Abstract
The overall use of antibiotics has increased significantly in recent years. Besides fighting infections, antibiotics also alter the gut microbiota. Commensal bacteria in the gastrointestinal tract are crucial to maintain immune homeostasis, and microbial imbalance or dysbiosis affects disease susceptibility and progression. We hypothesized that antibiotic-induced dysbiosis of the gut microbiota would suppress cytokine profiles in the host, thereby leading to changes in the tumor microenvironment. The induced dysbiosis was characterized by alterations in bacterial abundance, composition, and diversity in our animal models. On the host side, antibiotic-induced dysbiosis caused elongated small intestines and ceca, and B16-F10 melanoma and Lewis Lung carcinoma progressed more quickly than in control mice. Mechanistic studies revealed that this progression was mediated by suppressed TNF-α levels, both locally and systemically, resulting in reduced expression of tumor endothelial adhesion molecules, particularly intercellular adhesion molecule-1 (ICAM-1) and a subsequent decrease in the number of activated and effector CD8+ T-cells in the tumor. However, suppression of ICAM-1 or its binding site, the alpha subunit of lymphocyte function-associated antigen-1, was not seen in the spleen or thymus during dysbiosis. TNF-α supplementation in dysbiotic mice was able to increase ICAM-1 expression and leukocyte trafficking into the tumor. Overall, these results demonstrate the importance of commensal bacteria in supporting anticancer immune surveillance, define an important role of tumor endothelial cells within this process, and suggest adverse consequences of antibiotics on cancer control.
Keywords: Dysbiosis, tumor endothelial adhesion molecules, gut microbiome, ICAM-1, TNF-α, T cells, tumor microenvironment
Introduction
Bacteria colonize many parts of the body, and the crosstalk between the microbiota and the host is crucial to maintaining immune homeostasis. A growing body of literature supports the idea that microbial imbalance affects disease susceptibility and progression. For instance, intestinal dysbiosis has been associated with a growing list of diseases of inflammatory, autoimmune, allergic, metabolic, and psychologic/neurologic nature (1–3).
A common and significant influence on the microbiota in the gastrointestinal (GI) tract is the use of antibiotics. Oral antibiotics severely alter the bacteria in the GI tract by destroying beneficial bacteria as well as potentially pathogenic ones — producing a state of microbial imbalance called dysbiosis. The overall use of antibiotics has increased by more than 30% in recent years (4), and moreover many cancer patients are prescribed antibiotics during treatment, as infection is a frequent complication. A cohort study including over 3 million individuals showed that there is a positive correlation between antibiotic use and cancer risk (5). Specifically, individuals who received 2–5 prescriptions over two years had an increased relative risk (RR) of 1.27 for getting cancer (with a 95% confidence interval (CI) of 1.26–1.29), as compared to individuals who were prescribed one or fewer antibiotic treatments in that period. The cancer risk was even greater, RR (95% CI) of 1.37 times (1.34–1.40) for individuals with more than 6 prescriptions (5). Additionally, a growing body of evidence in murine studies indicates that dysbiosis of the GI tract affects local colon carcinogenesis due to the initial chronic inflammation and subsequent immune suppression that dysbiosis produces (6–8). However, whether commensal bacteria also play a role in stromal immune surveillance of distal tumors remains unclear.
The bidirectional communication between tumor endothelial cells and the immune system is increasingly appreciated, yet how leukocyte trafficking is impacted by dysbiosis is unknown. At the center of this interaction are adhesion molecules on endothelial cells, including intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule (VCAM-1), and selectins (9). These molecules promote rolling, adherence, and transmigration of leukocytes into tumor tissue, making tumors more vulnerable to host immunity. This communication is mediated by cytokines, the levels of which are controlled by cellular cross talk between host cells and commensal bacteria (10). We therefore examined the consequences of antibiotic-induced dysbiosis on stromal immune surveillance in distal tumors, as a mechanism by which GI-tract microbiota influence immune surveillance. Mechanistic studies revealed that this progression was mediated by suppressed tumor necrosis factor-alpha (TNF-α) levels, both locally and systemically, resulting in suppression of tumor endothelial adhesion molecules, and a subsequent decrease in the number of tumor-infiltrating activated CD8+ T cells.
Materials and Methods
Cell Lines and cell culture
B16-F10 (murine melanoma cell line; #CRL-6475) and LLC (murine lung carcinoma; #CRL-1642) were purchased from American Type Culture Collection (ATCC) and cultured according to the company’s instructions. All cell lines were cultured and maintained as previously described (11).
Mice and tumor mouse models
All mice (C57BL/6J, #0664; Foxn1−/−, #2019; ICAM-1−/−, #2867) were purchased from Jackson Laboratory and allowed to acclimatize to local conditions for at least 1 week. Animals were provided water and standard chow ad libitum and were maintained on a 12-hour light/dark cycle. For tumor cell inoculation, a 100μL solution of 2 × 105 of B16-F10 or 1 × 106 LLC was injected s.c. in the right rear leg of each mouse, as described previously (11). Mice (sex- and age-matched littermates) were randomly inoculated. Tumor volume was determined by measuring the diameters of tumors with calipers and calculated by the equation for volume of a spheroid: (a2 × b × π)/6, where a is the short axis and b is the long axis of the tumor. Dysbiosis was induced 2 weeks prior to the tumor inoculations by administering a cocktail of antibiotics, i.e. ampicillin (250 mg/L), vancomycin (125 mg/L), neomycin (250 mg/L), and metronidazole (250 mg/L) in their drinking water, available ad libitum during the experiment.(12) Mice receiving TNF-α were randomized on day 7 after tumor inoculation, and murine TNF-α treatment was initiated (q3dx4; 120 μg/kg in sterile PBS; i.p.). After euthanization, organs were promptly harvested, measured, and processed. Experiments were approved by the University of Arkansas for Medical Sciences Institutional Animal Care and Use Committee (IACUC Protocol #3610 and #3836).
Bacterial diversity analysis
Stool samples less than 6 hr old were collected from individual mice and stored at −80 °C (13). DNA extraction was performed using ZymoBiomics DNA Miniprep kit (#D4300; Zymo Research, Irvine, CA) according to the manufacturer’s instruction. Briefly, samples were suspended in lysis buffer and heated to 60 °C for 20 min prior to 20 min of horizontal vortexing with beads to homogenize the samples. Samples were centrifuged and the supernatant collected. From this several processing steps were performed to remove residual protein and the final DNA sample was eluted in 100 μL of nuclease-free H2O. The concentration and purity was determined by the A260/A280 value (Cytation 5; BioTek, Winooski VT, USA).
16S rRNA Gene Sequencing
The extracted sample DNA was sent to the ZymoBIOMICS® Targeted Sequencing Service for Microbiome Analysis (Zymo Research, Irvine, CA) and sequenced using the Quick-16S™ Primer set V3–V4 (Zymo Research, Irvine, CA) via the Illumina MiSeq v3 reagent kit using a 10% PhiX spike-in. Summary of the sequencing service: PCR products were quantified with qPCR fluorescence readings and pooled together based on equal molarity. The pooled library was cleaned using the Select-a-Size DNA Clean & Concentrator™ (Zymo Research, Irvine, CA), then quantified with TapeStation® Santa Clara, CA) (Agilent Technologies, (Thermo Fisher Scientific, Waltham, WA).
Data Processing and Analysis
Demultiplexed FASTQ files were received from ZymoBIOMICS® (GenBank BioProject Accession PRJNA561567) and processed with QIIME2 version 2019.1 and 2019.4 (14). Primer sequence is contained within the first 16 bp of the forward read and the first 24 bp of the reverse read. Due to difficulties with retaining high quality merged reads, we opted to only use the forward reads herein. Forward reads were denoised and converted to Amplicon Sequence Variants (ASVs) via DADA2 (15) through the q2-dada2 QIIME 2 plugin (all plugins are noted by q2-*). DADA2 was initiated by trimming the first 16 bp (to remove the proprietary ZymoBIOMICS® primer sequence), using the ‘pooled’ option for chimera detection and removal, and truncating the reads at 263 bp.
Taxonomy assignment was achieved by mapping against the QIIME formatted SILVA (v132) reference database (available at: https://www.arb-silva.de). To increase the robustness of taxonomy assignment the corresponding V3–V4 amplicon region was extracted from the clustered (99% similarity) SILVA reference alignment. This region corresponds to alignment positions 5,045 through 17,652 of the 99_alignment.fna file, provided through the QIIME formatted SILVA reference database. Alignment gaps were subsequently removed and the resulting sequences used as input to the q2-feature-classifier plugin, for both classifier training through fit-classifier-naive-bayes and taxonomy assignment via classify-sklearn to return 7-level consensus taxonomy. Any amplicon sequence variants (ASVs) that were classified as “Eukaryota”, “Chloroplast”, “Mitochondria”, and “Unclassified” were excluded. Only ASVs with at least a Phylum-level taxonomy assignment were retained.
The remaining ASVs were evaluated via q2-quality-control to assess possible ASV quality exclusion criteria based on visual inspection of the BLAST output of hits against the SILVA reference sequences via the evaluate-seqs method. After which, the plugin method exclude-seqs was used to remove ASVs with less than 95% sequence identity and 97% query coverage to any reference sequences contained within the SILVA reference set. Data was exported to R for analysis and plotting via phyloseq and ggplot and differential abundance analyses was performed via linear discriminant analysis effect size (LEfSe; 18).
Flow cytometry
Tumors were mechanically dissociated with shears until pieces were ~1 mm3. This was followed by enzymatic dissociation (1 mg/mL Collagenase Invitrogen #17101–015; 2.5 U/mL Dispase Invitrogen 17105–041 and 20 μg/mL DNaseI Sigma #D-4527) for 30 mins with continuous agitation by a MACSmix tube rotator in a 37 °C incubator. Subsequently the tissue suspensions were put on ice and 5 mL cold FACS buffer (sterile 2% FBS + 2mM EDTA in PBS) was added. Single cell suspensions were made by filtering through a 70-μm cell strainer (BD Falcon #352350) to remove undigested cell clumps followed by a second filtration step via a 40-μm cell strainer (BD Falcon #352340). After lysing the red blood cells (ACS lysis buffer Gibco; #A1049201), the cell suspension was washed and collected by centrifugation at 300 g for 5 min at 4 °C (11) followed by surface staining as described (19). For intracellular staining, cell suspensions were stimulated with cell activation cocktail (BD Biosciences; #423304) and 250 ng/mL anti-CD3e for 5 hr in a 37 °C tissue culture incubator. Subsequently the suspensions were fixed and permeabilized using the Perm/Wash™ kit, according to manufacturer’s instructions (BD Biosciences).
Anti-mouse antibodies targeting CD4 (#0041), CD8a (#0081), CD27 (#563365) CD31 (#0311), CD54 (#0541), CD44 (#0441), CD45 (#0451), CD69 (#0691), CD62E (#553751), CD62P (#0626), CD106 (#1061), CD146 (#1469), CD197 (#562675), TNF-α (#7321), INF-γ (#5945) and isotype control antibodies were purchased from Thermo Fisher Scientific (eBioscience), Biolegend, Invitrogen and BD Biosciences and used for fluorescence activated cell-sorting (FACS) analysis.
Samples were acquired by multiparameter flow cytometry on a LSR II flow cytometer (BD Biosciences) and analyzed using Flowjo software (Tree Star, Inc.). Singlets were gated on by doublet exclusion and dead cells were excluded from the analysis using Fixable Viability Dye (FVD; #0865 eBioscience). Fluorescence minus one (FMO) controls were utilized to determine gating strategy. Absolute numbers of cells are expressed per million cells in order to compare tumors of different sizes.
Histology
The small intestines and colons were stained with hematoxylin and eosin after being formalin-fixed, paraffin-embedded and cut in tissue 5 μm sections. Images of the sections and staining were acquired on Olympus IX71 microscope at 200× magnification and digitally analyzed and differentially quantified by morphometric analysis, as described earlier.(20) The size of the lumen was estimated by the formula to calculate the area of an ellipse: a × b × π.
ELISA
To obtain chemokine and growth factor levels ELISA kits for TNF-α (#MTA00B), INF-γ (#MIF00) and VEGF (#MMV00) were purchased from Biotechne and used according to the manufacturer’s instructions.
Statistical Analysis
Data are reported as mean +/− SEM unless otherwise stated and were analyzed by an unpaired two-tailed t-test. P values <0.05 were considered statistically significant.
Results
Tumorigenesis and antibiotics change the microbiome.
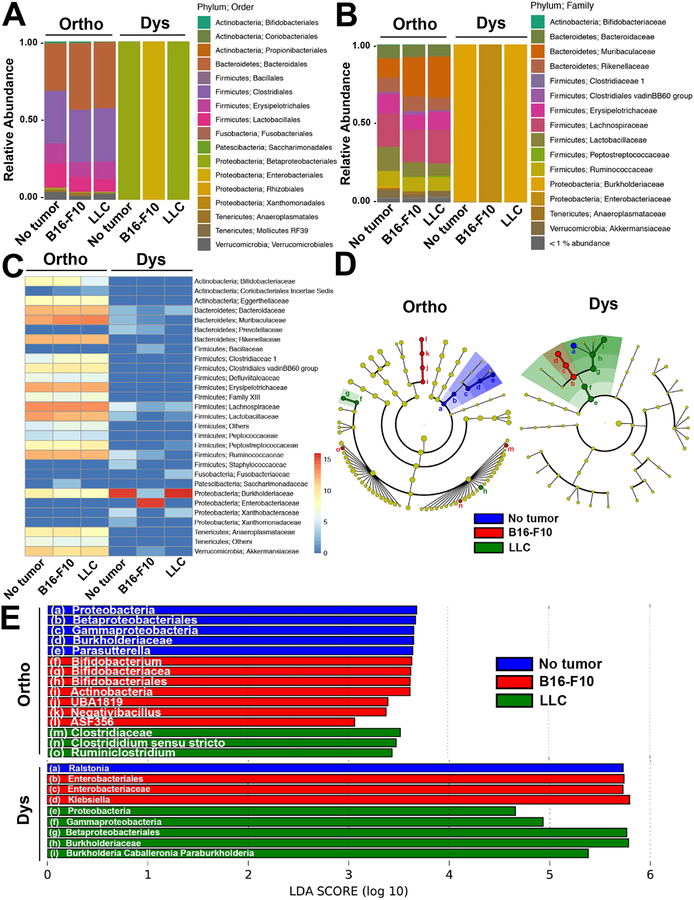
The microbial community composition among the sub-groups not treated with antibiotics displayed a prevalence of Bacteroidetes and Firmicutes over other groups of bacteria, whereas Proteobacteria were noticeably more abundant within the dysbiotic sub-groups (Figure 1). The antibiotics caused a substantial reduction in bacterial quantity and diversity and whereas the order Enterobacteriales was the most dominant in the dysbiotic mice bearing B16-F10 tumors, Betaproteobacteriales prevailed in the tumor free or LLC-bearing mice (Figure 1A). On the family level this manifested in Enterobacteriaceae in the dysbiotic mice bearing B16-F10 tumors, and Burkholderiaceae in the tumor free or LLC-bearing mice (Figure 1B and C). Linear discriminant analysis (LDA) revealed that there were several differentially enriched groups, as visualized though taxonomic hierarchy, of microbiota between the tumor types and treatment type (Figure 1D and E).
Figure 1. Microbial community composition in tumor free or mice bearing B16-F10 melanoma or LLC with or without dysbiosis.
Microbial composition in healthy controls (Ortho) and antibiotic exposed (Dys) mice with and without B16-F10 melanoma or LLC at the (A) Order and (B) Family levels. (C) Log2 abundance heatmap of microbial families. The relative abundance for the microbial families is indicated by hue. (D) Taxonomic cladograms obtained from LEfSe analysis of 16S rRNA sequences (blue = no tumor; red = B16-F10; green = LLC). Yellow circles represent non-significant differences in abundance between the groups. (E) LEfSe analysis revealing differentially enriched microbial groups across tumor types (blue = no tumor; red = B16-F10; green = LLC) within each of the Ortho and Dys groups. For A-C, independent samples (n = 4 each) were merged (i.e. ASV read counts summed) into their respective groups prior to visualization. The letters a-i in panel D refer to the microbial groups listed in panel E.
Both weighted and unweighted UniFrac distance matrices were produced by rarefying the data to 10,500 reads per sample. PERMANOVA of weighted UniFrac, revealed no differences in beta-diversity between the sub-groups not treated with antibiotics. However, there were significant differences between mice with LLC to both B16-F10-bearing mice (q ≤ 0.042) and tumor-free mice (q ≤ 0.042) when investigated via unweighted UniFrac. During dysbiotic conditions there were significant differences observed by weighted Unifrac between B16-F10 and LLC-bearing mice (q ≤ 0.023) and tumor-free mice (q ≤ 0.045), while no significant differences were observed by unweighted UniFrac. Through Kruskal-Wallis analysis of Faith’s phylodiversity (PD) we observed significant differences in alpha diversity between the orthobiotic controls and dysbiotic groups (p ≤ 0.0001). However, no significant differences of PD were observed within each of the orthobiotic or dysbiotic subgroups.
Dysbiosis alters the host’s GI tract and enhances tumor progression at distal sites.
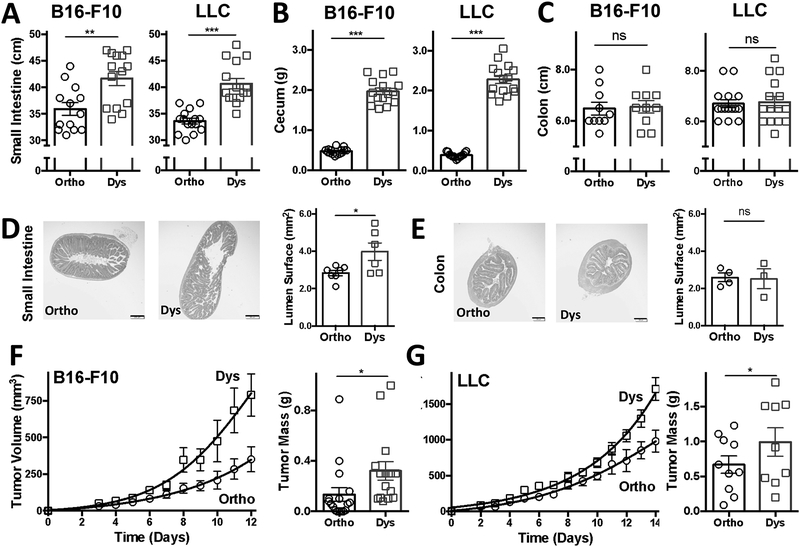
Antibiotic-induced dysbiosis caused the small intestine and cecum to enlarge and elongate, yet the colon was not affected (Figure 2). The length of the small intestines increased on average by 15% in the B16-F10 model from 35.9 ± 0.95 cm to 41.7 ± 1.11 cm (P < 0.01) during dysbiosis, and on average by 20% in the LLC model from 33.6 ± 0.54 cm to 40.7 ± 0.99 cm (P < 0.001) during dysbiosis (Figure 2A).
Figure 2. Dysbiosis alters the gastrointestinal tract and enhances melanoma and lung carcinoma progression.
The (A) small intestines, (B) ceca and (C) colons of mice bearing B16-F10 melanoma or Lewis lung carcinoma (LLC) with or without antibiotic-induced dysbiosis. Representative hematoxylin and eosin images of (D) small intestines and (E) colons in B16-F10 melanoma-bearing mice. Growth curves and masses of (F) B16-F10 melanoma and (G) LLC. Tumor growth curves are shown in mean volumes ± SEM with non-linear regression-fit lines and tumor weights are shown as mean mass ± SEM (n = 10–18 animals per group pooled from 2 or 3 individual experiments. Dysbiosis was induced by exposing mice to antibiotics (ampicillin, neomycin, metronidazole each at [250 mg/L]; vancomycin at [125 mg/L]). Scale bar = 200 μm. *P < 0.05, **P < 0.01, ***P < 0.05, two-sided t-test. ns = not significant. -◯- orthobiotic (Ortho) and -☐- dysbiotic (Dys) mice
In terms of relative enlargement, the ceca increased the most, on average by ~400% during dysbiosis. Namely, the ceca increased in weight in the B16-F10 model from 0.47 ± 0.02 g to 1.96 ± 0.07 g (P < 0.001) during dysbiosis, and on average by 615% in the LLC model from 0.38 ± 0.02 g to 2.34 ± 0.11 g (P < 0.001) during dysbiosis (Figure 2B). Images of representative ceca are shown in Supplemental Figure 1.
In contrast, the colons’ lengths were not significantly affected by dysbiosis. Namely, the colons were comparable in length in the B16-F10 model in the controls from 6.48 ± 0.18 cm vs. 6.55 ± 0.17 cm during dysbiosis, and in the LLC model from 6.70 ± 0.16 cm vs. 6.77 ± 0.23 cm during dysbiosis (Figure 2C).
The change in size of the small intestine was also reflected on a microscopic scale as assessed by histology. The lumen of the small intestines increased from 2.84 ± 0.14 mm2 in the controls to 3.98 ± 0.47 mm2 (P < 0.05) during dysbiosis (Figure 2D). The lumen of the colons, in contrast, changed only negligibly from 2.58 ± 0.23 mm2 in the controls to 2.51 ± 0.53 mm2 during dysbiosis (Figure 2E).
Tumor growth curves and tumor mass determinations indicated that both melanoma and lung carcinoma progressed roughly twice as fast, on average, in mice with antibiotic-induced dysbiosis as compared to control mice (Figure 2F and G). Namely, the tumor masses were increased by almost 250% in the B16-F10 model from 0.13 ± 0.05 g to 0.32 ± 0.07 g (P < 0.05) during dysbiosis, and by approximately 165% on average in the LLC model from 0.67 ± 0.13 g to 1.09 ± 0.2 g during dysbiosis (P < 0.05) on the day of sacrifice.
Dysbiosis did not affect body weights as mice bearing either B16-F10 or LLC tumors maintained their weights (Supplemental Figure 2A and B).
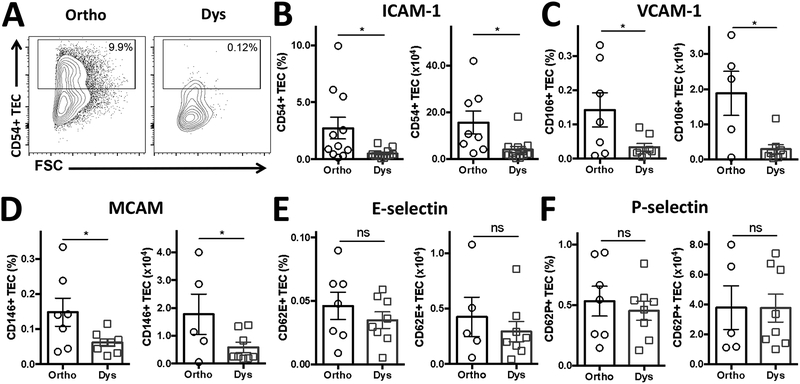
Dysbiosis suppresses tumor vascular adhesion molecules.
As tumors progressed more rapidly under dysbiotic conditions, we investigated the changes in vascular adhesion molecules, which play a crucial role in immune surveillance and leukocyte extravasation. Indeed, dysbiosis caused ICAM-1 (CD54), VCAM-1 (CD106), and MCAM (CD146) suppression on tumor endothelial cells (TEC; CD45- CD31+) in B16-F10 tumors (Figure 3). Namely, ICAM-1 expression reduced by 82% on average from 2.73 ± 0.95% to 0.49 ± 0.12%, VCAM by 79% from 0.14 ± 0.04% to 0.03 ± 0.01%, and MCAM by 60% from 0.15 ± 0.03% to 0.06 ± 0.01% during dysbiosis on TEC (all P-values < 0.05; Figure 3A–D). In addition, selectins also showed change during dysbiosis, namely E-selectin (CD62E) levels changed by 40% from 0.05 ± 0.01% to 0.03 ± 0.01% and P-selectin (CD62P) by 15% from 0.53 ± 0.10% to 0.45 ± 0.06% during dysbiosis on TEC (Figure 3E and F). However, these relative low expression levels and mild changes in selectins failed to attain statistical significance. Besides percentages, these same trends were seen for the number of TEC expressing these adhesion molecules in B16-F10 tumors (Figure 3B–F), and for the number and percentage of ICAM-1 positive TEC in LLC tumors (Supplemental Figure 3A–C).
Figure 3. Tumor vascular adhesion molecules are suppressed under dysbiotic conditions.
(A) Representative analysis plots for ICAM-1 (CD54) on tumor endothelial cells (TEC; CD45- CD31+). Quantification in percentage and absolute number of (B) ICAM-1 (CD54), (C) VCAM-1 (CD106), (D) MCAM (CD146), (E) E-selectin (CD62E) and (F) P-selectin (CD62P) expressing TEC. Data are the mean ± SEM (n = 5–11 B16-F10 tumors per group, pooled from 2–3 individual FACS experiments). *P < 0.05, two-sided t-test. ns = not significant. -◯- orthobiotic (Ortho) and -☐- dysbiotic (Dys) mice
This dysbiosis-induced suppression of ICAM-1 was not seen on established vasculature of tissues, such as the spleen and the thymus (Supplemental Figure 4A–I and Figure 5A–I). Additionally, integrin alpha L (CD11a), the alpha subunit of lymphocyte function-associated antigen-1 (LFA-1) and binding site of ICAM-1, was not suppressed on T cells in the spleen or thymus during dysbiosis (Supplemental Figures 4D–F and 5D–F, Supplemental Table 1). In concordance, dysbiosis did not affect the gross morphology and mass of the spleens in B16-F10-bearing mice (0.09 ± 0.01 g vs. 0.08 ± 0.01 g during dysbiosis), or LLC-bearing mice (0.12 ± 0.02 g vs. 0.10 ± 0.01 g during dysbiosis) as measured on the day of sacrifice (Supplemental Figure 6A and B).
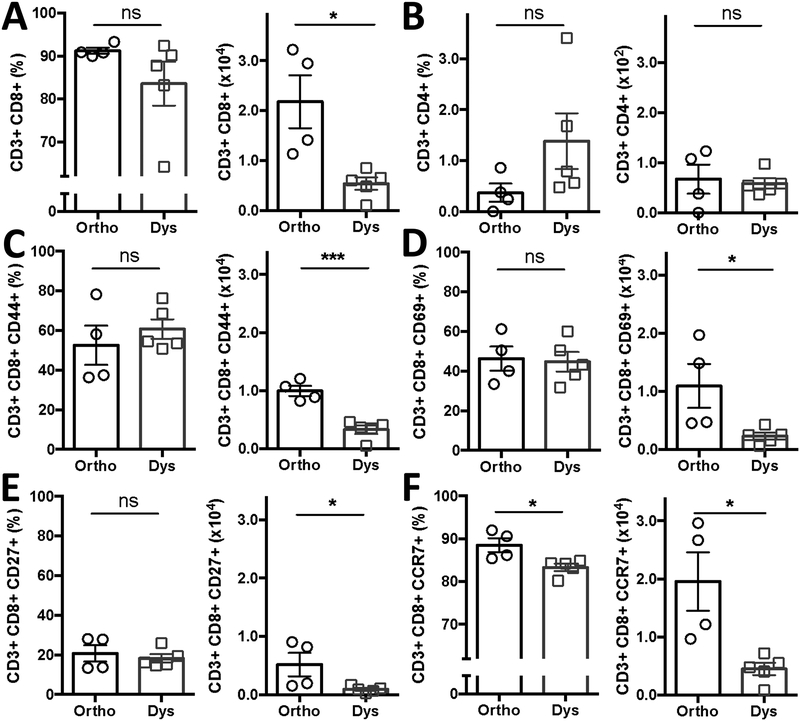
Dysbiosis decreases the number of activated and effector T-cells in tumors.
As tumor endothelial adhesion molecules are crucial for cytotoxic leukocyte trafficking and extravasation into the tumor, we subsequently investigated the tumor infiltrate (Figure 4). Dysbiotic conditions significantly decreased the average number of cytotoxic CD3+ CD8+ T cells that infiltrate tumors from 2.18 ± 0.53 [x104 cells] to 0.54 ± 0.12 [x104 cells] during dysbiosis (P < 0.05). The percentage of CD8+ T cells changed by fewer than 8 percentage points from 91.3% ± 0.7 to 83.6 ± 5.1% during dysbiosis (P < 0.23). Similarly, CD3+ CD4+ T cells did not change in percentage (0.37% ± 0.2 vs 1.4 ± 0.5% during dysbiosis), nor in total amount, which was 2-orders of magnitude lower to begin with (0.7 ± 0.3 [x102 cells] vs 0.6 ± 0.01 [x102 cells] during dysbiosis) as compared to the cytotoxic CD3+ CD8+ T-cells (Figure 4A and B).
Figure 4. Dysbiosis decreases the abundance of activated and effector T-cells in tumors.
B16-F10 infiltrated (A) CD3+ CD8+ T cells and (B) CD3+ CD4+ T cells with or without dysbiotic conditions. Percentage and amount of CD3+ CD8+ T cells expressing (C) CD44, (D) CD69, (E) CD27, (F) CCR7. Data presented as means ± SEM and is representative of 2 independent experiments n = 4–5 per group. *P < 0.05, ***P < 0.001, two-sided t-test. ns = not significant. -◯- orthobiotic (Ortho) and -☐- dysbiotic (Dys) mice
Next we assessed whether the T cells were active within the tumor microenvironment by evaluating activation markers hyaluronate receptor CD44, early activation antigen CD69, tumor necrosis factor receptor CD27 and effector function marker chemokine receptor type 7 (CCR7; CD197), (Figure 4C–F). Dysbiosis reduced the total number of, CD3+ CD8+ CD44+ T cells by 67% (from 1.0 ± 0.09 [x104 cells] to 0.33 ± 0.07 [x104 cells] during dysbiosis; P < 0.001), CD3+ CD8+ CD69+ T cells by 79% (from 1.10 ± 0.38 [x104 cells] to 0.23 ± 0.06 [x104 cells] during dysbiosis; P = 0.04), CD3+ CD8+ CD27+ T cells by by 83% (from 0.52 ± 0.20 [x104 cells] to 0.09 ± 0.02 [x104 cells] during dysbiosis; P < 0.05), and CD3+ CD8+ CCCR7+ T cells by 77% (from 1.95 ± 0.50 ([x104 cells] to 0.45 ± 0.11 [x104 cells] during dysbiosis; P = 0.01). Dysbiosis only significantly reduced the proportion of CD3+ CD8+ CCR7+ T cells by 6% (from 88.6% ± 1.6 to 83.3 ± 0.8% during dysbiosis; Figure 4D).
The overall decreased number of activated and effector CD8+ T cells in the tumor was not a result of a reduced number of T cells in the spleen or the thymus (Supplemental Figures 4A–C and 5A–C, Supplemental Table 1). Dysbiosis did not change overall cell viability in either tumor model (Supplemental Figure 7A and B).
TNF-α is suppressed under dysbiotic conditions.
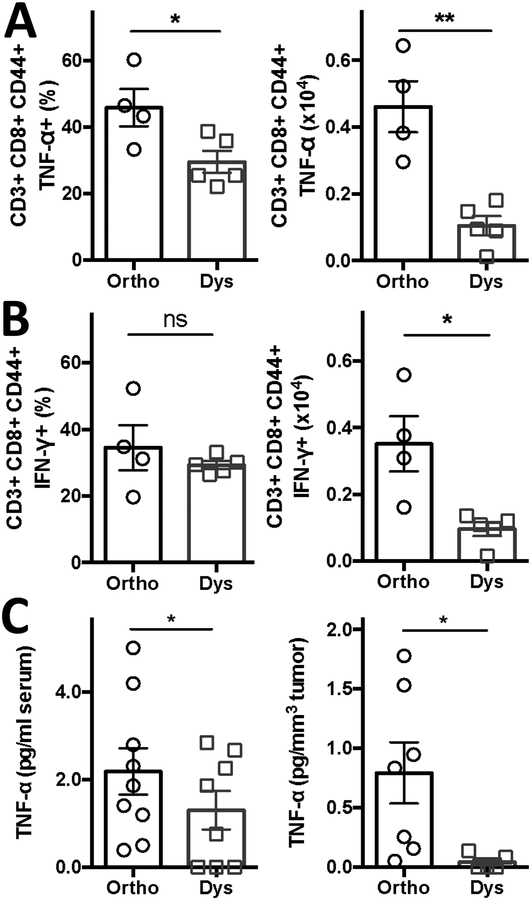
Next we measured effector molecules involved in ICAM-1 regulation, TNF-α and IFN-γ (Figure 5). Dysbiosis significantly reduced the total amount of CD3+ CD8+ CD44+ T cells expressing TNF-α by 78% (from 0.46 ± 0.08 [x104 cells] to 0.10 ± 0.03 [x104 cells] during dysbiosis; P < 0.002), and reduced total amount of CD3+ CD8+ CD44+ T cells expressing IFN-γ by 71% (from 0.35 ± 0.08 [x104 cells] to 0.10 ± 0.02 [x104 cells] during dysbiosis; P < 0.01). Dysbiosis only significantly reduced the proportion of TNF-α expressing T cells (from 45.9% ± 5.6 to 29.6 ± 3.3% during dysbiosis; P < 0.03) (Figure 5A and B).
Figure 5, TNF-α is suppressed under dysbiotic conditions.
Percentage and amount of (A) TNF-α producing or (B) IFN-y producing tumor-infiltrating CD3+ CD8+ CD44+ T cells and systemic (C) TNF-α serum levels under orthobiotic and dysbiotic conditions. Data presented as means ± SEM (n = 4–9 C57BL/6 mice bearing B16-F10 tumors per group pooled from 1–3 individual experiments). *P < 0.05, **P < 0.01, two-sided t-test. ns = not significant. -◯- orthobiotic (Ortho) and -☐- dysbiotic (Dys) mice
Additionally, serum levels of TNF-α also decreased from 2.2 ± 0.5 pg/ml to 1.3 ± 0.4 pg/ml, resulting in decreased TNF-α levels per tumor tissue from 0.8 ± 0.2 pg/mm3 to 0.04 ± 0.02 pg/mm3 during dysbiosis (Figure 5C). However, serum concentrations of IFN-γ and VEGF did not change significantly during dysbiosis (IFN-γ, from 24.5 ± 10.1 pg/ml to 14.7 ± 7.6 pg/ml and VEGF, from 156.5 ± 18.6 pg/ml to 155.8 ± 22.6 pg/ml; Supplemental Figure 8A and B).
Dysbiosis-induced tumor progression is ICAM-1 mediated.
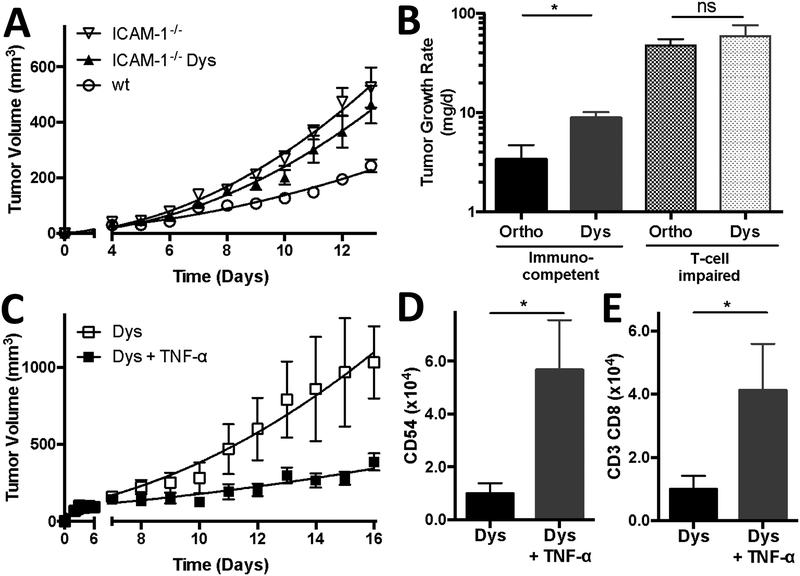
To confirm the importance of ICAM-1 in tumors during dysbiosis, we next grew melanoma in wild type and ICAM-1−/− mice (Figure 6). Tumors progressed up to twice as fast, on average, in ICAM-1−/− mice, as compared to wild type mice. However, dysbiosis did not enhance this tumor progression, indicating ICAM-1 involvement in dysbiotic-induced tumor progression (Figure 6A).
Figure 6, ICAM-1 and T cells are needed for dysbiotic-induced tumor progression and TNF-α supplementation rescues T cell trafficking under dysbiotic conditions.
(A) Tumor growth curves of B16-F10 melanoma in ICAM-1−/− mice with and without dysbiotic conditions. (B) Tumor growth rates of B16-F10 melanoma in immunocompetent (C57BL/6J) and T-cell impaired Foxn1−/− mice with and without dysbiotic conditions. (C) Tumor growth curves of B16-F10 melanoma during dysbiotic conditions with or without TNF-α supplementation (q3dx4; 120 μg/kg). (D) Quantification of ICAM-1 (CD54) expressing (CD45- CD31+) TEC and (E) tumor infiltrated CD3+ CD8+ T cells in B16-F10 tumors during dysbiotic conditions with or without TNF-α. Data presented as means ± SEM (n = 4 or 5 B16-F10 tumors per group). *P < 0.05, two-sided t-test. ns = not significant. Panel A: -▽- ICAM-1−/−, -▲- dysbiotic (Dys) ICAM-1−/−, and -◯- wild type (wt) mice; Panel C: -☐- dysbiotic (Dys) mice and -◼- dysbiotic mice supplemented with TNF-α (Dys + TNF-α)
TNF-α supplementation overcomes dysbiosis-induced tumor progression and enhances T-cell trafficking into the tumor.
To further validate the importance of T-cell trafficking and extravasation into tumors during dysbiosis, we injected B16-F10 cells simultaneously into immunocompetent (C57BL/6J) and T-cell deficient (Foxn1−/−) mice. We found that the growth rate of tumors was an order of magnitude lower in immunocompetent mice than in the T-cell deficient mice, and that dysbiosis only disrupted tumor growth in immunocompetent mice, indicating that dysbiosis-induced tumor progression is dependent on T-cell trafficking (Figure 6B).
Finally, based on the reduced levels of TNF-α under dysbiotic conditions, we hypothesized that the suppressed ICAM-1 expression and subsequent tumor-infiltrating T-cells, could be restored by TNF-α administration. Indeed, TNF-α supplementation (120 μg/kg in sterile PBS; q3dx4 initiated on day 7 after B16-F10 inoculation) resulted in an almost 6-fold increase on average in ICAM-1 expression on tumor endothelial cells (P < 0.02), and a 4-fold increase on average in CD8+ T-cell infiltrate in the tumor (P < 0.03), and a decrease of more than 60% in tumor growth (Figure 6C–E).
Discussion
The immune system can recognize and eliminate developing tumors, but many cancers ultimately escape immune surveillance. Studies have shown that a disruption of the microbiota can blunt anti-tumor immune response and the efficacy of cancer therapies by modulating circulating inflammatory chemo- and cytokines and thereby the host inflammatory response (21, 22). The microbiota of the GI tract is commonly perceived as diverse, robust and long-term stable to environmental disturbances (23). However, the individual bacterial communities within are dynamic in composition and susceptible to various external factors like antibiotics (24). Although the altered GI tract microbiome can be recovered and recolonized to a certain degree, loss of specific strains and overall diversity follows antibiotic selection pressure (25). Consequently, even after the completion of treatment the use of certain antibiotics may have direct and long-lasting deleterious effects on the host by altering the composition and functions of the microbiota (26). With the sharp rise of multi-regimen, high-dose antibiotics being prescribed worldwide, this is a particularly sobering possibility and our results and those of others suggest that much additional investigation is needed.
Here, we induced dysbiosis in mice with clinically relevant doses of antibiotics (27). Ampicillin and neomycin are both broad-spectrum antibiotics, while vancomycin is indicated for the treatment of Gram-positive bacteria. When given orally these antibiotics are very poorly absorbed, or not at all for neomycin, without any systemic effects (28, 29). Metronidazole is absorbed in the GI-tract but has no systemic effect as it only functions when it is reduced by anaerobic bacteria (30). In previous studies dysbiosis was induced with high doses of antibiotics (4 times as high) and/or acutely (1–3 days), resulting in changes in splenocytes and significant weight loss (> 30%), which are possible confounding variables (2, 31). The doses applied herein did not induce these alterations. While both approaches address clinically relevant aspects and considerations, one examines the acute effects of high-dose antibiotics, and the other assesses the continuing consequences of long-term or multiple courses of antibiotics on the microbiome and the host.
Cancer treatment is affected by changes of the GI tract microbiome as well, as microbiota can hinder the efficacy of conventional cancer therapy by modulating the host inflammatory response. For instance, Zitvogel et al. have shown that colon cancer only minimally responds to the immune-modulating anti-cancer drug cyclophosphamide during dysbiosis (21, 22). It was suggested that this effect is mediated by a decrease in leukocyte-derived circulating inflammatory cytokines. More recent clinical anti-cancer strategies are affected by the microbiome as well. Namely, a number of recent studies in mice and humans have shown the importance of GI tract microbiota and immunotherapy efficacy by immune checkpoint inhibitors (32). E.g. cytotoxic T-lymphocyte-associated protein 4 (CTLA-4; CD152) and programmed cell death protein-1 (PD-1) blockage only reduced tumor growth in mice harboring Bacteroides and Bifidobacterium species respectively (33, 34). Chaput et al. showed that patients with metastatic melanoma, treated with CTLA-4 inhibition, had longer progression free and overall survival when they were Faecalibacterium rich at the start of the treatment (35). Similarly, Routy et al. found that microbiota disruption with antibiotics in cancer patients immediately prior to checkpoint inhibitor treatment led to shorter progression free and overall survival (36). Wargo’s and Gajewski’s group reported that a more robust microbiota is associated with enhanced anti-PD-1 efficacy in melanoma patients (37, 38). Interestingly however, each group identified different ‘favorable’ bacteria, suggesting that either certain mechanisms are shared among bacterial species or that the overall bacterial abundance, composition and diversity is of importance rather than a specific bacterial strain. Along those lines, Honda et al. recently defined a commensal consortium of 11 strains (7 Bacteroidales and 4 non-Bacteroidales species) derived from human volunteers being able to induce effector IFN-γ+ CD8+ T cells in germfree mice. Moreover, these 11 strains were also able to enhance immune checkpoint inhibitors’ efficacy in murine tumor models, whereas a different 10-strain mix not associated with IFN-y+ CD8+ T cells failed to do so (39).
We noted that dysbiosis caused by this specific regimen of antibiotics resulted in a decline in bacterial amount and diversity, particularly inducing a change from Bacteroidetes and Firmicutes phyla dominance towards strains within the Proteobacteria phylum. This is in agreement with the above-mentioned human and murine studies. Moreover we found that dysbiosis also caused enlargement and elongation of the small intestine and cecum on a macro- and microscopic scale. The loss of anaerobic fusiform-shaped bacteria, which are abundant in the cecum and imbedded in the mucus layer of its epithelium, are likely at the center of this process as functionally they maintain the integrity of the water-transport mechanism (40). Losing these bacteria due to antibiotic treatment will disrupt this homeostasis and cause water and fluid retention in the cecum causing enlargement and elongation, which is also seen in germfree mice. Restoring the cecum to its normal anatomical and physiological state can be attained by reintroducing intestinal flora from unperturbed mice, but only to a certain degree as abnormalities of the cecum persist (40).
Several types of immunosuppression and tolerance, obstructing adequate T-cell effector function, have been described within the microenvironment during orthobiotic conditions. This includes co-inhibitory effects of cytokines on T-cell activation, proliferation and survival,(41, 42) interference of activated T-cell migration (43), or direct suppression of the effector function by iNOS (inducible nitric oxide synthase), TGF-β, and Tregs (44). Whether or not dysbiotic conditions exacerbate any or all of these immune suppression and escape mechanisms has yet to be elucidated. Our data supports the notion that dysbiosis negatively impacts T-cell trafficking, activation and effector function. Moreover, a low frequency of CD8+ CCR7+ effector T cells is a significant risk factor for disease recurrence in the clinic (45).
Here, we found that dysbiosis reduces the levels of TNF-α and IFN-γ in the tumor microenvironment, and TNF-α was suppressed systemically in the serum as well. TNF-α is a pleiotropic regulator of ICAM-1 (46), an essential adhesion molecule for leukocyte trafficking and extravasation into tumors (9). Whereas selectins are predominantly involved in the initial tethering, rolling and arrest of leukocytes on endothelial cells, it is particularly ICAM-1 that facilitates transcellular diapedesis into the tissue (47, 48). While dysbiosis enhanced tumor growth in ICAM-1+/+ and immunocompetent mice, it did not affect tumor growth in ICAM-1−/− or T-cell deficient mice, highlighting the influence and impact of dysbiosis on T-cell trafficking and extravasation. As expected from these observations, supplementing TNF-α during dysbiosis caused elevated ICAM-1 expression on the tumor vasculature, which increased the amount of effector T-cells in the tumor and inhibited tumor growth inhibition.
We focused on melanoma and lung carcinoma as exemplary immunogenic solid tumor models. Since dysbiosis enhanced progression in both models, our findings may have a broader impact for a variety of non-GI tumors, as it pertains to tumor vasculature modulation and thus could be pertinent for solid tumors in general. An important next step will be defining the commensal bacterial species, or mixture, and the metabolome responsible for maintaining a more competent immunogenic tumor microenvironment (39, 49, 50).
Overall, our work expands on the increasing appreciation of the importance of commensal bacteria in the GI tract in maintaining host immune homeostasis via conditioning of the tumor stroma. It indicates an additional deleterious effect of certain classes of antibiotics on the host’s ability to elicit an effective anti-tumor immune response.
Supplementary Material
Significance:
Antibiotic-induced dysbiosis enhances distal tumor progression by altering host cytokine levels resulting in suppression of tumor endothelial adhesion molecules and activated and effector CD8+ T-cells in the tumor.
Acknowledgements and funding
The study was supported by P20GM103625: the Center for Microbial Pathogenesis and Host Inflammatory Responses grant through the NIH National Institute of General Medical Sciences Centers of Biomedical Research Excellence to R.P.M. Dings. This work was also supported in part by a grant from the Arkansas Biosciences Institute and the Winthrop P. Rockefeller Cancer Institute to R.P.M. Dings. K.B. Vang received grant NSF OIA-1457888 through the Center for Advanced Surface Engineering.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NSF or UAMS/WPRCI.
Abbreviations:
- ACT
alignment, classification and tree
- ASV
amplicon sequence variant
- BIC
Bayesian information criterion
- CI
confidence interval
- EAM
endothelial adhesion molecules
- GI tract
gastrointestinal tract
- ICAM-1
intercellular adhesion molecule 1
- LDA
linear discriminant analysis
- LEfSe
Linear discriminant analysis effect size
- LFA-1
lymphocyte function-associated antigen-1
- PD
phylodiversity
- QIIME
Quantitative insights into microbial ecology
- TEC
tumor endothelial cells
- TME
tumor microenvironment
- TNF-α
tumor necrosis factor-alpha
- VCAM-1
vascular cell adhesion molecule
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Erturk-Hasdemir D, Kasper DL. Resident commensals shaping immunity. Curr Opin Immunol. 2013;25:450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, et al. Microbiota Modulate Tumoral Immune Surveillance in Lung through a gammadeltaT17 Immune Cell-Dependent Mechanism. Cancer Res. 2014;74:4030–41. [DOI] [PubMed] [Google Scholar]
- 3.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nature reviews Microbiology. 2011;9:233–43. [DOI] [PubMed] [Google Scholar]
- 4.CDC. Antibiotic use in the United States, 2017: Progress and Opportunities. Atlanta, GA: US department of Health and human Services; 2017. [Google Scholar]
- 5.Kilkkinen A, Rissanen H, Klaukka T, Pukkala E, Heliovaara M, Huovinen P, et al. Antibiotic use predicts an increased risk of cancer. Int J Cancer. 2008;123:2152–5. [DOI] [PubMed] [Google Scholar]
- 6.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. 2014;157:776–83. [DOI] [PubMed] [Google Scholar]
- 7.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–14. [DOI] [PubMed] [Google Scholar]
- 10.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dings RP, Vang KB, Castermans K, Popescu F, Zhang Y, Oude Egbrink MG, et al. Enhancement of T-cell-mediated antitumor response: angiostatic adjuvant to immunotherapy against cancer. Clin Cancer Res. 2011;17:3134–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins SV, Vang KB, Gies A, Griffin RJ, Jun SR, Nookaew I, et al. Sample storage conditions induce post-collection biases in microbiome profiles. BMC Microbiol. 2018;18:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature methods. 2017;14:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 19.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dings RP, Chen X, Hellebrekers DM, van Eijk LI, Zhang Y, Hoye TR, et al. Design of nonpeptidic topomimetics of antiangiogenic proteins with antitumor activities. J Natl Cancer Inst. 2006;98:932–6. [DOI] [PubMed] [Google Scholar]
- 21.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez-Baeza Y, Callewaert C, Debelius J, Hyde E, Marotz C, Morton JT, et al. Impacts of the Human Gut Microbiome on Therapeutics. Annu Rev Pharmacol Toxicol. 2018;58:253–70. [DOI] [PubMed] [Google Scholar]
- 25.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becattini S, Taur Y, Pamer EG. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends in molecular medicine. 2016;22:458–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uptodate.com. Drug information - Dosing. 2018. [cited; Available from:
- 28.Ferrier L, Berard F, Debrauwer L, Chabo C, Langella P, Bueno L, et al. Impairment of the intestinal barrier by ethanol involves enteric microflora and mast cell activation in rodents. Am J Pathol. 2006;168:1148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moellering RC, Jr. Pharmacokinetics of vancomycin. The Journal of antimicrobial chemotherapy. 1984;14 Suppl D:43–52. [DOI] [PubMed] [Google Scholar]
- 30.Lofmark S, Edlund C, Nord CE. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin Infect Dis. 2010;50 Suppl 1:S16–23. [DOI] [PubMed] [Google Scholar]
- 31.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroemer G, Zitvogel L. Cancer immunotherapy in 2017: The breakthrough of the microbiota. Nat Rev Immunol. 2018;18:87–8. [DOI] [PubMed] [Google Scholar]
- 33.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368–79. [DOI] [PubMed] [Google Scholar]
- 36.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- 37.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–5. [DOI] [PubMed] [Google Scholar]
- 40.Savage DC, Dubos R. Alterations in the mouse cecum and its flora produced by antibacterial drugs. J Exp Med. 1968;128:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. [DOI] [PubMed] [Google Scholar]
- 42.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lizee G, Cantu MA, Hwu P. Less yin, more yang: confronting the barriers to cancer immunotherapy. Clin Cancer Res. 2007;13:5250–5. [DOI] [PubMed] [Google Scholar]
- 44.Sakaguchi S Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol. 2005;6:345–52. [DOI] [PubMed] [Google Scholar]
- 45.Czystowska M, Gooding W, Szczepanski MJ, Lopez-Abaitero A, Ferris RL, Johnson JT, et al. The immune signature of CD8(+)CCR7(+) T cells in the peripheral circulation associates with disease recurrence in patients with HNSCC. Clin Cancer Res. 2013;19:889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnoor M, Alcaide P, Voisin MB, van Buul JD. Crossing the Vascular Wall: Common and Unique Mechanisms Exploited by Different Leukocyte Subsets during Extravasation. Mediators of inflammation. 2015;2015:946509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celli L, Ryckewaert JJ, Delachanal E, Duperray A. Evidence of a functional role for interaction between ICAM-1 and nonmuscle alpha-actinins in leukocyte diapedesis. J Immunol. 2006;177:4113–21. [DOI] [PubMed] [Google Scholar]
- 49.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, et al. Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Scientific reports. 2018;8:14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.