Abstract
A consequence of the intratumor heterogeneity (ITH) of glioblastoma (GB) is the susceptibility to treatment driven evolution. To determine the potential of radiotherapy to influence GB evolution we used orthotopic xenografts initiated from CD133+ GB stem-like cells (GSCs). Towards this end, orthotopic xenografts grown in nude mice were exposed to a fractionated radiation protocol that resulted in a significant increase in animal survival. Brain tumors from control and irradiated mice were then collected at morbidity and compared in terms of growth pattern, clonal diversity and genomic architecture. In mice that received fractionated radiation, tumors were less invasive with more clearly demarcated borders and tumor core hypercellularity as compared to controls suggesting a fundamental change in tumor biology. Viral integration site analysis indicated a reduction in clonal diversity in the irradiated tumors implying a decrease in intratumor heterogeneity. Changes in clonal diversity were not detected after irradiation of GSCs in vitro suggesting that the radiation-induced reduction in intratumor heterogeneity was dependent on the brain microenvironment. Whole exome sequencing revealed differences in mutation patterns between control and irradiated tumors, which included modifications in the presence and clonality of driver mutations associated with GB. Moreover, changes in the distribution of mutations as a function of subpopulation size between control and irradiated tumors were consistent with subclone expansion and contraction, i.e. subpopulation evolution. Taken together, these results indicate that radiation drives the evolution of the GSC-initiated orthotopic xenografts and suggest that radiation-driven evolution may have therapeutic implications for recurrent GB.
Introduction
Glioblastoma (GB) is the most common form of malignant brain cancer; despite the combination of surgery, radiotherapy, and temozolomide, the median survival time is only about 14 months (1). Whereas the mechanisms mediating this consistent therapeutic resistance have not been defined, the clonal diversity and evolutionary dynamics inherent to GB is considered a major obstacle in the development of effective treatment (2–6). Along these lines, comparison of genomic data generated from glioma tissue obtained at initial surgery and at recurrence revealed an altered mutational profile, an effect that was attributed to temozolomide treatment (7). The implication of such studies is that the temozolomide driven evolution results in the emergence of resistant clones. Consistent with studies of clinical specimens, temozolomide treatment of mice bearing brain tumor xenografts initiated from GB primary cultures suggested the expansion of drug resistant clones (8). Given that GBs regrow after initial treatment, understanding the consequences of treatment-driven evolution may not only generate insight into the fundamental biology of recurrent GBs but also suggest novel therapeutic strategies.
While studies to date have focused on temozolomide (7,9,10), a role for radiotherapy as an independent driver of GB evolution has not been investigated. Towards this end, orthotopic xenografts initiated from CD133+ GB stem-like cells (GSCs) would appear to provide a model system for testing the potential of radiation to influence GB evolution. GSCs represent a clonogenic subpopulation considered to be critical in the development, maintenance and treatment response of GBs (11–13). Moreover, orthotopic xenografts grown from GSCs replicate the genotype, phenotype and in vivo growth pattern of GB (14). With respect to GB evolution, we have previously shown that after the initial implant of 100% CD133+ cells, xenografts at the time of morbidity are comprised of a variety of cell subpopulations including those expressing GFAP or βIII tubulin (15), which is consistent with tumor cells that have differentiated, at least partially, along astrocytic and neuronal pathways, respectively. In addition, there continued to be a small subpopulation (approximately 10%) of tumor cells expressing CD133, suggesting the presence of GSCs. Finally, based on analysis of ɣH2AXand 53BP1 foci, CD133+ cells were less radiosensitive than CD133− tumor cells (16). Thus, the GSC xenograft model exhibits the intratumor heterogeneity and evolutionary dynamics that may simulate that of a GB in situ.
To investigate the potential of radiotherapy to influence GB evolution, in the study described here we defined the consequences of a fractionated radiation protocol on the growth pattern, clonal diversity and genomic architecture of GSC-initiated orthotopic xenografts. The data presented show that tumors that regrow after irradiation were less invasive and had different mutational signatures as compared to untreated tumors. In addition, based on viral integration site analysis (VISA), radiation exposure resulted in a reduction in intratumor heterogeneity (clonal diversity), an effect that was dependent on the brain microenvironment. These results indicate that radiation drives the evolution of the GSC-initiated orthotopic xenografts.
Materials and Methods
Glioblastoma Cell Lines
GSC lines NSC11 and NSC20 (provided by Dr. Frederick Lang, MD Anderson Cancer Center in 2008 as frozen stocks) were grown as neurospheres in stem cell medium and CD133+ GSC cells were isolated by FACS as reported previously (17). The U251 human GB cell line was obtained from the Division of Cancer Treatment and Diagnosis Tumor Repository (DCTD), National Cancer Institute (NCI) and grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS (Invitrogen). All cell lines were cultured less than 2 months after resuscitation; each tested negative for mycoplasma by PCR. U251 cells were authenticated in July 2019 by STR analysis (Idexx BioAnalytics); GSCs were authenticated by routine morphologic and growth analysis. All lines were transduced with lentivirus (LVpFUGQ-UbC-ffLuc2-eGFP2) at an MOI of 1 (16). For in vitro experiment, GSC neurospheres were disaggregated into single cells (17) and seeded onto poly-L-ornithine (Invitrogen)/laminin (Sigma-Aldrich) coated tissue culture dishes in stem cell media. Under these conditions, single-cell GSCs attach and grow as an adherent monolayer maintaining CD133 expression and stem-like characteristics (18). Radiation was delivered in vitro using a 320 kV X ray machine (Precision X-Ray Inc.) (19).
Xenografts
CD133+ GSCs (1.0 × 105) or U251 (2.5 × 105) transduced to express luciferase and GFP were implanted into the right striatum of 6-week-old athymic female nude mice (NCr nu/nu; NCI Animal Production Program) (16). Bioluminescent imaging (BLI) and local irradiation were all performed as described (16). On day 6 (U251) or day 21 (NSC11, NSC20) after implantation mice were randomized according to BLI signal into two groups: control and irradiated (RT). For GSC tumors, 5Gy was delivered for 3 consecutive days; for U251 tumors 3Gy was delivered on 3 consecutive days. Radiation was delivered using an X-Rad 320 X-irradiator (Precision X-Rays, Inc.), dose rate of 2.9 Gy/minute. Mice were monitored daily until the onset of neurologic symptoms (morbidity). BLI was performed weekly after irradiation until the first mouse of the group was lost. GraphPad Prism 7 (GraphPad Software) was used to generate Kaplan–Meier survival curves. To generate leg tumors 1.0 × 106 U251 cells were subcutaneously (SC) injected into the right hind leg. When tumors were 150–200 mm3, mice were randomized; radiation was delivered locally with animals restrained in a custom designed jig. SC tumors were collected for analysis at 1500 mm3 or 2 cm in any dimension. All experiments were performed as approved by the NIH Guide for Care and Use of Animals and conducted in accordance with the Institutional Animal Care and Use Committee.
Immunohistochemistry
At the initial signs of morbidity, mice were euthanized and perfused with chilled PBS then formalin via cardiac puncture. Brains were removed, placed in 10% buffered formalin before sectioning, embedded in paraffin, and cut into 6 µm thick slices. After heat-induced epitope retrieval and blocking slides were incubated with human SOX2 primary antibody (anti-rabbit, Cell signaling) followed by HRP secondary (anti-rabbit, Vector Labs). Slides were exposed to DAB reagent (Vector Labs), counterstained with hematoxylin and visualized with an AxioScan imager (Zeiss).
Viral Integration Site Analysis (VISA)
VISA was performed by the Center for Cancer Research Genomics Technology Laboratory (20,21). Briefly, genomic DNA was sheared to an average size of 400bp and subjected to linker-mediated, nested PCR using a combination of long terminal repeat (lentivirus) and linker-specific primers. Illumina sequencing adaptors were added at the same time. The library was sequenced on Illumina MiSeq using 2×150bp PE reads. Integration site junctions were mapped to hg19 human reference genome. Insertion sites are expressed as a percentage of the total reads. To compare samples within an experiment, integration sites detected in at least 2 of the samples were subjected to unsupervised hierarchical cluster analysis using R to visualize relative changes in clonal diversity.
Whole Exome Sequencing and Variant Analysis
Genomic DNA was subjected to WES, which was performed by the Center for Cancer Research Genomics Technology Laboratory. Extracted DNA underwent library prep according to the Agilent SureSelect XT (All Exon V5 +UTR) protocol and sequenced on Illumina HiSeq4000 device using paired-end sequencing to an average sequencing depth of > 180x. Mouse reads were removed as described (22). Alignment and tumor-only variant calling was performed with the Center for Cancer Research Collaborative Bioinformatics Resource (CCBR) pipeline (https://github.com/CCBR/Pipeliner). Germline variants were excluded using a panel of normals (23). Only protein-altering variants were retained (24). COSMIC mutational signature analysis (https://cancer.sanger.ac.uk/cosmic/signatures_v2) was performed with the R package YAPSA R package (v1.8.0; http://bioconductor.org/packages/release/bioc/vignettes/YAPSA/inst/doc/YAPSA.html) as described (25,26). Tumor subpopulations were further defined using EXPANDS algorithm and R package (v2.1.2) with default parameters (27). EXPANDS determines cell-frequency probabilities by using copy number variation (CNV) and single nucleotide variants (SNV) allele frequencies to estimate the fraction of cells harboring a SNV (SNVs and CNVs are filtered to remove variants on the X and Y chromosomes) (27). For COSMIC and EXPANDS analyses variants were included when they were protein altering, present in at least 2 out of 3 replicates, and had vaf > 5%. Sequence data have been deposited in NCBI’s BioProject database and are accessible through ID PRJNA576782 (http://www.ncbi.nlm.nih.gov/bioproject/576782).
Results
In vivo models of radioresponse
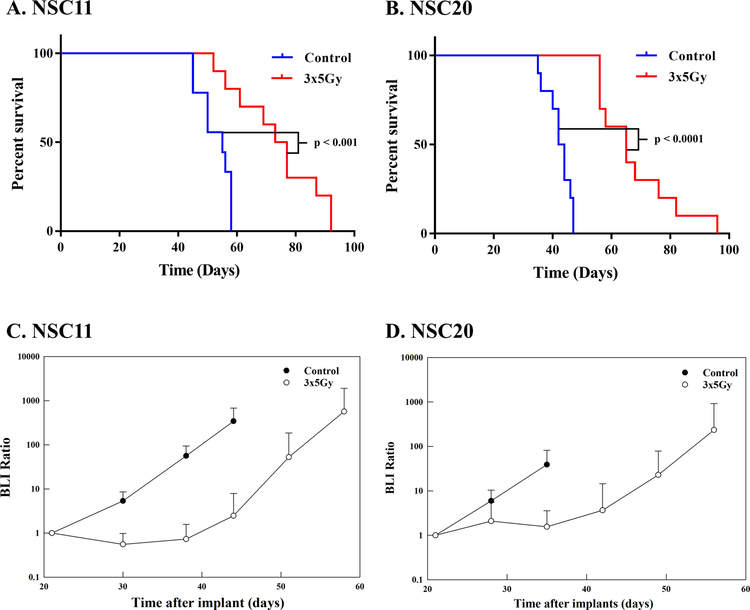
As a model system for investigating the impact of radiotherapy on GB evolution, we used orthotopic xenografts initiated from CD133+ GSC lines. GSCs were transduced with a lentivirus containing a bimodal expression vector fused with the bioluminescent protein ffLuc2 and fluorescent protein eGFP2 under UbC promoter control (LVpFUGQ-UbC-ffLuc2-eGFP2) (16). After in vitro expansion, cells were implanted into the right striatum of nude mice. At 21 days post-implant, upon reaching a size consistently detectable by BLI, tumor-bearing mice were randomized into treatment groups, control (mock) and 3×5Gy. As shown in figures 1A–B, this fractionated irradiation protocol resulted in significant survival increases for NSC11 or NSC20 tumor-bearing mice. This survival advantage was consistent with the delay in growth rate reflected by BLI as a function of time after irradiation (Figs. 1C–D). Given that 3×5Gy initiated at day 21 post-implant significantly delays tumor growth yet does not achieve curative effects, a situation not unlike that typically observed in clinic, we used this treatment protocol to test the hypothesis that radiation drives the evolution of GSC-initiated brain tumor xenografts.
Figure 1.
Radioresponse of GSC-initiated xenografts. On day 21 post-implant mice were randomized and treatment initiated the following day as described in text (3×5Gy). Kaplan–Meier survival curves were generated for A) NSC11 and B) NSC20 tumor-bearing mice (n=10 mice per group). Tumor growth defined by BLI ratio as a function of time after irradiation for C) NSC11 and D) NSC20
Radiation-induced changes in morphology and histology
As an initial assessment of the potential for radiation to influence GB evolution, the growth patterns of control and irradiated GSC xenografts were compared. GSCs engineered to express GFP can be visualized in tumor-bearing brains using a stereomicroscope. Representative images (Fig. 2A) of control mice demonstrate GFP signal from NSC11 or NSC20 cells extends anteriorly and posteriorly throughout most of the right hemisphere, well beyond the right striatum implantation site, reflecting a highly diffuse tumor (corresponding bright field images are shown in Figure 2B). However, in irradiated mice, GFP fluorescence was more restricted and limited primarily to the anterior portion of the hemisphere. To determine whether these observations extended to the histological level, right hemisphere sagittal sections from control and irradiated NSC11 and NSC20 bearing brains were evaluated for human SOX2 staining, which is highly expressed in both lines. As shown in Figure 2C and Supplemental Figure S1, GSCs in control mice were diffusely distributed with poorly defined margins and grey and white matter infiltration. In mice that received fractionated radiation, tumors were less infiltrative with more clearly demarcated borders as compared to controls. While indicative of an altered growth pattern, in broader terms these results suggest that the fundamental biology of GSC-initiated tumors that regrow after irradiation diverges from that of untreated tumors. As a possible explanation, we hypothesized that morphological/histological modifications detected in recurrent xenografts reflected the radiation-induced selection of tumor cell subpopulations, i.e., radiation-driven evolution.
Figure 2.
Morphology and histology of NSC11 and NSC20 tumors. A) Representative GFP fluorescence images of control and irradiated (3×5Gy) tumor-bearing brains at morbidity. B) Corresponding bright field images. Photos were taken on a stereoscope at 2.5x magnification. C) Right hemisphere sagittal sections at plane of injection site stained for SOX2 (magnification 20x).
Viral Integration Site Analysis
To directly address the question as to whether radiation modifies the clonal diversity (i.e., intratumoral heterogeneity) of GSC-initiated xenografts, viral integration site analysis (VISA) was used (20). Facilitating the application of VISA, our in vivo studies have centered on GSCs transduced with the lentivirus LVpFUGQ-UbC-ffLuc2-eGFP2, which, as discussed above, allows for in vivo tumor growth monitoring and ex vivo visualization. In addition, the stable integration of the lentivirus into the genome at essentially random sites provides a unique tag for each cell in the transduced population, which is inherited by subsequent daughter cells. Because the integration site is largely within a transcriptional unit, sequencing of PCR amplified genomic DNA surrounding the lentiviral sequence allows for assigning each site to a single location/gene or nearest gene. Identifying the gene thus merely provides a method for easily cataloging the integration site. The percent reads for a given gene/site corresponds to the size of the clone within the transduced population.
For this experiment CD133+ NSC11 cells were transduced with the described lentivirus; the culture was expanded and used for orthotopic implantation or collected for in vitro (IV) VISA. At 21 days post-implant, as for the survival studies, mice were randomized according to BLI into 2 groups: control (ic) and irradiated (icRT, 3×5Gy). At morbidity, mice were euthanized, GFP-expressing brain tissue grossly dissected, and DNA was extracted for VISA. The % reads detected at a given integration site (clonal frequency) were visualized for the individual xenografts and in vitro cultures in the heat map shown in figure 3A. As compared to NSC11 cells grown in vitro, there was a reduction in the number of clones detected (unique integration sites) in each of the 4 untreated xenografts (ic), suggesting a selection for clones that preferentially grow under in vivo orthotopic conditions. Moreover, as compared to the untreated tumors (ic) there was a further reduction in the total number of individual clones detected in the 4 xenografts that had been irradiated (icRT).
Figure 3.
Viral Integration Site Analysis (VISA) comparing GSCs in vitro to those grown as intracerebral xenografts that had received mock treatment (ic) or 3×5Gy (icRT). Treatment protocol is as described in text. Unsupervised hierarchical cluster analysis of integration sites detected in A) NSC11 and B) NSC20. The absence of color indicates the clone was not detected in a given sample (≤0.001% of total reads). The number of unique integration sites (mean ± standard error) detected by VISA in C) NSC11 and D) NSC20 samples. Because there was overlap of sites within each group, sites shared between samples of a given group were counted only once. * p < 0.05
VISA and unsupervised hierarchical cluster analysis of clonal frequency were also performed on NSC20 samples (Fig. 3B). Although there was variability between xenografts in the control (ic) and irradiated (icRT) groups, as compared to NSC20 in vitro, there was a reduction in clonal diversity in each of the orthotopic xenografts, with a further reduction evident in the irradiated tumors, similar to NSC11. The VISA results were also expressed in figures 3C–D as the average number of unique integration sites detected in each group. As shown, the largest number of integration sites was detected under in vitro conditions, which was reduced in orthotopic xenografts and further reduced after irradiation of xenografts. These results suggest that only a subset of the NSC11 and NSC20 cells proliferating in vitro will form a tumor after orthotopic implantation. Moreover, of those clones that grow orthotopically, after irradiation there is an additional reduction in clonal diversity, which is consistent with radiation-driven tumor evolution.
To investigate the role of the microenvironment in the radiation-induced reduction in clonal diversity, VISA was performed on NSC11 cells irradiated in vitro. Twenty-four hours after plating, cultures received either mock (control), 4Gy (single dose), or 3×2Gy. Cells were harvested for VISA when they reached 70–80% confluency, which corresponded to 7, 19 and 21 days for control, 4Gy and 3×2Gy, respectively. Clonogenic survival analysis showed that the surviving fraction of NSC11 cell exposed to 4Gy and 3×2Gy was 0.008±0.004 and 0.039±0.031 (mean±SEM, n=3), respectively. As shown in Figure 4A, cultures did not cluster according to treatment group. Moreover, there was no significant difference in the average number of integration sites among the 3 groups (Fig. 4B). These results suggest that, in contrast to the orthotopic model, in vitro irradiation had no detectable effect on clonal diversity. These data suggest that the radiation-induced reduction in clonal diversity is dependent on the in vivo microenvironment.
Figure 4.
Viral Integration Site Analysis of NSC11 in vitro and U251 cells grown in vitro (IV) and as subcutaneous (SC) and intracerebral (ic) xenografts. A) Unsupervised hierarchical cluster analysis of integration sites detected in NSC11 cells that were untreated (control) or irradiated with a single dose of 4Gy or 3 daily doses of 2Gy (3×2Gy) B) Unsupervised hierarchical cluster analysis of U251 cells. U251 xenografts (sc and ic) received 3×3Gy as described in text. IV samples were initiated from the same pool of cells used for sc and ic implantation and collected when 70–80% confluent. C) Number of unique integration sites (n=3, mean ± standard error) detected in NSC11 in vitro. D) Number of unique integration sites (n=3, mean ± standard error) detected in U251 cells. * p < 0.05
To determine whether radiation-driven evolution was limited to xenografts initiated from GSCs and to further interrogate the role of the brain microenvironment, VISA was applied to the established glioma cell line U251. In this experiment, lentivirus transduced U251 cells were grown as intracerebral (ic) xenografts and as subcutaneous (sc) leg tumors. At 6 days after ic (when tumors were detectable by BLI) and 18 days after sc implantation (150–200 mm3), U251 tumors received 3Gy for 3 consecutive days (3×3Gy). Irradiated and control ic tumors were collected at morbidity and sc tumors when the volume exceeded 1500mm3. Unsupervised hierarchical cluster analysis of the clonal frequency of the tumor samples as compared to U251 grown in vitro is shown in figure 4C; the average number of integration sites in each group is shown in figure 4D. Whereas there was no consistent difference between control ic and sc tumors, the number of integration sites were reduced in each compared to U251 in vitro, although not to the degree as observed for the GSC ic tumors. In response to irradiation, the number of integration sites in sc tumors is reduced suggesting a decrease in clonal diversity. However, in ic U251 tumors irradiation induced a greater reduction in integration sites, suggesting that the radiation-mediated reduction in clonal diversity is not limited to xenografts initiated from GSCs. Moreover, these results suggest that the brain microenvironment plays a critical role in radiation-driven evolution of GB xenografts.
Whole exome sequencing
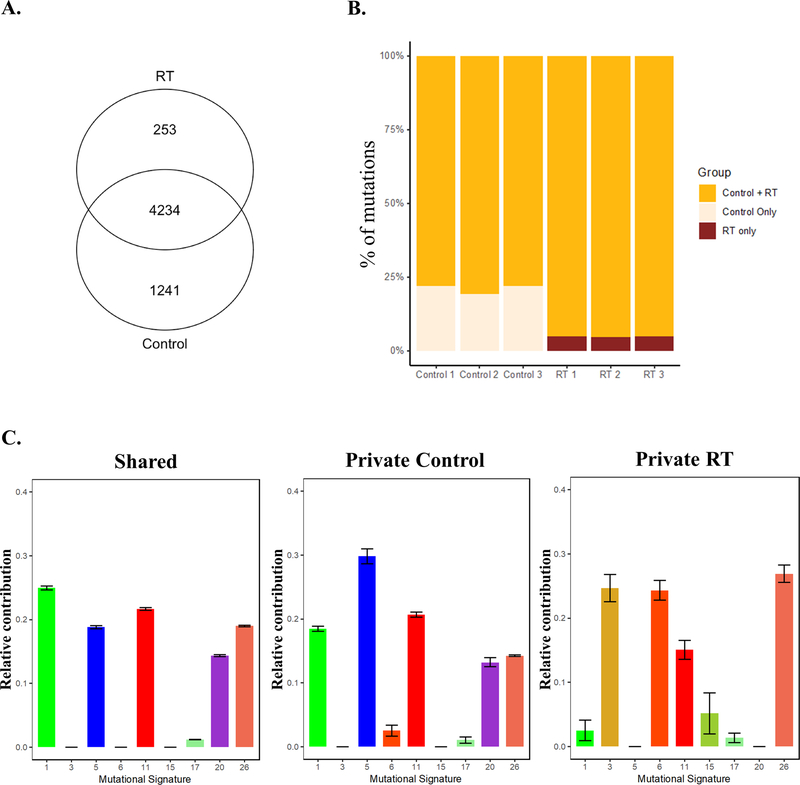
Because investigations of tumor evolution typically apply WES to describe genetic ITH, we used this approach to evaluate the potential for radiation-driven evolution of GSC-initiated orthotopic xenografts. Genomic DNA samples extracted from morbid tumors treated as in the VISA experiments shown in figure 3 were utilized for WES. Over 5500 SNVs + indels were detected in NSC11 tumors of which the majority were shared between control and irradiated tumors (Fig. 5A). Variants unique to control or irradiated tumors (private mutations) were consistently detected among the 3 samples in each group (Fig. 5B). The shared and private mutations in control and irradiated tumors were classified according to the mutational signatures generated from patient characteristics within large cancer cohorts using COSMIC mutational signature profiling (25,28,29). As shown in Figure 5C, the shared mutations distributed to mutational signatures 1 (Deamination of 5-methylcytosine), 5 (unspecified etiology), 11 (TMZ treatment), 20 (Concurrent POLD1 mutation and mismatch repair deficiency) and 26 (Defective DNA mismatch repair). Mutations unique to control tumors associated with similar signature profiles. However, private mutations found in irradiated tumors did not include signatures 1, 5, and 20, but mapped to signatures 3 (Defective HR DNA repair: BRCA1/2 mutation), 6 (Defective DNA mismatch repair), and 15 (Defective DNA mismatch repair), which were not associated with the unirradiated tumors. These results suggest the emergence of different subpopulations in irradiated samples as compared to control NSC11 tumors.
Figure 5.
The influence of radiation on mutations detected in NSC11 xenografts. On day 21 post-implant, brain tumors (n=3) were exposed to 3×5Gy (RT) or mock irradiated (control) and collected for WES at morbidity. A) Venn diagram comparing the number of mutations between treatment groups. B) The relative percentage of shared or private variants for each tumor (control and treated). C) The relative contribution of shared and private mutations in control and irradiated tumors to pre-defined COSMIC signature profiles (25,29). Signatures were defined as positive when they had a cumulative normalized contribution over signature-specific cutoffs. In A and C, variants were included if they were protein-altering, were present in at least 2 out of 3 replicates, and had vaf > 5%.
These analyses were also performed on WES data generated from control and irradiated NSC20 tumors (figure 6). In NSC20 tumors over 6000 SNVs + indels were detected with most of the variants shared between control and irradiated tumors with a similar number of private mutations detected only in control or irradiated NSC20 tumors (figures 6A–B). Using the shared mutations between control and irradiated NSC20 tumors the mutational signatures identified were similar to those in NSC11 tumors (signature #s 1,5,11,20,26) (figure 6C). In contrast, the signatures defined using the private mutations found in irradiated NSC20 tumors corresponded to increases in signatures 5 (unspecified etiology) and 19 (unspecified etiology) and decrease in signature 11 (TMZ treatment). Whereas these signatures were substantially different from those identified for irradiated NSC11 tumors, as for NSC11, these analyses suggest that irradiation modified the subpopulations comprising NSC20 tumors, consistent with radiation-driven evolution.
Figure 6.
The influence of radiation on mutations detected in NSC20 xenografts. Treatment as in Figure 5. A) Venn diagram comparing the number of mutations between treatment groups. B) The relative percentage of shared or private variants for each tumor (control and treated). C) The relative contribution of shared and private mutations for control and irradiated tumors to pre-defined COSMIC signature profiles. Signatures were defined as positive when they had a cumulative normalized contribution over signature-specific cutoffs.
To investigate the effects of radiation on xenograft subpopulation heterogeneity in more detail, the bioinformatics tool EXPANDS was applied to the WES data set. EXPANDS utilizes VAFs and copy number data to estimate cell-frequency probabilities (27). For this analysis, variants that were detected in at least 2 of the 3 replicates per group (control and RT) were included. Figure 7A depicts the 10 predicted subpopulations found in control and irradiated NSC11 tumors with the largest subpopulation (blue) at the bottom of the graph; each black dot corresponds to a variant predicted to be in a subpopulation. The lines indicate whether a specific variant in control tumors appeared in a subpopulation of the same ranking in the irradiated tumors or whether it had moved to another ranking. Whereas most variants are conserved between the largest subpopulations of control and irradiated tumors (depicted by straight lines), there was also a considerable amount of shifting from control tumors to a subpopulation of a different prevalence in irradiated tumors, which is especially apparent in comparisons of the smaller subpopulations (top of figure). Figure 7B shows 13 predicted subpopulations for NSC20 tumors. Again, the largest, most prevalent subpopulation (blue) is at the bottom of the graph. Similar to NSC11, while the majority of variants are conserved within the largest subpopulations, there is still substantial subpopulation shifting for variants between control and irradiated tumors.
Figure 7.
Influence of radiation on subpopulation dynamics. Orthotopic xenografts were treated and collected for WES as described in previous figures. Overall variant shifts within defined subpopulations of control and irradiated A) NSC11 and B) NSC20 tumors. The size of the subpopulations corresponds to its position on the y-axis, with the largest subpopulation is at the bottom. The black circles correspond to individual variants. GB driver mutations in control and irradiated C) NSC11 and D) NSC20 tumors. Clonal variants were those in the sample’s largest subpopulation; subclonal variants were those in any of a sample’s smaller subpopulations. Gray boxes represent mutations other than autosomal SNVs (indels, X or Y chromosome SNVs), which are not included in EXPANDS. GB driver gene mutation shifts within the defined subpopulations of control and irradiated E) NSC11 and F) NSC20 tumors.
These analyses were extended to driver gene mutations previously associated with GB (30–33). Initially, as shown in figures 7C–D, variants in driver genes were identified in each of 3 control and 3 irradiated GSC xenografts and classified as clonal (present in largest subpopulation), subclonal (present in smaller subpopulation), or both (multiple mutations for a given gene were individually found in both clonal and subclonal populations). In NSC11 tumors (Fig. 7C), multiple driver gene mutations were consistently lost in irradiated tumors, including AKAP9, FN1, SOX2, and SPTA1. There were also mutations that shifted in clonal status from control to irradiated tumors such as MAP4K3. Although there was more variability between individual tumors within control and irradiated groups, irradiation also resulted in changes in driver gene mutations detected in NSC20 tumors (figure 7D). Lost consistently after irradiation of NSC20 tumors were mutations in PIK3CA and CARM1. These data suggest that irradiation influenced the presence and clonality of GB driver mutations.
The shifts of these GB driver gene mutations between the previously defined subpopulations were then assessed (only subpopulations containing a GB driver gene variant were included). As in the overall WES data (Fig. 7A), the majority of NSC11 GB driver gene mutations were conserved within the largest subpopulations in control and irradiated samples (Fig. 7E). However, there were several mutations that became less prevalent after irradiation (e.g., lines from control subpopulation 1 to RT subpopulations 2 and 4). There was also one mutation that become more prevalent after irradiation (line from control subpopulation 5 to RT subpopulation 1), suggesting that this subclone may have had a better survival rate after radiotherapy. Figure 7F displays similar subpopulation dynamics for NSC20 tumors, although more variant shifts between subpopulations were noted for NSC20 compared to NSC11, including at least 3 variants that increased in prevalence after treatment. Changes in the distribution of mutations as a function of subpopulation size between control and irradiated tumors are consistent with subclone expansion and contraction, i.e. subpopulation evolution. Taken together, WES data support the radiation-driven evolution of orthotopic xenografts initiated from GSCs.
Discussion
The goal of this study was to test the hypothesis that radiation drives the evolution of orthotopic xenografts initiated from GSCs. Given that ITH is a critical parameter mediating tumor evolution, VISA was used to assess the changes in clonal diversity existing in cell lines and tumor xenografts. In these analyses, in the absence of any radiation exposure, comparison of the GSC lines grown in vitro and grown as brain tumors showed a significant reduction in the clonal diversity with intracerebral growth. These results imply that, although isolated by CD133 expression, the GSC cultures are comprised of a heterogeneous population, which is consistent with previous reports (34–37) and that only selected subpopulations were capable of proliferation under orthotopic conditions. As compared to untreated tumors, there was a dramatic additional reduction in clonal diversity in GSC-initiated tumors that had been irradiated. These results are suggestive of radiation-induced selection of subpopulations within GSC xenografts with the ultimate consequence of a reduction in ITH. The radiation-induced reduction in clonal diversity was also detected in orthotopic xenografts initiated from the long-established GB cell line U251 indicating that the process is not unique to the GSC model and suggesting that radiation-driven evolution may be applicable to brain tumors in general.
Further analyses indicated that the reduction in ITH was dependent on the brain microenvironment. No change was detected in clonal diversity when VISA was performed on NSC11 cells irradiated in vitro. Moreover, comparison of U251 sc and ic xenografts showed that while there was a reduction in clonal diversity after irradiation of sc tumors, the effect was considerably more pronounced in ic U251 tumors, a reduction comparable to those detected in the GSC tumors. The implication of this reduction in ITH is that the brain microenvironment imposes the selection of subpopulations of GSC and U251 cells. Such a subpopulation selective effect could be attributed to normal tissue influence dependent on intrinsic characteristics of the cells implanted (e.g., genotype) or localization into a niche independent of tumor cell type. Whereas the mechanisms remain to be determined, the results presented emphasize the need to account for the brain microenvironment when investigating GB radioresponse.
As an alternative approach to evaluating radiation-driven evolution, studies were extended to WES, which has been the standard approach to evaluating tumor evolution. Comparison of untreated GSC tumors and tumors that had regrown after irradiation revealed a consistently different set of mutations as well as mutational signatures. There also appeared to be a radiation induced shift in the genetic composition of the ranked sub-populations comprising the NSC11 and NSC20 xenografts. In evaluating the driver genes associated with GB (30–33), there was a reduction in the number of mutations in irradiated NSC11 tumors, which would be consistent with a reduction in ITH. In NSC20 tumors, although there was also a reduction in the number GB associated gene mutations, the number of losses as well as the specific genes affected were different as compared to NSC11. The different mutation spectrums observed for NSC11 and NSC20 tumors with and without RT suggest that whereas radiation drives the evolutionary process, the specific events along with the functional consequences may be tumor dependent. Thus, whereas WES results are indicative of radiation-driven evolution, they do not provide any clear insight into the mechanisms involved or the characteristics of the surviving tumor cells. Of note, the reduction in mutations detected in the irradiated xenografts contrasts with the hypermutation reported for tumors treated with temozolomide (5,7,38,39). Whether temozolomide, a component of GB standard of care, influences radiation-driven evolution remains to be determined.
If radiation alters GB evolution, then the fundamental biology of the tumors that recur after an initially effective course of radiotherapy should be altered. An example appears to be the morphology/histology of the NSC11 and NSC20 xenografts that regrow after the fractionated radiation protocol as compared to untreated tumors. As shown, each tumor exhibited a more restricted growth pattern in response to irradiation. Although in conflict with reports showing that radiation increases the invasion propensity of glioma cells in vitro (40,41), these results agree with clinical observations indicating that > 80% of GB recur in the initial radiation treatment volume (42,43). The studies described here show that radiation alone drives the evolution of GSC orthotopic xenografts affecting their ITH, mutation profile and growth pattern. If a similar process is operative in a clinical setting, then therapeutic targets in a primary, untreated GB may differ from those in the recurrent tumor, as previously suggested (44). GSC xenograft tumors that regrow after an initial radiation exposure may thus provide a model system for testing therapies for recurrent GB. This would include re-irradiation alone as well as in combination with potential radiosensitizers.
Supplementary Material
Statement of significance:
Radiation drives the evolution of glioblastoma orthotopic xenografts, when translated to the clinic this may have therapeutic implications for recurrent tumors.
Acknowledgements
Financial Support provided by Division of Basic Sciences, Intramural Program, National Cancer Institute (Z1ABC011372) to P.J. Tofilon. This project has also been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
The authors declare no potential conflicts of interest
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine 2005;352:987–96 [DOI] [PubMed] [Google Scholar]
- 2.Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America 2013;110:4009–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JK, Wang J, Sa JK, Ladewig E, Lee HO, Lee IH, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet 2017;49:594–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (New York, NY) 2014;344:1396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome research 2015;25:316–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 2015;28:318–28 [DOI] [PubMed] [Google Scholar]
- 7.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science (New York, NY) 2014;343. [DOI] [PMC free article] [PubMed]
- 8.Lan X, Jorg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 2017;549:227–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clinical cancer research : an official journal of the American Association for Cancer Research 2009;15:4622–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clinical cancer research : an official journal of the American Association for Cancer Research 2007;13:2038–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer research 2004;64:7011–21 [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396–401 [DOI] [PubMed] [Google Scholar]
- 13.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60 [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell 2006;9:391–403 [DOI] [PubMed] [Google Scholar]
- 15.Jamal M, Rath BH, Williams ES, Camphausen K, Tofilon PJ. Microenvironmental regulation of glioblastoma radioresponse. Clinical cancer research : an official journal of the American Association for Cancer Research 2010;16:6049–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamal M, Rath BH, Tsang PS, Camphausen K, Tofilon PJ. The brain microenvironment preferentially enhances the radioresistance of CD133(+) glioblastoma stem-like cells. Neoplasia (New York, NY) 2012;14:150–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCord AM, Jamal M, Williams ES, Camphausen K, Tofilon PJ. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clinical cancer research : an official journal of the American Association for Cancer Research 2009;15:5145–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, et al. Glioma Stem Cell Lines Expanded in Adherent Culture Have Tumor-Specific Phenotypes and Are Suitable for Chemical and Genetic Screens. Cell Stem Cell 2009;4:568–80 [DOI] [PubMed] [Google Scholar]
- 19.Wahba A, Rath BH, O’Neill JW, Camphausen K, Tofilon PJ. The XPO1 Inhibitor Selinexor Inhibits Translation and Enhances the Radiosensitivity of Glioblastoma Cells Grown In Vitro and In Vivo. Mol Cancer Ther 2018;17:1717–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science 2003;300:1749–51 [DOI] [PubMed] [Google Scholar]
- 21.De Ravin SS, Wu X, Moir S, Anaya-O’Brien S, Kwatemaa N, Littel P, et al. Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med 2016;8:335ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khandelwal G, Girotti MR, Smowton C, Taylor S, Wirth C, Dynowski M, et al. Next-Generation Sequencing Analysis and Algorithms for PDX and CDX Models. Molecular Cancer Research 2017;15:1012–6 [DOI] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 2011;12:745–55 [DOI] [PubMed] [Google Scholar]
- 25.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D H, Gu Z, Schlesner M. YAPSA: Yet Another Package for Signature Analysis. 2019. [DOI] [PubMed]
- 27.Andor N, Harness JV, Müller S, Mewes HW, Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics 2014;30:50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Körber V, Yang J, Barah P, Wu Y, Stichel D, Gu Z, et al. Evolutionary Trajectories of IDHWT Glioblastomas Reveal a Common Path of Early Tumorigenesis Instigated Years ahead of Initial Diagnosis. Cancer Cell 2019;35:692–704.e12 [DOI] [PubMed] [Google Scholar]
- 29.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng AW, Boot A, et al. The Repertoire of Mutational Signatures in Human Cancer. bioRxiv 2018:322859
- 30.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science (New York, NY) 2008;321:1807–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;174:1034–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2017;171:1029–41 e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orzan F, De Bacco F, Crisafulli G, Pellegatta S, Mussolin B, Siravegna G, et al. Genetic Evolution of Glioblastoma Stem-Like Cells From Primary to Recurrent Tumor. Stem cells (Dayton, Ohio) 2017;35:2218–28 [DOI] [PubMed] [Google Scholar]
- 35.Piccirillo SG, Combi R, Cajola L, Patrizi A, Redaelli S, Bentivegna A, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene 2009;28:1807–11 [DOI] [PubMed] [Google Scholar]
- 36.Piccirillo SGM, Colman S, Potter NE, van Delft FW, Lillis S, Carnicer M-J, et al. Genetic and functional diversity of propagating cells in glioblastoma. Stem cell reports 2015;4:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stieber D, Golebiewska A, Evers L, Lenkiewicz E, Brons NH, Nicot N, et al. Glioblastomas are composed of genetically divergent clones with distinct tumourigenic potential and variable stem cell-associated phenotypes. Acta Neuropathol 2014;127:203–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 2006;66:3987–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016;48:768–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wild-Bode C, Weller M, Rimner A, Dichgans J, Wick W. Sublethal irradiation promotes migration and invasiveness of glioma cells: implications for radiotherapy of human glioblastoma. Cancer Res 2001;61:2744–50 [PubMed] [Google Scholar]
- 41.Goetze K, Scholz M, Taucher-Scholz G, Mueller-Klieser W. The impact of conventional and heavy ion irradiation on tumor cell migration in vitro. Int J Radiat Biol 2007;83:889–96 [DOI] [PubMed] [Google Scholar]
- 42.Minniti G, Amelio D, Amichetti M, Salvati M, Muni R, Bozzao A, et al. Patterns of failure and comparison of different target volume delineations in patients with glioblastoma treated with conformal radiotherapy plus concomitant and adjuvant temozolomide. Radiother Oncol 2010;97:377–81 [DOI] [PubMed] [Google Scholar]
- 43.Narayana A, Yamada J, Berry S, Shah P, Hunt M, Gutin PH, et al. Intensity-modulated radiotherapy in high-grade gliomas: clinical and dosimetric results. Int J Radiat Oncol Biol Phys 2006;64:892–7 [DOI] [PubMed] [Google Scholar]
- 44.Campos B, Olsen LR, Urup T, Poulsen HS. A comprehensive profile of recurrent glioblastoma. Oncogene 2016;35:5819–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.