Abstract
Chronic inflammation is an established risk factor for colorectal cancer (CRC). To study reactive products of gut inflammation and redox signaling on CRC development we used untargeted adductomics to detect adduct features in pre-diagnostic serum from the EPIC-Italy cohort. We focused on modifications to Cys34 in human serum albumin (HSA), which is responsible for scavenging small reactive electrophiles that might initiate cancers. Employing a combination of statistical methods, we selected seven Cys34 adducts associated with CRC, as well as BMI (a well-known risk factor). Five adducts were more abundant in CRC cases than controls and clustered with each other, suggesting a common pathway. Since two of these adducts were Cys34 modifications by methanethiol, a microbial-human co-metabolite, and crotonaldehyde, a product of lipid peroxidation, these findings further implicate infiltration of gut microbes into the intestinal mucosa and the corresponding inflammatory response as causes of CRC. The other two associated adducts were Cys34 disulfides of homocysteine that were less abundant in CRC cases than controls and may implicate homocysteine metabolism as another causal pathway. The selected adducts and BMI ranked higher as potentially causal factors than variables previously associated with CRC (smoking, alcohol consumption, physical activity and total meat consumption). Regressions of case-control differences in adduct levels on days to diagnosis showed no statistical evidence that disease progression, rather than causal factors at recruitment, contributed to the observed differences. These findings support the hypothesis that infiltration of gut microbes into the intestinal mucosa and the resulting inflammation are causal factors for CRC.
Introduction
Colorectal cancer (CRC) is a major cause of human mortality, accounting for about nine percent of all cancer deaths (1); however, the etiology of CRC is poorly understood. Since studies of families and twins have shown that heritable genetics contribute less than 15% to CRC incidence (2,3), non-genetic factors must be important. Indeed, many studies have implicated diet and lifestyle factors with CRC risks (reviewed by (4)). Interestingly, some associations pointed to increased risks – notably, consumption of fat and red meat, smoking and alcohol use - while others suggested reduced risks, namely, consumption of fish, fish oil and fiber, plus regular exercise and intake of vitamin D, calcium and aspirin. Since most of these risk factors implicate dietary exposures, recent interest has focused on the interplay between the diet and gut microbiota as contributors to CRC (5,6). In particular, evidence is accumulating that the shift away from fiber-rich foods in the ‘Westernized diet’ has discouraged gut fermentation that enhances colonic health.
An emerging theme from this collection of risk factors is the hypothesis that CRC results from chronic promotion of gut dysbiosis “… creating a microclimate that promotes inflammation, proliferation and neoplastic progression” (5). Certainly, chronic colonic inflammation is a hallmark of inflammatory bowel disease and colitis-associated cancer, and is an established risk factor for CRC. A critical adjunct to gut inflammation is production of reactive oxygen species (ROS) by neutrophils and macrophages that are mobilized in response to infiltration of microbiota into the intestinal mucosa. Reactive oxygen species can damage DNA and thereby initiate tumors; they can react with polyunsaturated fatty acids to produce reactive carbonyl species (RCS) that modify proteins and promote cancers; and they are important modulators of redox-signaling pathways that are activated by gut inflammation (7).
Despite their potential importance to cancer causation, ROS, RCS and other reactive electrophilic products of human and microbial metabolism cannot generally be measured in vivo. This has motivated investigators to study the dispositions of reactive metabolites by monitoring adducts of these species with abundant proteins, particularly hemoglobin (Hb) and human serum albumin (HSA). Although most assays have targeted particular modifications of Hb and HSA selected a priori (8), recent work has explored untargeted avenues for characterizing adductomes at particular nucleophilic loci (9–11). Our laboratory developed an adductomics pipeline to investigate modifications at the highly nucleophilic Cys34 residue of HSA (11). We focused on Cys34, not only because it efficiently scavenges small reactive electrophiles (12), but also because its oxidation by ROS generates a host of reversible sulfoxidations that act as redox switches in homeostatic processes (13–16). Indeed, oxidation of HSA-Cys34 to the reactive sulfenic acid (Cys34-SOH) serves as an intermediate in formation of mixed Cys34-disulfides that are also sentinels of redox biology during the one-month residence time of HSA (17).
Given evidence that reactive products of gut inflammation and modulation of redox signaling pathways are potential contributors to CRC, we conducted Cys34 adductomics with archived serum from incident CRC cases and matched controls from the European Prospective Investigation into Cancer and Nutrition (EPIC) (18). This exploratory study is intended to discover discriminating adducts that can motivate hypotheses and follow-up of potentially important exposures or pathways leading to CRC. Results point to CRC associations with several adducts, some of which further implicate the gut microbiota and redox biology as potential causes.
Materials and Methods
CRC Cases and controls
Serum samples were obtained at recruitment from 95 pairs of incident CRC cases and matched controls (68 male pairs and 27 female pairs), collected between 1993 and 1997 from subjects in Turin, Italy as part of the EPIC cohort study (18). Written informed consent was obtained from all participants and the study was conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule). The study protocol was approved by an institutional review board of the Human Genetics Foundation (Turin, Italy). Controls were sampled from within the cohort (a sample of the general population) and matched by age, gender and enrollment year and season. The cohort was regularly followed up and, at diagnosis of CRC, cases were confirmed by colonoscopy and biopsy; matched controls were healthy and with few exceptions, did not undergo colonoscopies. Information related to the diet, body mass index (BMI) and lifestyle factors were obtained by questionnaire (19). Serum samples were obtained in cryostraws from the central biorepository of the International Agency for Research on Cancer (IARC; Lyon, France) where they had been stored in liquid nitrogen prior to shipment to our laboratory with further storage at −80°C for approximately two years prior to analysis. Upon processing of the serum, 59 samples had a gelled consistency, which was traced to an additive in the cryostraws (20). Because these gelled samples affected adductomic profiles, they were excluded, as were two subjects with large percentages of missing adducts, leaving 129 samples for downstream statistical analysis (57 cases and 72 controls), including 47 matched case-control pairs. Table 1 provides summary statistics for these subjects and relevant covariates (smoking, physical activity, consumption of alcohol and meat and BMI). Out of these covariates, BMI was the most different between cases and controls (nominal p-value = 0.026 from a two-sample t-test), with cases having a higher average BMI.
Table 1.
Descriptive statistics of human subjects matched by age and gender.
| Total n= | CRC cases n=57 | Controls n=72 | p-value* | |
|---|---|---|---|---|
| Gender | Male | 39 | 49 | |
| Female | 18 | 23 | ||
| Age at enrollment (y) | mean | 55.30 | 55.05 | |
| median | 57.02 | 56.4 | ||
| min | 35.48 | 35.46 | ||
| max | 64.68 | 63.58 | ||
| mean | 6.86 | - | ||
| Years to diagnosis | median | 6.99 | - | |
| min | 0.02 | - | ||
| max | 14.41 | - | ||
| BMI (kg/m2) | mean | 27.06 | 25.52 | 0.026 |
| median | 26.71 | 25.01 | ||
| min | 19.68 | 18.73 | ||
| max | 40.68 | 33.57 | ||
| Smoking status | current | 10 | 17 | |
| former | 25 | 26 | ||
| never | 17 | 25 | ||
| NA | 5 | 4 | ||
| Alcohol consumption (mL/day) | mean | 21.94 | 19.78 | 0.585 |
| median | 13.47 | 11.77 | ||
| min | 0.0 | 0.0 | ||
| max | 80.57 | 93.54 | ||
| Physical activity† | active | 9 | 11 | |
| moderately active | 10 | 20 | ||
| moderately inactive | 23 | 20 | ||
| inactive | 10 | 17 | ||
| NA | 5 | 4 | ||
| Total meat consumption(g/day) | mean | 80.24 | 72.76 | 0.386 |
| median | 75.30 | 63.45 | ||
| min | 2.60 | 0.0 | ||
| max | 189 | 201.3 | ||
| Total vegetable consumption¶ (g/day) | mean | 259.1 | 255.6 | 0.849 |
| median | 227.9 | 241.5 | ||
| min | 74.5 | 80.7 | ||
| max | 739.7 | 593.6 |
NA – not available
Nominal p-values from a two-sided t-test.
For definitions and validation see (50).
Sum of: leafy vegetables (raw and cooked), other vegetables, tomatoes (raw and cooked), root vegetables, cabbages, mushrooms, onion, garlic, mixed salad, mixed vegetables, and legumes.
Chemicals and reagents
With the following exceptions, all of the chemicals used in this study were the same as described previously (11). For the current investigation, sodium thiomethoxide (≥ 95%) and iodine (≥ 99%), were from Sigma-Aldrich (St. Louis, MO), and hydrogen peroxide (30 wt. % aqueous solution) and formic acid (Optima, LCMS grade), were from Fisher Scientific (Pittsburgh, PA).
Sample processing and nLC-HRMS data acquisition
Sample processing and analysis by nano-liquid chromatography-high resolution mass spectrometry (nLC-HRMS) were performed as previously described (11). The order of analyses was randomized except that each case-control pair was analyzed on the same day, also with random order. Briefly, HSA was purified (≥ 75 %) by precipitating other serum proteins and residual Hb with 60% methanol. HSA was digested with trypsin at 37°C with high-pressure cycling for 30 min (NEP2320, Pressure Biosciences Inc., South Easton, MA) and without prior reduction of disulfide bonds. Adducts were located on the triply charged ‘T3 peptide’ (21ALVLIAFAQYLQQC34PFEDHVK41, m/z 811.7593). Prior to nLC-HRMS, 1 μL of an internal standard, consisting of the isotopically labeled T3 peptide modified at Cys34 with iodoacetamide (IAA-iT3, 20 pmol/μL), was added to normalize data for instrument performance. One microliter of each digest was injected into the nLC-HRMS, consisting of a Dionex Ultimate® 3000 nanoflow LC system equipped with a Dionex monolithic column (100 μm i.d. × 25 cm) and connected via a Flex Ion nano-ESI source to an LTQ Orbitrap XL hybrid mass spectrometer (Thermo Scientific, Sunnyvale, CA) that was operated in positive-ion mode. After duplicate injections of a sample, a blank sample was injected to reduce carryover effects, and after analysis of three samples the LC column was washed with 1 μL of a solution containing 80% acetonitrile, 10% acetic acid, 5% DMSO, and 5% water to stabilize the chromatography.
Adducts were located on the T3 peptide based on the monoisotopic mass (MIM) within 10 ppm as described previously (11). By performing nLC-HRMS in data-dependent mode, the MS2 spectra for all triply charged precursor ions were first interrogated for b+- and y2+-series ions that are signatures of the T3 peptide and its modifications. Spectra displaying the requisite fragment ions were designated as putative T3 modifications. The corresponding precursor ions were then extracted from the total ion chromatogram (TIC) to obtain a MIM for each adduct feature. To normalize peak areas for the amount of HSA in each tryptic digest, the MIM was also extracted for the doubly charged HSA peptide (42LVNEVTEFAK51, m/z, 575.3111) adjacent to T3 and referred to as the ‘housekeeping peptide’ (HKP). As shown previously (11) the peak area ratio (PAR), representing the ratio of the adduct-peak abundance to the HKP peak abundance is a robust measure of the adduct concentration. Peaks representing the selected ion chromatogram (SIC) for the internal standard (IAA-iT3) were used to normalize for instrument performance. Peak picking and integration were performed using the Xcalibur Processing Method (version 3.0, Thermo Fisher Scientific, Waltham, MA) based on the average MIMs and retention times. Peak integration employed the Genesis algorithm after normalizing for instrument performance via iT3-IAA. Added masses relative to the Cys34 thiolate ion were estimated as Madduct = (m/zadduct-m/zT3-peptide)*3 + 1.0078, where m/zadduct and m/zT3-peptide are the observed m/z values for the triply-charged MIMs of a given precursor ion for an adduct and the unmodified T3 peptide, respectively, and 1.0078 is the mass of a hydrogen atom. All data processing utilized in-house software written in R.
Synthesis of reference standards
The identities of several adducts were verified by synthetic reference standards that had been prepared previously (11,21,22). A new reference standard for the Cys34 S-methanethiol adduct was prepared as follows. Two microliters of 25 mM sodium thiomethoxide were diluted with 0.25 mL of water with and without 10 μL of 1 mM hydrochloric acid and incubated at room temperature for 1 h. Purified HSA from 15 μL of serum from a volunteer subject was diluted with 0.2 mL of digestion buffer and mixed with the thiomethoxide solution plus 1 μL of 30 % H2O2 and 0.5 μL of 35 mM iodine. A negative control was also prepared with HSA from an additional 15 μL of serum that was processed with all reagents except sodium thiomethoxide. After incubation at room temperature with constant agitation for 24 h the reagents were removed with 30K MWCO spin columns (Millipore Sigma, MA) and the modified HSA was digested with trypsin and analyzed by nLC-HRMS as described above. The monoisotopic mass for the Cys34 S-methanethiol adduct (m/z 827.0890) was extracted from TICs using a mass tolerance of 5 ppm.
Statistical analysis
Duplicate injections were averaged for each adduct peak, ignoring missing values, in order to reduce technical variation. Eight adducts were detected in only one or two serum samples and were excluded from further analyses. Using a cutoff of 15% for missing values across adducts, two subjects were excluded. Missing values were imputed using the k-nearest-neighbor method (23), with k = 5 adduct neighbors. Data were normalized using the Bioconductor R package ‘scone’ (24), which employs linear regression models on scaled and logged feature abundances to adjust for various combinations of factors of unwanted variation (25). The ‘scone’ package then evaluates each candidate normalization scheme with metrics that gauge the removal of unwanted variation and retention of wanted variation (e.g., case-control status) to help users select an appropriate normalization scheme. The top-ranking normalization scheme according to ‘scone’ used DESeq scaling (26) and adjusted for unwanted variation due to digested HSA and instrument performance. Here, ‘digested HSA’ was quantified by the abundance of the HKP and ‘instrument performance’ was indicated by the drift in abundance of the internal standard (iT3-IAA) peak over time. All quantified T-3 peptides were clustered using the partitioning around medoids (PAM) method (k=6, ‘pam’ function in R) using Spearman correlations on the normalized abundances.
A combination of regression and classification methods was used to select adducts that were associated with CRC cases and controls. Since BMI was greater on average in CRC cases than controls (Table 1), this variable was also investigated. (Due to missing values of BMI, 5 cases and 7 controls were excluded, leaving 117 subjects for analysis). First, the following multivariate linear regression model was fitted:
| (1) |
where Yij represents logged and DESeq-scaled abundances for the jth adduct (or BMI) in the ith subject Xcase and Xsex are binary indicators, Xage is a continuous variable, XHKP is the vector of housekeeping peptide abundances, XIS is the vector of internal standard abundances, and εi is a random error term for the ith subject. The nominal p-value corresponding to the coefficient β1 was used to rank each variable by its association with case-control status. The mean case/control fold-change in adduct levels was calculated as exp(β1), and β1 was used to represent the difference in average BMI between cases and controls, adjusting for sex and age.
Second, a regularized logistic regression (LASSO) (27) of CRC case-control status on normalized adduct abundances and BMI along with sex and age (matching variables), was performed to find groups of variables associated with CRC. The logistic LASSO regression was performed on 500 bootstrapped datasets to provide stability (28), using the number of times a given adduct was selected in 500 iterations as a measure of its importance. A concordance plot was used to evaluate agreement between ranked p-values from Model (1) and the bootstrapped LASSO variable importance measures. Variables that were top-ranking for both methods were selected.
Finally, variable importance measures from random forest classification of CRC case-control status were used to provide a nonlinear index of association (29). A random forest of 500 trees was used to predict case-control status based on adduct abundances and BMI, and all variables were ranked in importance based on their mean decrease in Gini index (30,31). Variables with large increases in random forest variable importance were also considered for addition to the list of selected variables.
To investigate factors that could potentially drive relationships between selected adducts and CRC status, the covariates BMI (kg/m2), smoking (current vs. former/never), alcohol consumption (g/d), physical activity (active/moderately active vs. moderately inactive/inactive) and total meat consumption (g/d) were evaluated because these variables have been implicated as risk factors for CRC (4). First, to determine whether the above covariates might have influenced selection of adducts in our ensemble variable-selection method, a random forest classifier was used to rank all selected adducts and covariates by their importance in classifying CRC case status (31–33). Then, to obtain additional information about potential associations between adducts and covariates - regardless of case-control status - random forest classifiers were used to rank all measured adducts in terms of their predictive power for each particular covariate.
Because CRC cases and matched controls were evaluated more than 14 years after recruitment, we tested associations between adduct abundances and days (from recruitment) to diagnosis to discern whether they represent potentially causal effects or reactive effects of disease progression (34). If a significant linear trend in the log fold-change for a given feature were detected with increasing days to diagnosis, the adduct would be regarded as potentially reactive.
Results
Adducts detected
A total of 55 modifications to the T3 peptide were detected in CRC cases and controls (Supplementary Table S1). Peak abundances covered a 2,250-fold range (PARx1000: 0.09 – 203). Based on ANOVA of duplicate injections across blood specimens for the 46 adducts with sufficient data (Supplementary Table S2), the median intraclass correlation coefficient (ICC) was 0.777 (range: 0.345 – 0.982), indicating that technical variation typically accounted for 23% of the total variance of adduct abundances. Coefficients of variations (CVs) across duplicate injections for adducts ranged from 0.134 to 0.758 with a median value of 0.283, consistent with previous applications of the assay (11).
Accurate masses for 51 adducts led to reasonable elemental compositions added to the Cys34-S− ion within 3 ppm of theoretical values from −46 Da to 510 Da (negative added masses refer to deletions and truncations). A subset of 30 modifications to the T3 peptide was annotated including: truncations [e.g. 796.43 (Cys34→Gly) and 805.76 (Cys34→oxoalanine or formylglycine)], Cys34 sulfoxidation products (816.42, 822.42 and 827.76, representing addition of 1, 2 and 3 oxygens to Cys34, respectively), RCS (i.e., crotonaldehyde, 835.11) and a host of mixed Cys34 disulfides, notably those of methanethiol (827.09), Cys (851.43), homocysteine (hCys, 856.10), CysGly (870.44), GluCys (894.44) and glutathione (913.45). About one third of the T3 adducts were unannotated, including several whose MS2 spectra indicated T3 modifications at sites other than Cys34, including methylation (816.43).
We had previously detected 43 of these adducts in at least one of four studies with serum/plasma from diverse populations (11,22,35,36), and 17 of these adducts were common to all four studies. This points to a pool of modifications of the T3-peptide that arises from a set of precursor molecules, including ROS, RCS and small thiols from metabolic pathways involving common nutrients. Twelve adducts were unique to the current study, and none of these modifications was annotated (Supplementary Table S1). The MS2 spectra and SICs/MS1 spectra of these 12 new adducts are reproduced in Supplementary Figures S1 and S2, respectively.
Adducts associated with CRC
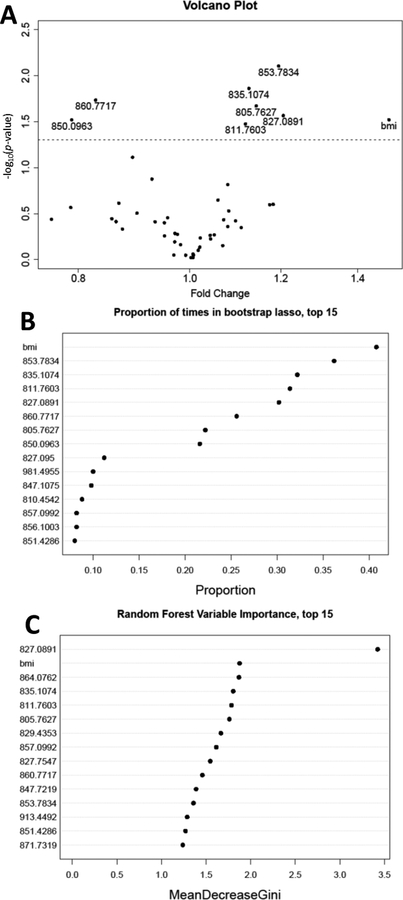
Results of our variable selection strategy are summarized in Figure 1. The concordance of linear regression (Model 1) and bootstrapped LASSO logistic regression was 100% for the 8 highest ranked variables, including 7 adducts and BMI (Figure 1 A and 1B). In addition to BMI, five of the selected adducts were present at higher levels in CRC cases, namely, 853.78 (unknown), 835.11 (crotonaldehyde), 805.76 (Cys34→oxoalanine or formylglycine), 827.09 (S-methanethiol) and 811.76 (a ‘T3-labile adduct’ detected with the same MIM as the T3 peptide but a different retention time, suggesting truncation of the adduct in the ESI source), while two adducts representing hCys disulfides were more abundant in controls, namely, 860.77 [S-hCys (+CH3)] and 850.10 S-hCys (-H2O)]. Many of the adducts and BMI that had been selected by both linear regression and LASSO logistic regression were among the top ranked variables determined by random forest (Figure 1C). In fact, the S-methanethiol adduct was the only adduct to demonstrate a marked increase in Gini index by random forest (Figure 1C).
Figure 1.
Regression and classification methods used to measure associations between CRC status and modifications to the T3 peptide or BMI in 57 CRC cases and 72 controls. A) Volcano plot of nominal p-values for case-control status in multivariate linear regression of each adduct and BMI in Model (1) (the dashed line represents a nominal p-value of 0.05); B) proportion of times that a given adduct or BMI was selected by regularized logistic regression (LASSO) of CRC case-control status; C) ranked variable importance measures from random forest classification of case-control status (top 15 variables).
Clusters of adducts and BMI resulting from the PAM algorithm are shown in Supplementary Figure S3 (k=6 resulted in the highest average silhouette width among k=2,…, 8). Of the six clusters identified, the most informative was cluster 2, which included all of the five adducts that were more abundant in CRC cases than controls (from Figure 1A). Other adducts in this cluster included the Cys34 sulfonic acid (827.75), the T3 dimer (811.42) and two unknowns (847.77 and 815.44). The two selected adducts that were less abundant in CRC cases were disulfides of hCys that had been either methylated (860.77) or dehydrated (850.10). These adducts were grouped with each other and with the parent hCys disulfide (856.10) in cluster 5 of Supplementary Figure S3. Interestingly, BMI did not cluster with any of the seven adducts associated with CRC. Spearman correlations between these selected adducts and BMI were ≤ |0.11| except for the dehydration product of hCys (850.10), which was −0.21.
Effects of covariates on associations between adducts and CRC
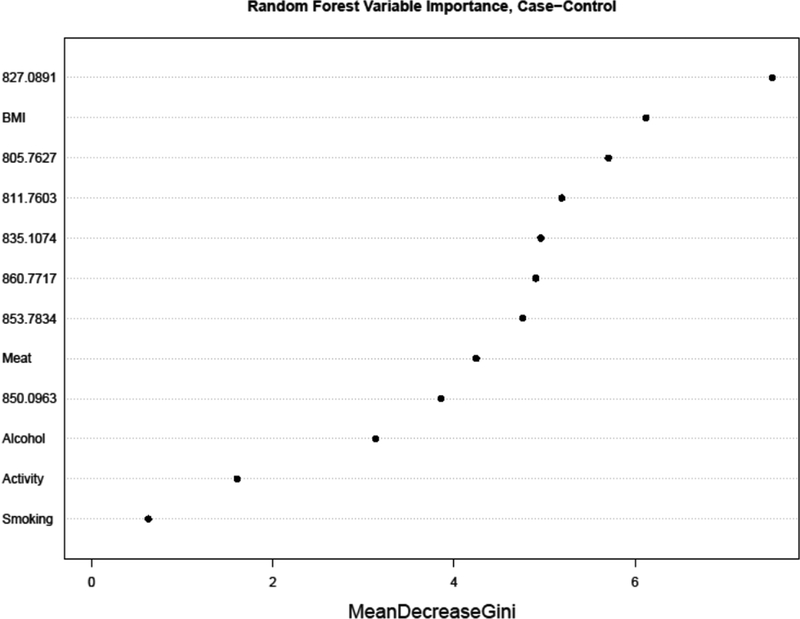
Variables ranked for importance by a random forest classification of case-control status are shown in Figure 2 to compare the selected adducts (Figure 1) and covariates previously associated with CRC (BMI, smoking, alcohol consumption, physical activity and total meat consumption). Aside from BMI (second ranked), the other covariates ranked below the selected adducts as classifiers of CRC case status, suggesting that these covariates are probably not responsible for the potential associations shown in Figure 1. When random forest models were constructed to investigate the variable importance of all 46 adducts as predictors of each covariate individually, the seven selected adducts were not top-ranking for most of the covariates, further suggesting that the covariates are not driving the associations shown in Figure 1. Nonetheless, there were some interesting results (Supplementary Figure S4). For example, among smokers the variables with greatest importance were adducts of acrylonitrile (829.43) and ethylene oxide (826.43) (Supplementary Figure S4–A), both of which had been previously associated with smoking in our adductomics pipeline (11). Also, the top three variables for BMI were Cys34 sulfoxidation products [(-H2+O), 816.42; (+CH3O2), 827.10; (+HO2), 822.42] (Supplementary Figure S4–B). Top-ranking adducts for total meat consumption included two unknowns (847.77 and 815.44) and the Cys34→Gly truncation (796.43) (Supplementary Figure S4–C); top-ranking adducts for physical activity included three unknowns (981.50, 894.13 and 879.13) plus the T-3 labile adduct (811.76) and S-Cys (NH2→OH) (851.76) (Supplementary Figure S4–D); and top-ranking adducts for alcohol consumption included S-Cys (NH2→OH) (851.76), S-hCys (856.10) and S-glutathione (913.45) (Supplementary Figure S4–E). Thus, of the seven adducts selected as associated with CRC (Figure 1) only the T-3 labile adduct (811.76) had high-ranking variable importance for any of the tested covariates (i.e., physical activity and alcohol consumption), further suggesting that the underlying CRC associations were largely free of confounding by these variables.
Figure 2.
Ranking of adducts and covariates by random forest variable importance measures for CRC status in 57 CRC cases and 72 controls. (‘Activity’ represents physical activity and ‘Meat’ represents total meat consumption).
Discussion
This is the first study to apply our adductomics pipeline to prospective analysis of CRC or cancer generally. We had validated our adductomics methodology with archived serum/plasma from healthy smoking and nonsmoking subjects (11) and subsequently applied it to populations with and without high exposures to indoor combustion products (36) or benzene (22), and to subjects with and without lung or heart disease (35). This led to measurement of over 75 adducts, several of which were significantly associated with particular exposures or diseases, notably Cys34 modifications of reactive oxygen and carbonyl species and disulfides of small thiols derived from redox processes. This combination of results points to separate windows that Cys34 modifications provide for viewing exposure-specific electrophiles and global characteristics of the redox proteome (14).
Using an ensemble of regression and classification methods developed initially for untargeted metabolomics (20,31), we selected seven adducts as potentially associated with CRC in 57 cases and 72 control subjects from the EPIC cohort. The fact that our adductomics pipeline had previously detected all seven of the selected adducts in various human populations (11,22,35,36) suggests that the reactive precursors of these modifications represent common exposures of everyday life that are modulated by pathways leading to CRC.
Using results from two large cohort studies, Morikawa et al. argued that high BMI increased CRC risk through a combination of obesity, insulin and insulin-like growth factor-1 that modulated the Wnt-β-catenin signaling pathway in unspecified ways (37). Based on a review of recent literature, Liu et al. concluded that the Wnt-β-catenin signaling pathway was modulated by production of ROS (38). Thus, it is interesting that the top-ranking adducts in terms of random forest variable importance for BMI were all sulfoxidation products of Cys34 (Supplementary Figure S4–B) that are formed by Cys34 reactions with ROS. However, since these Cys34 sulfoxidation products were not among the seven adducts selected as associated with CRC (Figure 1), it appears that multiple pathways are involved in the etiology of this cancer.
Five of the seven adducts associated with CRC incidence were more abundant in cases than controls, with fold-changes between 1.11 and 1.20 (Figure 1A). Interestingly, all five of these adducts clustered with each other (Figure 2, cluster 2), suggesting a common pathway. Yet, the sources and biochemistry underlying production of these adducts are varied. Perhaps the most informative of these five adducts is the S-methanethiol modification of Cys34 (827.09) that was also observed in two previous studies (11,36). This modification results from oxidation of Cys34 to the sulfenic acid (Cys34-SOH) which subsequently binds with circulating methanethiol, with loss of H2O, to form the corresponding Cys34 disulfide (17). Methanethiol is a product of microbial-human co-metabolism that is mediated by the gut microbiota via catabolism of methionine and/or methylation of hydrogen sulfide (39). Interestingly, methanethiol was found to be more abundant in feces from CRC patients compared to controls (40). Thus, we speculate that the Cys34 adduct of methanethiol is a biomarker of human enteric bacteria and that the increased abundance of this adduct in CRC cases further implicates the gut microbiota as a risk factor, consistent with formal hypotheses (5,6). It is also worth noting that Bae et al. reported a positive association between CRC risk in postmenopausal women and plasma trimethylamine-N-oxide (TMAO), which is another human/microbial cometabolite (41).
Another Cys34 adduct with mechanistic significance is the crotonaldehyde modification (835.11). Crotonaldehyde is a reactive α, β-unsaturated aldehyde produced by ROS oxidation of membrane lipids (42). We had previously shown that workers exposed to high levels of benzene – a strong promoter of ROS - had elevated serum levels of this crotonaldehyde adduct compared to controls (22). In their review of redox biology and CRC, Liu et al. linked lipid peroxidation with COX2 expression and two subsequent pathways towards CRC, one involving production of prostaglandins and the other involving reduced degradation of β-catenin (38). The fact that the crotonaldehyde adduct (835.11) clustered with the S-methanethiol adduct (827.09) (cluster 2 of Supplementary Figure S3), lends credibility to the hypothesis that invasion of gut microbiota into the intestinal mucosa initiates a chain of events involving an inflammatory response followed by production of ROS, RCS and subsequent damage to DNA and proteins as well as modulation of redox-signaling pathways (7).
Potential origins of the other three adducts in cluster 2 (Supplementary Figure S3) that are associated with CRC are more difficult to characterize. Adduct 805.76 represents conversion of Cys34 to oxoalanine or formylglycine with a mass loss of 18 Da. While oxidative cleavage of the sulfhydryl group from protein cysteine residues to produce dehydroalanine (−34 Da) and serine (−16 Da) has been reported (43,44), we have not found evidence of modifications yielding the observed mass shift of −18 Da. It also seems unlikely that 805.76 represents conversion to Cys34 to formylglycine by human or microbial sulfatase metabolism because Cys34 is not embedded in the sequence motif (CXPXR) recognized by sulfatases (45). Regarding unknown adduct 853.78, we had previously detected this modification in two studies and suspected that it was a Cys34 disulfide of a small thiol because it disappeared after treatment of HSA with TCEP, a reagent that selectively cleaves disulfide bonds (11,36). However, none of the putative elemental compositions that include a sulfur atom (C7H11S, C5H7N2S or C6H9NS) resulted in a plausible added mass relative to that observed (127.077 Da). Based on analysis of MS2 fragmentation spectra it appears that the same precursor ion (853.7834) can generate two different sets of fragment ions that suggest rearrangement during collision induced dissociation in the mass spectrometer. And finally, the T3-labile adduct (811.76) appears to represent a T3 modification(s) that is cleaved in the ESI source to yield the unadducted T3 peptide, albeit with a different retention time. Although this modification has been observed in all previous studies, we have no information regarding its identity.
The other two adducts potentially associated with CRC in our samples were Cys34 disulfides of hCys that were either methylated (860.77) or dehydrated (850.10) at another site on the T3 peptide. Unlike the other five associated adducts, these hCys modifications were less abundant in CRC cases than controls (Figure 1A) and clustered with each other and the unmodified Cys34-hCys disulfide (856.10) (cluster 5 of Supplementary Figure S3). As a key intermediate in one-carbon metabolism, hCys is remethylated to produce methionine, and subsequently S-adenosylmethionine (SAM), which plays an important role in DNA methylation that has been linked to CRC and other cancers (46). However, recent meta-analyses of many case-control studies (46,47) and a combination of case-control and cohort studies (48) point to CRC risks that increase with hCys blood concentrations, which is the reverse of what we observed. However, estimated effect sizes were smaller in cohorts than case-control studies, and numerous dietary and lifestyle factors increased CRC risks (reduced intake of fiber, methionine, vitamin B9 or folate, and vitamin B6, and increased intake of B12 intake, alcohol, and smoking) (48). Also, hCys levels have been shown to increase with age greater (but not less than) 65 y (49) and the mean age across 16 studies that linked CRC with increasing hCys by Xu et al. (47) was 61.4 y (SD = 3.7 y). This indicates that many subjects in the meta-analysis (47) were greater than 65 y and this may have contributed to increased levels of hCys. In contrast, the mean age at phlebotomy of the 57 cases in our study was 55.3 y. Thus, although it is difficult to entirely reconcile our findings regarding adducts 860.77 and 850.10 with the current epidemiologic literature, we cannot rule out their connections to a potentially causal pathway involving hCys metabolism.
Finally, to determine whether modulation in levels of selected adducts was the result of disease progression rather than a causal factor, we examined the relationships between log-fold-changes of adduct abundances of CRC case-control pairs and days (from recruitment) to diagnosis (34). Results are presented in Supplementary Figure S5 as individual plots for the selected adducts. In each case, the p-value for slope of the linear relationship was large indicating that there is little statistical evidence supporting the notion that disease progression (reverse causality) rather than a causal pathway(s) lead to differential adduct abundances between cases and matched controls.
Our study had several limitations. The initial sample size was small (95 cases and matched controls) and then was reduced due to exclusion of gelled samples from cryostraw-storage and missing information about BMI in some subjects. Also, we had no information regarding aspirin use and histories of CRC in families of cases, two factors that have been associated with CRC (4). The storage of biological specimens for decades can lead to artifacts but in our study all specimens were collected within four years and cases and controls were matched by year of enrollment to minimize potential effects of sample storage on case-control differences. Three of the seven adducts selected for associations with CRC were unannotated and, therefore, of limited utility in discovery of causal factors. Another limitation was our inability to examine possible connections between adducts and advanced neoplasms (precursors of CRC) and advanced stage vs. early stage cancers.
In summary, we used untargeted adductomics to detect 51 adduct features in HSA from incident cases and controls from the EPIC cohort of which seven were found to be associated with CRC (Figure 1). Two adducts were more abundant in CRC cases than controls and represent Cys34 modifications by methanethiol and crotonaldehyde that jointly implicate infiltration of gut microbes into the intestinal mucosa and the corresponding inflammatory response as potential causes of CRC. Two other associated adducts were disulfides of hCys that were both less abundant in CRC cases than controls and may implicate hCys metabolism as a contributor to CRC. These adducts should be targeted for validation in independent samples of CRC cases and controls and should motivate mechanistic hypotheses regarding the underlying causal exposures and pathways. For example, the methanethiol/crotonaldehyde adducts could be measured in CRC cases and controls in conjunction with metagenomics of fecal samples to determine whether particular strains of microbiota may be responsible for the observed effects. It would also be interesting to determine whether there are associations between Cys34 adducts and DNA adducts or mutations in oncogenes or tumor suppressor genes in CRC cases.
Supplementary Material
Statement of significance.
Infiltration of gut microbes into the intestinal mucosa and the resulting inflammation are causal factors for colorectal cancer.
Acknowledgements
Financial support for this work was provided from the U.S. National Institutes of Health through grants R33CA191159 from the National Cancer Institute (S. Rappaport, C. Schiffman, S. Dudoit) and P42ES04705 from the National Institute for Environmental Health Sciences (S. Rappaport) and grant agreement 308610-FP7 from the European Commission (Project Exposomics) (P. Vineis, S. Rappaport). The authors appreciate the assistance of Kelsi Perttula and William Edmands, who extracted serum from the cryostraws, Anthony Iavarone who assisted with mass spectrometry, and Carlotta Sacerdote who provided information regarding covariates.
Abbreviations list.
- BMI
body mass index
- CRC
colorectal cancer
- EPIC
European Prospective Investigation into Cancer and Nutrition
- ESI
electrospray ionization for mass spectrometry
- Hb
hemoglobin
- HSA
human serum albumin
- HKP
housekeeping peptide (adjacent to T3)
- IAA
iodoacetamide
- IAA-iT3
carboxyamidomethylated Cys34 adduct of iT3
- iT3
isotopically-modified T3
- IARC
International Agency for Research on Cancer
- LASSO
least absolute shrinkage and selection operator
- MIM
monoisotopic mass
- nLC-HRMS
nano-liquid chromatography high resolution mass spectrometry
- PAR
ratio of the adduct-peak abundance to the HKP peak abundance
- PAM
partitioning around medoids method
- RCS
reactive carbonyl species
- ROS
reactive oxygen species
- SAM
S-adenosylmethionine
- SIC
selected ion chromatogram
- T3
third largest peptide of HSA in tryptic digests
- TCEP
tris(2-carboxyethyl)phosphine
- TIC
total ion chromatogram
- TMAO
trimethylamine-N-oxide
Footnotes
Conflict of interest disclosure statement: The authors declare no potential conflicts of interest.
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66(4):683–91 doi 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Hemminki K, Czene K. Attributable risks of familial cancer from the Family-Cancer Database. Cancer Epidemiol Biomarkers Prev 2002;11(12):1638–44. [PubMed] [Google Scholar]
- 3.Rappaport SM. Genetic Factors Are Not the Major Causes of Chronic Diseases. PLoS One 2016;11(4):e0154387 doi 10.1371/journal.pone.0154387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet 2016;7(3):105–14. [PMC free article] [PubMed] [Google Scholar]
- 5.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 2016;13(12):691–706 doi 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vipperla K, O’Keefe SJ. Diet, microbiota, and dysbiosis: a ‘recipe’ for colorectal cancer. Food & function 2016;7(4):1731–40 doi 10.1039/c5fo01276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guina T, Biasi F, Calfapietra S, Nano M, Poli G. Inflammatory and redox reactions in colorectal carcinogenesis. Ann N Y Acad Sci 2015;1340:95–103 doi 10.1111/nyas.12734. [DOI] [PubMed] [Google Scholar]
- 8.Rubino FM, Pitton M, Di Fabio D, Colombi A. Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom Rev 2009;28(5):725–84 doi 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, et al. Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Mol Cell Proteomics 2011;10(3):M110 004606 doi 10.1074/mcp.M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlsson H, von Stedingk H, Nilsson U, Tornqvist M. LC-MS/MS screening strategy for unknown adducts to N-terminal valine in hemoglobin applied to smokers and nonsmokers. Chem Res Toxicol 2014;27(12):2062–70 doi 10.1021/tx5002749. [DOI] [PubMed] [Google Scholar]
- 11.Grigoryan H, Edmands W, Lu SS, Yano Y, Regazzoni L, Iavarone AT, et al. Adductomics Pipeline for Untargeted Analysis of Modifications to Cys34 of Human Serum Albumin. Anal Chem 2016;88(21):10504–12 doi 10.1021/acs.analchem.6b02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabbioni G, Turesky RJ. Biomonitoring Human Albumin Adducts: The Past, the Present, and the Future. Chem Res Toxicol 2017;30(1):332–66 doi 10.1021/acs.chemrestox.6b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Go YM, Jones DP. Redox biology: interface of the exposome with the proteome, epigenome and genome. Redox biology 2014;2:358–60 doi 10.1016/j.redox.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Go YM, Jones DP. The redox proteome. J Biol Chem 2013;288(37):26512–20 doi 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R. Sulfenic acid in human serum albumin. Amino acids 2007;32(4):543–51. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe H, Imafuku T, Otagiri M, Maruyama T. Clinical Implications Associated With the Posttranslational Modification-Induced Functional Impairment of Albumin in Oxidative Stress-Related Diseases. Journal of pharmaceutical sciences 2017;106(9):2195–203 doi 10.1016/j.xphs.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Nagumo K, Tanaka M, Chuang VT, Setoyama H, Watanabe H, Yamada N, et al. Cys34-cysteinylated human serum albumin is a sensitive plasma marker in oxidative stress-related chronic diseases. PLoS One 2014;9(1):e85216 doi 10.1371/journal.pone.0085216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr 2002;5(6B):1113–24 doi 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 19.Chajes V, Jenab M, Romieu I, Ferrari P, Dahm CC, Overvad K, et al. Plasma phospholipid fatty acid concentrations and risk of gastric adenocarcinomas in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Am J Clin Nutr 2011;94(5):1304–13 doi ajcn.110.005892 [pii] 10.3945/ajcn.110.005892. [DOI] [PubMed] [Google Scholar]
- 20.Perttula K, Schiffman C, Edmands WMB, Petrick L, Grigoryan H, Cai X, et al. Untargeted lipidomic features associated with colorectal cancer in a prospective cohort. BMC cancer 2018;18(1):996 doi 10.1186/s12885-018-4894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigoryan H, Li H, Iavarone AT, Williams ER, Rappaport SM. Cys34 adducts of reactive oxygen species in human serum albumin. Chem Res Toxicol 2012;25(8):1633–42 doi 10.1021/tx300096a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigoryan H, Edmands WMB, Lan Q, Carlsson H, Vermeulen R, Zhang L, et al. Adductomic signatures of benzene exposure provide insights into cancer induction. Carcinogenesis 2018;39(5):661–8 doi 10.1093/carcin/bgy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, et al. Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17(6):520–5. [DOI] [PubMed] [Google Scholar]
- 24.Cole M, Risso D. Scone: Single Cell Overview of Normalized Expression data.
- 25.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32(9):896–902 doi 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106 doi 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibshirani R Regression Shrinkage and Selection via the Lasso. J R Stat Soc Ser B 1996;58:267–88. [Google Scholar]
- 28.Bach FR. BoLASSO: Model Consistent Lasso Estimation through the Bootstrap. New York, NY: ACM Press; 2008. [Google Scholar]
- 29.Liaw A, Wiener M. Classification and Regression by Random Forest. R News 2002;2:18–22. [Google Scholar]
- 30.Calle ML, Urrea V. Letter to the Editor: Stability of Random Forest importance measures. Briefings in bioinformatics 2011;12:86–9 doi 10.1093/bib/bbq011. [DOI] [PubMed] [Google Scholar]
- 31.Petrick LM, Schiffman C, Edmands WMB, Yano Y, Perttula K, Whitehead T, et al. Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatric acute lymphoblastic leukemia and potential effects of early-life nutrition. Cancer letters 2019;452:71–8 doi 10.1016/j.canlet.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneeweiss S, Eddings W, Glynn RJ, Patorno E, Rassen J, Franklin JM. Variable Selection for Confounding Adjustment in High-dimensional Covariate Spaces When Analyzing Healthcare Databases. Epidemiology 2017;28(2):237–48 doi 10.1097/EDE.0000000000000581. [DOI] [PubMed] [Google Scholar]
- 33.Lu M, Sadiq S, Feaster DJ, Ishwaran H. Estimating Individual Treatment Effect in Observational Data Using Random Forest Methods. J Comput Graph Stat 2018;27(1):209–19 doi 10.1080/10618600.2017.1356325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perttula K, Edmands WM, Grigoryan H, Cai X, Iavarone AT, Gunter MJ, et al. Evaluating Ultra-long-Chain Fatty Acids as Biomarkers of Colorectal Cancer Risk. Cancer Epidemiol Biomarkers Prev 2016;25(8):1216–23 doi 10.1158/1055-9965.EPI-16-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S, Grigoryan H, Edmands WMB, Dagnino S, Sinharay R, Cullinan P, et al. Cys34 Adductomes Differ between Patients with Chronic Lung or Heart Disease and Healthy Controls in Central London. Environmental science & technology 2018;52(4):2307–13 doi 10.1021/acs.est.7b05554. [DOI] [PubMed] [Google Scholar]
- 36.Lu SS, Grigoryan H, Edmands WM, Hu W, Iavarone AT, Hubbard A, et al. Profiling the Serum Albumin Cys34 Adductome of Solid Fuel Users in Xuanwei and Fuyuan, China. Environmental science & technology 2017;51(1):46–57 doi 10.1021/acs.est.6b03955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morikawa T, Kuchiba A, Lochhead P, Nishihara R, Yamauchi M, Imamura Y, et al. Prospective analysis of body mass index, physical activity, and colorectal cancer risk associated with beta-catenin (CTNNB1) status. Cancer research 2013;73(5):1600–10 doi 10.1158/0008-5472.CAN-12-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Liu X, Zhang C, Zhu H, Xu Q, Bu Y, et al. Redox Imbalance in the Development of Colorectal Cancer. J Cancer 2017;8(9):1586–97 doi 10.7150/jca.18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He X, Slupsky CM. Metabolic fingerprint of dimethyl sulfone (DMSO2) in microbial-mammalian co-metabolism. J Proteome Res 2014;13(12):5281–92 doi 10.1021/pr500629t. [DOI] [PubMed] [Google Scholar]
- 40.Ishibe A, Ota M, Takeshita A, Tsuboi H, Kizuka S, Oka H, et al. Detection of gas components as a novel diagnostic method for colorectal cancer. Ann Gastroenterol Surg 2018;2(2):147–53 doi 10.1002/ags3.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bae S, Ulrich CM, Neuhouser ML, Malysheva O, Bailey LB, Xiao L, et al. Plasma choline metabolites and colorectal cancer risk in the Women’s Health Initiative Observational Study. Cancer research 2014;74(24):7442–52 doi 10.1158/0008-5472.CAN-14-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aldini G, Dalle-Donne I, Facino RM, Milzani A, Carini M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Med Res Rev 2007;27(6):817–68 doi 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]
- 43.Jeong J, Jung Y, Na S, Jeong J, Lee E, Kim MS, et al. Novel oxidative modifications in redox-active cysteine residues. Mol Cell Proteomics 2011;10(3):M110 000513 doi 10.1074/mcp.M110.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HJ, Ha S, Lee HY, Lee KJ. ROSics: chemistry and proteomics of cysteine modifications in redox biology. Mass Spectrom Rev 2015;34(2):184–208 doi 10.1002/mas.21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Appel MJ, Bertozzi CR. Formylglycine, a post-translationally generated residue with unique catalytic capabilities and biotechnology applications. ACS Chem Biol 2015;10(1):72–84 doi 10.1021/cb500897w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang D, Wen X, Wu W, Guo Y, Cui W. Elevated homocysteine level and folate deficiency associated with increased overall risk of carcinogenesis: meta-analysis of 83 case-control studies involving 35,758 individuals. PLoS One 2015;10(5):e0123423 doi 10.1371/journal.pone.0123423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, Zhao X, Sun S, Ni P, Li C, Ren A, et al. Homocysteine and Digestive Tract Cancer Risk: A Dose-Response Meta-Analysis. J Oncol 2018;2018:3720684 doi 10.1155/2018/3720684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiao SPK, Lie A, Yu CH. Meta-analysis of homocysteine-related factors on the risk of colorectal cancer. Oncotarget 2018;9(39):25681–97 doi 10.18632/oncotarget.25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao Y, Gao LJ, Zhou Y, Zhao JH, Lv Q, Dong JZ, et al. Effect of advanced age on plasma homocysteine levels and its association with ischemic stroke in non-valvular atrial fibrillation. J Geriatr Cardiol 2017;14(12):743–9 doi 10.11909/j.issn.1671-5411.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cust AE, Smith BJ, Chau J, van der Ploeg HP, Friedenreich CM, Armstrong BK, et al. Validity and repeatability of the EPIC physical activity questionnaire: a validation study using accelerometers as an objective measure. Int J Behav Nutr Phys Act 2008;5:33 doi 10.1186/1479-5868-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.