected (Table 1) (28). Previously, TEIPPs have been described in cells that lack TAP or are low in HLA surface expression (28-30, 57). Here we instead showed that in cells that have increased levels of HLA and TAP proteins, TEIPPs were presented, suggesting an alternate mechanism for their appearance.
Table 1:
HLA-A*02:01 TEIPP peptides found after RET inhibitor treatment
| TIEPP sequence | DMSO | AST487 | Cabozantinib |
|---|---|---|---|
| ALFSFVTAL | 0 | 0 | 0 |
| FLGPWPAAS | 0 | 1 | 1 |
| FLSELQYYL | 0 | 0 | 0 |
| FLYPFLSHL | 0 | 0 | 0 |
| ILEYLTAEV | 0 | 0 | 0 |
| LLALAAGLAV | 0 | 0 | 0 |
| LLLDVPTAAV | 0 | 0 | 1 |
| LLLSAEPVPA | 0 | 0 | 0 |
| LLWGRQLFA | 0 | 0 | 0 |
| LSEKLERI | 0 | 0 | 0 |
| LTLLGTLWGA | 0 | 0 | 0 |
| SVLWLGALGL | 0 | 0 | 0 |
| TLLGASLPA | 0 | 0 | 0 |
| VIIKPLVWV | 0 | 0 | 0 |
| VLAVFIKAV | 0 | 0 | 0 |
| VLLDHLSLA | 0 | 0 | 0 |
The 16 known TEIPP peptides reported for HLA-A*02 and the drug treatment groups in which those TEIPP peptides were found. Number indicates runs TEIPP peptide were found in (n=3).
PBMC viability and HLA levels were not affected by the inhibitors, suggesting specificity of the drugs for cells with these altered pathways (Supplemental Figure S9). In addition, only the canonical HLA molecules were affected, while noncanonical molecules like HLA-E were unchanged (Supplemental Fig. S10).
Unmasked antigens after RET inhibition enhanced cellular cytotoxicity against HLA complexes
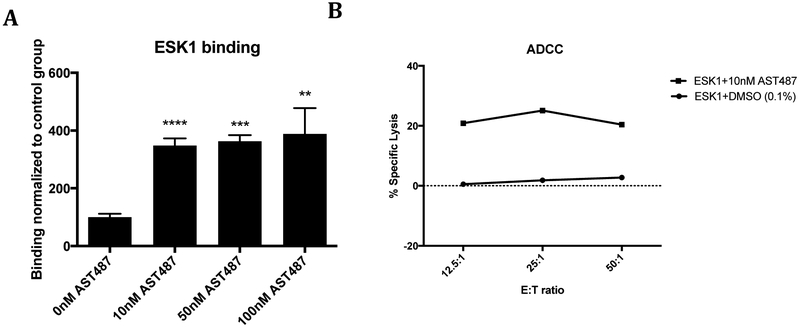
TCR mimic monoclonal antibodies (TCRm) recognize peptide/HLA complex epitopes in a manner similar to that of a TCR, but have the advantageous pharmacological properties of an antibody (52). ESK1 is a TCR mimic antibody that reacts with the RMFPNAPYL peptide derived from WT1 as well as several other peptides with similar sequences, when complexed with HLA-A*02:01(53). Although, ESK1 bound minimally to naïve TPC1 cells, increased ESK1 binding was seen following RET inhibition (Figure 6A). Analysis of the mass spectrometry data show that TPC1 cells do not present known epitopes that could be bound by ESK1 before treatment; however, after RET inhibition, the off target peptide (RMFPGEVAL) is present and allows binding of ESK1 (54). Hence, we used ESK1 as a tool to show that the increased HLA expression and presentation of new peptides following RET inhibition resulted in improved antibody-dependent cellular cytotoxicity (ADCC) activity when TPC1 cells were pre-incubated with the RET inhibitor, AST487 (Figure 6B). Thus, the ability to unmask new antigens has the potential to enhance TCR-based recognition and increase T-cell mediated lysis of target cells.
Figure 6: Unmasked antigen led to lysis of TPC1 cells by a TCR mimic antibody.
(A) ESK1, a TCR mimic antibody, was fluorescently labeled to probe binding after 72 hours RET inhibitor treatment by flow cytometry. (B) Chromium-51 labeled TPC1 cells were incubated with ESK1 and human PBMCs for 5hrs at 37°C and percent specific lysis was calculated for DMSO (control) and AST487 treated groups.
DISCUSSION
With the plethora of effective T cell-based therapies for cancer, the ability to safely increase HLA levels could have a profound impact across a wide variety of treatments. The minimal, or absence of, ALK and RET expression on normal cells make these RTKs appealing targets for selective HLA upregulation on cancer cells that express their activated forms, with little risk for side effects. Whereas we and others have previously shown that MEK inhibitors also upregulated HLA, these drugs also adversely affect T cell function (14,19,55,56). ALK and RET inhibitors act upstream of other pathways and if more than one pathway affecting HLA is inhibited, this could lead to an additive effect on HLA upregulation.
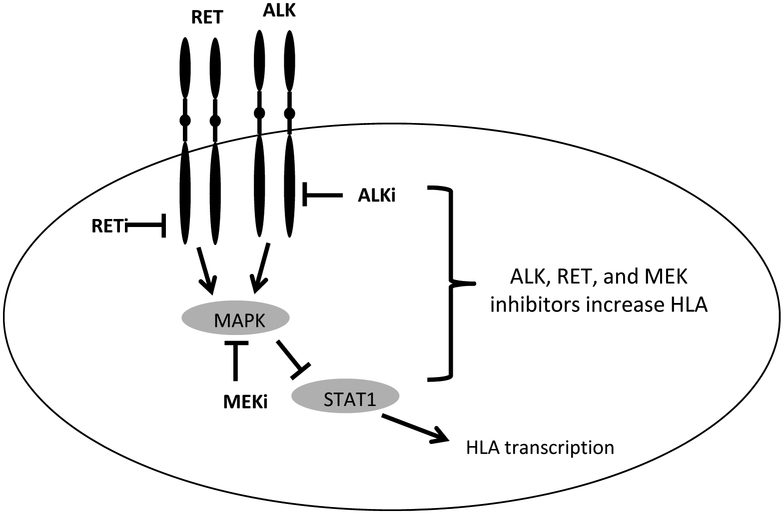
We have shown here consistently, with multiple drugs and RNAi, using multiple cell lines, that inhibition of ALK and RET led to substantial increases in the surface levels of HLA-A,B,C. Figure 7 summarizes our proposed model on the signaling pathway for HLA upregulation 7. Antigen processing machinery transcript and proteins in cells that contain the respective target mutant oncogenes also increased with inhibition. These large changes in HLA complex quantities in cancer cells have wide implications for T cell immunosurveillance. The amount of HLA complexes, but not the amount of peptides present in the ER is the limiting factor in HLA ligand presentation (57). Increases in HLA complex capacities, as were demonstrated in this study, not only provides the opportunity to display more of the same peptides, but also rarely presented HLA ligands. This hypothesis was confirmed by multiple mass spectrometry experiments with TKI inhibition, resulting in large changes in the quantity and quality of the immunopeptidome, with potentially hundreds of new epitopes displayed on the cell surface. As escape from the immune system is a hallmark of cancer survival and progression, these data provide another mechanism by which oncogenesis is promoted, by downregulating antigen presentation during the oncogenic process (58). Thus, ALK and RET inhibitors could be used in combination with T cell immunotherapies by making cancer cells more susceptible to T recognition (59).
Figure 7: Simplified schema of signaling pathway for HLA upregulation.
MEK, ALK or RET positively regulate the output of the MAPK pathway, which in turn downregulates STAT1, which leads to reduced HLA. Inhibitors of these kinases, reverse the process.
The appearance of the numerous new peptide epitopes may be a consequence of several mechanisms: 1. increased detection rate in mass spectrometry experiments due to presentation of the same peptides in higher numbers, 2. altered gene expression due to pathway inhibition, or 3. altered protein processing and new cleavage patterns of new and existing proteins. First, the several fold increases in cell surface HLA molecules (perhaps hundreds of thousands of additional HLA molecules per cell), which in concert with the increased antigen processing machinery, could lead to large increases in total presented peptides and thus the increased sensitivity of T cells to recognize the rarer epitopes. It is possible that the increase in epitopes detected by mass spectrometry may be in part due to the increase in absolute number of the same peptides that were already present, but now presented at higher frequencies due to increased HLA expression. However, the potential for increased cell surface HLA expression to bias the sensitivity of detection of rare peptides does not seem to be a sufficient explanation for their detection, since results of the overlap of the ligandome from three individual experiments showed not only a disappearance of many HLA ligands found in the control group, which otherwise should still be detected, but also identified two distinct new groups after two RET inhibitor treatments. There was only 30% overlap of HLA ligands appearing after either TKI treatment; we hypothesize that this proportion would have to be much higher to argue that their appearance is only mediated exclusively by increase of detection rates after upregulating HLA levels. Thus, in addition to the increase in HLA expression, the ALK and RET inhibitors could also alter the cell’s protein repertoire independent of the effects on antigen presentation pathway throughput, thus providing potential new antigens. The inhibited ALK and RET kinases are upstream of multiple signaling pathways that control expression of multiple target genes. This could lead to the appearance of the new peptides found in the drug treated groups, which could potentially include tumor-associated antigens.
The generation of new peptides could have resulted from altered proteasomal cleavage patterns of proteins that normally need to be degraded for HLA presentation, as has been seen with interferon gamma (60,61). However, motif analysis of the A*02 9-mer peptides in each treatment group yielded mostly identical frequencies of amino acids over all positions, arguing against the changes in proteasomal cleavage as a major explanation for the detection of many new HLA ligands after TKI treatment.
Analysis of RNA-Seq data showed the 32 genes were upregulated at least two-fold in the AST487 treated cells and 50 genes in the cabozantinib treated cells. However, altered gene expression as an effect of TKI treatment was not sufficient to fully explain changes in the HLA ligand repertoire since only a very small fraction (four for AST487 and five for Cabozantinib, 1.2% of total new peptides) of newly identified HLA ligands in the treatment groups showed at least a 2-fold increase in mRNA levels. Overall, this indicates that most of the new peptides might have been detected because of the overexpression of HLA in combination with MAPK pathway alterations after TKI treatment. This notion was further supported in the comparison between RET inhibitor altered peptides present in all three experiments and control peptides present in at least one of the experiments (Supplemental Fig. S8G). Almost all of the most robustly expressed peptides that were presented in all three experiments after RET inhibition, were found at least once in the control cells as well (3365 total peptides.) Despite this overlap in presentation, there were more than two dozen peptides regularly revealed by the inhibitors that were never found presented in the control cells. Looking at the 2 RET inhibitor revealed peptides found in at least one run, the number of new peptides was broadened to 1818 and 1642 peptides, for the 2 drugs respectively, that were never present in the control treatment group. These data lead us to conclude that the peptides displayed on the cancer cell at a given time are derived from a large pool (for instance, 195 control peptides were seen in all three runs, compared to the 3365 control peptides that were seen at least one time). RET inhibitors caused potential convergence and better detection of the peptides displayed. RET inhibition leads to new peptides with a minimum of 25 found in all three runs to a maximum of a few thousand peptides found at least once in the three experiments.
From the pool of new peptides, the presence of TEIPPs further expands the therapeutic potential of RET inhibition by presenting a set of known neoantigens that have detectable frequencies of CD8+ cognate T cells and to which CD8+ T cells have shown reactivity (28,30). It is generally assumed that TEIPPs are found primarily in TAP deficient cells, in which peptides from the cytoplasm are limited, and there is excess HLA capacity. Here, we showed an alternative mechanism for TEIPP presentation, in which markedly increasing the total HLA carrying capacity led to presentation of these unusual TEIPP epitopes by changing the ratio of available HLA molecules to available peptides. Based on our RNA seq and motif data, we do not think there is generation of new TEIPPs through transcription or cleavage. TEIPPs are not normally presented due to the large abundance of processed peptides with favorable binding characteristics from which the limited HLA molecules can choose. Instead, here, by increasing HLA abundance we allowed for these normally unselected TEIPP peptides to be loaded into HLA (57). These data again underlie the ability of kinase inhibition to shift the peptide repertoire to produce immunogenic peptides. Because a random sampling of the new peptides that appeared after RET inhibition were capable of stimulating an immune response in human T cells, we hypothesize that the new peptide repertoire may enhance the immunogenic potential of cancer cells in the setting of inhibitor therapy.
In conclusion, we identified a new strategy for upregulating the expression of HLA in ALK and RET mutated cancers in vitro and in vivo by using ALK and RET tyrosine kinase inhibitors. This increase in the expression of HLA in cancer cells can make those cells preferable targets for T-cell based immunotherapies. We demonstrated that this increase in HLA binding capacities gave rise to a distinct new repertoire of HLA ligands, which were capable of eliciting CD8+ T cell–responses and can mediate ADCC through TCR mimic antibodies, as a surrogate for T cell killing. The detection of TEIPPs in this new repertoire gives new insights into the biology of these rare HLA ligands and expands the list of potential tumor-specific targets which are induced through RET and ALK inhibitor treatment (28,30). When these effects are taken together, ALK and RET inhibitors provide a method to increase HLA expression in cancer cells and simultaneously unmask hundreds of treatment induced HLA ligands capable of inducing T cell responses. This opens up the potential for combinatorial therapies of ALK and RET inhibition and subsequent TCR-based immunotherapy.
Supplementary Material
ACKNOWLEDGEMENTS
Blueprint Medicines generously provided BLU6864. We thank the ImCORE network at Roche and Genentech for helpful discussions. We would also like to thank James Fagin, Neal Rosen and Virginia Seijas for helpful discussions.
FUNDING
This research was funded by the National Cancer Institute Training Grant T32 CA062948, R01 CA55349, P30 CA008748, the Thyroid SPORE grant P50 CA172012-01A1, the Lymphoma Foundation, Memorial Sloan Kettering Cancer Center’s Experimental Therapeutics Center, the German Research Foundation KL3118/1-1, and the Doris Duke Charitable Foundation. Further, this imCORE Network project was funded in part by Genentech, Inc.
Footnotes
DISCLOSURES MSK has filed for patent protection for compositions and methods related to their use, for certain work discussed in this paper on behalf of some of the authors as inventors. DAS and TD are consultants to Eureka Therapeutics. DAS is a consultant to, and/or has equity in: SLS, KLUS, IOVA, PFE, Actinium, Oncopep.
REFERENCES
- (1).Thorsby E, “The role of HLA in T cell activation,” Hum. Immunol, vol. 9, no. 1, pp. 1–7, 1984. [DOI] [PubMed] [Google Scholar]
- (2).Rosenberg SA, “A new era for cancer immunotherapy based on the genes that encode cancer antigens,” Immunity, vol. 10, no. 3 pp. 281–287, 1999. [DOI] [PubMed] [Google Scholar]
- (3).Blum JS, Wearsch PA, and Cresswell P, Pathways of antigen processing., vol. 31 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Andersen MH, Schrama D, Thor Straten P, and Becker JC, “Cytotoxic T cells,” Journal of Investigative Dermatology, vol. 126, no. 1 pp. 32–41, 2006. [DOI] [PubMed] [Google Scholar]
- (5).Demanet C et al. , “Down-regulation of HLA-A and HLA-Bw6, but not HLA-Bw4, allospecificities in leukemic cells: An escape mechanism from CTL and NK attack?,” Blood, vol. 103, no. 8, pp. 3122–3130, 2004. [DOI] [PubMed] [Google Scholar]
- (6).Hicklin DJ, Marincola FM, and Ferrone S, “HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story,” Molecular Medicine Today, vol. 5, no. 4 pp. 178–186, 1999. [DOI] [PubMed] [Google Scholar]
- (7).Gejman R et al. , “Rejection of immunogenic tumor clones is limited by clonal fraction,” Elife, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Campoli M and Ferrone S, “HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance,” Oncogene, vol. 27, no. 45 pp. 5869–5885, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mcgranahan N, Rosenthal R, Hiley CT, Herrero J, and Swanton C, “Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution,” Cell, pp. 1–13, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mendez R et al. , “HLA and melanoma: Multiple alterations in HLA class i and II expression in human melanoma cell lines from ESTDAB cell bank,” Cancer Immunology, Immunotherapy, vol. 58, no. 9 pp. 1507–1515, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cabrera T et al. , “HLA class I expression in bladder carcinomas,” Tissue Antigens, vol. 62, no. 4, pp. 324–327, 2003. [DOI] [PubMed] [Google Scholar]
- (12).Maleno I et al. , “Distribution of HLA class I altered phenotypes in colorectal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21.,” Immunogenetics, vol. 56, no. 4, pp. 244–53, 2004. [DOI] [PubMed] [Google Scholar]
- (13).Garrido C et al. , “MHC class I molecules act as tumor suppressor genes regulating the cell cycle gene expression, invasion and intrinsic tumorigenicity of melanoma cells,” Carcinogenesis, vol. 33, no. 3, pp. 687–693, 2012. [DOI] [PubMed] [Google Scholar]
- (14).Brea EJ et al. , “Kinase Regulation of Human MHC Class I Molecule Expression on Cancer Cells,” Cancer Immunol. Res, vol. 4, no.11, pp936–947, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gobin SJSJP, van Zutphen M, Woltman AMA, van den Elsen PJ, van Zutphen M, and van den Elsen PJ, “Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element.,” J. Immunol, vol. 163, no. 3, pp. 1428–34, 1999. [PubMed] [Google Scholar]
- (16).Vella LJ et al. , “MEK inhibition, alone or in combination with BRAF inhibition, impairs multiple functions of isolated normal human lymphocytes and dendritic cells,” J. Immunother. Cancer, vol. 1, no. Suppl 1, p. P93, 2013. [DOI] [PubMed] [Google Scholar]
- (17).Ebert PJR et al. , “MAP Kinase Inhibition Promotes T Cell and Anti-tumor Activity in Combination with PD-L1 Checkpoint Blockade,” Immunity, vol. 44, no. 3, pp. 609–621, 2016. [DOI] [PubMed] [Google Scholar]
- (18).D’Souza WN, Chang C-F, Fischer AM, Li M, and Hedrick SM, “The Erk2 MAPK Regulates CD8 T Cell Proliferation and Survival,” J. Immunol, vol. 181, no. 11, pp. 7617–7629, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dushyanthen S et al. , “Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer,” Nat. Commun, vol. 8, no. 1, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Menicali E, Moretti S, Voce P, Romagnoli S, Avenia N, and Puxeddu E, “Intracellular signal transduction and modification of the tumor microenvironment induced by RET/ptcs in papillary thyroid carcinoma,” Front. Endocrinol. (Lausanne)., vol. 3, no. MAY, pp. 1–23, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Knauf JA and Fagin JA, “Role of MAPK pathway oncoproteins in thyroid cancer pathogenesis and as drug targets,” Curr. Opin. Cell Biol, vol. 21, no. 2, pp. 296–303, 2009. [DOI] [PubMed] [Google Scholar]
- (22).Santoro M, Carlomagno F, Melillo RM, Billaud M, Vecchio G, and Fusco A, “Molecular mechanisms of RET activation in human neoplasia,” Journal of Endocrinological Investigation, vol. 22, no. 10 pp. 811–819, 1999. [DOI] [PubMed] [Google Scholar]
- (23).Knauf JA, Kuroda H, Basu S, and Fagin JA, “RET/PTC-induced dedifferentiation of thyroid cells is mediated through Y1062 signaling through SHC-RAS-MAP kinase,” Oncogene, vol. 22, no. 28, pp. 4406–4412, 2003. [DOI] [PubMed] [Google Scholar]
- (24).Gainor JF and Shaw AT, “Novel Targets in Non-Small Cell Lung Cancer: ROS1 and RET Fusions,” Oncologist, vol. 18, no. 7, pp. 865–875, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hallberg B and Palmer RH, “Mechanistic insight into ALK cancer biology,” Nat. Rev. Cancer, vol. 13, pp. 685–700, 2013. [DOI] [PubMed] [Google Scholar]
- (26).Webb TR et al. , “Anaplastic lymphoma kinase: role in cancer pathogenesis and small-molecule inhibitor development for therapy.,” Expert Rev. Anticancer Ther, vol. 9, no. 3, pp. 331–56, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).George SK, Vishwamitra D, Manshouri R, Shi P, and Amin HM, “The ALK inhibitor ASP3026 eradicates NPM-ALK+ T-cell anaplastic large-cell lymphoma in vitro and in a systemic xenograft lymphoma model.,” Oncotarget, vol. 5, no. 14, pp. 5750–63, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Marijt KA et al. , “Identification of non-mutated neoantigens presented by TAP-deficient tumors,” J. Exp. Med., vol. 215, no. 9, p. 2325 LP–2337, September 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kiessling R, “TAP-ing into TIEPPs for cancer immunotherapy,” Journal of Clinical Investigation, vol. 126, no. 2 pp. 480–482, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lampen MH, Verweij MC, Querido B, van der Burg SH, Wiertz EJHJ, and van Hall T, “CD8+ T Cell Responses against TAP-Inhibited Cells Are Readily Detected in the Human Population,” J. Immunol, vol. 185, no. 11, pp. 6508–6517, 2010. [DOI] [PubMed] [Google Scholar]
- (31).Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, and Mann M, “Mass Spectrometry of Human Leukocyte Antigen Class I Peptidomes Reveals Strong Effects of Protein Abundance and Turnover on Antigen Presentation,” Mol. Cell. Proteomics, vol. 14, no. 3, pp. 658–673, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Awad MM and Shaw AT, “ALK inhibitors in non-small cell lung cancer: Crizotinib and beyond,” Clin. Adv. Hematol. Oncol, vol. 12, no. 7, pp. 429–439, 2014. [PMC free article] [PubMed] [Google Scholar]
- (33).Sullivan I and Planchard D, “ALK inhibitors in non-small cell lung cancer : the latest evidence and developments,” pp. 32–47, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Shaw AT et al. , “Ceritinib in ALK -Rearranged Non–Small-Cell Lung Cancer,” N. Engl. J. Med, vol. 370, no. 13, pp. 1189–1197, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kodama T et al. , “Alectinib shows potent antitumor activity against RET-rearranged non-small cell lung cancer.,” Mol. Cancer Ther, vol. 13, no. 12, pp. 2910–8, 2014. [DOI] [PubMed] [Google Scholar]
- (36).Akeno-Stuart N et al. , “The RET kinase inhibitor NVP-AST487 blocks growth and calcitonin gene expression through distinct mechanisms in medullary thyroid cancer cells,” Cancer Res, vol. 67, no. 14, pp. 6956–6964, 2007. [DOI] [PubMed] [Google Scholar]
- (37).Grülich C, “Cabozantinib: A MET, RET, and VEGFR2 tyrosine kinase inhibitor,” Recent Results Cancer Res, vol. 201, pp. 207–214, 2014. [DOI] [PubMed] [Google Scholar]
- (38).Matsubara D et al. , “Identification of CCDC6-RET Fusion in the Human Lung Adenocarcinoma Cell Line, LC-2/ad,” J. Thorac. Oncol, vol. 7, no. 12, pp. 1872–1876, 2012. [DOI] [PubMed] [Google Scholar]
- (39).Carlomagno F et al. , “Point mutation of the RET proto-oncogene in the TT human medullary thyroid carcinoma cell line,” Biochem. Biophys. Res. Commun, vol. 207, no. 3, pp. 1022–1028, 1995. [DOI] [PubMed] [Google Scholar]
- (40).Min W, Pober JS, and Johnson DR, “Kinetically coordinated induction of TAP1 and HLA class I by IFN-γ: The rapid induction of TAP1 by IFN-γ is mediated by Stat1α,” J. Immunol, vol. 156, no.9, pp. 3174–83. 1996. [PubMed] [Google Scholar]
- (41).Girdlestone J, Isamat M, Gewert D, and Milstein C, “Transcriptional regulation of HLA-A and -B: differential binding of members of the Rel and IRF families of transcription factors.,” Proc. Natl. Acad. Sci, vol. 90, no. 24, pp. 11568–11572, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Srivastava RM et al. , “STAT1-Induced HLA Class I Upregulation Enhances Immunogenicity and Clinical Response to Anti-EGFR mAb Cetuximab Therapy in HNC Patients,” Cancer Immunol. Res, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Pollack BP, Sapkota B, and Cartee TV, “Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes,” Clin. Cancer Res, vol.17, no. 13, 2011. [DOI] [PubMed] [Google Scholar]
- (44).Marzi L et al. , “FOXO3a and the MAPK p38 are activated by cetuximab to induce cell death and inhibit cell proliferation and their expression predicts cetuximab efficacy in colorectal cancer,” Br. J. Cancer, vol.115, no.10, pp. 1223–1233, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Piotrowska Z et al. , “Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion,” Cancer Discov, vol.8,no.12, pp. 1529–1539, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wu C et al. , “STAT1 is phosphorylated and downregulated by the oncogenic tyrosine kinase NPM-ALK in ALK-positive anaplastic large-cell lymphoma,” Blood, vol. 126, no.3, pp. :336–45, 2015. [DOI] [PubMed] [Google Scholar]
- (47).Liu B et al. , “Inhibition of Stat1-mediated gene activation by PIAS1,” Proc. Natl. Acad. Sci, vol.95, no.18, pp.10626–31, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Zhang Y, Chen Y, Liu Z, and Lai R, “ERK is a negative feedback regulator for IFN-γ/STAT1 signaling by promoting STAT1 ubiquitination,” BMC Cancer, vol. 18, no. 1, p613, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Vanhatupa S, Ungureanu D, Paakkunainen M, and Silvennoinen O, “ MAPK-induced Ser 727 phosphorylation promotes SUMOylation of STAT1,” Biochem. J, vol. 409, no. 1 pp. 179–85, 2008. [DOI] [PubMed] [Google Scholar]
- (50).Schumacher TN and Schreiber RD, “Neoantigens in cancer immunotherapy,” Science (80-. )., vol. 348, no. 6230, pp. 69–74, 2015. [DOI] [PubMed] [Google Scholar]
- (51).Yarchoan M, Johnson BA, Lutz ER, Laheru DA, and Jaffee EM, “Targeting neoantigens to augment antitumour immunity,” Nature Reviews Cancer, vol. 17, no. 4 pp. 209–222, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Dubrovsky L et al. , “A TCR-mimic antibody to WT1 bypasses tyrosine kinase inhibitor resistance in human BCR-ABL + leukemias.” Blood, vol. 123, no. 21 pp. 3296–304, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Veomett N et al. , “Therapeutic efficacy of an Fc-enhanced TCR-like antibody to the intracellular WT1 oncoprotein,” Clin. Cancer Res, vol. 20, no. 15, pp. 4036–4046, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Gejman R et al. , “Prospective identification of cross-reactive human peptide-MHC ligands for T cell receptor based therapies,” Manuscr. Submitt. Publ. [Google Scholar]
- (55).Liu L et al. , “The BRAF and MEK inhibitors dabrafenib and trametinib: Effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4,” Clin. Cancer Res, vol. 21, no. 7, pp. 1639–1651, 2015. [DOI] [PubMed] [Google Scholar]
- (56).Mimura K et al. , “The MAPK Pathway Is a Predominant Regulator of HLA-A Expression in Esophageal and Gastric Cancer,” J. Immunol, vol. 191, no. 12, pp. 6261–6272, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Komov L, Kadosh DM, Barnea E, Milner E, Hendler A, and Admon A, “Cell Surface MHC Class I Expression Is Limited by the Availability of Peptide-Receptive ‘Empty’ Molecules Rather than by the Supply of Peptide Ligands,” Proteomics, vol. 18, no. 12, 2018. [DOI] [PubMed] [Google Scholar]
- (58).Hanahan D and Weinberg RA, “Hallmarks of cancer: The next generation,” Cell, vol. 144, no. 5 pp. 646–674, 2011. [DOI] [PubMed] [Google Scholar]
- (59).French JD, “Revisiting Immune-Based Therapies for Aggressive Follicular Cell–Derived Thyroid Cancers,” Thyroid, vol. 23, no. 5, pp. 529–542, 2013. [DOI] [PubMed] [Google Scholar]
- (60).Chang AY et al. , “A therapeutic T cell receptor mimic antibody targets tumor-associated PRAME peptide/HLA-I antigens,” J. Clin. Invest, vol. 127, no. 7, pp. 2705–2718, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Gravett AM, Trautwein N, Stevanović S, Dalgleish AG, and Copier J, “Gemcitabine alters the proteasome composition and immunopeptidome of tumour cells,” Oncoimmunology, vol. 7, no. 6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.