Abstract
The potent immunosuppression induced by glioblastoma (GBM) is one of the primary obstacles to finding effective immunotherapies. One hallmark of the GBM-associated immunosuppressive landscape is the massive infiltration of myeloid-derived suppressor cells (MDSCs) and, to a lesser extent, regulatory T cells (Tregs), within the tumor microenvironment. Here, we showed that regulatory B cells (Bregs) are a prominent feature of the GBM microenvironment in both preclinical models and clinical samples. Forty percent of GBM patients (n=60) scored positive for B-cell tumor infiltration. Human and mouse GBM-associated Bregs were characterized by immunosuppressive activity towards activated CD8+ T cells, the overexpression of inhibitory molecules PD-L1 and CD155, and production of immunosuppressive cytokines TGFβ and IL10. Local delivery of B-cell depleting anti-CD20 immunotherapy improved overall animals’ survival (IgG vs. anti-CD20 mean survival: 18.5 vs. 33 days, P=0.0001), suggesting a potential role of Bregs in GBM progression. We unveiled that GBM-associated MDSCs promoted regulatory B-cell function by delivering microvesicles transporting membrane-bound PD-L1, able to be up-taken by tumoral B cells. The transfer of functional PD-L1 via microvesicles conferred Bregs the potential to suppress CD8+ T-cell activation and acquisition of an effector phenotype. This work uncovered the role of B cells in GBM physiopathology and provides a mechanism by which the GBM microenvironment controls B cell–mediated immunosuppression.
Keywords: Glioblastoma, regulatory B cells, myeloid-derived suppressive cells, PD-L1, microvesicles
Introduction
Glioblastoma (GBM), an incurable malignant brain cancer, is characterized by its ability to build an immunosuppressive environment, which blocks antitumor immunity and promotes tumorigenesis. There are many drivers of GBM-induced immunosuppression, including the production of immunoregulatory secreted factors such as cytokines (TGFβ1, IL10), enzymes (IDO), or prostaglandins (1–3). Cellular components such as regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs), and tumor-associated macrophages (TAMs) are also known to mediate GBM-associated immunosuppression (4–6). MDSCs are known to act as major immunosuppressive cells within the tumor vicinity. They have also been found in peripheral blood of GBM patients, suggesting a systemic influence on immunity (5). Inhibitory molecules expressed on the surface of GBM and myeloid suppressive cells, such as programmed death ligand 1 (PD-L1) or CD155 (poliovirus receptor), are known to inhibit the effector function of T cells (7,8), and it has been suggested that the GBM-derived hypoxic environment might contribute to MDSC and Treg function (9,10).
Although immunotherapy remains a promising treatment for GBM (11), a better understanding of how GBM controls immunity to promote its survival, progression, and immune escape is necessary to optimize current immunotherapeutic approaches. Our group and others have reported the importance of GBM-associated immunosuppressive factors such as MDSCs or, to a lesser extent, Tregs as fundamental immunosuppressive players in GBM (12,13). Other players of the immune system, such as B cells, have also been reported to be part of the GBM immune landscape (14). However, whether tumor-infiltrating B cells play a relevant role in GBM progression remains to be elucidated.
This present work provides a comprehensive phenotype and functional profile of GBM regulatory B cells (Bregs). We showed that human and murine GBM-associated B cells exhibit an immunosuppressive phenotype characterized by the presence of inhibitory molecules PD-L1 and CD155 and production of immunoregulatory cytokines TGFβ and IL10. Bregs represented ∼10% of bone marrow–derived infiltrating immune cells in two different orthotopic brain tumor models (GL261 and CT2A), and 40% of GBM patients that were screened scored positive for B-cell tumor infiltration. GBM-associated B cells showed an immunosuppressive function toward activated CD8+ T cells, and their pathophysiological relevance was highlighted by extended animal survival after local delivery of B cell–depleting immunotherapy. We showed that the tumor microenvironment, and more precisely, MDSCs, played a fundamental role in promoting immunosuppressive B cells. MDSCs mediated transfer of membrane-bound PD-L1 to B cells, resulting in the promotion of B cell–mediated immunosuppression. Overall, the present study provides an extensive characterization of GBM-associated B cells and unveiled a mechanism of intercellular communication between MDSCs and B cells to expand MDSC immunosuppressive effects.
Materials and methods
Human samples
All human samples (tumor and peripheral blood) and frozen tissue were collected by the Nervous System Tumor Bank of the Northwestern University (NSTB) under the IRB protocol N° STU00202003. All patients signed a written consent. Only samples from GBM patients with greater than 50% tumor cellularity, as determined by H&E examination, were included in the study. The study was conducted in accordance to U.S. Common Rule of ethical standards.
GBM sample immunohistochemistry (IHC)
A total of 60 GBM patients were analyzed for the presence of CD20+ B cells by IHC. Formalin-fixed paraffin-embedded tumor blocks of GBM patients (see Table 1 for patients’ characteristics) were double-labeled with rabbit anti-human CD20 (1:2 dilution, clone EP459Y, Thermo Fischer) and mouse anti-human CD8 (1:100 dilution, clone 4B11, Leica). Anti-rabbit and anti-mouse HRP secondary antibodies (both from Abcam) were used at 1:500 dilution. Twenty consecutive high-power fields were evaluated on each tumor by a board-certified neuropathologist from the Neurosurgical Department at Northwestern University. Images were captured on a Leica DMi8 inverted microscope.
Table 1. GBM patients’ characteristics and tumors analysis for B-cell infiltration.
A total of 60 GBM patients’ tumor samples were tested for the presence of CD20+ B cells by IHC. Patients were stratified (variable) by age, gender, newly diagnosed or recurrent, isocitrate dehydrogenase (IDH) mutation and O[6]-methylguanine-DNA methyltransferase (MGMT) methylation status, CD8+ T-cell infiltration and location. These variables were analyzed for statistical correlation with presence or not of B cells (p-value).
| Variable | B-cell present N=25 |
B-cell NOT present N=37 |
P-value |
|---|---|---|---|
| Age at diagnosis (yrs) | |||
| N | 25 | 37 | 0.1778 |
| Mean(SD) | 52.72 (15.31) | 57.78 (13.66) | . |
| Median(Q1,Q3) | 54 (40,66) | 62 (51,68) | . |
| Minimum, Maximum | 23–77 | 27–86 | . |
| Gender | |||
| MALE | 19 (76%) | 25 (67.57%) | 0.5740 |
| FEMALE | 6 (24%) | 12 (32.43%) | . |
| New or Recurrence | |||
| New | 18 (72%) | 25 (67.57%) | 0.7840 |
| Recurrence | 7 (28%) | 12 (32.43%) | . |
| IDH1 Mutation | 0.4683 | ||
| Yes | 5 (20%) | 6 (16.22%) | . |
| No | 20 (80%) | 30 (81.08%) | . |
| Unknown | 0 | 1 (2.7%) | . |
| MGMT Methylation | 0.3762 | ||
| Positive | 9 (36%) | 20 (54.05%) | . |
| Negative | 15 (60%) | 16 (43.24%) | . |
| N/A | 1 (4%) | 1 (2.71%) | . |
| Associated with CD8 | |||
| Yes | 25 (100%) | 0 (0%) | <0.0001 |
| No | 0 (0%) | 37 (100%) | . |
| Location | |||
| Perivascular | 23 (92%) | 0 (0%) | . |
| Perivascular intraparenchymal | 2 (8%) | 0 (0%) | . |
Cd20 gene (MS4A1) expression analysis in GBM patients by TCGA database
MS4A1 gene expression was analyzed in grade II, III and IV gliomas using the TCGA-GBMLGG dataset. The data show the analysis of a total of 620 patients (grade II = 226, grade III = 244 and grade IV = 150). MS4A1 gene expression (Microarray HG-U133A platform) and overall GBM (grade IV) patient’s survival comparison was assessed using the TCGA-GBM dataset. High (n=254) and low (n=271) MS4A1 gene expression was determined by median of gene expression. Survival curves were compared by the log-rank test.
Isolation of GBM infiltrating immune cells and PBMCs
Freshly resected tumor samples were diced using a razor blade and incubate for 30 minutes at 37°C in a Petri dish with digestion buffer, consisting of 4 mL of Hank’s balanced salt solution (HBSS, Gibco) supplemented with 8 mg of collagenase D (Sigma-Aldrich), 80 μg DNaseI (Sigma-Aldrich), and 40 μg TLCK (Sigma-Aldrich) per approximately 2 grams of tumor. The sample was mixed by pipetting up and down several times every 10 minutes. Then, the cell suspension was mechanically dissociated using a tissue homogenizer (Potter-Elvehjem PTFE pestle) in HBSS. Cells clumps were removed using a 70 μm cell strainer (Thermo Fischer). Red blood cells, myelin, and debris were removed by 30/70 Percoll (GE Healthcare) gradient separation (30min, 1000 x g at room temperature). Peripheral blood samples from GBM patients were collected in EDTA tubes. Peripheral blood mononuclear cells (PBMC) were isolated using the Ficoll (GE Healthcare) gradient. Tumor cells and PBMCs were immediately put in complete RPMI media [RPMI + 10% heat-inactivated FBS, 10 mM HEPES–sodium Pyruvate, 1 mM sodium pyruvate, 0.01% 2-mercaptoethanol, 2 mM L-glutamine, penicillin (100 U/mL), and streptomycin (100 μg/mL); all reagents from Thermo Fischer].
GBM B cell-mediated T-cell suppression assay
The assay was performed in an autologous manner, and thus, B cells (from tumor and PBMCs) and T cells (PBMCs) were from the same patient. B cells from tumor and PBMCs were obtained using the EasySep™ Human CD19 Positive Selection Kit II (StemCell Technologies). PBMC T cells were isolated using the EasySep™ Human T Cell Isolation Kit (StemCell Technologies) and labeled with 10 μM of the eBioscience™ cell proliferation dye eFluor 450 (Thermo Fischer). Cells were activated with T-cell activator anti-CD3/CD28 beads (Dynabeads, Invitrogen, Thermo Fischer) at 1:3 beads:T-cell ratio supplemented with IL2 (50 U/mL; Peprotech) and cocultured at a 1:1 ratio with tumor-infiltrating or PBMC CD19+ B cells for 72 hours. CD4+ and CD8+ T-cell proliferation (eFluor450 dilution) and activation status [intracellular granzyme B (GzmB) and IFNγ expression] were analyzed using flow cytometry (Supplementary Table S1).
Tumor-infiltrating CD163+ cells isolation and microvesicle uptake
Tumor cells and PBMCs were obtained as described above, and CD163+ macrophages were isolated using an anti-human CD163 biotin (clone GHI/61, BioLegend) and the anti-biotin Microbeads (Miltenyi Biotec). Cells were magnetically isolated using LS columns (Miltenyi Biotec). CD163+ cells were labeled with the lipophilic dye Cell Trace Violet (CTV, Invitrogen, Thermo Fischer) and placed in the upper chamber of a 0.4 μm transwell system in complete RPMI. In the lower chamber, PBMCs from the same donor were placed at 106 cells/mL in complete RPMI. After 24 hours, cells from the bottom chamber were harvested and tested by flow cytometry the acquisition of the CTV dye by B cells, CD4+Foxp3+ Tregs, and CD33+ myeloid cells by flow cytometry. See Supplementary Table S2 for antibody information.
Mice
C57BL/6, CD45.1 C57BL/6, B cell–deficient (μMT, BKO), and IL10-deficient (IL10 KO) mice were from The Jackson Laboratory. Animals were 6 to 8 weeks-old at the time of the experiment initiation. All animal experimentation protocols are approved by the Institutional Animal Care and Use Committee (IACUC) under the protocol # IS00002459 at the Northwestern University. All animals were housed in an SPF animal facility at Northwestern University.
Cell lines
GL261 cells were obtained from the National Cancer Institute (NCI), and CT2A cells were a gift from Pr. Tom Seyfried (Boston College). The GL261 cell line identity and purity were evaluated annually using short tandem repeats (STR) profiling performed by at Northwestern sequencing facility. All cell lines were routinely tested for Mycoplasma contamination every 2 months using the Universal Mycoplasma Detection Kit (ATCC® 30–1012K™). Both murine syngeneic glioma cell lines were maintained in DMEM (Corning) with 10% fetal bovine serum (FBS, HyClone), penicillin (100 U/mL), and streptomycin (100 mg/mL; Corning) and incubated at 37 in 5% CO2.
Brain tumor injection
A total of 2 × 105 GL261 or CT2A cells were intracranially (i.c.) implanted as previously described (2). Mice were anesthetized through intraperitoneal administration of a stock solution containing ketamine (100 mg/kg) and xylazine (10 mg/kg). The surgical site was shaved and prepared with a swab of povidone-iodine followed by a 70% ethanol. The swabbing procedure was performed three times in total. An incision was made at the midline for access to the skull. A 1-mm-diameter burr hole was drilled 2 mm posterior to the coronal suture and 2 mm lateral to the sagittal suture. Mice were then placed in a stereotaxic frame, and tumor cells were injected in a total volume of 2.5 μL using a Hamilton syringe fitted with a 26-gauge blunt needle at a depth of 3 mm. The incision was then stapled closed.
Cannula implantation
Briefly, mice were anesthetized and a skin incision ~10 mm in length was made over the middle frontal to the parietal bone to expose the surface of the skull. A 26-gauge sterile guide cannula for mice (Plastics One) was installed into the mouse brain at 2 mm depth through the burr hole generated during tumor implantation as described above. Tissue glue was applied around the burr hole and secure the protrusion of the cannula for long term stable positioning. The scalp was closed with surgical glue around the implantation site. A protection dummy cannula was used to secure the protrusion end during the post-op recovery and following observation period. For anti-CD20 injection, a 33-gauge sterile syringe was inserted into the guide cannula. The syringe can be covered with a sleeve designed to extend 1 mm beyond the tip of the guide cannula. Diluted anti-CD20 (25 μg/mouse/injection) in sterile 0.9% saline was injected into the brain (final volume of 2.5 μL/injection). After injection, the cannula was covered using a 33-gauge dummy cannula for mice.
B-cell rescue and animal survival
Survival experiments were performed according to the IACUC guidelines. Mice were monitored daily after tumor implantation. If endpoint was reached, mice were euthanized. For B-cell rescue experiments, B cell–deficient mice (μMT, BKO) were injected intracranially with 2×105 CT2A cells as described above. Ten days later, mice received 5×106 splenic B cells negatively isolated from C57BL/6 mice or IL10–deficient mice using EasySep™ Mouse B-cell Isolation Kit (StemCell Technologies). In all cases, B-cell purity was >98%. B cells were injected intravenously.
In vivo B-cell depletion
Systemic B-cell depletion was achieved by intraperitoneal injection of CD20 depleting antibody (Ab; clone 5D2, Genentech, 250 μg/mouse/injection) 7 and 10 days after tumor implantation. IgG2a (clone C1.18 Mab, BioXCell, 250 μg/mouse/injection) was used as control. After 3 days of the last CD20 Ab injection, mice were bled retro-orbitally to check for B-cell depletion by measuring circulating CD19+ cells (clone 1D3/CD19, BioLegend) by flow cytometry. Alternatively, intracranial administration of CD20 depleting Ab was performed using a cannula implantation system 7 and 10 days after tumor implantation. Briefly, a 33-gauge sterile syringe was inserted into the guide cannula. The syringe was covered with a sleeve designed to extend 1 mm beyond the tip of the guide cannula. The diluted anti-CD20 in sterile 0.9% saline was injected into the brain (25μg/mouse/injection; final volume of 2.5μl/injection). After injection, the cannula was covered using a 33-gauge dummy cannula for mice.
Brain, lymphoid tissue, and blood single-cell suspensions
Mice were bled retro-orbitally and blood samples were collected in heparinized-PBS solution (1mg/ml, Sigma Aldrich). Red blood cells were lysed using an ACK lysing solution (Gibco, Thermo Fischer). After blood collection, mice were euthanized in a CO2 chamber and intracardially perfused with chilled PBS. Brain single-cell suspensions were obtained by mechanical dissociation using a manual tissue homogenizer (Potter-Elvehjem PTFE pestle, Sigma Aldrich) in HBSS. Myelin and debris were removed by Percoll gradient separation. Leukocytes from deep and superficial cervical lymph nodes were obtained by mechanical tissue dissociated using a 70μm cell strainer and a syringe plunger. Brain, blood, and lymph nodes cells were collected in complete RPMI media. Cells were used for immunophenotype analysis or ex vivo functional assays as described below.
Flow cytometry and immunophenotype analysis
Immunophenotype analysis of immune cells from tumor-bearing mice was performed a different time points (0, 7, 14, and 21 days after tumor injection) as described below. In some experiments, the immunophenotype analysis was perform at one time point defined in the Results section. After collection single-cell suspensions, cells were counted and washed with staining buffer (5% bovine serum albumin, 0.001% sodium azide in PBS). Cells were incubated with 1 μL Fc receptor blocking Ab (anti-CD16/32, clone 93, BioLegend) per 106 cells in 100 μL staining buffer for 5 minutes at room temperature. For surface staining, cells were incubated with 1 μL Ab per 106 cells for 30 minutes at 4°C. Cells were washed twice with cold PBS. Cells were stained with Fixable Viability Dye eFluor™ 780 (eBioscience, Thermo Fischer) for 30 minutes at 4°C. Cells were washed twice with staining buffer. For intracellular staining, cells were fixed and permeabilized using the eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set (Invitrogen, Thermo Fischer) for 90 minutes at room temperature. Cells were washed twice with the Permeabilization Buffer (provided in the permeabilization/fixation buffer kit) and incubated with 1μl Ab for 1 hour at 4°C. Cells were washed twice with staining buffer. To evaluate cytokines expression, cells were stimulated for 5 hours at 37°C with the eBioscience™ Cell Stimulation Cocktail plus protein transport inhibitors (500x, Thermo Fischer) prior staining. The list of antibodies used to analyze the different immune cells population can be found in the Supplementary Tables S1, S3, and S4. All antibodies were from BioLegend. Data was acquired with BD FACS Symphony analyzer and analyzed with FlowJo software. Dead cells and debris were excluded using the Live/Dead staining (Viability Dye eFluor™ 780). B cells were identified as CD45+CD11b− CD19+ cells. CD4+ and+ CD8 T cells were identified as CD45+CD11b−CD4+ cells or CD45+CD11b−CD8+ cells, respectively. Monocytic MDSCs were identified as CD45+CD11b+CD11C−Ly6C+Ly6Glow cells. Polymorphonucleated (PMN) MDSCs were identified as CD45+CD11b+CD11C−Ly6C−Ly6G+ cells. All Ab details are found in Supplementary Tables S1, S3, and S4.
Human B-cell phenotype was assessed from tumor cells and PBMCs obtained as described above. Cells were processed using the same method as for murine cells. B cells were defined as CD45+CD11b−CD19+CD20+ cells, and expression of PD-L1, CD155, IL10 and LAP (TGFβ) were assessed. All Abs were from BioLegend unless otherwise specified. Details are found in the Supplementary Table S2.
In vitro murine MDSC generation
GBM-associated MDSCs were generated as in previous reports (13,15). Bone marrow (BM) cells from C57BL/6 mouse were flushed out from femurs with complete RPMI using a 10 mL syringe and 25-gauge needle into complete RPMI (Corning). BM cells were centrifuged (10 minutes, 1500 RPM, at 4°C), and red blood cells were lysed using ACK lysing buffer (Sigma) for 5 minutes at RT. Cells were washed (5 minutes, 1500 RPM, at 4°C) with complete RPMI, counted, and plated into 24-well plates (Corning) at a density of 2.5×105 cells per well with 50% complete RPMI, 50% conditioned media (8mL DMEM (Corning) supernatant that was collected from original seeding of 2×106 CT2A cells after 72 hours of culture), and GM-CSF (40 ng/mL; PeproTech). After 3 days of culture, old media was removed by aspiration and new media (same as aforementioned) was added to the cells for an additional 3 days of incubation. Six days after BM isolation, cells were lifted by pipetting up and down, washed (10 minutes, 300 x g, at 4°C), and stained for characteristic flow cytometric analysis (CD11b+Ly6C+PD-L1+arginase-1+, Supplementary Table S3 and further functional assays as described below.
In vitro human MDSCs generation
Human monocytes were isolated from buffy coat samples using the Human Monocyte Isolation Kit (StemCell technologies). Cells were counted and plated in a 24-well plate with the density of 106 per well with 50% complete RPMI (20% FBS), 50% conditioned media (8ml DMEM (2% FBS) supernatant that was collected from original seeding of 2×106 GBM6, GBM43, GBM12 and MES83 cells after confluency of the cells, human GM-CSF (80 ng/m), and human IL6 (80ng/mL; both cytokines from PeproTech). Old media was removed and replaced with new media (same as aforementioned) on day 3 and 6 of culture. After 9 days of culture, cells were lifted, washed (5 minutes, 1500 RPM at RT) and tested for the expression of arginase-1 and PD-L1 by flow cytometry (Supplementary Table S2).
Murine and human Breg conversion assay by MDSCs
Splenic B-cells from naïve mice were isolated using EasySep™ Mouse B-cell Isolation Kit (StemCell Technologies, Vancouver, Canada). To test the ability of MDSCs to convert naïve B-cells into Bregs, cells were cultured for 48 hours at 1:1 ratio. Then, B-cells were analyzed for their expression of PD-L1, CD155, TGFβ (LAP) and IL10 (Supplementary Table S3 for Ab details). Alternatively, cells were cultured in a 0.4μm membrane transwell system (Corning, Lifesciences). MDSCs were placed in the upper compartment and naïve B-cells supplemented with 100nM murine BAFF (BioLegend) in the lower compartment, at 1:1 ratio. Coculture was maintained for 12, 24, 48 or 72 hours unless otherwise specified. For human Breg conversion using human MDSCs, B-cells were isolated using the EasySep™ Human B-cell Isolation Kit (StemCell Technologies, Vancouver, Canada). B-cells were incubated with 100mM recombinant human BAFF (Peprotech).
In vivo BLZ954 treatment and Breg conversion
B cell–deficient mice (μMT, BKO) were injected intracranially with 2×105 CT2A cells, as described above. The CSF-R1 inhibitor BLZ954 (Novartis) was dissolved in Captisol® and administrated daily by gavage (200 mg/kg) from day 3 to day 7 after tumor inoculation. B cells from CD45.1+ C57BL/6 mouse spleens were isolated using the EasySep™ Mouse B-cell Isolation Kit (StemCell Technologies, purity >97%) in injected intravenously (retro-orbital injection) 24 hours after BLZ954 treatment termination. Forty-eight hours after, CD45.1+ B cells (clone A20, BioLegend) were assessed by flow cytometry for their expression of CD155 and PD-L1 (Supplementary Table S4 for Ab details).
In vivo PD-L1 blockade and Breg conversion
B cell–deficient mice (μMT, BKO) were injected intracranially with 2×105 CT2A cells. Mouse brains were irradiated with 3 Gy for three consecutive days (days 8–10) using a Gammacell 40 Exactor (Best Theratronics). Three days after, mice received 5 μL of blocking anti-PD-L1 (clone 10F.9G2, BioXCell, 5 μg/injection) intracranially via cannula. Two days after, mice received intracranially 5×106 splenic B cells, negatively isolated from CD45.1+ C57BL/6 spleens using the EasySep™ Mouse B-cell Isolation Kit (StemCell Technologies, purity ≥ 97%). Forty-eight hours after the adoptive transfer, mice were sacrificed, and B cells were isolated from tumor-bearing brains using the EasySep™ Mouse CD19 Positive Selection Kit II (StemCell Technologies). B-cell purity (≥ 96%) was evaluated by the congenic marker CD45.1 (clone A20, BioLegend). B cells were tested for their suppressive function against activated splenic CD8+ T-cells as described below.
Murine CD8+ T-cell suppression assay
CD8+ T cells were isolated from spleens of naïve mice using EasySep™ Mouse CD8+ T-Cell Isolation Kit (StemCell Technologies,). Cells were labeled with the eBioscience™ cell proliferation dye eFluor 450 (Thermo Fischer). To test B-cell ability to suppress activated CD8+ T-cell proliferation, cells were mixed at 1:1 B: T ratio. CD8+ T-cell activation was assessed using the anti-CD3/CD28 T-cell activating beads used at 1:3 beads:CD8+ T-cell ration (Invitrogen, Thermo Fischer) plus IL2 (50 U/mL, Peprotech) for 3 days in complete RPMI. T-cell proliferation (eFluor450 dye dilution) and effector T-cell factors such as granzyme B and IFNγ (Supplementary Table S1) were analyzed by flow cytometry. Anti-IL10 (Thermo Fischer), anti-TGFβ (R&D), anti-TIGIT (BioLegend), and anti–PD-L1 (BioXcell) were added every day in the culture at 10 μg/mL. Alternatively, expression of TGFβ (LAP), IL10, and IFNγ by B cells while cocultured with CD8+ T-cells was examined after 36 hours (Supplementary Table S4).
MDSCs-derived microvesicles isolation, quantification, labeling uptake by B cells
Microvesicles (MVs) were prepared from MDSCs conditioned media (CM) as previously described (16,17). Briefly, MDSC-CM was collected after initial centrifugation to pellet life cells at 300 x g, 5 minutes at room temperature. Supernatants were cleared of dead cells, cell debris, and large vesicles by centrifugation at 2000 x g, 30 minutes at 4°C. Apoptotic bodies were removed by ultracentrifugation 10,000 x g for 30 minutes at 4°C. Remaining supernatant was ultracentrifuged at 100,000 x g for 90 minutes at 4°C. Pellets containing vesicles were resuspended in cold PBS and ultracentrifuged at 100,000 x g for 90 minutes at 4°C. MV pellets were resuspended in 50–100 μL of ice-cold PBS and used for a downstream application. The particle size distribution of microvesicles was measured by dynamic light scattering (DLS) using a Zetasizer Nano ZSP (Malvern Panalytical) and presented as diameter in nm and polydispersity index (PDI). The surface charge of MV was determined by zeta-potential using a Zetasizer Nano ZSP (Malvern Panalytical).
To label MDSC-derived MVs, MDSC were labeled with the lipophilic dye Cell Trace Violet (CTV; Invitrogen) for 5 minutes at room temperature. Dye excess was removed by washing cells 3 times with complete RPMI. MDSCs were cultured for 4 days in complete RPMI using exosome-free FBS (Gibco, Thermo Fischer) protected from light. MVs were isolated from supernatants as described above. To test the MV uptake by B cells, CTV-labeled MDSC were cocultured with splenic B cells isolated from naïve mice using the EasySep™ Mouse B-cell Isolation Kit (StemCell Technologies) in a 0.4 μm transwell system (Corning, Lifescience). B cells were harvested at different time points depending on the experiment and evaluated for the acquisition of the CTV fluorescence by flow cytometry. In some experiments, MDSCs were incubated for 2 hours at 37°C with the MV release inhibitor GW4869 (20 μM, Cayman Chemicals). Cells were washed 3 times with complete RPMI and cocultured in a 0.4 μm transwell system with B cells. Alternatively, MVs were fluorescently labeled with PKH67 green fluorescent cell linker kit (Sigma-Aldrich) following manufacturer’s instruction. Briefly, 1 μL of PKH67 dyes in 150 μL of Diluent C solution were mixed with the equal volume of MV (from 106 cells/mL) at room temperature for 5 minutes. To remove unlabeled PKH67 dyes, 1.5 mL of 0.971 M sucrose solution was carefully added into the bottom of the tube following 8 mL of serum-free media was added to PKH67-labed MVs. After centrifugation at 200,000 x g for 2 hours, unlabeled PKH67 dyes located in the interface layer were aspirated. PKH67-labeled MVs located in the bottom were resuspended with PBS. MV content was normalized by their protein content using a standard Bradford method before incubation with B cells. A range of 5–15 μg of MV protein was used across the experiments to convert B cells into Bregs. However, the same protein content was used among groups within the same experiment.
PD-L1 knockdown using siRNA
Lipid nanoparticles (LNPs) for siRNA transfection were formulated by 1,2-dioleoyl-3-trimethylammonium-propane, cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (Avanti Polar Lipids), and synthesized using a thin-film rehydration and sonication method. pd-l1 siRNA (Sigma) was encapsulated into LNP by co-incubation in HEPES buffer for 30 minutes at room temperature. MDSCs were lifted and re-plated at 106 cells/mL/well in 24-well plates and were allowed to adhere overnight. MDSCs were incubated with siRNA (pd-l1 or scramble)/LNP complex in Opti-MEM at siRNA concentration of 100 nM for 4 hours at 37 °C. The medium was replaced by complete RPMI and the cells were cultured for 24 hours at 37 °C. RNA was isolated from siRNA treated MDSCs using the RNEasy Plus Mini Kit (Qiagen). Following chloroform extraction, samples were loaded onto RNeasy Plus columns (Qiagen) and processed according to the manufacturer’s protocol. Total RNA was quantified using a Nanodrop apparatus (Thermo Scientific) and then converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad). pd-l1 and control β-actin transcripts were analyzed by using the 2–ΔΔCT method. Murine pd-l1 primers: forward TGCTGCATAATCAGCTACGG; reverse CCACGGAAATTCTCTGGTTG (18). Murine β-actin primers: forward TTGCTGACAGGATGCAGAAG; reverse ACATCTGCTGGAAGGTGGAC (19).
Membrane-cytosol fractionation and immunoblotting
MDSCs and MVs were lysed using Mem-PER™ Plus Kit (Thermo Scientific #89842) according to manufacturer instructions. A total of 10 μg of protein/samples/fraction was resolved by SDS-PAGE (Bio-Rad), transferred to Immobilon-P PVDF membranes (Millipore), then probed with primary Abs anti-CD45 (1/1000, clone 72787, Cell Signaling), anti–PD-L1 (1/1000, clone BE0101, Bio X Cell), and anti-GAPDH (1/15000, clone MAB374, Millipore). The GE Healthcare Amersham™ ECL™ anti-rabbit HRP (Thermo Fischer) and the Goat Anti-Mouse IgG (H + L)-HRP (Bio-Rad) were used as secondary Abs at 1:5000 dilution. After reaction with Amersham ECL detection reagent (GE Healthcare), blots were visualized for the indicated proteins using autoradiography.
Cryo-electronic microscopy
MVs were imaged by cryo-electron microscopy using the software package Digital Micrograph (Gatan, Inc., Warrendale, PA) employing a JEOL 3200FS Transmission Electron Microscope (JEOL USA, Inc. Peabody, MA) equipped with an in-column energy filter (omega filter) operating at 300 kV at a magnification of 30,000X. Images were recorded using a K2 Summit Direct Electron Detection camera (Gatan, Inc.) in counting mode, and motion corrected using tools in Digital Micrograph. Sample preparation utilized 400 mesh Lacey Carbon copper grids (LC400-CU, Electron Microscopy Sciences, Hatfield, PA), glow discharged at either 5 watts, 10 watts or 25 watts for 10s, with a Pelco easiGlow 91000 Glow Discharge Cleaning System (Ted Pella, Inc., Redding, CA). Grids were vitrified by plunging into liquid ethane. 4 μL of sample were applied to each prepared grid and incubated for 30s under >95% humidity at 4◦C using a Vitrobot Mark IV cryo system (FEI/ThermoFisher). Blot forces of 0.0 and 1.0 and a range of blot times, from 1s-4s were tested.
Microvesicle uptake inhibition assay
MDSCs were incubated for 2 hours with endocytosis inhibitors. Supplementary Table S5 describes the type of inhibitor and the concentration used. MVs were derived from MDSCs fluorescently labeled with the lipophilic dye (CTV) described above. Cells were then incubated with MVs (10μg of protein) in complete RPMI for 12 hours. MVs uptake was assessed by flow cytometry by percentage of CTV positive fluorescence CD19+ B cells (clone 1D3/CD19, BioLegend). The Viability Dye eFluor™ 780 (eBioscience, Thermo Fischer) was used to exclude dead cells and debris.
Statistical analysis
Data are shown as mean±SD for a continuous variable and number (percentage) for a categorical variable. Differences between two groups were analyzed by Student’s t-test or Wilcoxon Rank-sum test as appropriate. Differences among multiple groups were evaluated using one-way ANOVA with post hoc Tukey’s test, or Kruskal-Wallis H Tests followed by post hoc Dunn’s multiple tests as appropriate. Survival curves were generated via the Kaplan-Meier method and compared by log-rank test and multiple comparisons were adjusted using Bonferroni method. Categorical variables were analyzed using Fisher’s exact tests or Chi-square tests as appropriate. All the tests are two-sided and p-values or Benjamini-Hochberg adjusted false discovery rates less than 0.05 were considered as significant. Statistical analyses were performed using SAS9.4 and GraphPad Prism7.03.
Results
Functional and immunophenotypical characterization of GBM-infiltrating B cells
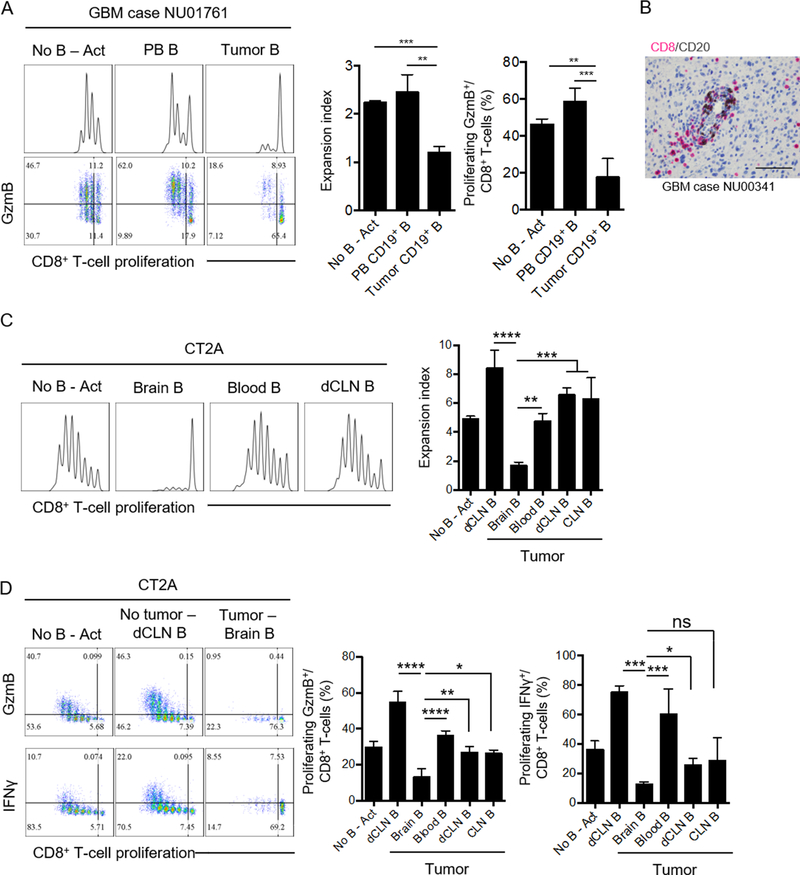
We first evaluated whether infiltration of B cells occurred in tumors of GBM patients. A total of 60 GBM tissue sections were analyzed for the expression of CD20. 40% of the samples scored positive for CD20, and B-cell infiltration showed a predominant perivascular distribution (Table 1). Our data showed that B-cell recruitment did not significantly correlate with age or gender of the patient, nor the IDH mutation or MGMT methylation status of the tumor (Table 1). CD20 gene (MS4A1) expression was found higher in grade IV gliomas only when compared to grade II gliomas (p=0.05, Supplementary Fig. S1A, TCGA database). This data supported the lack of correlation between GBM patient survival and CD20 gene expression in tumors (Supplementary Fig. S1B, TCGA database). In the context of GBM, B cells have been reported to exhibit immunosuppressive function towards activated CD8+ T cells (20,21) in vitro, suggesting a potential role in pro-tumorigenic immune responses. We observed that B cells isolated from freshly resected tumors suppressed activated autologous CD8+ T-cell proliferation and expression of the cytotoxic factor granzyme B (GzmB, Fig. 1A). No significant effect was observed in CD4+ T-cell activation (Supplementary Fig. S1C). Our histopathological evaluation showed that B-cell infiltrates were in close proximity to CD8+ T-cells in the perivascular areas of the tumor (Fig. 1B, Table 1).
Figure 1. Glioma-associated B cells suppress CD8+ T-cell activation.
(A) Representative experiment (n=3 patients) showing the immunosuppressive activity of tumor-infiltrating B cells in GBM patients against autologous CD8+ T cells. Freshly resected tumor and peripheral blood (PB) were collected from patient NU01761. B cells (from tumor and PB) and CD8+ T cells (from PB) were magnetically isolated and mixed at 1:1 ratio (+anti-CD3/CD28 beads and recombinant human IL2). Proliferation was assessed by dilution of cell proliferation dye eFluoer450, and measured as expansion index, after 72h. Effector CD8+ T-cell expansion was measured by the intracellular detection of granzyme B (GzmB) in proliferating cells. The experiment was performed in triplicates. A total of 3 independent experiments from 3 different tumor/PBMC donors was performed. (B) CD8+ (pink) and CD20+ (brown) cells were detected in paraffin embedded GBM sections. Shown is a representative section (40x magnification, NU00341) of a total of 60 different GBM sections. (C) B cells from tumors (brain), blood, and deep cervical lymph nodes (dCLNs) were magnetically isolated and mixed at 1:1 ratio with splenic CD8+ T cells in the presence of anti-CD3/CD28 activating beads and recombinant mouse IL2. B cells from dCLN of a naïve mouse were used as controls (dCLN B). Representative data from 2 independent experiments performed with triplicates. (D) Intracellular expression of granzyme B (GzmB) and IFNγ in the different tissues compared to control groups. Representative data from 2 independent experiments performed with triplicates (n=4 mice/group). All histograms mean±SD. For all experiments, differences among multiple groups were evaluated using one-way ANOVA with post hoc Tukey’s test, ns: no statistically significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
The analysis of the immune landscape in GBM orthotopic GL261 and CT2A experimental models revealed that B cells infiltrated the tumor. Absolute counts of B cells in the brain were significantly increased after tumor implantation (Supplementary Fig. S1D–E). Presence of B cells in the tumor correlated with an overall B-cell lymphopenia in the deep cervical lymph nodes (dCLN) and circulation (Supplementary Fig. S1D-E). B cells from the brain, blood, dCLN and superficial CLN from either CT2A or GL261 tumor-bearing mice were isolated and tested for their ability to suppress splenic CD8+ T-cell activation (Fig. 1C, Supplementary Fig. S2A). Activated CD8+ T cells without B cells (No B Act) were used as a positive control for proliferation. B cells from the dCLN of naïve mice were used as an internal control as our data showed that these B cells functioned as an enhancer of CD8+ T-cell activation (dCLN B, Fig. 1C). Tumor-infiltrating B cells from CT2A (Fig. 1C–D) or GL261 (Supplementary Fig. S2A) significantly suppressed activated CD8+ T-cell proliferation. Circulating B cells and B cells from the dCLN and the superficial CLN from both CT2A- or GL261-bearing mice did not affect CD8+ T-cell expansion, suggesting that B cells’ suppressive function is a unique feature of the B cells located in the tumor microenvironment. Tumor-associated B cells also inhibited the acquisition of effector phenotype by CD8+ T-cells, as seen by the inhibition of GzmB and IFNγ expression (Fig. 1D, Supplementary Fig. S2A). These data showed that GBM-infiltrating B cells had the capability to suppress proliferation and expansion of effector CD8+ T cells.
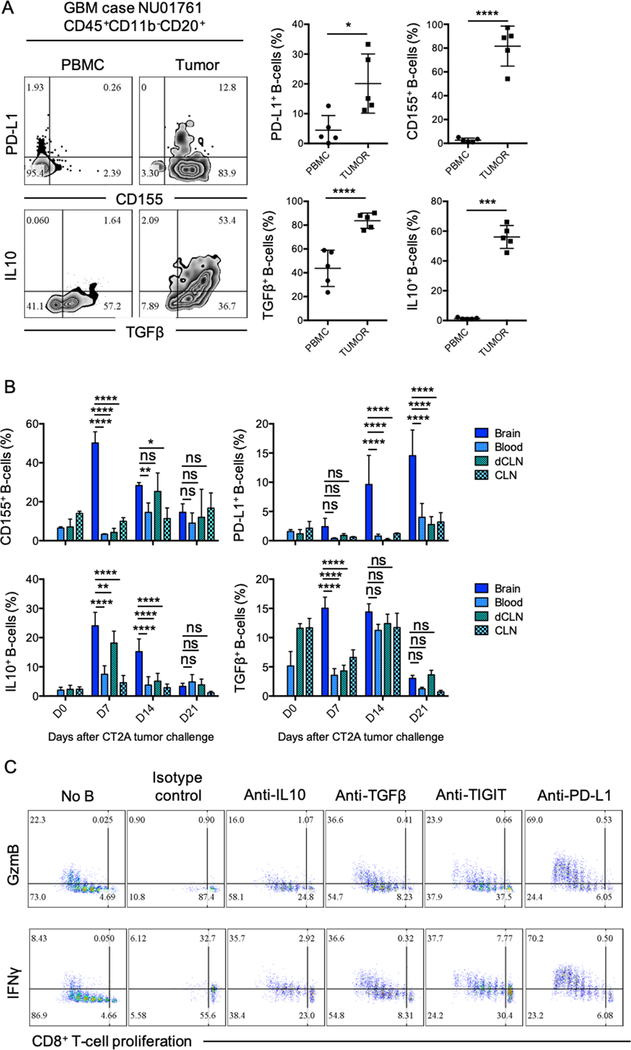
To evaluate the possible mechanisms controlling B cell–mediated CD8+ T-cell suppression, we analyzed the immunophenotype of GBM-associated B cells. Tumor-infiltrating B cells from 5 different GBM patients showed overexpression of inhibitory molecules PD-L1, the poliovirus receptor (PVR) CD155, and immunosuppressive cytokines IL10 and TGFβ (LAP) (Fig. 2A). The GBM-associated immunosuppressive phenotype observed in humans was also found in in both CT2A (Fig. 2B) and GL261 (Supplementary Fig. S2B) murine models. The overexpression of CD155, IL10, and TGFβ was predominant during the earlier stages of the tumor progression (Day 7 and 14 post-tumor cell injection), whereas PD-L1 expression was at its highest at late stages (Day 14 and 21 post-tumor implantation). Overall our data suggested that B cells infiltrating the tumor vicinity were regulatory B cells (Bregs) that could inhibit CD8+ T-cell activation and acquisition of effector cytotoxic properties.
Figure 2. Glioma-associated B-cells phenotype.
(A) Five paired tumor and PBMC samples from newly diagnosed GBM patients were analyzed for the presence of B cells and their phenotype by flow cytometry. PD-L1, CD155, TGFβ (LAP), and IL10 were assessed (n=4 patients). Difference between tumor and PBMC B-cell phenotype was assessed by unpaired T-test. (B) CD155, PD-L1, IL10, and TGFβ (LAP) expression were analyzed in B cells from tumor (brain), blood, deep cervical lymph nodes (dCLNs), and superficial cervical lymph nodes (CLNs) at 7, 14, and 21 days after CT2A tumor implantation. n=4 mice/group in 2 independent experiments. For comparisons of the mean differences between groups based on two factors (time and tissue), ordinary two-way ANOVA with Turkey’s multiple comparison test were used. (C) B cells from CT2A tumors (n=5/experiment) were tested for suppression of CD8+ T-cell activation after treatment with blocking anti-IL10, anti-TGFβ, anti-TIGIT, or anti-PD-L1. Abs were added every day throughout the experiment (72 hours). CD8+ T-cell activation was assessed by cell proliferation and expression of intracellular GzmB and IFNγ. Representative experiment of 3 independent experiments performed in triplicates. All histograms mean±SD of triplicates. In all experiments, ns: no statistically significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
The collection of molecules expressed by GBM-associated B cells are well known to play an important role in cancer-mediated immunosuppression and immunosurveillance escape and are under investigation as potential targets for the development of immunotherapies (22–28). Supporting this current knowledge, neutralization of soluble IL10 and TGFβ, blockade of TIGIT (ligand for CD155) on CD8+ T-cells, and PD-L1 on tumor-infiltrating B cells using inhibitory Abs rescued CD8+ T-cell proliferation and expression of GzmB and IFNγ (Fig. 2C), with anti-PD-L1 being the most effective regimen. Taken together, our results suggested that GBM Bregs negatively regulated activation of CD8+ T cells by expression of membrane inhibitory molecules PD-L1 and CD155 and production of immunosuppressive soluble factors such as IL10 and TGFβ.
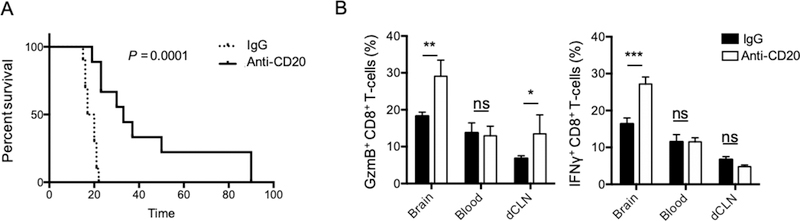
Anti-CD20 depleting immunotherapy provides an extended animal survival
To evaluate the pathophysiological relevance of GBM Bregs in tumor progression, we tested the effects of B-cell depleting immunotherapy using anti-CD20. Because the immunosuppressive function of GBM Bregs appeared to be restricted to the tumor and not the periphery, we aimed to preferentially deplete B cells in the brain. To do so, we delivered the CD20 depleting Ab intracranially via cannula after 7 days of CT2A tumor implantation. We observed that animals receiving the B-cell depleting immunotherapy survived significantly longer than the IgG control group (IgG vs anti-CD20 mean survival: 18.5 vs 33 days, P=0.0001, Fig. 3A). In an independent experiment, we confirmed local injections of B-cell depleting Ab only partially depleted peripheral B cells (blood, spleen and dCLN, Supplementary Fig. S3A). The extended animal survival after local B-cell depleting immunotherapy correlated with increased tumor-infiltrating effector CD8+ T cells expressing GmzB and IFNγ (Fig. 3B). However, no survival benefit was observed in the animals receiving the immunotherapy systemically (intraperitoneal injection, Supplementary Fig. S3B–C).
Figure 3. Local B cell–depletion immunotherapy provides an animal survival benefit.
(A) CT2A-bearing mice received intracranially 25μg/mouse (5μL/injection) of depleting anti-CD20 (anti-CD20) or IgG isotype control (IgG) 7 and 9 days after tumor inoculation (n=10mice/group). Survival of treated and control-treated mice shown. Survival curves were generated via the Kaplan-Meier method and compared by log-rank test and multiple comparisons were adjusted using Bonferroni method. (B) CD8+ T cells were analyzed for the expression of granzyme B (GzmB) and IFNγ in the tumor (brain), blood, and deep cervical lymph nodes (dCLN). Ordinary Two-way ANOVA with Turkey’s multiple comparison test were used. Histograms are shown as mean±SD of n=5mice/group. In all experiments, statistical significance is depicted as ns: no statistically significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
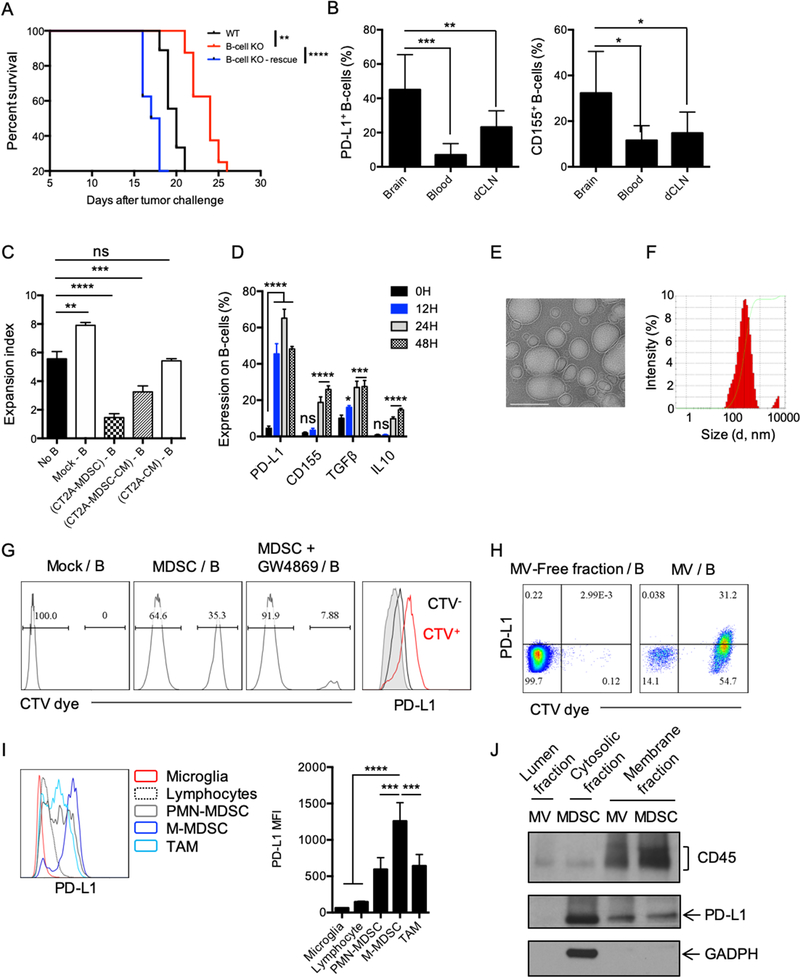
Supporting our observations using B-cell depletion, B cell–deficient μMT (B-cell KO) mice survived significantly longer than wild-type (WT) controls (Fig. 4A), and this correlated with increased expression of GzmB by CD8+ T cells (Supplementary Fig. S4A). Adoptive transfer of WT B cells into B cell–deficient mice (B-cell KO–rescue group, Fig. 4A) showed a significant reduction in overall survival compared to the B-cell KO group, including compared to the WT group. B-cell immunophenotypic analysis revealed that adoptively transferred naïve B cells exhibited significantly higher expression of both inhibitory molecules CD155 and PD-L1 (Fig. 4B) in the tumor compared to those in the circulation (blood) and dCLN. These data suggested that naïve B cells acquired a Breg phenotype within the tumor microenvironment. IL10-deficient B cells did not show any animal survival improvement (Supplementary Fig. S4B), suggesting that unlike other Breg populations (29), GBM-associated Bregs do not utilize IL10 as the main immunoregulatory mechanism.
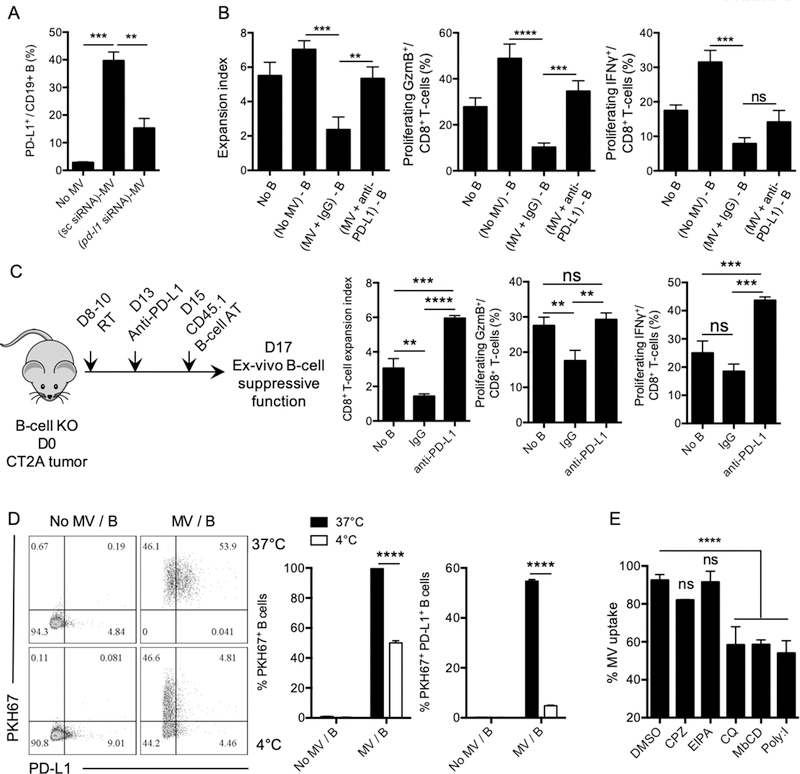
Figure 4. MDSC converts naïve B-cells into Bregs via microvesicles.
(A) Survival of CT2A-bearing, B cell–deficient mice (B-cell KO) vs. C57BL/6 (WT) mice. 5×106 naïve B-cells were adoptively transferred into B-cell KO mice (B-cell KO–rescue) and B-cell KO or WT groups (n=10mice/group). Survival curves were compared by log-rank test and multiple comparisons were adjusted using Bonferroni method. (B) B cell–deficient mice were injected with 5×106 B cells 7 days after tumor implantation and B-cell phenotype was analyzed 3 days after. Percentage of PD-L1+ and CD155+ B cells in the brain, blood, or in the deep cervical lymph nodes (dCLNs) were assessed in n=4 mice/group. Differences were assessed by one-way ANOVA with Tukey’s multiple comparison test. (C) MDSCs were generated using CT2A–conditioned media (CM) for 6 days in the presence of GM-CSF (CT2A-MDSC). CT2A-MDSCs were then cultured at 1:1 ratio with naïve B cells ((CT2A-MDSC)-B). CT2A-MDSCs were cultured alone for 4 days. Supernatants were collected (CT2A-MDSC-CM). Naïve B cells were incubated with ½ CT2A-MDSC-CM + ½ complete RPMI ((CT2A-MDSC-CM) – B). Naïve B cells were incubated with ½ CT2A-CM + ½ complete RPMI ((CT2A-CM) – B). All B cells were incubated for 48 hours. B cells alone (Mock B) was also kept 48h as control. B cells were evaluated for suppression of CD8+ T-cell activation. CD8+ T-cell proliferation was measured as expansion index. Differences were assessed by one-way ANOVA with Tukey’s multiple comparison test. (D) CT2A-MDSCs (upper chamber) and B cells (lower chamber) were cultured for different timepoints (12, 24, 48, and 72h) and expression of PD-L1, CD155, TGFβ (LAP), and IL10 was analyzed. The experiment was performed in triplicates and repeated in 2 independent experiments. Each gene expression difference over the time was statistically assessed using one-way ANOVA with Tukey’s multiple comparison test. (E-F) CT2A-MDSCs were cultured in microvesicle (MV)-free media for 4 days at 106 cells/mL. Supernatants were harvested and ultracentrifugation used to eliminate cell debris and apoptotic bodies. MVs were (E) imaged by cryo-electronic microscopy (scale bar 100 nm) and (F) the size distribution of the MVs evaluated by dynamic light scattering (DLS) analysis. (G) CT2A-MDSCs labeled with the lipophilic dye Cell Trace Violet (CTV; upper chamber, MDSCs) and naïve B cells (lower chamber). 48 hours after, B cells were harvested and evaluated for the acquisition of the CTV dye and PD-L1 by flow cytometry. In parallel, CT2A-MDSCs were treated with an extracellular vesicle release inhibitor GW4869 for 5h prior transwell culture (MDSC+GW4869). GW4869 was washed 3 times with complete RPMI before setting the coculture with B cells. (H) CT2A-MDSCs were labeled with the CTV dye and cultured in MV-free media. Supernatants were collected 4 days after. MVs were obtained after ultracentrifugation. The MV-free fraction (last step of centrifugation) was also collected. B cells were incubated with MV-free and MVs for 12 hours. B cells were harvested and evaluated for the acquisition of the CTV dye and PD-L1 by flow cytometry. (I) Flow cytometry analysis of PD-L1 expression by different immune cell populations in CT2A tumors (n= 4 mice). (J) PD-L1 detection in the MV membrane and lumen fractions versus MDSC membrane and cytosol fraction by Western blot. All histograms mean±SD of triplicates. In all experiments, ns: no statistically significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
GBM-associated Breg generation is an active process that requires MDSCs
Myeloid cells, especially monocytic MDSCs, are a prominent component of the GBM microenvironment (12,30), and might play an important role in shaping the adaptive immune response to promote immune escape. We tested the ability of tumor cell–derived factors and MDSCs to promote Breg differentiation and function by culturing B cells with CT2A-conditioned media (CT2A-CM) or immunosuppressive MDSCs generated from BM cells using CT2A-CM (CT2A-MDSCs, Supplementary Fig. S4C–D, Fig 4C). Supernatants from CT2A-MDSCs (CT2A-MDSC-CM) were also tested. After 48 hours, B cells were collected and evaluated for their CD8+ T-cell suppressive capability (Fig. 4C). Naïve B cells (Mock B) showed an enhancing effect in CD8+ T-cell activation (No B vs Mock-B, p<0.005). B cells cocultured with MDSCs or MDSC-CM showed a significant suppressive effect compared to activated CD8+ T cells alone [No B vs (CT2A-MDSC)-B or (CT2A-MDSC-CM)-B; p<0.00001]. However, B cells incubated with CT2A-CM were not suppressive [No B vs (CT2A-CM)-B, p 0.67] nor an activator of CD8+ T-cells [Mock-B vs (CT2A-CM)-B, p<0.005]. Because both CT2A-MDSCs and CT2A-MDSC-CM conferred regulatory function to B cells, we next evaluated whether this interaction required cell-to-cell contact. B-cell immunophenotype and function were analyzed in a MDSC/B-cell transwell system. We assured that no contaminating MDSCs crossed the transwell 0.4μm pores membrane (Supplementary Fig. S4E). We observed that the presence of PD-L1 on the surface of B cells was significantly increased after only 12 hours of culture, followed by increased expression of CD155, TGFβ (LAP), and IL10 at later time points (Fig 4D, Supplementary Fig. S4F). These observations might suggest that the rapid acquisition of PD-L1 by B cells might mark the transition from naïve B cells to Bregs.
GBM-associated MDSCs induce Bregs via transfer of membrane-bound PD-L1
MDSCs can modulate the tumor microenvironment via cell-to-cell contact (i.e. PD-L1, FasL) by altering the surrounding microenvironment nutrient and metabolic profile (i.e. amino acid deprivation via arginase, ROS production) and by the production of immunosuppressive cytokines such as TGFβ or IL10 (31–33). In the context of cancer, MDSCs are known to secrete extracellular vesicles that can recapitulate many aspects of their immunosuppressive factors, mainly by transporting both membrane proteins and cytosolic factors to the target T cells (34–36). We observed that GBM MDSCs produced a significant amount of extracellular vesicles (Fig. 4E–F) able to cross the transwell 0.4μm pore membrane. These extracellular vesicles were imaged by cryo-electronic microscopy (Fig. 4E), and the dynamic light scattering (DLS) analysis revealed the presence of vesicles with an average size of 188nm (Fig. 4F). These microvesicles (MVs) might correspond to ectosomes (100n-1000nm) rather than exosomes (40–100nm)(37,38). To test whether naïve B cells could uptake the MDSC membrane-derived MVs, MDSCs were labeled with a lipophilic dye (CTV), and we repeated the transwell experiment. After 48 hours, B cells were harvested and evaluated for the acquisition of the CTV dye by flow cytometry. We observed that B cells acquired MDSC membrane-derived structures (CTV dye positive), which correlated with the presence of PD-L1 (Fig. 4G). Pretreatment of MDSCs with an inhibitor of MV release (GW4869) reduced CTV dye uptake by the B cells (Fig. 4G).
We next confirmed that B cells’ uptake shown in Fig. 4G was due to MDSC-derived MVs because the majority of naïve B cells (>85%) directly incubated with MVs derived from MDSCs acquired the CTV signal after 12 hours, and >30% were also positive for PD-L1 (Fig. 4H).This suggested that the “rapid” increase of PD-L1 on B cells (Fig. 4D) was likely due to the uptake by the B-cell membrane of MDSC-derived MVs carrying PD-L1. MV uptake by B cells slightly increased expression levels of CD155 and TGFβ. No significant changes were observed for IL10 (Supplementary Fig. S4G). In CT2A-bearing mouse brains, monocytic MDSCs showed the highest PD-L1 expression in the tumor microenvironment (Fig. 4I), and MDSC-derived MVs carried PD-L1 on their membranes (not in the lumen, Fig. 4J). To verify this phenomenon, we knocked down PD-L1 in MDSCs using positively charged lipid nanoparticles (LNP) to deliver siRNA. After 24 hours post-transfection, MDSCs were harvested and tested for their PD-L1 transcripts using quantitative PCR (Supplementary Fig. S5A). MVs from transfected MDSCs (scramble siRNAs and pd-l1 siRNA) were harvested from supernatants and cultured with naïve B cells for 12 hours. Presence of membrane PD-L1 on B cells was tested by flow cytometry. Our data showed that pd-l1 siRNA–transfected MDSC MVs transferred significantly reduced amounts of PD-L1 to B cells compared to the control scramble siRNA group (Fig. 5A). We confirmed that B cells cultured with MDSC-derived MVs suppressed CD8+ T-cell activation. However, blockade of PD-L1 on MVs before coculture with B cells significantly inhibited B-cell suppressive effects on CD8+ T-cell expansion and effector phenotype (Fig. 5B). Altogether these data suggested that MDSCs transferred PD-L1 to B cells via MVs, which conferred regulatory properties to B cells.
Figure 5. MDSCs transfer PD-L1 to B cells via microvesicles.
(A) PD-L1 was knocked down using lipid nanoparticles (LNP)+siRNA for 24 hours. MDSC-derived MVs from scramble (sc) siRNA or pd-l1 siRNA were isolated by ultracentrifugation after 4 days in culture with MV-free complete RPMI. Naïve B cells were then cultured with sc siRNA–treated MV or pd-l1 siRNA–treated MVs for 12 hours. PD-L1 on the surface of B cells was assessed by flow cytometry. Performed in 2 independent experiments in triplicates. (B) B cells treated with MVs carrying PD-L1 were evaluated for their suppressive effect on activated CD8+ T cells. MVs were pretreated with blocking anti-PD-L1 (MV+antiPD-L1) or isotype control (MV+IgG) prior incubation with B cells. CD8+ T-cell proliferation was measured as expansion index 72 hours after. The experiment was performed in triplicates. (C) B cell–deficient (KO) mice were challenged intracranially with 105 CT2A glioma cells. Mouse brains were irradiated with 3Gy three consecutive days (D8–10) to upregulate PD-L1. Three days after, mice were injected intracranially with anti-PD-L1 (5μL, 5.5μg/injection) via cannula. 48h after, CD45.1+ B cells were adoptively transferred intracranially. CD45.1+ B cells were magnetically isolated after 2 days and tested for their ex vivo suppressive function toward activated CD8+ T-cells (+anti-CD3/CD28+IL2). CD8+ T-cell activation was measured by cell proliferation (expansion index) and expression of granzyme B (GzmB) and IFNγ. (D) Microvesicles (MVs) were isolated from in vitro–generated glioma-associated MDSCs. MVs were labeled with PKH67 dye. Splenic naïve B cells were incubated with PKH67+ MVs at 4°C or 37°C. MV uptake and PD-L1 expression were assessed by flow cytometry. n=3 independent experiments. (E) Splenic naïve B cells were treated with different endocytosis inhibitors for 2 hours prior addition of PKH67-labed MVs. MV uptake by B cells was assessed by flow cytometry as PKH67+ B cells (n=2 independent experiments). All histograms mean±SD of triplicates. In all experiments, statistical significance was assessed using the one-way ANOVA with Tukey’s multiple comparison test; ns: no statistically significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Brain penetrant CSF-1R inhibitor (BLZ945) is known to reduce numbers of cancer-associated myeloid cells (39,40), inhibit GBM-associated myeloid cells’ immunosuppressive functions (41–43), and reduced PD-L1 expression by tumor-associated myeloid cells (39)Supplementary Fig. S5B). Our data showed that naïve B cells transferred to B cell–deficient mice treated with BLZ945 (prior to B-cell injection) failed to acquire PD-L1 (Supplementary Fig. S5C). In support, in vivo blockade of PD-L1 upon radiotherapy (known to upregulate PD-L1 in myeloid cells, (44)) prevented the acquisition of regulatory properties by adoptively transferred B cells (Fig. 5C).
MVs can be acquired by B cells in an ATP-independent (4°C culture, passive fusion) or -dependent manner (37°C, active endocytosis) (45). We observed that optimal MV uptake and PD-L1 transfer involved an ATP-dependent mechanism (37°C; Fig 5D). An endocytosis inhibitor study showed that caveolae-mediated endocytosis could be a major mechanism of MV uptake by B cells. These results implied that PD-L1 on MVs was probably transferred to B cells via receptor-mediated endocytosis followed by endocytic recycling toward the plasma membrane of B cells (Fig. 5E, (45)).
Finally, the interaction of GBM-infiltrating Bregs with effector CD8+ T cells promoted overexpression of B cell–derived TGFβ and IL10 (Supplementary Fig. S5D). This might reflect Bregs attempting to maintain the immunosuppressive environment by restraining CD8+ T-cell activation. Blockade of PD-L1 on tumor Bregs significantly reduced the expression of both TGFβ and IL10 by B cells. This observation highlights the magnitude of PD-L1 acquisition by B cells in controlling their own immune functions and the relevant role of MDSCs in inducing B cell–mediated immunosuppression.
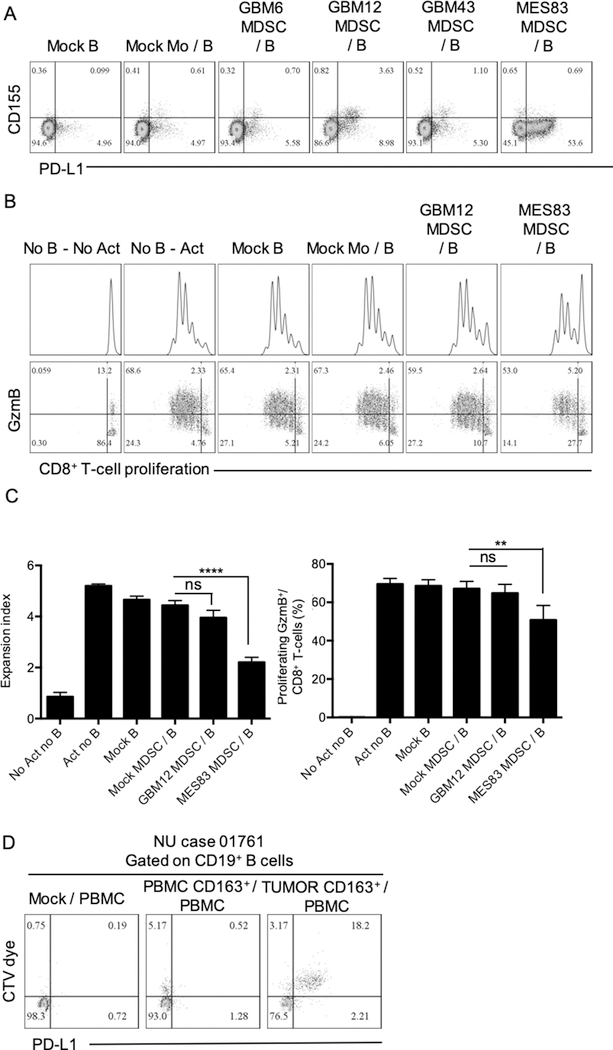
Next, we aimed to elucidate whether human MDSCs and B cells could interact via transfer of MV-bound PD-L1. Human MDSCs were generated from peripheral blood monocytes using CM from patient-derived GBM xenograft lines (46): a classical subtype (GBM6), a proneural subtype (GBM12 and 43) and a mesenchymal subtype (MES83). We observed that MDSCs generated with CM from GBM12 and MES83 expressed high PD-L1 (Supplementary Fig. S6A). This observation correlated with acquisition of PD-L1 by B cells upon transwell coculture for 24hrs (Fig. 6A). B cells cultured with GBM12 and MES83-MDSCs were tested for their ability to suppress autologous CD8+ T cells in transwell experiments. Coculture with MES83-MDSCs provided the highest B-cell PD-L1 expression and significant immunosuppression (Fig. 6B–C). Because tumor-associated macrophages also express high PD-L1 and are also able to secrete MVs (47), we investigated whether CD163+ infiltrating macrophages from GBM patients’ tumors are able to transfer PD-L1 to the target cells. CD163+ infiltrating macrophages from freshly resected GBM tumors were isolated and tested for their ability to promote PD-L1+ Bregs. CD163+ macrophages labeled with the CTV dye (upper chamber) and PBMCs (lower chamber) were cocultured in a transwell system as in previous experiments. After 4 days, different immune cells populations were evaluated for the acquisition of the CTV dye and the presence of PD-L1. Foxp3+ Tregs and B cells showed the highest uptake compared to CD8+ T cells and myeloid cells (Fig. 6D, Supplementary Fig. S6B). CTV dye acquisition correlated with the presence of membrane PD-L1. CD33+CD14+ myeloid cells expressed high PD-L1, but no sign of uptake (CTV signal) was observed (Supplementary Fig. S6B). Altogether, our observations suggest that GBM-associated MDSCs produced MVs that carry membrane-bound PD-L1, which are then taken up by target cells such as Bregs. Therefore, MDSCs may promote GBM by sharing their immunosuppressive factors with other cells prone to their reception.
Figure 6. GBM-associated human MDSCs transfer PD-L1 to B cells.
(A) Monocytes from peripheral blood were incubated with GM-CSF, IL6, and 50% conditioned media from different patient-derived GBM cell lines: GBM6, GBM12, GBM43, and MES83. MDSCs were transwell cultured with autologous circulating B cells for 24 hours. PD-L1 and CD155 were evaluated in B cells by flow cytometry. Representative flow cytometry plot of 2 independent experiments. Mock B: no MDSC; Mock Mo/B: Control monocytes (upper chamber) cultured with B cells (lower chamber); GBM6 MDSC/B: MDSC generated from GBM6 conditioned media (upper chamber) cultured with B cells (lower chamber); GBM12 MDSC/B: MDSC generated from GBM12 conditioned media (upper chamber) cultured with B cells (lower chamber); GBM43 MDSC/B: MDSC generated from GBM43 conditioned media (upper chamber) cultured with B cells (lower chamber); MES83 MDSC/B: MDSC generated from MES83 conditioned media (upper chamber) cultured with B cells (lower chamber). (B-C) B cells obtained after transwell culture with MDSCs generated with GBM12 and MES83 conditioned media were tested for their ability to inhibit CD8+ T-cell proliferation and expression of granzyme B (GzmB) after 72 hours. Experiment was performed in triplicates and repeated in 2 independent experiments. (D) CD163+ myeloid cells from tumor and peripheral blood from donor NU01761 were magnetically isolated and labeled with CTV dye. Total PBMCs were placed in the lower chamber. After 24 hours, CD19+ B cells in the lower chamber (PBMC) were tested for the acquisition of CTV and PD-L1 by flow cytometry. All histograms mean±SD of triplicates. In all experiments, statistical significance was assessed using the one-way ANOVA with Tukey’s multiple comparison test; ns: no statistically significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Discussion
In the present study, we investigated the phenotype and function of GBM-infiltrating B cells. Whereas the role of other components of the tumor microenvironment such as MDSCs and Tregs are relatively well-studied (4,48–51), the involvement of B cells in GBM biology has not been thoroughly examined, limited only to the observation of B-cell infiltration in GBMs (14). Here, we demonstrated that B cells harvested from patients’ GBM tumors possessed a suppressive effect on activated CD8+ T cells, shown by the inhibition of CD8+ T-cell proliferation and further acquisition of an effector phenotype. Immunosuppressive functions of Bregs in cancer are also extended in the CD4+ T-cell compartment (52,53). We observe a mild effect of glioma Bregs in suppressing activated CD4+ T cells. However, further studies should be pursued to deeply elucidate the role of glioma Bregs in CD4+ T-cell polarization and differentiation.
GBM-associated B cells showed a definite immunoregulatory phenotype as judged by expression of PD-L1, CD155, TGFβ, and IL10. These observations were also obtained in GL261 and CT2A orthotopic models of GBM. This functional and immunophenotypic profile was exclusive to tumor-infiltrating B cells. We observed that local intratumoral depletion of B cells resulted in the improvement of animal survival. The therapeutic effectiveness was accompanied by increased intratumor GzmB+ CD8+ T cells, suggesting that depletion of GBM-associated Bregs favored CD8+ T-cell activation and promoted their effector function. We did not observe significant animal survival benefit when B cells were depleted systemically. These results reinforce the hypothesis that B cells might present dual functions depending on their localization: pro-tumorigenic functions in the tumor (Breg) vs antitumor functions in the periphery. Candolfi and colleagues (54) reported similar observations when using systemic anti-CD20 depleting therapy in the GL261 GBM model. The authors observed that B cells played an important role in promoting antitumor responses by antigen-presentation rather than Ab production. Our observations also contradict a computational analysis of GBM tumor-infiltrating immune cells, where the high magnitude of B cells predicted overall increased patient survival (55). Nevertheless, ex vivo tumor-infiltrating B cells from GBM tumor samples showed an immunosuppressive effect toward the CD8+ T-cell activation in agreement with previous observations (20,21). Based on these data, we hypothesize that GBM and its microenvironment actively promote the conversion of B cells infiltrating the tumor into Bregs.
The evaluation of GBM-bearing, B cell–deficient mice contributes to the understanding that the acquisition of the regulatory phenotype is an active process. In fact, adoptive transfer of naïve B cells not only rescued the survival phenotype but also revealed that only tumor infiltrating B cells showed high PD-L1 and CD155. These data suggest that a dynamic process might take place in the tumor microenvironment that converts naïve B cells into GBM-associated Bregs. This hypothesis correlates with a current view of Breg differentiation, a biological mechanism that requires local immunological activation (56). The GBM vicinity is an ideal microenvironment to attract and promote survival of potent immunoregulatory cells such as MDSCs or Tregs (12), and possibly Bregs. GBM cells are known to release Breg-inducing factors (20,21). Other components of the tumor microenvironment such as myeloid cells can promote activation of naïve B cells and their differentiation toward an immunosuppressive functional profile. For instance, breast cancer-associated MDSCs have been reported to convert naïve B cells into immunosuppressive B cells able to inhibit T-cell responses (57). In GBMs, monocytic Ly6C-expressing MDSCs are the main component of the GBM microenvironment and represent approximately 40% of the total tumor mass (12,13). We observed that B cells exhibited an immunoregulatory phenotype when cultured in the presence of GBM-associated MDSCs or MDSC supernatants, as they became capable of abrogating the CD8+ T-cell activation. The fast upregulation of MDSC-induced PD-L1 (unlike CD155, TGFβ or IL10) on B cells after transwell culture with MDSCs led us to hypothesize that the initial increase of PD-L1 on B cells membrane might be partially due to a protein transfer from MDSCs. Chen and colleagues (58) report that tumor-derived exosomes, which are extracellular vesicles of endosomal origin, can carry PD-L1 on their membrane surface. Authors observed that these “nanobodies” could suppress CD8+ T cells via direct PD-L1/PD1 interaction remotely from the tumor site. This mechanism has been reported in GBM patients, where tumor-derived extracellular vesicles carrying PD-L1 on their surface can inhibit CD8+ T-cell activity (59). Accordingly, we observed that GBM-associated MDSCs secreted ∼200nm MVs carrying PD-L1 on their membrane. Our experiments showed that these MV shared cytoplasmic membrane as their cellular source, which are MDSCs. Naïve B cells were readily prone to uptake these shedding vesicles. We hypothesize that PD-L1 is carried by MDSC-derived MV membranes (since no detectable PD-L1 was observed in the MV lumen fraction) and is transferred to B cells. This hypothesis is supported by work reporting that MDSC-derived extracellular vesicles carry biologically relevant proteins (34,35,60). Accordingly, downregulation of PD-L1 expression on MDSCs significantly reduced PD-L1 presence on the surface of B cells and subsequently reduced Breg immunosuppressive effects on activated CD8+ T cells. This indicated that MDSC-derived MVs carrying PD-L1 are, at least in part, responsible for the presence of PD-L1 on B cells. This phenomenon might drive B cell–mediated immunosuppressive function in brain tumors.
Based on our results, one hallmark of GBM-associated Bregs is the overexpression of TGFβ and IL10 upon interaction with activated CD8+ T-cells. This phenomenon might reflect an attempt of Bregs to maintain a sustained immunosuppressive environment to prevent further CD8+ T-cell activation upon initial inhibitory contact. We are currently unable to determine whether MDSC-MV-bound PD-L1 transfer to B cells mediates TGFβ and IL10 upregulation, as it is unclear whether the PD-L1 downstream pathway is fully functional. Nevertheless, blockade of PD-L1 on tumor Bregs upon interaction with CD8+ T cells abrogated the overexpression of both TGFβ and IL10 by Bregs. One possible explanation might be the inability of CD8+ T cells to produce IFNγ upon contact with GBM Bregs. The lack of IFNγ might be the result of PD-1–mediated inhibition (61) regulated by Bregs. Accordingly, PD-1/PD-L1 interaction blockade promoted CD8+ T cell–derived IFNγ expression, which was associated with reduced expression of TGFβ and IL10 by B cells.
The mechanisms by which B cells infiltrate glioblastoma are unknown, and, albeit, Breg function might be relevant only in a reduced group of GBM patients. The overall study highlights a mechanism of MDSC cellular interaction with the adaptive immune system. The potential effect of GBM-associated myeloid cells in promoting Bregs via transfer of membrane-bound PDL1 might also occur in human myeloid cells. Both in vitro–generated MDSC-like cells (using MES83 mesenchymal cell line CM) and tumor-infiltrating CD163+ macrophages promoted increased PD-L1 in autologous B cells in transwell culture settings. CD163+ myeloid cell-derived MVs were preferentially up-taken by both B cells and Foxp3+ Tregs, suggesting the transfer of PD-L1–carrying MVs might represent a universal process of intercellular communication between regulatory cells.
In summary, we demonstrated that B cells infiltrating human and murine GBMs acquire the regulatory phenotype in the tumor milieu. This dynamic process was mediated, at least in part, by communication of MDSCs with B cells via MV-bound PD-L1 transfer from MDSCs. Our data suggest that Bregs play an important role in GBM evasion from the surveillance of cytotoxic CD8+ T cells, further highlighting the complexity of immunosuppressive GBM network.
Supplementary Material
Acknowledgments
We would like to thank Katy McCortney and Rodrigo Javier at the Nervous System Tumor Bank (Feinberg School of Medicine, Northwestern University) for managing the interface between the clinic and the laboratory regarding GBM patients’ biological samples. Lastly, we are extremely grateful for the patients who consented to donate their biological samples.
Funding
This study was supported by the following grants from the NIH: P50CA221747, R35CA197725, R01NS093903, R01 NS087990 (MSL). CLC received a SPORE Career Enhancement Program (P50CA221747). We acknowledge support from the Structural Biology Facility at Northwestern University, the Robert H Lurie Comprehensive Cancer Center of Northwestern University and NCI CCSG P30 CA060553. The Gatan K2 direct electron detector was purchased with funds provided by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust.
Footnotes
The authors declare no potential conflicts of interest
References
- 1.Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology 2015;4(7):e1016700 doi 10.1080/2162402X.2015.1016700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wainwright DA, Balyasnikova IV, Chang AL, Ahmed AU, Moon KS, Auffinger B, et al. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clin Cancer Res 2012;18(22):6110–21 doi 10.1158/1078-0432.CCR-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vleeschouwer S, Spencer Lopes I, Ceuppens JL, Van Gool SW. Persistent IL-10 production is required for glioma growth suppressive activity by Th1-directed effector cells after stimulation with tumor lysate-loaded dendritic cells. J Neurooncol 2007;84(2):131–40 doi 10.1007/s11060-007-9362-y. [DOI] [PubMed] [Google Scholar]
- 4.Raychaudhuri B, Rayman P, Ireland J, Ko J, Rini B, Borden EC, et al. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro Oncol 2011;13(6):591–9 doi 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Andaloussi A, Lesniak MS. CD4+ CD25+ FoxP3+ T-cell infiltration and heme oxygenase-1 expression correlate with tumor grade in human gliomas. J Neurooncol 2007;83(2):145–52 doi 10.1007/s11060-006-9314-y. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs DI, Liu Y, Gabrusiewicz K, Tsavachidis S, Armstrong GN, Zhou R, et al. Germline polymorphisms in myeloid-associated genes are not associated with survival in glioma patients. J Neurooncol 2018;136(1):33–9 doi 10.1007/s11060-017-2622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol 2015;11(9):504–14 doi 10.1038/nrneurol.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li XY, Das I, Lepletier A, Addala V, Bald T, Stannard K, et al. CD155 loss enhances tumor suppression via combined host and tumor-intrinsic mechanisms. J Clin Invest 2018;128(6):2613–25 doi 10.1172/JCI98769. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Wei J, Wu A, Kong LY, Wang Y, Fuller G, Fokt I, et al. Hypoxia potentiates glioma-mediated immunosuppression. PLoS One 2011;6(1):e16195 doi 10.1371/journal.pone.0016195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei J, Barr J, Kong LY, Wang Y, Wu A, Sharma AK, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther 2010;9(1):67–78 doi 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol 2018;15(7):422–42 doi 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 12.Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. CCL2 Produced by the Glioma Microenvironment Is Essential for the Recruitment of Regulatory T Cells and Myeloid-Derived Suppressor Cells. Cancer Res 2016;76(19):5671–82 doi 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamran N, Kadiyala P, Saxena M, Candolfi M, Li Y, Moreno-Ayala MA, et al. Immunosuppressive Myeloid Cells’ Blockade in the Glioma Microenvironment Enhances the Efficacy of Immune-Stimulatory Gene Therapy. Mol Ther 2017;25(1):232–48 doi 10.1016/j.ymthe.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingues P, Gonzalez-Tablas M, Otero A, Pascual D, Miranda D, Ruiz L, et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun 2016;53:1–15 doi 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Wu C, Muroski ME, Miska J, Lee-Chang C, Shen Y, Rashidi A, et al. Repolarization of myeloid derived suppressor cells via magnetic nanoparticles to promote radiotherapy for glioma treatment. Nanomedicine 2019;16:126–37 doi 10.1016/j.nano.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3 22 doi 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 17.Pituch KC, Moyano AL, Lopez-Rosas A, Marottoli FM, Li G, Hu C, et al. Dysfunction of platelet-derived growth factor receptor alpha (PDGFRalpha) represses the production of oligodendrocytes from arylsulfatase A-deficient multipotential neural precursor cells. J Biol Chem 2015;290(11):7040–53 doi 10.1074/jbc.M115.636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram JR, Dougan M, Rashidian M, Knoll M, Keliher EJ, Garrett S, et al. PD-L1 is an activation-independent marker of brown adipocytes. Nat Commun 2017;8(1):647 doi 10.1038/s41467-017-00799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miska J, Lee-Chang C, Rashidi A, Muroski ME, Chang AL, Lopez-Rosas A, et al. HIF-1alpha Is a Metabolic Switch between Glycolytic-Driven Migration and Oxidative Phosphorylation-Driven Immunosuppression of Tregs in Glioblastoma. Cell Rep 2019;27(1):226–37 e4 doi 10.1016/j.celrep.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye ZP, He HY, Wang H, Li WS, Luo L, Huang ZC, et al. Glioma-derived ADAM10 induces regulatory B cells to suppress CD8+ T cells. PLoS One 2014;9(8):e105350 doi 10.1371/journal.pone.0105350. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Han S, Feng S, Ren M, Ma E, Wang X, Xu L, et al. Glioma cell-derived placental growth factor induces regulatory B cells. Int J Biochem Cell Biol 2014;57:63–8 doi 10.1016/j.biocel.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A 2002;99(19):12293–7 doi 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Wu X, Guo C, Zhang K, Xu J, Li Z, et al. Development of Inhibitors of the Programmed Cell Death-1/Programmed Cell Death-Ligand 1 Signaling Pathway. J Med Chem 2018. doi 10.1021/acs.jmedchem.8b00990. [DOI] [PubMed] [Google Scholar]
- 24.Brown MC, Holl EK, Boczkowski D, Dobrikova E, Mosaheb M, Chandramohan V, et al. Cancer immunotherapy with recombinant poliovirus induces IFN-dominant activation of dendritic cells and tumor antigen-specific CTLs. Sci Transl Med 2017;9(408) doi 10.1126/scitranslmed.aan4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massague J TGFbeta in Cancer. Cell 2008;134(2):215–30 doi 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554(7693):544–8 doi 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oft M IL-10: master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol Res 2014;2(3):194–9 doi 10.1158/2326-6066.CIR-13-0214. [DOI] [PubMed] [Google Scholar]
- 28.DiLillo DJ, Matsushita T, Tedder TF. B10 cells and regulatory B cells balance immune responses during inflammation, autoimmunity, and cancer. Ann N Y Acad Sci 2010;1183:38–57 doi 10.1111/j.1749-6632.2009.05137.x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, et al. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature 2012;491(7423):264–8 doi 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The Immune Landscape of Cancer. Immunity 2018;48(4):812–30 e14 doi 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol 2009;182(9):5693–701 doi 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar V, Gabrilovich DI. Hypoxia-inducible factors in regulation of immune responses in tumour microenvironment. Immunology 2014;143(4):512–9 doi 10.1111/imm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009;9(3):162–74 doi 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke M, Choksawangkarn W, Edwards N, Ostrand-Rosenberg S, Fenselau C. Exosomes from myeloid-derived suppressor cells carry biologically active proteins. J Proteome Res 2014;13(2):836–43 doi 10.1021/pr400879c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zoller M Janus-Faced Myeloid-Derived Suppressor Cell Exosomes for the Good and the Bad in Cancer and Autoimmune Disease. Front Immunol 2018;9:137 doi 10.3389/fimmu.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng Z, Rong Y, Teng Y, Zhuang X, Samykutty A, Mu J, et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene 2017;36(5):639–51 doi 10.1038/onc.2016.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 2010;78(9):838–48 doi 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 38.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200(4):373–83 doi 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao Y, Eissler N, Blanc KL, Johnsen JI, Kogner P, Kiessling R. Targeting Suppressive Myeloid Cells Potentiates Checkpoint Inhibitors to Control Spontaneous Neuroblastoma. Clin Cancer Res 2016;22(15):3849–59 doi 10.1158/1078-0432.CCR-15-1912. [DOI] [PubMed] [Google Scholar]
- 40.Beffinger M, Tallon de Lara P, Tugues S, Vermeer M, Montagnolo Y, Ohs I, et al. CSF1R-dependent myeloid cells are required for NKmediated control of metastasis. JCI Insight 2018;3(10) doi 10.1172/jci.insight.97792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med 2013;19(10):1264–72 doi 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 2016;352(6288):aad3018 doi 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha D, Martuza RL, Rabkin SD. Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell 2017;32(2):253–67 e5 doi 10.1016/j.ccell.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng L, Liang H, Burnette B, Beckett M, Darga T, Weichselbaum RR, et al. Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J Clin Invest 2014;124(2):687–95 doi 10.1172/JCI67313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017;549(7670):101–5 doi 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar R, de Mooij T, Peterson TE, Kaptzan T, Johnson AJ, Daniels DJ, et al. Modulating glioma-mediated myeloid-derived suppressor cell development with sulforaphane. PLoS One 2017;12(6):e0179012 doi 10.1371/journal.pone.0179012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan J, Sun L, Xu F, Liu L, Hu F, Song D, et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res 2019;79(1):146–58 doi 10.1158/0008-5472.CAN-18-0014. [DOI] [PubMed] [Google Scholar]
- 48.Wintterle S, Schreiner B, Mitsdoerffer M, Schneider D, Chen L, Meyermann R, et al. Expression of the B7-related molecule B7-H1 by glioma cells: a potential mechanism of immune paralysis. Cancer Res 2003;63(21):7462–7. [PubMed] [Google Scholar]
- 49.Fecci PE, Mitchell DA, Whitesides JF, Xie W, Friedman AH, Archer GE, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res 2006;66(6):3294–302. [DOI] [PubMed] [Google Scholar]
- 50.El Andaloussi A, Lesniak MS. An increase in CD4+CD25+FOXP3+ regulatory T cells in tumor-infiltrating lymphocytes of human glioblastoma multiforme. Neuro Oncol 2006;8(3):234–43 doi 10.1215/15228517-2006-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Andaloussi A, Han Y, Lesniak MS. Prolongation of survival following depletion of CD4+CD25+ regulatory T cells in mice with experimental brain tumors. J Neurosurg 2006;105(3):430–7 doi 10.3171/jns.2006.105.3.430. [DOI] [PubMed] [Google Scholar]
- 52.Flores-Borja F, Bosma A, Ng D, Reddy V, Ehrenstein MR, Isenberg DA, et al. CD19+CD24hiCD38hi B cells maintain regulatory T cells while limiting TH1 and TH17 differentiation. Sci Transl Med 2013;5(173):173ra23 doi 10.1126/scitranslmed.3005407. [DOI] [PubMed] [Google Scholar]
- 53.Shen P, Fillatreau S. Antibody-independent functions of B cells: a focus on cytokines. Nat Rev Immunol 2015;15(7):441–51 doi 10.1038/nri3857. [DOI] [PubMed] [Google Scholar]
- 54.Candolfi M, Curtin JF, Yagiz K, Assi H, Wibowo MK, Alzadeh GE, et al. B cells are critical to T-cell-mediated antitumor immunity induced by a combined immune-stimulatory/conditionally cytotoxic therapy for glioblastoma. Neoplasia 2011;13(10):947–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17(1):174 doi 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauri C, Menon M. Human regulatory B cells in health and disease: therapeutic potential. J Clin Invest 2017;127(3):772–9 doi 10.1172/JCI85113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen M, Wang J, Yu W, Zhang C, Liu M, Wang K, et al. A novel MDSC-induced PD-1(−)PD-L1(+) B-cell subset in breast tumor microenvironment possesses immuno-suppressive properties. Oncoimmunology 2018;7(4):e1413520 doi 10.1080/2162402X.2017.1413520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560(7718):382–6 doi 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ricklefs FL, Alayo Q, Krenzlin H, Mahmoud AB, Speranza MC, Nakashima H, et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv 2018;4(3):eaar2766 doi 10.1126/sciadv.aar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geis-Asteggiante L, Belew AT, Clements VK, Edwards NJ, Ostrand-Rosenberg S, El-Sayed NM, et al. Differential Content of Proteins, mRNAs, and miRNAs Suggests that MDSC and Their Exosomes May Mediate Distinct Immune Suppressive Functions. J Proteome Res 2018;17(1):486–98 doi 10.1021/acs.jproteome.7b00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen S, Lee LF, Fisher TS, Jessen B, Elliott M, Evering W, et al. Combination of 4–1BB agonist and PD-1 antagonist promotes antitumor effector/memory CD8 T cells in a poorly immunogenic tumor model. Cancer Immunol Res 2015;3(2):149–60 doi 10.1158/2326-6066.CIR-14-0118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.