Abstract
A key hallmark of cancer, unlimited replication, requires cancer cells to evade both replicative senescence and potentially lethal chromosomal instability induced by telomere dysfunction. The majority of cancers overcome these critical barriers by up-regulating telomerase, a telomere-specific reverse transcriptase. However, a subset of cancers maintains telomere lengths by the telomerase-independent Alternative Lengthening of Telomeres (ALT) pathway. The presence of ALT is strongly associated with recurrent cancer-specific somatic inactivating mutations in the ATRX-DAXX chromatin remodeling complex. Here, we generate an ALT-positive adenocarcinoma cell line following functional inactivation of ATRX and telomerase in a telomerase-positive adenocarcinoma cell line. Inactivating mutations in ATRX were introduced using CRISPR-cas9 nickase into two prostate cancer cell lines, LAPC-4 (derived from a lymph node metastasis) and CWR22Rv1 (sourced from a xenograft established from a primary prostate cancer). In LAPC-4, but not CWR22Rv1, abolishing ATRX was sufficient to induce multiple ALT-associated hallmarks, including the presence of ALT-associated PML bodies (APBs), extrachromosomal telomere C-circles, and dramatic telomere length heterogeneity. However, telomerase activity was still present in these ATRXKO cells. Telomerase activity was subsequently crippled in these LAPC-4 ATRXKO cells by introducing mutations in the TERC locus, the essential RNA component of telomerase. These LAPC-4 ATRXKO TERCmut cells continued to proliferate long-term and retained ALT-associated hallmarks, thereby demonstrating their reliance on the ALT mechanism for telomere maintenance.
Keywords: ATRX, TERC, Telomeres, Alternative lengthening of telomeres, Cancer, Prostate cancer, telomerase, gene knockout, CRISPR/Cas
INTRODUCTION
Telomeres are nucleoprotein complexes that consist of a repetitive hexameric DNA sequence, 5’-TTAGGG/3’-AATCCC, that are bound by the shelterin protein complex. Located at the ends of eukaryotic chromosomes, telomeres maintain genomic stability and integrity by preventing exonucleolytic degradation and recognition of the chromosomal ends as double stranded breaks by DNA damage response (DDR) proteins (1). Following each round of cell division, telomeres progressively shorten as a direct result of incomplete DNA replication in the lagging strand (2). Once telomeres reach a critical length, normal cells undergo p53-dependent replicative senescence or apoptosis (3). While there are rare exceptions without a known telomere maintenance mechanism (4,5), telomere maintenance is required for most cancers to maintain unlimited replication while evading replicative senescence or apoptosis (6). The vast majority of cancers up-regulate telomerase, a telomere-specific reverse transcriptase (7); however, 10–15% of cancers lack telomerase activity (7,8).
A subset of telomerase-negative cancers maintains their telomere lengths by a telomere-specific mechanism of homology directed repair called Alternative Lengthening of Telomeres (ALT) (9–11). The mechanism underlying ALT generates unique molecular characteristics that readily distinguish ALT-positive and ALT-negative cancers. For example, ALT-positive cancers display dramatic telomere length heterogeneity, manifesting both as cell-to-cell variation and intracellular heterogeneity (9). ALT-positive cancers also have ALT-associated promyelocytic leukemia (PML) bodies, called APBs. These unique nuclear structures contain donut-shaped PML protein bodies that colocalize with telomeric DNA, the shelterin components TRF1 and TRF2, and proteins involved in DNA synthesis and recombination (12,13). Additionally, ALT-positive cancers are enriched for C-circles, extrachromosomal circular DNA comprised of single stranded C-rich telomere DNA sequence partially overlapped by linear complementary G-rich telomere DNA (14). Sequencing studies of molecularly characterized ALT-positive cancers from our group, and others, identified a robust association between ALT and mutations in α-thalassemia/mental retardation X-linked protein (ATRX) and death domain-associated protein (DAXX) in multiple tumor types, including pancreatic neuroendocrine tumors (PanNETs), sarcomas, and tumors of the central nervous system (15–17). As a complex, ATRX/DAXX recognizes H3K9me3 in chromatin and deposits histone variant H3.3 at repetitive sequences. This chromatin remodeling function of ATRX/DAXX is important for heterochromatin maintenance at repetitive sequences, including telomeres (18,19). More recently, inactivating mutations in the ATP-dependent annealing helicase, SMARCAL1, were also found to be tightly associated with ALT in a subset of ATRX and DAXX wildtype glioblastomas (20).
The majority of established ALT-positive cancer cell lines have inactivated ATRX, although there are some recent exceptions (20–23). Previous attempts to induce ALT through knockdown or knockout of ATRX have largely been unsuccessful (21,23–25). However, in a context dependent manner, genetic knockout of ATRX or SMARCAL1 in some telomerase-positive glioma cell lines has induced multiple hallmarks of ALT (20,26). Thus, a constellation of genetic and epigenetic changes may be gatekeepers for permitting ALT. Interestingly, the combination of knocking down ATRX, knocking out DAXX, and introducing a mutation in TPP1 (that suppresses telomerase activity and induces telomere-specific DNA damage) was sufficient to activate ALT in the telomerase-positive fibrosarcoma cell line HTC75 (27). Strategies to cripple telomerase via knocking out TERC in telomerase-positive cell lines have been successful in activating ALT at extremely low frequency in the spontaneously immortalized human lung fibroblast cell line SW39 and the lung carcinoma cell line H1299 (28).
We previously identified a prostate cancer case where a chromosomal inversion disrupting ATRX was found in multiple distant metastases, but not in the primary cancer, and these metastatic lesions showed evidence of ALT. Intriguingly, these findings suggest that the ATRX loss-of-function mutation was advantageous in the lethal metastases (29). Here, we have introduced mutations in ATRX in two different telomerase-positive, ALT-negative, prostate cancer cell lines. CWR22Rv1, originally derived from a primary tumor (30), did not develop hallmarks of ALT following ATRX knockout. However, LAPC-4, which was derived from a lymph node metastasis (31), displayed multiple hallmarks consistent with ALT following ATRX knockout. Pathway analysis of the transcriptome of LAPC-4 ATRXKO cells was consistent with previous reports of ALT-positive cancers, particularly the down-regulation of MYC target genes (32). Furthermore, when telomerase was crippled in LAPC-4 ATRXKO, these cells were able to continue proliferating long term and maintain their telomere length. These telomerase-independent cells displayed increased telomere heterogeneity, increased number of APB-positive cells, and increased C-circle levels. Here, we demonstrate bona fide ALT following functional inactivation of ATRX and telomerase in a telomerase-positive adenocarcinoma cell line.
MATERIALS AND METHODS
Cell culture
LAPC-4 was cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma), 1% of mixture of Penicillin 10,000 units/mL and Streptomycin 10,000 ug/mL (Pen/Strep, Quality Biological), and 1 nM of R1881. CWR22Rv1 and PC3 were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS and 1% Pen/Strep. U2OS was cultured in Dulbecco’s Modified Eagle Medium, high glucose (DMEM, Gibco) supplemented with 10% FBS and 1% Pen/Strep. All cell lines were submitted to the Genetic Resources Core Facility at Johns Hopkins for mycoplasma detection and cell line authentication by short tandem repeat (STR) profiling using the GenePrint 10 kit (Promega, June 2018).
ATRX CRISPR genome editing
As previously described, two CRISPR Cas9 nickase guide RNAs (Table S1) were designed to target exon 9 of ATRX using CRISPR Design (crispr.mit.edu, Figure S2). The gRNAs were cloned into the GFP-expressing Cas9n plasmid, PX461, a gift from Feng Zhang (Addgene #48140) (33). Lipofectamine 3000 (ThermoFisher) was used to transfect either empty vector PX461 or co-transfect both ATRX gRNA1-PX461 and ATRX gRNA 2-PX461 into LAPC-4 and CWR22Rv1 cells. GFP positive cells were sorted by FACS after 48 hours and 1000 cells were plated in 150 mm dishes. Cell colonies were isolated using cloning cylinders (Sigma) and screened preliminarily for ATRX protein by immunostaining. Promising clones were subsequently validated by western blotting and Sanger sequencing.
CRISPR genome editing of the TERC locus
A CRISPR plasmid cloned with the TERC gRNA-1 (Table S1) cloned into plasmid PX458 was a kind gift from Jaewon Min and colleagues (34). The original vector was a gift from Feng Zhang (Addgene #48138) (35). Lipofectamine 3000 (ThermoFisher) was used to transfect TERC1-gRNA-PX458 in LAPC-4 ATRXKO cells. GFP positive cells were sorted by FACS after 48 hours and 1000 cells were plated in 150 mm dishes. In another approach, CRISPR-mediated homology directed repair with a donor plasmid containing dsRed was used to knock-out TERC and knock-in dsRed in LAPC-4 ATRXKO1. TERC gRNA 2 and TERC gRNA 3 (Table S1) were individually cloned into PX458. The donor plasmid was constructed with a left TERC locus homology arm–loxP site–CMV enhancer and promoter–dsRed–Sv40 Poly A tail–right TERC locus homology arm. Lipofectamine 3000 (ThermoFisher) was used to co-transfect TERC gRNA 2 PX458, TERC gRNA 3 PX458, and donor plasmid in the LAPC-4 ATRXKO1 cell line. Cells positive for both RFP and GFP were sorted by FACS after 48 hours and 1000 cells were plated in 150 mm dishes. Cell colonies from both CRISPR approaches were isolated using cloning cylinders (Sigma) and screened preliminarily for TERC expression by RT-qPCR. Promising clones were subsequently validated by the TRAP assay, Sanger sequencing and RNA CISH.
Telomere repeated amplification protocol (TRAP assay)
The TRAP assay was performed as previously published with minor modifications (36). Frozen cell pellets (1×106 cells) were resuspended in 400 μL of NP-40 lysis buffer and incubated on ice for 30 minutes. The equivalent of 2500 cells (1 μL of crude lysate) was combined with 49 μL of 1X TRAP buffer, 200 μM dNTPs, 2 units of DNA polymerase (Platinum™ Taq DNA Polymerase, Invitrogen), 340 nM TS oligonucleotide conjugated with Alexa Fluor 488, 170 nM ACX oligonucleotide, 20 aM (2×10−17 M) TSNT oligonucleotide, and 340 nM NT oligonucleotide (Table S1 for oligo sequences). For RNase treated lysate, 1 μL of RNase A (Qiagen) diluted to 2.5 mg/mL was combined with 8 μL of lysate and 15 μL of nuclease-free water. The reaction was incubated at room temperature for 5 minutes and 2500 cell equivalents was used in the TRAP reaction. In a thermocycler, the following conditions were used to initiate TRAP reaction: 1 cycle at 30°C for 30 minutes, 1 cycle at 94°C for 10 minutes, 26 cycles of 94°C for 30 seconds, 50°C for 30 seconds, 72°C for 45 seconds, and ending with a hold at 4°C. To visualize the TRAP products using a non-radioactive detection method, 10 μL of 6X nucleic acid loading buffer (Thermo Scientific) was added to each TRAP reaction. An aliquot of 40 μL of the reaction was loaded into a well of a 20% Novex® TBE Gel (Invitrogen), and run for 125 minutes at 200 Volts in 0.5XTBE Buffer. TRAP products labeled with Alexa Fluor 488 were visualized using the ChemiDoc™ Imaging System (Bio-Rad).
Western blot
Frozen cell pellets collected from T-75 flasks were resuspended in 100 μL of RIPA buffer (Cell Signaling) and incubated on ice for 30 minutes. Cell lysates were centrifuged for 30 minutes at 13,000 RPM for 15 minutes at 4°C. Supernatant was collected and protein concentration was determined using the BCA Protein Assay Kit (Pierce). An equal volume of 2X Laemmli Buffer (Bio-Rad) containing 5% 2-mercaptoethanol (MilliporeSigma) was added to the cell lysate and incubated for 10 minutes at 95°C. Denatured lysate was cooled on ice and 30 μg of cell lysate was loaded into well of NuPAGE™ 3–8% Tris-Acetate Protein Gel (Invitrogen). Samples were run for 1–1.5 hours at 150 volts in NuPAGE™ tris-acetate SDS running buffer (Invitrogen). Electrophoretically separated proteins were transferred to a nitrocellulose membrane (Bio-Rad) in Tris-Glycine transfer buffer (Bio-Rad) containing 5% methanol (MilliporeSigma) for 3 hours at 325 mA at 4°C. Membranes were blocked in 5% milk (Bio-Rad) in tris buffered saline with Tween 20 (TBST, MilliporeSigma) for one hour, and then incubated in primary antibody overnight at 4°C in 5% milk in TBST. The following dilutions in 5% milk were prepared for each primary antibody: 1:20,000 GAPDH (D16H11, Cell Signaling), 1:10,000 beta-actin (8H10D10, Cell Signaling), or 1:2000 DAXX (HPA008736, Sigma). ATRX antibody (D1N2E, Cell Signaling) was diluted 1:1000 in 5% BSA in TBST. Membranes were washed in TBST and incubated in secondary antibody for one hour in 5% milk in TBST, either 1:10,000 HRP-linked anti-rabbit IgG (7074, Cell Signaling) or 1:20:000 HRP-linked anti-mouse IgG (7076, Cell Signaling). Signal was detected using Clarity ECL (Bio-Rad). Blots were either exposed to film or developed using the ChemiDoc™ Imaging System (Bio-Rad).
Telomere-specific FISH, PML immunofluorescence, and APB scoring
Freshly prepared cell pellets from confluent T-175 flasks were fixed in 10% phosphate-buffered formalin (MilliporeSigma) for 48 hours and paraffin-embedded. Telomere fluorescence in situ hybridization (FISH) was performed as previously described (37). FFPE cell pellets of 5 μM sections were deparaffinized, rehydrated, treated for 1 minute in 1% Tween 20, and steamed for 30 minutes in citrate buffer, pH 6 (Vector Laboratories). Slides were incubated with 333 ng/mL of telomere probe, a cy3-labeled peptide nucleic acid (PNA) oligonucleotide (N-CCCTAACCCTAACCCTAA-C) in PNA buffer (10 mM Tris pH 7.5, 70% formamide) for 5 minutes at 84°C, followed by an incubation for 2 hours or overnight at room temperature. Excess probe was washed off at room temperature with PNA buffer. For immunofluorescence staining, slides were blocked for 10 minutes in serum-free protein block (Agilent Dako), washed in phosphate buffered saline with Tween 20 (PBST), and subsequently incubated for two hours in 1:100 PML antibody (A301–167A, Bethyl) diluted in antibody diluent buffer (Ventana). Excess antibody was washed in PBST, and stained with secondary antibody, anti-rabbit Alexa Fluor 647 (Invitrogen) diluted in PBS. Slides were stained with DAPI and cured in ProLong™ Gold Antifade reagent (Invitrogen). Stained slides were scanned using the TissueFAXS Plus (Tissue Gnostics) automated microscopy workstation equipped with a Zeiss Z2 Axioimager microscope with high quality optics. Scanned 400X images were exported as TIF images using TissueQuest software (Tissue Gnostics), and manually scored for cell count and co-localization of PML with ALT-associated telomeric foci (APBs) with ImageJ/Fiji cell count tool (38).
C-circle assay
A previously described C-circle dot blot assay was followed with some modifications (14). Genomic DNA was extracted from frozen cell pellets harvested from a confluent T-75 flask using the DNeasy Blood and Tissue Kit (Qiagen). DNA less than 10 kilobases were enriched using the QiaQuick PCR Purification Kit (Qiagen). C-circle signal was amplified using rolling circle replication, combining 150 ng of DNA with 2 units of phi29 DNA polymerase (New England Biolabs) in 20 μL of C-circle buffer (1x Phi29 reaction buffer, 200 ng/mL BSA, 0.1% Tween 20, 1 mM dATP, 1 mM dGTP, 1 mM dTTP). Reaction products were blotted onto a positively-charged nylon membrane (Roche) and hybridized to Digoxigenin (DIG) conjugated telomere probe (CCCTAACCCTAACCCTAACCCTAA-DIG, Integrated DNA Technologies) in DIG Easy Hyb™ buffer (Roche) overnight at 42°C. Excess probe was washed using 2X saline-sodium citrate (SSC) buffer with 0.1% SDS at room temperature, followed by 0.2X SSC 0.1% SDS at 50°C. Membranes were blocked in 5% milk TBST for at least 30 minutes at room temperature, and subsequently incubated with 1:10,000 anti-Digoxigenin-AP, Fab fragments (Roche) for at least 30 minutes. Excess antibody was washed and amplified C-circle products were detected using CDP-Star kit (Roche). C-circle signals were quantified by densitometry using the ChemiDoc™ Imaging System (Bio-Rad).
Telomere length assay - Telomere Restriction Fragment (TRF) Southern blotting
A previous published protocol for genomic DNA extraction and TRF Southern Blotting was followed with some modifications (39). Cells were resuspended in SNET buffer (20 mM Tris pH 8.0, 5 mM EDTA, 400 mM NaCl, 1% SDS) containing 400 ug/mL of proteinase K (P8107S, NEB), and incubated at 55oC with gentle agitation overnight. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, pH 8, MilliporeSigma) was added. The aqueous layer was isolated, and DNA was precipitated with equal volume of isoproponal. DNA pellet was washed twice 70% ethanol and airdried. Isolated genomic DNA was digested overnight with 20 units of MseI (R0525, NEB) and 0.07 units of RNase A (Qiagen) in 1XCutSmart buffer at 37°C. Digested DNA was supplemented with 20 units of AluI (R0137, NEB) and 10 units of MboI (R0147, NEB). DNA Molecular Weight Marker II, DIG-labeled (Roche) and digested DNA (2–20 ug, empirically determined input) was combined with 6XDNA loading dye (NEB) and run on 0.7% agarose TAE gel for 12 hours at 47 V. Gel was post-stained in 0.5 ug/mL ethidium bromide, and UV treated for 60 mJ using a Stratalinker (40). The gel was subsequently treated for 30 minutes in Denaturation Solution (0.5 M NaOH/1.5 M NaCl, Millipore), 30 minutes in Neutralization Buffer (1.5 M NaCl/0.5 M Tris-HCl pH 7.4), and 30 minutes in 20X SSC buffer. DNA was transferred to positively-charged nylon membrane (Roche) overnight using the Turboblotter transfer system (GE), and subsequently UV cross-linked using a Stratalinker. Transferred TRFs were hybridized to Digoxigenin (DIG) conjugated telomere probe (CCCTAACCCTAACCCTAACCCTAA-DIG, Integrated DNA Technologies) in DIG Easy Hyb™ buffer (Roche) overnight at 42°C. Excess probe was washed using 2X saline-sodium citrate (SSC) buffer with 0.1% SDS at room temperature, followed by 0.2X SSC 0.1% SDS at 50°C. Membranes were blocked in 5% milk TBST for at least 30 minutes at room temperature, and subsequently incubated with 1:10,000 anti-Digoxigenin-AP, Fab fragments (Roche) for at least 30 minutes. Excess antibody was washed and amplified TRFs were detected using CDP-Star kit (Roche). TRF sizes were determined using the ChemiDoc™ Imaging System (Bio-Rad), and telomere length distribution was recorded based on densitometry reads with the plot profile tool on ImageJ/Fiji (38).
Microarray
Total RNA was isolated from cell pellets using TriZol reagent (Invitrogen) followed by RNeasy mini kit with DNase on-column digestion (Qiagen). RNA was quantified with NanoDrop ND-1000 followed by quality assessment with 2100 Bioanalyzer (Agilent Technologies) according to manufacturer’s protocol. Sample amplification and labeling procedures were carried out by using Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies), with minor modifications. Briefly, 0.4 ug total RNA was reverse-transcribed into cDNA by MMLV-RT using an oligo dT primer (System Biosciences) that incorporates a T7 promoter sequence. The cDNA is then used as a template for in vitro transcription in the presence of T7 RNA polymerase and Cyanine-3 labeled CTP (Perkin Elmer). The labeled cRNA is purified using RNeasy mini kit (Qiagen). RNA spike-in controls (Agilent Technologies) are added to RNA samples before amplification and labeling according to manufacturer’s protocol. Agilent human GE 4× 44K v2 microarrays (G4845A) were used, which contain 41,000 unique probes targeting 27,958 Entrez gene RNAs. 0.825 ug of each Cy3-labeled samples was used for hybridization at 65°C for 17 hours in a hybridization oven with rotation. After hybridization, microarrays are washed and dried according to Agilent microarray processing protocol in a walk-in ozone-controlled enclosure. Microarrays were scanned using an Agilent G2565A Scanner controlled by Agilent Scan Control 7.0 software. Data were extracted with Agilent Feature Extraction 9.5.3.1 software.
Differential Expression Analysis
All data analysis for DNA was carried out using the R statistical software suite using both standard and customized routines (41). Agilent expression array data was imported and quantile normalized using functions included in the Limma Bioconductor software package (42). Finally, expression values were transformed to a log-base-2 scale. Gene annotations were provided by the array manufacturer.Differential expression was calculated using Empirical Bayes linear models as implemented in the limma package from Bioconductor (42). When both cell lines were used together in an analysis, expression values were standardized within cell line, by subtracting the parental transcriptional profile from each ATRX knockout replicate. Gene set analysis was performed based on curated data sets from MSigDB (http://software.broadinstitute.org/gsea/msigdb/index.jsp) to identify altered pathways, using Wilcoxon tests as implemented in the limma package.
Statistics
All statistical tests, including T-tests, were performed using Graphpad Prism 7 software. P-values < 0.05 were considered statistically significant.
RESULTS
LAPC-4 and CWR22RV1 rely on telomerase for telomere maintenance
LAPC-4 (31) and CWR22Rv1 (30) are two well-characterized, established human prostate cancer cell lines that rely on telomerase for telomere maintenance, as measured by Telomerase Repeated Amplification Protocol (TRAP) analysis (Figure S1A), and do not display dramatic telomere length heterogeneity as assessed by Telomere Restriction Fragment (TRF) southern blotting (Figure S1B). Additionally, LAPC-4 and CWR22Rv1 also lack ultrabright telomeric foci, APBs, and C-circles, features consistent with ALT, in contrast to U2OS, a well-characterized ALT-positive osteosarcoma cell line (43) (Figure S1C and D). In addition, LAPC-4 and CWR22Rv1 express full-length ATRX and DAXX proteins (Figure 1A and B). Functional inactivation in either of these chromatin remodeling proteins, ATRX or DAXX, typically resulting in loss of protein, is enriched in ALT-positive cancers (15,16).
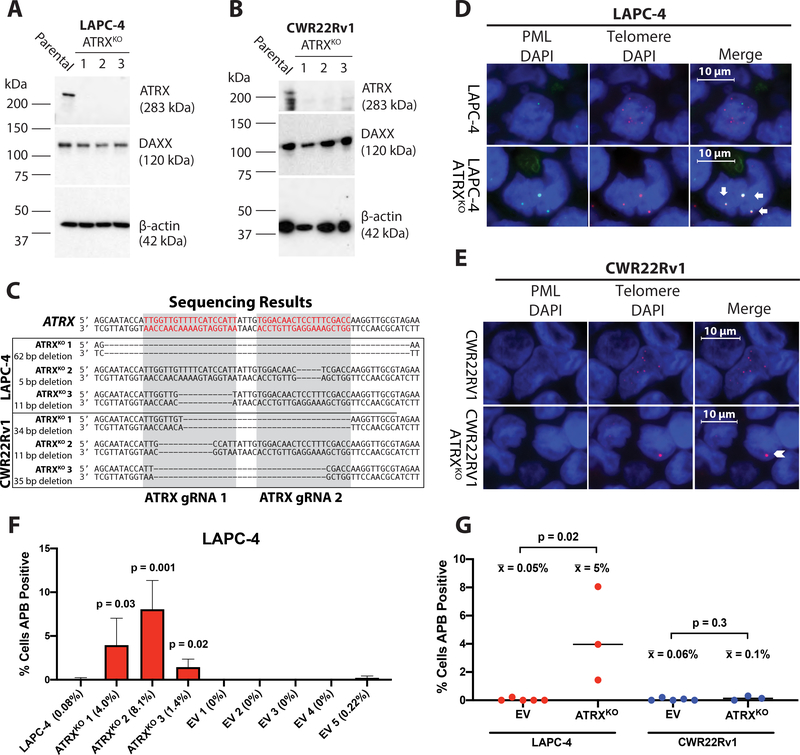
Figure 1.
ATRX frameshift mutations lead to protein disruption and the formation of APBs. Immunoblotting demonstrates the retention of DAXX protein expression, whereas ATRX protein expression is lost in the (A) LAPC-4 ATRXKO clones and (B) CWR22Rv1 ATRXKO clones. (C) Sanger sequencing of all LAPC-4 and CWR22Rv1 ATRXKO clones confirmed frameshift deletions overlapping the intended target region. Representative telomere FISH (red) and PML immunolabeling (green) in (D) LAPC-4 and (E) CWR22Rv1 ATRXKO clones. APBs (arrows) or ultrabright telomeric foci that do not co-localize with PML (chevron) are shown. Cells (N>1200) were scored for APBs in parental, ATRXKO, and EV clones for (F) LAPC-4 and CWR22Rv1 (not shown). P-values were determined using an unpaired t-test comparing the parental cell line with each ATRXKO or EV clone, as well as (G) EV clones with ATRXKO clones for % cells positive for APBs. Only p-values < 0.05 are indicated.
Introducing ATRX frameshift mutations disrupts protein expression
We used the CRISPR Cas9 nickase system to generate multiple ATRXKO clones in LAPC4 and CWR22Rv1. The aspartate-to-alanine (D10A) mutation in the catalytic domain of Cas9 results in a nickase mutant, requiring two gRNAs to generate the desired double strand break and initiate error-prone repair, thus decreasing the incidence of unwanted mutations in non-specific targets (35). We used previously designed gRNAs that targeted the ATRX-DNMT3-DNMT3L (ADD) domain of ATRX (26) (Figure S2A). We generated six independent isogenic KO clones: LAPC-4 ATRXKO 1–3 and CWR22Rv1 ATRXKO 1–3, with sequence-confirmed frameshift mutations in ATRX (Figure 1C) and confirmed disrupted ATRX protein expression (Figure 1A and B).
Recently, it has been reported that siRNA knockdown of ATRX is associated with a reduction in PML protein (44). In our ATRXKO clones, we did not observe a reduction of PML protein compared to parental cells (Figure S2B–C). However, baseline PML levels in both parental LAPC-4 and CWR22Rv1 cells were relatively low compared to U2OS, an ALT-positive cell line known to express PML but lacks ATRX (45).
Functional loss of ATRX induces APBs in LAPC-4, but not CWR22Rv1
In order to determine the effect of ATRX loss and the emergence of ALT-associated hallmarks in LAPC-4 and CWR22Rv1, we assessed our ATRX KO clones for the presence of APBs. All of the LAPC-4 ATRX KO clones displayed APBs, unlike the parental and empty vector (EV) clones (Figure 1D, F, and G). In contrast, APBs were rarely, if ever, observed in the CWR22Rv1 ATRX KO clones (Figure 1E and G). Although ultrabright telomere DNA foci that do not co-localize with PML are another marker of ALT (15), these too were rare in the CWR22Rv1 ATRX KO clones. Quantification of the percentage of cells positive for APBs (N>1200 cells assessed per cell line) showed that the percentage increased from 0.08% in parental LAPC-4 cells to more than 17-fold in all three LAPC-4 ATRX KO clones (range: 1.4–8.1%); whereas the percentage of cells positive for APBs in CWR22Rv1 ATRX KO clones were less than < 0.3% (range: 0–0.3%), comparable to the parental CWR22Rv1 and EV clones (Figure 1F and G). In general, ALT-positive cancers have more than 1% of cells displaying the ALT-associated ultrabright telomeric foci (15,29).
Functional loss of ATRX induces telomere length heterogeneity in LAPC-4
In telomere restriction fragment (TRF) assays, telomerase-positive cancer cell lines display a discernable mean telomere length with telomeres typically ranging a few kilobases from the mean value. In contrast, ALT-positive cells display a broad and extremely heterogeneous distribution of telomere lengths ranging from very short to greater than 50 kb (46). While parental LAPC-4 and EV clones have well-defined mean telomere lengths of ~3.5 kb (Table S2), the LAPC-4 ATRXKO clones have an increase in telomere length heterogeneity similar to that observed in ALT-positive cell lines and tissues (Figure S3A and B). In contrast, the telomere length distribution did not significantly change in the CWR22Rv1 ATRXKO clones compared to their parental and EV clones after more than 50 population doublings (Figure S3C and D). Within each cell line group, LAPC-4 or CWR22Rv1, the mean telomere lengths of parental, ATRXKO clones, and EV clones were not significantly different (Table S2, Figure S3E–F).
Functional loss of ATRX induces C-circles in LAPC-4
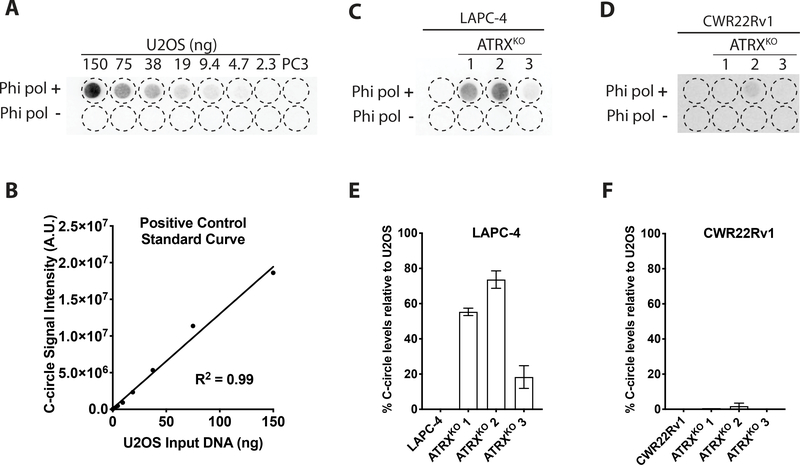
ALT-positive cancers are enriched for C-circles, extrachromosomal DNA containing circular single-stranded C-rich telomeric sequence. Using the processive phi29 DNA polymerase, C-circles can be preferentially amplified since C-circles are partially double stranded, and the G-rich telomeric DNA can serve as a primer for rolling circle amplification (14). Relative to an ALT-positive cell line (U2OS), C-circle analysis revealed the absence of C-circle production in the LAPC-4 parental line, whereas the LAPC-4 ATRXKO clones produced appreciable C-circles, with levels ranging from 18–74% of those observed in U2OS (Figure 2A–E). In contrast, all CWR22Rv1 ATRXKO clones were comparable to the parental counterpart, with all cell lines displaying <2% C-circle levels relative to U2OS (Figure 2F).
Figure 2.
Assessment of C-circle levels of LAPC-4 ATRXKO and CWR22Rv1 ATRXKO clones. (A) Blot of C-circle assay generated from U2OS, an ALT-positive cell line. C-circles are absent in PC3 (ALT-negative prostate cancer cell line). (B) C-circle assays were performed with a U2OS (positive control) standard curve to determine relative C-circle levels in LAPC-4 (C&E) and CWR22Rv1 (D&F).
Abolishing ATRX does not disrupt telomerase activity in LAPC-4 and CWR22Rv1
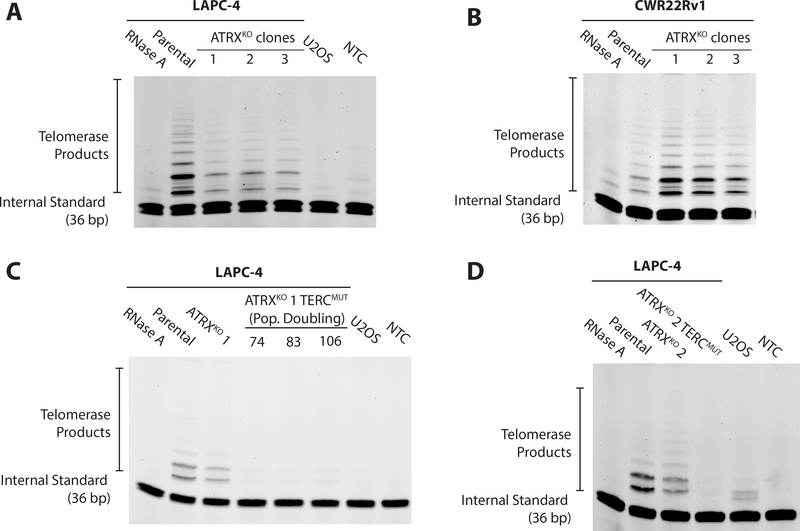
The majority of telomerase-deficient cancers rely exclusively on ALT for telomere maintenance (9). However, telomerase and ALT can co-exist in the same cell line when hTERT, the catalytic subunit of telomerase, is overexpressed in an ALT-positive cell line (47). Similarly, TRAP analysis demonstrated telomerase activity remained in all LAPC-4 ATRXKO and CWR22Rv1 ATRXKO clones (Figure 3A and B). While TERT expression was reduced in LAPC-4 ATRXKO clones, the difference was not statistically significant compared to EV clones (Figure S4A–B).
Figure 3.
Assessment of telomerase activity in ATRXKO clones. Telomere repeated amplification protocol (TRAP) assay was performed on (A) parental and LAPC-4 ATRXKO clones, (B) parental CWR22Rv1 ATRXKO clones, (C) LAPC-4 ATRXKO 1;TERCmut and (D) LAPC-4 ATRXKO 2;TERCmut. U2OS was used a telomerase-negative control. Additionally, a no template control (NTC) and RNase treated LAPC-4 or CWR22Rv1 cell lysate demonstrate lack of telomerase activity. PD = population doubling.
Introducing mutations in the TERC locus to inhibit telomerase activity in LAPC-4 ATRXKO clones
Unlike CWR22Rv1 ATRXKO clones, LAPC-4 ATRXKO clones displayed multiple characteristics consistent with ALT – formation of APBs, telomere length heterogeneity, and the generation of C-circles; however, LAPC-4 ATRXKO clones maintained telomerase activity. To assess if LAPC-4 ATRXKO clones could rely exclusively on ALT for telomere maintenance (i.e. in the absence of telomerase), we used multiple CRISPR Cas9 genome editing strategies to cripple telomerase. Initial attempts targeting TERT, the gene encoding the catalytic subunit of telomerase, were unsuccessful. However, targeting TERC, the gene encoding the essential RNA template of telomerase, in parallel approaches, generated two LAPC-4 ATRXKO clones deficient for telomerase activity. In one approach, homology-directed genome editing with two CRISPR Cas9 gRNAs targeting sequences flanking TERC were co-transfected with a donor plasmid containing dsRed (48) (Figure S5A). This strategy generated the LAPC-4 ATRXKO 1;TERCmut clone, in which all 3 copies of TERC were disrupted, resulting in a loss of telomerase activity (Figure 3C). TERC expression in LAPC-4 ATRXKO 1;TERCmut clone was <1% expression compared to LAPC-4 parental, as measured by RT-qPCR and validated by chromogenic in situ RNA detection (Figure S4C and E). In this clone, telomerase activity was crippled through over 100 population doublings of continuous growth in culture.
In another approach, wild type CRISPR Cas9 was coupled with a gRNA targeted to the core domain of TERC (34) (Figure S5B). Out of 3,000 LAPC-4 ATRXKO2 cells positively sorted for the GFP-expressing CRISPR plasmid, 21 colonies were established, one of which was telomerase-negative. Sequencing of the clone that was telomerase-negative revealed that 2 of the 3 copies of TERC were mutated. Although TERC expression of these LAPC-4 ATRXKO 2;TERCmut cells was retained, (Figure S4D and E), the telomerase activity was nonetheless undetectable (Figure 3D), suggesting that mutations in the core domain of TERC was sufficient to cripple telomerase activity. Using the same strategies to target TERC in the parental LAPC-4 cells, we were unable to recover any TERCmut clones, suggesting that crippling telomerase was lethal. In rare surviving clones that grew into colonies (N=24), all clones maintained TERC expression comparable to parental cells (data not shown).
Crippling telomerase in LAPC-4 ATRXKO clones increases ALT-associated hallmarks
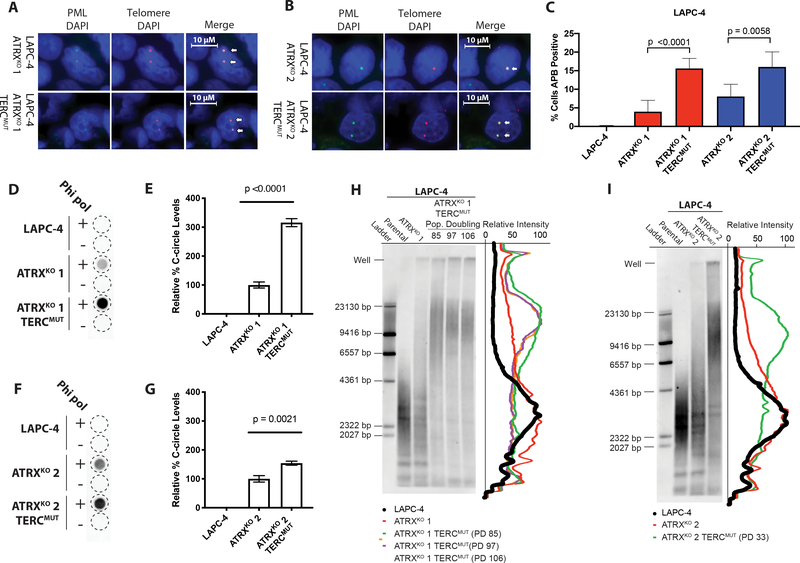
Hallmarks associated with ALT were evaluated in LAPC-4 ATRXKO clones with crippled telomerase. Cells (N>1200) were assessed for the presence or absence of nuclear APBs via combined PML-IF/telomere-specific FISH and quantitative image analysis (Figure 4A–C). Both LAPC-4 ATRXKO 1;TERCmut and LAPC-4 ATRXKO 2;TERCmut had more than two-fold increase in percent cells positive for APBs when compared to LAPC-4 ATRXKO 1 and LAPC-4 ATRXKO 2, the cell lines from which the TERCmut sub-lines were derived, respectively (Figure 4C). Similarly, there were significant increases in C-circle levels in both LAPC-4 ATRXKO 1;TERCmut and LAPC-4 ATRXKO 2;TERCmut (Figure 4D and F). A 2.2-fold increase was observed in C-circle levels in LAPC-4 ATRXKO 1;TERCmut compared to parental LAPC-4 ATRXKO 1 (Figure 4E). Likewise, LAPC-4 ATRXKO 2;TERCmut displayed a 54% increase in C-circle levels compared to LAPC-4 ATRXKO 2, from which it was derived (Figure 4G).
Figure 4.
APBs, C-circles, and telomere lengths increase in LAPC-4 ATRXKO clones lacking telomerase activity. Representative images of telomere FISH (red) and PML immunoblotting (green) from (A) LAPC-4 ATRXKO 1 and LAPC-4 ATRXKO 1;TERCmut; (B) LAPC-4 ATRXKO 2 and LAPC-4 ATRXKO 2;TERCmut. Arrows indicate examples of ALT-associated PML Bodies (APBs). Cells were scored for APBs in (C) LAPC-4 ATRXKO 1;TERCmut and LAPC-4 ATRXKO 2;TERCmut. P-values were determined using two-sided t-tests. C-circle assay of (D) LAPC-4 ATRXKO 1;TERCmut and (F) LAPC-4 ATRXKO 2;TERCmut. The relative quantity of C-circle was assessed from blots of (E) LAPC-4 ATRXKO 1;TERCmut and (G) LAPC-4 ATRXKO 2;TERCmut. P-values were determined using unpaired two-sided t-tests. Representative telomere restriction fragment (TRF) southern blots of (H) LAPC-4 ATRXKO 1;TERCmut and (I) LAPC-4 ATRXKO 2;TERCmut. Intensity traces of the distribution of TRFs is shown. PD = population doubling.
Long telomeres are maintained during serial passaging in ATRXKO cells in which telomerase has been inactivated
In these LAPC-4 ATRXKO clones lacking telomerase activity, overall telomere length was assessed by TRF Southern blot analysis. When TERC is abolished, in the absence of another telomere maintenance mechanism, telomeres will progressively shorten over multiple population doublings due to the end replication problem (49). In fact, when TERC expression is the limiting factor in telomerase activity, TERC haploinsufficiency is sufficient to induce telomere shortening (50). However, in LAPC-4 ATRXKO 1;TERCmut, there is a notable shift to increased telomere lengths as compared to both LAPC-4 and LAPC-4 ATRXKO 1, and retention of telomeric DNA content in gel-wells, indicative of recombination intermediates observed in ALT-positive cells (Figure 4H). Importantly, this shift to increased telomere length is maintained beyond 100 population doublings. Similarly, the overall telomere length distribution shifts to longer telomere lengths in LAPC-4 ATRXKO 2;TERCmut after 33 population doublings (the longest timepoint measured) in comparison to LAPC-4 and LAPC-4 ATRXKO 2 (Figure 4I). Additionally, both LAPC-4 ATRXKO clones with inactivated telomerase activity appear to maintain telomere heterogeneity, with a bulk of the telomere length increasing from ~ 3 kb to > 20 kb. These results demonstrate that LAPC-4 ATRXKO clones rely on ALT for telomere maintenance when telomerase activity is compromised.
Genes associated with cell cycle progression and DNA repair pathways are differentially expressed in ATRXKO clones
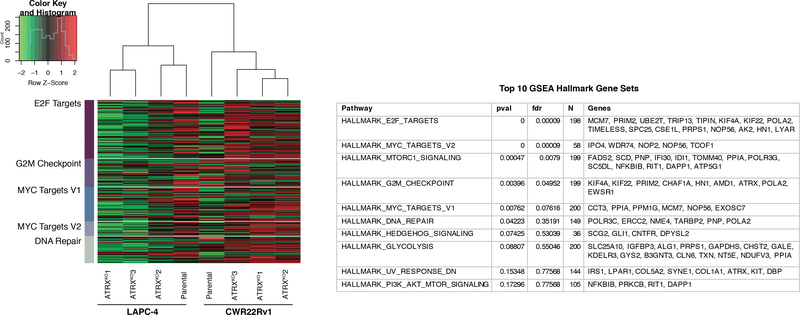
The gene expression of parental LAPC-4 and CWR22Rv1, and associated ATRXKO clones, were profiled using a transcriptome microarray (GEO submission GSE129448). A gene set analysis based on curated datasets from Molecular Signatures Database (MSigDB) revealed that several differentially expressed genes were cell cycle targets of E2F transcription factors, involved in the G2/M checkpoint, regulated by MYC, and involved in DNA repair (Figure 5). In general, transcript levels for genes in our pathway analysis were down-regulated in LAPC-4 ATRXKO clones and up-regulated in CWR22Rv1 ATRXKO clones, suggesting that differences in cell cycle progression, DNA repair, and/or MYC activity may contribute to the emergence of ALT-associated hallmarks in LAPC-4, and not in CWR22Rv1, following loss of ATRX. Intriguingly, TIMELESS and TIPIN were a couple of the top genes affected in the E2F target genes in our pathway analysis. TIMELESS is known to form a complex with TIPIN, and is important in telomere length maintenance (51). RT-qPCR analysis of TIMELESS and TIPIN revealed that ATRXKOs consistently had higher expression than EV clones (Figure S6A). Finally, unsupervised hierarchical clustering of parental and ATRXKO clones shows that isogenic clones cluster based on cell line (Figure 5).
Figure 5.
Gene set analysis of parental (LAPC-4 and CWR22Rv1) and accompanying ATRXKO clones. Based on curated data sets from Molecular Signatures Database (MSigDB), gene set analysis reveals down-regulation of E2F targets, MYC targets, G2/M checkpoint genes, and DNA repair genes in LAPC-4 ATRXKO clones compared to parental LAPC-4 cell line. In contrast, the targets of these pathways are up-regulated in CWR22Rv1 ATRXKO clones compared to parental CWR22Rv1 cell line.
Of the top 10 differentially expressed genes comparing parental ATRX wild-type cells and ATRXKO cells, ranked by false discovery rate (FDR)-adjusted p-values, all four microarray probes that target ATRX were present, as expected (Table S3). A subset of genes found to be most differentially expressed in ATRXKO clones compared to parental ATRX wild-type cells (Table S3), or most differentially expressed in LAPC-4 ATRXKO clones compared to CWR22Rv1 ATRXKO clones (Table S4) were evaluated by RT-qPCR with additional EV clones included (Figure S6B–C). GPRC5C was found to be significantly down-regulated in both LAPC-4 ATRXKO clones and CWR22Rv1 ATRXKO clones compared to their EV counterparts, suggesting that loss of ATRX results in a down-regulation of GPRC5C expression (Figure S6). KIF1A was found to be significantly down-regulated in CWR22Rv1 ATRXKO clones, while KIF1A was up-regulated in CWR22Rv1 EV clones, LAPC-4 EV clones, and LAPC-4 ATRXKO clones (Figure S6). However, no statistically significant changes were observed in POLE2, HOXB4, PRSS23 and C2orf54 when ATRXKO and EV clones were compared (Figure S6).
DISCUSSION
The ALT telomere maintenance mechanism is strongly associated with the functional inactivation of ATRX or DAXX, and infrequently, the ATP-dependent annealing helicase SMARCAL1 (15–17,20). While the majority of ALT-positive cancers harbor mutations in ATRX, demonstrating experimentally that loss-of-function mutations in ATRX can activate ALT in cancer cells has been elusive. Here, we have shown that the telomerase-positive, ALT-negative human prostate cancer cell line LAPC-4 acquires multiple ALT-associated hallmarks following ATRX knockout, and more importantly, can continue to proliferate long term (> 100 population doublings) and maintain telomere length when telomerase activity is subsequently crippled – the most rigorous definition of ALT. The findings of these studies demonstrate that telomerase-independent, ALT-positive adenocarcinoma lines can be established from a telomerase-positive cell line following ATRX inactivation.
While ATRX acts as a suppressor of ALT in vivo (25,52), prior attempts to induce ALT in vitro through knockdown or knockout of ATRX have mostly been unsuccessful (21,23–25). However, in a context dependent manner, genetic knockout of ATRX or SMARCAL1 in some telomerase-positive glioma lines induces multiple hallmarks of ALT, specifically the presence of APBs and C-circle production (20,26). A notable limitation of these studies is that it is unknown whether cells that acquire ALT-associated hallmarks following ATRX knockout can rely exclusively on ALT for telomere maintenance (i.e. in the absence of telomerase activity).
In a stepwise manner, we have demonstrated here that LAPC-4 prostate cancer cells can acquire ALT-associated hallmarks following ATRX knockout. Furthermore, telomerase-deficient LAPC-4 TERCmut survivors were only recovered in LAPC-4 ATRXKO clones, and not in the parental LAPC-4, where ATRX remains intact. More importantly, LAPC-4 ATRXKO TERCmut clones continued to proliferate and maintain telomere lengths when telomerase activity was crippled. With approximately 50 bp loss of telomere content per division (53), LAPC-4 cells (~3 kb mean telomere length) lacking telomerase would be expected to reach critical lengths following roughly 40 population doublings. However, LAPC-4 ATRXKO TERCmut cells were able to continue to proliferate in the absence of telomerase well beyond this point. Furthermore, we observed increases in telomere length distribution, frequency of APBs, and C-circles levels in LAPC-4 ATRXKO TERCmut compared to LAPC-4 ATRXKO clones. Perhaps both telomere maintenance pathways, telomerase and ALT, were employed in LAPC-4 ATRXKO. However, if so, it is unclear whether both telomere maintenance pathways are operating in the same individual cell, or mutually exclusive as a mixed population. An alternative, and equally plausible, possibility is that the elimination of telomerase may have instigated telomere crisis either driving LAPC-4 ATRXKO cells fully to ALT, or selecting for sub-clones poised for full ALT conversion. Notably, a prior approach to cripple telomerase via knocking out TERC in a telomerase-positive adenocarcinoma cell line (H1299) successfully activated ALT in one out of more than one hundred million cells in crisis (frequency of 0.9 × 10−8); an extremely rare event (28). In contrast, using the same CRISPR construct for targeting TERC, we successfully established an ALT-positive, telomerase-negative clone from 3,000 LAPC-4 ATRXKO cells (frequency of 0.3 × 10−3). This represents several orders of magnitude higher success rate and supports the notion that a significantly larger fraction of LAPC-4 ATRXKO cells were poised for full ALT conversion.
In this study, multiple isogenic ATRXKO clones were used to investigate the activation of ALT in prostate cancer cells. We introduced frameshift mutations in the ATRX gene in two prostate adenocarcinoma cell lines, LAPC-4 and CWR22Rv1. Following ATRX knockout, LAPC-4 activated multiple hallmarks of ALT as assessed by APBs, telomere heterogeneity, C-circles, and telomerase-independence. In contrast, CWR22Rv1 did not. Interestingly, one CWR22Rv1 ATRXKO clone with shorter telomeres had very low levels of C-circles (Figure S3D and Figure 2D). However, the same clone did not display telomere length heterogeneity, and less than 1% of the cells were APB positive. Thus, in a cell context dependent manner, functional loss of ATRX is capable of activating multiple hallmarks of ALT in human prostate cancer cells. Of particular significance, we also show that LAPC-4 ATRXKO TERCmut cells with crippled telomerase activity have truly activated ALT. These results confirm and extend what we and others have previously shown – specifically, that ATRX inactivation, alone, is not sufficient to activate ALT in telomerase-positive cancer cells. Thus, additional key genetic and/or epigenetic changes must coincide with ATRX inactivation to induce ALT (24,25).
It is unclear why ALT hallmarks arose after ATRX knockout in LAPC-4 and not CWR22Rv1. Although no firm conclusions can be made, the transcriptome pathway analysis of LAPC-4 and CWR22Rv1 ATRXKO clones may provide some insight. Genes encoded by cell cycle targets of E2F transcription factors, involved in the G2/M checkpoint, regulated by MYC, and involved in DNA repair were found to be down-regulated in LAPC-4 ATRXKOs, but up-regulated in CWR22Rv1 ATRXKOs (Figure 5). These pathways highlight distinct differences between the ATRXKO cell lines, and interestingly, are consistent with previous reports describing attributes of ALT-positive cancers. For example, ALT-positive cancer cells have defects in the G2/M checkpoint and are slow to repair double DNA breaks (21), which may, in part, explain why genes in both pathways are down-regulated in LAPC-4 ATRXKO cells. While MYC has been implicated in the initiation and progression of prostate cancer (54), transcriptome analysis of ALT-positive cancers have reported reduced expression of MYC target genes (32), similar to our LAPC-4 ATRXKOs. MYC is tightly linked to cell cycle progression, and the induction of E2F transcription factors are important in the transition from G1 to S phase (54). However, it is worthwhile noting that the top genes affected in the E2F target genes in our pathway analysis include TIMELESS and TIPIN. The TIMELESS/TIPIN complex has also been reported to suppress telomere clustering, mitotic DNA synthesis, and APB formation in the ALT-positive SAOS-2 cell line (34). Perhaps in our ATRXKO clones, upon loss of ATRX, TIMELESS and TIPIN are induced to prevent fork collapse due to increased instability at telomeres.
Differences in frequency of PML foci between LAPC-4 and CWR22Rv1 may also partially account for whether or not ALT hallmarks arise following ATRX loss of function. Although PML protein levels are low in both LAPC-4 and CWR22Rv1 (Figure S2B), immunostaining of PML indicated that LAPC-4 had stronger nuclear PML foci than CWR22Rv1 (Figure S1D, Figure 1D and F). In LAPC-4 ATRXKOs, almost all of the large telomeric foci co-localized with PML (>97%). PML is thought to be an important component of the ALT mechanism, especially since PML bodies are associated with many proteins involved in DDR and homology directed repair (12). While large telomeric DNA foci stained by telomere FISH do not always co-localize with PML to form APBs, PML levels have been shown to directly affect hallmarks of ALT. Knockdown of PML in the ALT-positive cancer cell line, U2OS, resulted in decreased telomere length and C-circle levels (55). Thus, the lack of PML nuclear bodies in CWR22Rv1 may potentially contribute to the lack of ALT hallmarks following loss of ATRX.
Finally, perhaps the genetic and epigenetic alterations differ in primary and metastatic prostate cancer, thereby contributing to whether ALT will be activated following functional loss of ATRX. Previously, an inversion on chromosome X disrupting the ATRX gene was identified in several distant lethal metastases in a patient with prostate cancer, and these metastases showed loss of ATRX protein and the presence of ALT-associated hallmarks. Interestingly, the ALT-negative primary tumor that gave rise to the metastases did not show this inversion in chromosome X (29), consistent with previous observations that ALT has not been reported in primary prostate adenocarcinomas (56). LAPC-4 was derived from a lymph node metastasis of a prostate cancer patient (31). In contrast, CWR22Rv1 was originally sourced from CWR22, a xenograft established from a primary prostate cancer (30). However, in the absence of a clear understanding of the genetic and epigenetic changes co-occurring with ALT, it is unclear if tumor site is a relevant factor.
Taken together, these studies demonstrate the induction of bona fide ALT in adenocarcinoma cells following ATRX inactivation and subsequent loss of telomerase activity, and re-emphasize the importance of ATRX loss in the activation of ALT. Additionally, the results highlight a possible resistance mechanism in cancers treated with telomerase-targeted therapies (8). Under selective pressure due to loss of telomerase activity, inactivation of key ALT suppressors such as ATRX can switch telomere maintenance from telomerase to ALT, given the right genetic and/or epigenetic context. Importantly, the cell lines generated from these studies will be useful tools for the development and testing of therapies specifically targeting ALT using approaches such as genomic and small molecule screens.
Supplementary Material
Implications.
These prostate cancer cell line models provide a unique system to explore the distinct molecular alterations that occur upon induction of ALT, and may be useful tools to screen for ALT-specific therapies.
Acknowledgements
This work was supported by National Institutes of Health [5T32CA009110–38 to M. Graham, F32CA213742 to M. Graham, 5R01CA172380–05 to A. Meeker, P30 CA006973 to the Sidney Kimmel Comprehensive Cancer Center]; and the Prostate Cancer Foundation [2014 Bonnie Pfeifer Evans Young Investigator Award to C. Heaphy]. We wish to thank the staff at the Flow Cytometry and Cell Sorting Core Facility at Johns Hopkins School of Public Health, and the Oncology Tissue Core and Microarray Core at Johns Hopkins Sidney Kimmel Comprehensive Cancer Center.
Footnotes
Conflicts of Interest: None
REFERENCES
- 1.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 2010;11(3):171–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 1973;41(1):181–90 [DOI] [PubMed] [Google Scholar]
- 3.Vaziri H Critical telomere shortening regulated by the ataxia-telangiectasia gene acts as a DNA damage signal leading to activation of p53 protein and limited life-span of human diploid fibroblasts. A review. Biochemistry (Mosc) 1997;62(11):1306–10 [PubMed] [Google Scholar]
- 4.Dagg RA, Pickett HA, Neumann AA, Napier CE, Henson JD, Teber ET, et al. Extensive Proliferation of Human Cancer Cells with Ever-Shorter Telomeres. Cell Rep 2017;19(12):2544–56 [DOI] [PubMed] [Google Scholar]
- 5.Viceconte N, Dheur MS, Majerova E, Pierreux CE, Baurain JF, van Baren N, et al. Highly Aggressive Metastatic Melanoma Cells Unable to Maintain Telomere Length. Cell Rep 2017;19(12):2529–43 [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144(5):646–74 [DOI] [PubMed] [Google Scholar]
- 7.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer 1997;33(5):787–91 [DOI] [PubMed] [Google Scholar]
- 8.Shay JW, Reddel RR, Wright WE. Cancer. Cancer and telomeres--an ALTernative to telomerase. Science 2012;336(6087):1388–90 [DOI] [PubMed] [Google Scholar]
- 9.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 1997;3(11):1271–4 [DOI] [PubMed] [Google Scholar]
- 10.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016;539(7627):54–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JM, Yadav T, Ouyang J, Lan L, Zou L. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep 2019;26(4):955–68 e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 1999;59(17):4175–9 [PubMed] [Google Scholar]
- 13.Min J, Wright WE, Shay JW. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev 2019;33(13–14):814–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 2009;27(12):1181–5 [DOI] [PubMed] [Google Scholar]
- 15.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011;333(6041):425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482(7384):226–31 [DOI] [PubMed] [Google Scholar]
- 17.Makinen N, Aavikko M, Heikkinen T, Taipale M, Taipale J, Koivisto-Korander R, et al. Exome Sequencing of Uterine Leiomyosarcomas Identifies Frequent Mutations in TP53, ATRX, and MED12. PLoS Genet 2016;12(2):e1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev 2010;24(12):1253–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010;140(5):678–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diplas BH, He X, Brosnan-Cashman JA, Liu H, Chen LH, Wang Z, et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun 2018;9(1):2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 2012;8(7):e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason-Osann E, Dai A, Floro J, Lock YJ, Reiss M, Gali H, et al. Identification of a novel gene fusion in ALT positive osteosarcoma. Oncotarget 2018;9(67):32868–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol 2014;21(2):167–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eid R, Demattei MV, Episkopou H, Auge-Gouillou C, Decottignies A, Grandin N, et al. Genetic Inactivation of ATRX Leads to a Decrease in the Amount of Telomeric Cohesin and Level of Telomere Transcription in Human Glioma Cells. Mol Cell Biol 2015;35(16):2818–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Napier CE, Huschtscha LI, Harvey A, Bower K, Noble JR, Hendrickson EA, et al. ATRX represses alternative lengthening of telomeres. Oncotarget 2015;6(18):16543–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosnan-Cashman JA, Yuan M, Graham MK, Rizzo AJ, Myers KM, Davis C, et al. ATRX loss induces multiple hallmarks of the alternative lengthening of telomeres (ALT) phenotype in human glioma cell lines in a cell line-specific manner. PLoS One 2018;13(9):e0204159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, Shi G, Zhang L, Li F, Jiang Y, Jiang S, et al. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci Rep 2016;6:32280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min J, Wright WE, Shay JW. Alternative lengthening of telomeres can be maintained by preferential elongation of lagging strands. Nucleic Acids Res 2017;45(5):2615–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest 2013;123(11):4918–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sramkoski RM, Pretlow TG 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim 1999;35(7):403–9 [DOI] [PubMed] [Google Scholar]
- 31.Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med 1997;3(4):402–8 [DOI] [PubMed] [Google Scholar]
- 32.Lafferty-Whyte K, Cairney CJ, Will MB, Serakinci N, Daidone MG, Zaffaroni N, et al. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene 2009;28(43):3765–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154(6):1380–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min J, Wright WE, Shay JW. Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol Cell Biol 2017;37(20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8(11):2281–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mender I, Shay JW. Telomerase Repeated Amplification Protocol (TRAP). Bio Protoc 2015;5(22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meeker AK, Gage WR, Hicks JL, Simon I, Coffman JR, Platz EA, et al. Telomere length assessment in human archival tissues: combined telomere fluorescence in situ hybridization and immunostaining. Am J Pathol 2002;160(4):1259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9(7):676–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura M, Stone RC, Hunt SC, Skurnick J, Lu X, Cao X, et al. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat Protoc 2010;5(9):1596–607 [DOI] [PubMed] [Google Scholar]
- 40.Lee H, Birren B, Lai E. Ultraviolet nicking of large DNA molecules from pulsed-field gels for southern transfer and hybridization. Anal Biochem 1991;199(1):29–34 [DOI] [PubMed] [Google Scholar]
- 41.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: URL http://www.R-project.org/. R Foundation for Statistical Computing; 2019. [Google Scholar]
- 42.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheel C, Schaefer KL, Jauch A, Keller M, Wai D, Brinkschmidt C, et al. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene 2001;20(29):3835–44 [DOI] [PubMed] [Google Scholar]
- 44.Han M, Napier CE, Frolich S, Teber E, Wong T, Noble JR, et al. Synthetic lethality of cytolytic HSV-1 in cancer cells with ATRX and PML deficiency. J Cell Sci 2019;132(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol 2010;84(8):4026–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 1995;14(17):4240–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol 2001;21(12):3862–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 2016;34(3):339–44 [DOI] [PubMed] [Google Scholar]
- 49.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 1997;91(1):25–34 [DOI] [PubMed] [Google Scholar]
- 50.Goldman F, Bouarich R, Kulkarni S, Freeman S, Du HY, Harrington L, et al. The effect of TERC haploinsufficiency on the inheritance of telomere length. Proc Natl Acad Sci U S A 2005;102(47):17119–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leman AR, Dheekollu J, Deng Z, Lee SW, Das MM, Lieberman PM, et al. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 2012;11(12):2337–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clynes D, Jelinska C, Xella B, Ayyub H, Scott C, Mitson M, et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun 2015;6:7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol 1992;225(4):951–60 [DOI] [PubMed] [Google Scholar]
- 54.Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell 2001;8(1):105–13 [DOI] [PubMed] [Google Scholar]
- 55.Osterwald S, Deeg KI, Chung I, Parisotto D, Worz S, Rohr K, et al. PML induces compaction, TRF2 depletion and DNA damage signaling at telomeres and promotes their alternative lengthening. J Cell Sci 2015;128(10):1887–900 [DOI] [PubMed] [Google Scholar]
- 56.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 2011;179(4):1608–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.