The solution of the crystal structure of the Rep domain of Wheat dwarf virus can help inform the development of protein–ssDNA binding tags.
Keywords: crystal structure, Wheat dwarf virus, Rep domain, HUH motif, HUH-tag, ssDNA, engineered protein–ssDNA complexes
Abstract
The Rep domain of Wheat dwarf virus (WDV Rep) is an HUH endonuclease involved in rolling-circle replication. HUH endonucleases coordinate a metal ion to enable the nicking of a specific ssDNA sequence and the subsequent formation of an intermediate phosphotyrosine bond. This covalent protein–ssDNA adduct makes HUH endonucleases attractive fusion tags (HUH-tags) in a diverse number of biotechnological applications. Solving the structure of an HUH endonuclease in complex with ssDNA will provide critical information about ssDNA recognition and sequence specificity, thus enabling rationally engineered protein–DNA interactions that are programmable. The structure of the WDV Rep domain reported here was solved in the apo state from a crystal diffracting to 1.24 Å resolution and represents an initial step in the direction of solving the structure of a protein–ssDNA complex.
1. Introduction
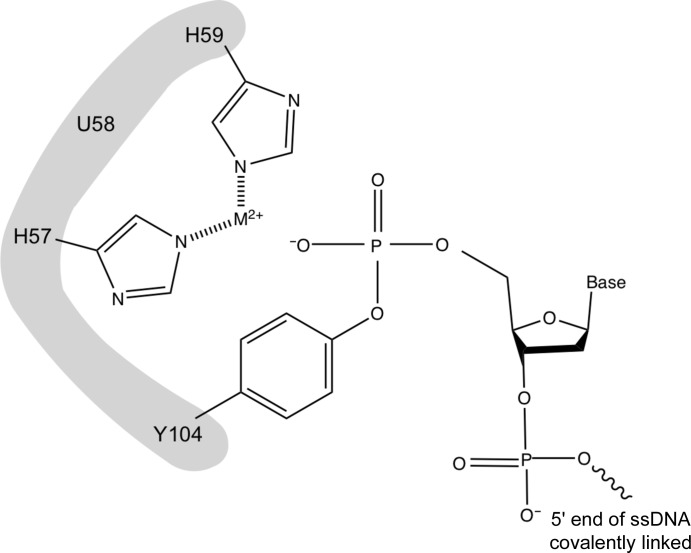
The Rep domain of Wheat dwarf virus (WDV), a type of geminivirus, is an HUH endonuclease (HUH-tag; Heyraud-Nitschke et al., 1995 ▸; Lovendahl et al., 2017 ▸). These proteins have been identified in both viral (Eisenberg et al., 1977 ▸) and bacterial (Ilyina & Koonin, 1992 ▸) genomes and play an important role in processes such as rolling-circle replication (Chandler et al., 2013 ▸). The histidine–nonpolar residue–histidine motif (aka HUH), from which the protein family derives its name, allows the coordination of a metal ion necessary for endonuclease functionality (Fig. 1 ▸). After binding a specific and conserved ssDNA sequence, the protein introduces a 5′ nick and a subsequent phosphotyrosine linkage (Fig. 1 ▸). This allows the initiation of replication of circular ssDNA (Chandler et al., 2013 ▸) by sequestering the 5′ end of the DNA.
Figure 1.
A model of the covalent phosphotyrosine intermediate between a tyrosine residue in an HUH-tag and the 5′ end ssDNA.
The existence of an intermediate protein–ssDNA covalent complex has made HUH-tags (Lovendahl et al., 2017 ▸) candidates in many molecular technologies. Protein–DNA complexes have already been utilized in a number of technologies, including DNA-guided protein localization and function (Derr et al., 2012 ▸; Engelen et al., 2016 ▸), single-molecule manipulation of DNA-tethered proteins (Halvorsen et al., 2011 ▸), cellular imaging (Jungmann et al., 2014 ▸) and cellular barcoding (Mali et al., 2013 ▸). However, HUH-tags are especially attractive because the protein–ssDNA complex is specific, covalent and inexpensive compared with linkages which rely on modified bases (Keppler et al., 2003 ▸; Los et al., 2008 ▸). We have demonstrated that HUH-tags can be used as fusion tags to tether a diverse number of proteins to ssDNA without disruption of function, including nuclear, cytoplasmic and cell-surface target proteins (Lovendahl et al., 2017 ▸). One particularly interesting application involves using HUH-tags in fusion with Cas9 to tether ssDNA donor repair templates, which resulted in increased homology-directed repair efficiency (Aird et al., 2018 ▸).
To improve upon these applications, it is desirable to utilize multiple HUH-tags in a single system and perform multiplex experiments (Lovendahl et al., 2017 ▸). This presents a challenge owing to the overlapping reactivity between HUH-tags and the conserved DNA sequence(s), particularly in HUH-tags derived from viruses (Lovendahl et al., 2017 ▸). Using structural information, the binding sites of various HUH-tags could be engineered to specifically react with unique oligonucleotides; however, this information is limited. Crystal structures of some circoviral (Porcine circovirus type 2; PDB entry 5xor; Luo et al., 2018 ▸) and nanoviral (Faba bean necrotic yellows virus; PDB entry 6h8o; Moncalian & Gonzalez-Mones, unpublished work) HUH-tags have been solved, and NMR structures have been determined of the geminivirus counterparts. However, there are neither crystal nor NMR structures of viral HUH-tags bound to ssDNA. A solved structure of a viral HUH-tag bound to ssDNA would provide information on the nature of the covalent interaction between the HUH-tag and the oligonucleotide. While attempted here, we were unable to obtain a structure of the HUH-tag bound to ssDNA. Instead, we report the first structure of the Rep domain from the geminivirus WDV, describe the strategies used for crystallization and make comparisons with existing geminivirus HUH-tag structures.
2. Materials and methods
2.1. Protein production and purification
A gene block containing the sequence of the WDV Rep domain, flanked by 15 bases homologous to the parent vector, pTD68/His6-SUMO, was ordered from Integrated DNA Technologies. The parent vector was digested using BamHI and XhoI restriction enzymes (New England Biolabs), and the gene block was directly cloned into the plasmid via the In-Fusion HD Cloning Kit (Takara) following the manufacturer’s protocol such that WDV Rep would have a His6-SUMO tag upon expression. The recombinant plasmid was transformed into competent Escherichia coli Stellar cells. The transformed cells were plated onto 100 µg ml−1 ampicillin plates and were cultured at 310 K overnight. A miniprep (Qiagen) of the colony was performed, and the purified DNA was sent to the University of Minnesota Genomics Center for sequencing in order to confirm that cloning had been successful. The recombinant plasmid was then cloned into E. coli BL21(DE3) competent cells (Agilent). Transformed cells were cultured in 2 l Erlenmeyer flasks in 1 l LB medium with 100 µg ml−1 ampicillin at 310 K until an OD600 nm of 0.6–1 was attained. Protein expression was induced with isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.5 mM. The cells were grown at 291 K for approximately 20 h.
The cells were harvested by centrifugation at 4000 rev min−1 and 277 K for 30 min. The pellets were then resuspended in lysis buffer (50 mM Tris pH 7.5, 250 mM NaCl, 1 mM EDTA). EDTA was present throughout purification to prevent metal binding. A protease-inhibitor cocktail was added to prevent any degradation, and lysis was carried out via sonication at 277 K. The resulting homogenate was centrifuged at 24 000g and 277 K for 20 min. The decanted supernatant was added to 4 ml Ni-IMAC bead slurry equilibrated with wash buffer (50 mM Tris pH 7.5, 250 mM NaCl, 1 mM EDTA, 30 µM imidazole) for batch binding on a rotator at 277 K. The homogenate was poured into a gravity-flow column and allowed to flow through. Approximately 30 ml of wash buffer was added and allowed to flow through, followed by approximately 7 ml of elution buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 250 µM imidazole). Fractions during each step of purification were saved and analyzed by SDS–PAGE to verify that the elution fractions contained a single band at 29 kDa, the molecular weight of the desired construct. The fractions containing a single band at approximately 29 kDa were then pooled for concurrent dialysis and His/SUMO-tag cleavage. DTT was added to the pooled fractions to a concentration of 1 mM, and 5 µl of ULP1 (1 U µl−1) was added. The pooled SUMO cleavage solution was transferred to a dialysis bag and left in dialysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) for approximately 18 h. Another round of Ni-IMAC purification was carried out; however, the desired product was now expected to be in the flowthrough. This was again verified using SDS–PAGE. The fractions containing a single band at approximately 16 kDa were pooled for concentration using a Vivaspin 6 (3 kDa molecular-weight cutoff) until a concentration of 7 mg ml−1 was reached. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | Wheat dwarf virus |
| DNA source | Integrated DNA Technologies |
| Forward primer | GAGAACAGATTGGTGGATCCATGGCAAGCAGCAGCA |
| Reverse primer | AAGCTTATTACTCGAGTTAATCTGCATCACGATCTTTACGACCCG |
| Expression vector | pTD68 |
| Expression host | E. coli |
| Complete amino-acid sequence of the construct produced | MASSSTPRFRVYSKYLFLTYPQCTLEPQYALDSLRTLLNKYEPLYIAAVRELHEDGSPHLHVLVQNKLRASITNPNALNLRMDTSPFSIFHPNIQAAKDCNQVRDYITKEVDSDVNTAEWGTFVAVSTPGRKDRDAD |
2.2. Crystallization
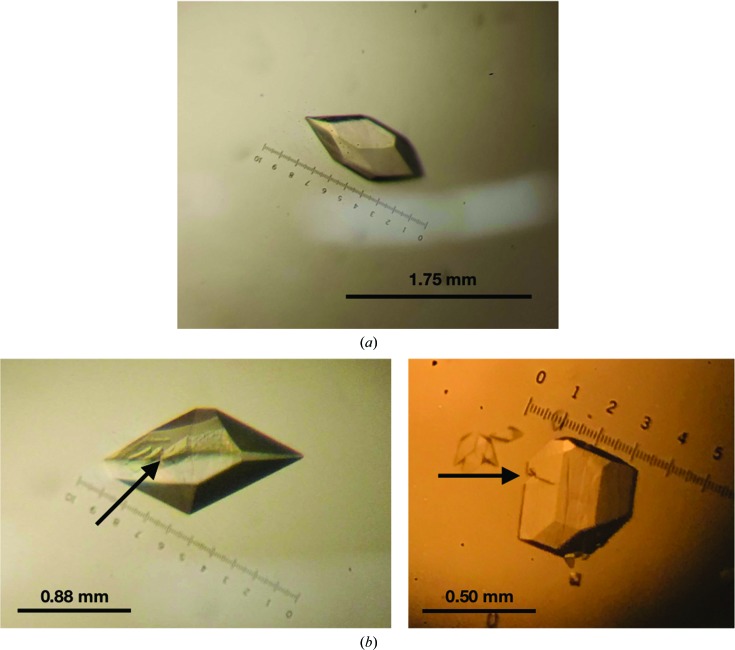
Initial crystallization conditions were determined by replicating the conditions resulting in crystals of the Rep domain from Porcine circovirus type 2 (Luo et al., 2018 ▸) using the hanging-drop vapor-diffusion method. Drops were prepared by mixing 1 µl protein solution and 1 µl well solution and were equilibrated against 500 µl well solution. The initial well condition consisted of 0.1 M HEPES buffer pH 7.5, 0.2 M sodium citrate, 30% (±)-2-methyl-2,4-pentanediol at 298 K, which produced crystals within 24 h. Sodium citrate and (±)-2-methyl-2,4-pentanediol kits were obtained from Hampton Research, while HEPES buffers were prepared using Milli-Q water (Milli-Q Academic, Millipore). After subsequent screening around this condition, varying the sodium citrate and (±)-2-methyl-2,4-pentanediol concentrations while maintaining 0.1 M HEPES pH 7.5, the most robust crystals were found to grow in a well solution consisting of solely 0.1 M HEPES buffer pH 7.5 at 298 K. These robust crystals were eventually used to collect a data set. The average size of the crystals was 1.0 × 0.5 mm. These crystals were fully developed and stopped growing within the first hour (Table 2 ▸, Fig. 2 ▸ a). However, secondary crystals and fractures were observed throughout growth (Fig. 2 ▸ b). The first data set was collected at room temperature and without any soaking using our in-house generator, but subsequent crystals were soaked for 40 s in a drop containing the mother liquor, 1 mM MgCl2, 1 mM 8 nt DNA oligonucleotide (ATATTACC) and 20% glycerol. When soaked, the crystals degraded quickly. Crystals were removed from the soaking solution when cracking was observed. All solutions were brought to final concentrations using Milli-Q water (Milli-Q Academic, Millipore).
Table 2. Crystallization.
| Method | Hanging drop |
| Plate type | 24-well plate, Hampton Research |
| Temperature (K) | 298 |
| Protein concentration (mg ml−1) | 7 |
| Buffer composition of protein solution | 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT |
| Composition of reservoir solution | 0.1 M HEPES buffer pH 7.5 |
| Volume and ratio of drop | 2 µl; 1:1 ratio |
| Volume of reservoir (µl) | 500 |
Figure 2.
Crystals imaged prior to handling or soaking. All crystals were grown in drops consisting of 1 µl well solution (0.1 M HEPES buffer pH 7.5) and 1 µl protein solution. (a) The WDV Rep crystal resulting in the deposited structure measured 1.23 × 0.53 mm. Note the lack of fracturing and attached secondary crystals. (b) WDV Rep crystals with fracture lines indicated by arrows. Note the secondary crystals attached to the bottom of the crystal on the right.
2.3. Data collection, processing and structure refinement
The first data set was collected without soaking and at room temperature on an in-house Rigaku MicroMax-007 HF rotating-anode copper source using a Rigaku Saturn 944+ detector at the Kahlert Structural Biology Center, University of Minnesota. Using molecular replacement with the minimized average NMR structure of Rep from Tomato yellow leaf curl virus (TYLCV) with 40% sequence identity (PDB entry 1l2m; Campos-Olivas et al., 2002 ▸), trimmed to Cβ atoms using PyMOL (DeLano, 2002 ▸), a model was built to 2.6 Å resolution. The final data set was collected on beamline 24-ID-C at the Advanced Photon Source, Argonne National Laboratory using a Dectris PILATUS3 6M-F detector. The initial model was then used for molecular replacement, resulting in the final 1.24 Å resolution model. XDS (Kabsch, 2010 ▸) was used to process data and Phaser (McCoy et al., 2007 ▸) in Phenix (Liebschner et al., 2019 ▸) was used for molecular replacement and refinement. Manual model building for refinement was performed in Coot (Emsley et al., 2010 ▸), with subsequent refinement performed in Phenix. As a final pass, anisotropic refinement was performed on all atoms except waters, improving the R work and R free statistics significantly. Through structure refinement, it was determined that the ssDNA soaks did not result in an ssDNA-bound structure. The final refinement and data-collection statistics are shown in Tables 3 ▸ and 4 ▸. The final model was deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank with PDB code 6q1m.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| X-ray source | Beamline 24-ID-C, APS |
| Wavelength (Å) | 0.97910 |
| Detector | PILATUS3 6M-F |
| Exposure time (s) | 0.197 |
| Crystal-to-detector distance (cm) | 17.000 |
| Angle increment (°) | 0.2000 |
| Resolution range (Å) | 46.92–1.24 (1.284–1.240) |
| Space group | P41212 |
| a, b, c (Å) | 49.57, 49.57, 145.7 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Matthews coefficient (Å3 Da−1) | 2.87 |
| Solvent content (%) | 57.07 |
| Total reflections | 337460 |
| Unique reflections | 52013 (5098) |
| Multiplicity | 6.5 |
| Mosaicity (°) | 0.09 |
| Completeness (%) | 98.83 (99.43) |
| 〈I/σ(I)〉 | 18.8 |
| Wilson B factor (Å2) | 14.96 |
| R merge | 0.050 |
| R meas | 0.054 |
| R p.i.m. | 0.028 |
| CC1/2 | 0.999 |
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Reflections used in refinement | 51949 (5122) |
| Reflections used for R free | 2556 (250) |
| R work | 0.158 (0.253) |
| R free | 0.166 (0.287) |
| No. of non-H atoms | |
| Total | 1198 |
| Macromolecules | 1065 |
| Ligands | 24 |
| Solvent | 109 |
| No. of protein residues | 119 |
| R.m.s.d, bonds (Å) | 0.006 |
| R.m.s.d, angles (°) | 0.88 |
| Ramachandran favored (%) | 94.87 |
| Ramachandran allowed (%) | 5.13 |
| Ramachandran outliers (%) | 0.00 |
3. Results and discussion
3.1. Crystallization and structure determination
WDV Rep was expressed in E. coli BL21(DE3) cells and was purified using nickel affinity to a final concentration of 7 mg ml−1. WDV Rep crystallized in one hour using a hanging drop consisting of 1 µl protein solution and 1 µl well solution equilibrated against 500 µl well solution (0.1 M HEPES buffer pH 7.5 and no precipitant) at 298 K. Interestingly, the crystals had varying degrees of high mosaicity. The initial 2.6 Å resolution structure was solved by collecting data from multiple crystals until an outlier crystal with a lower mosaicity of 1.29° was found. The final crystal (Fig. 2 ▸ a) had a mosaicity of 0.09°. Crystals were soaked for 40 s in a drop consisting of the mother liquor, 1 mM MgCl2, 1 mM 8 nt DNA oligonucleotude (ATATTACC) and 20% glycerol.
A data set was collected to 1.24 Å resolution. The crystal belonged to space group P41212, with unit-cell parameters a = 49.57, b = 49.57, c = 145.7 Å and one molecule per asymmetric unit. The structure was refined to R work and R free values of 0.158 and 0.166, respectively. Cryoprotectant glycerol molecules were added to the model. During refinement, there was electron density near the C-terminal α-helix that appeared to have a peptide-like structure. However, multiple attempts to fill this density with both DNA and amino-acid residues were unsuccessful. Electron density was not observed near the histidine residues to support metal-ion coordination or near the active-site tyrosine to support DNA linkage. The ssDNA soak could have been ineffective owing to the 40 s soak time being insufficient for equilibration. However, the crystals appeared to fracture after 40 s and to salvage them they were removed from solution. Attempts to co-crystallize both WDV and ssDNA through mass screens have so far been ineffective.
3.2. Structural analysis
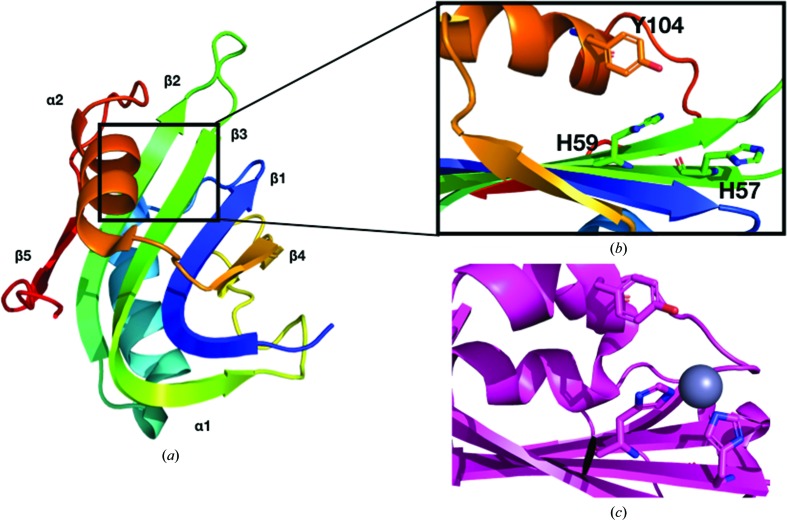
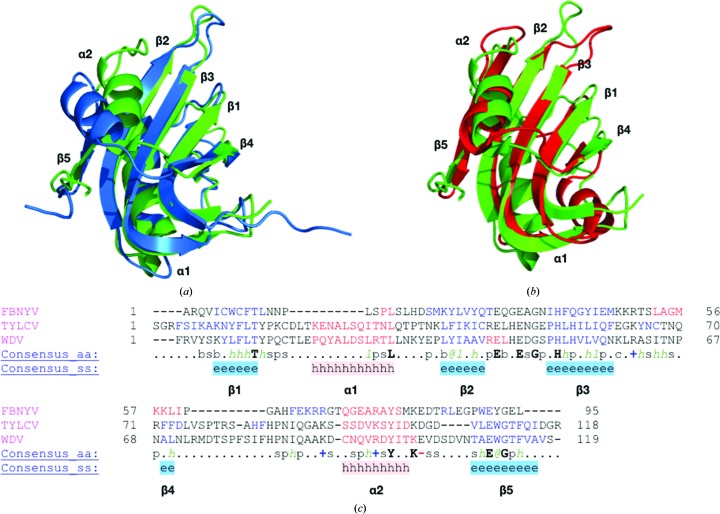
The secondary structure of WDV Rep is β1–α1–β2–β3–β4–α2–β5 from the N-terminus to the C-terminus (Fig. 3 ▸ a). The antiparallel β-sheet is ordered β5–β2–β3–β1–β4 from left to right in Fig. 3( ▸ a). The tyrosine residue of the HUH motif is located on α2 and the histidine residues are located on β3 (Fig. 3 ▸ a). The nonpolar residue of the HUH motif is a leucine. The solved WDV Rep structure was superimposed with the TYLCV Rep NMR structure used for molecular replacement using Secondary Structure Matching or Superimpose (Krissinel & Henrick, 2004 ▸) in CCP4 (Winn et al., 2011 ▸; Fig. 4 ▸ a). The r.m.s.d. of all atoms from superimposition is 1.6 Å. TYLCV Rep and WDV Rep were structurally similar apart from the positioning of α2. This difference could be owing to the more dynamic nature of NMR structures. Sequence alignment and structural alignment revealed that the sequence identity is 40%, and the secondary structure is highly conserved in these two proteins (Fig. 4 ▸ c).
Figure 3.
Structural comparison of the active sites of WDV Rep and Adeno-associated virus (AAV) type 5 Rep (PDB entry 1m55; Hickman et al., 2002 ▸). (a) The ribbon structure of the WDV protein is colored from the N-terminus (blue) to the C-terminus (red). (b) The active site of WDV Rep. Note the orientation of the histidine pair of the HUH motif. (c) The active site of AAV type 5 Rep. The histidine pair of the HUH motif is coordinated by a zinc ion (gray). Note the shifted orientation of the histidine pair towards the zinc ion and the catalytic tyrosine.
Figure 4.
Structural and sequence comparison of WDV, TYLCV and FBNYV Reps. Metal ions and ssDNA are absent from all structures. (a) Superimposition of the WDV Rep (green) and TYLCV Rep (blue) structures. The r.m.s.d. of superimposition was 1.6 Å. (b) Superimposition of the WDV Rep (green) and FBNYV Rep (red) structures. The r.m.s.d. of superimposition was 2.3 Å. (c) Sequence and structural alignment of the TYLCV and WDV Rep proteins in PROMALS3D (Pei et al., 2008 ▸). Secondary-structure alignment of β-strands is shown in blue and that of α-helices is shown in red. Consensus amino-acid symbols are as follows: conserved amino acids are in bold upper case letters; l, aliphatic; @, aromatic; h, hydrophobic; o, alcohol; p, polar; t, tiny; s, small; b, bulky; +, positively charged; −, negatively charged; c, charged.
The solved WDV Rep structure was also superimposed and aligned with the crystal structure of Rep from Faba bean necrotic yellows virus (FBNYV; PDB entry 6h8o; Moncalian & Gonzalez-Mones, unpublished work). The r.m.s.d. value for superimposition of all atoms is 2.3 Å and the sequence identity is 24%. The secondary structure is very similar between FBNYV Rep and WDV Rep, apart from the structure of α1, which is a loop in FBNYV Rep, and an additional α-helix that is present between β3 and β4 in FBNYV Rep (Fig. 4 ▸ b). The HUH motif in FBYNV contains Gln43 instead of a histidine (Fig. 4 ▸ c).
The three structures do not contain the bound divalent metal ion needed for cleavage (Fig. 3 ▸ b). Divalent metal ions coordinate to the two histidine residues of the HUH motif (Chandler et al., 2013 ▸). This causes a change in the active site, with the two histidine residues pointing inwards towards the catalytic tyrosine residue (see Fig. 3 ▸ c; Hickman et al., 2002 ▸).
Supplementary Material
PDB reference: Rep domain of Wheat dwarf virus, 6q1m
Acknowledgments
The structural data set was collected on beamline 24 (NE-CAT) at the Advanced Photon Source, Argonne National Laboratory.
Funding Statement
This work was funded by National Institute of General Medical Sciences grants GM119483 and NIGMS R35-GM118047.
References
- Aird, E. J., Lovendahl, K. N., St Martin, A., Harris, R. S. & Gordon, W. R. (2018). Commun. Biol. 1, 54. [DOI] [PMC free article] [PubMed]
- Campos-Olivas, R., Louis, J. M., Clerot, D., Gronenborn, B. & Gronenborn, A. M. (2002). Proc. Natl Acad. Sci. USA, 99, 10310–10315. [DOI] [PMC free article] [PubMed]
- Chandler, M., de la Cruz, F., Dyda, F., Hickman, A. B., Moncalian, G. & Ton-Hoang, B. (2013). Nat. Rev. Microbiol. 11, 525–538. [DOI] [PMC free article] [PubMed]
- DeLano, W. (2002). PyMOL. http://www.pymol.org.
- Derr, N. D., Goodman, B. S., Jungmann, R., Leschziner, A. E., Shih, W. M. & Reck-Peterson, S. L. (2012). Science, 338, 662–665. [DOI] [PMC free article] [PubMed]
- Eisenberg, S., Griffith, J. & Kornberg, A. (1977). Proc. Natl Acad. Sci. USA, 74, 3198–3202. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Engelen, W., Janssen, B. M. G. & Merkx, M. (2016). Chem. Commun. 52, 3598–3610. [DOI] [PMC free article] [PubMed]
- Halvorsen, K., Schaak, D. & Wong, W. P. (2011). Nanotechnology, 22, 494005. [DOI] [PubMed]
- Heyraud-Nitschke, F., Schumacher, S., Laufs, J., Schaefer, S., Schell, J. & Gronenborn, B. (1995). Nucleic Acids Res. 23, 910–916. [DOI] [PMC free article] [PubMed]
- Hickman, A. B., Ronning, D. R., Kotin, R. M. & Dyda, F. (2002). Mol. Cell, 10, 327–337. [DOI] [PubMed]
- Ilyina, T. V. & Koonin, E. V. (1992). Nucleic Acids Res. 20, 3279–3285. [DOI] [PMC free article] [PubMed]
- Jungmann, R., Avendaño, M. S., Woehrstein, J. B., Dai, M., Shih, W. M. & Yin, P. (2014). Nat. Methods, 11, 313–318. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H. & Johnsson, K. (2003). Nat. Biotechnol. 21, 86–89. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen,V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Los, G. V., Encell, L. P., McDougall, M. G., Hartzell, D. D., Karassina, N., Zimprich, C., Wood, M. G., Learish, R., Ohana, R. F., Urh, M., Simpson, D., Mendez, J., Zimmerman, K., Otto, P., Vidugiris, G., Zhu, J., Darzins, A., Klaubert, D. H., Bulleit, R. F. & Wood, K. V. (2008). ACS Chem. Biol. 3, 373–382. [DOI] [PubMed]
- Lovendahl, K. N., Hayward, A. N. & Gordon, W. R. (2017). J. Am. Chem. Soc. 139, 7030–7035. [DOI] [PMC free article] [PubMed]
- Luo, L., Zhu, X., Lv, Y., Lv, B., Fang, J., Cao, S., Chen, H., Peng, G. & Song, Y. (2018). J. Virol. 92, e00724-18. [DOI] [PMC free article] [PubMed]
- Mali, P., Aach, J., Lee, J.-H., Levner, D., Nip, L. & Church, G. M. (2013). Nat. Methods, 10, 403–406. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Pei, J., Kim, B.-H. & Grishin, N. V. (2008). Nucleic Acids Res. 36, 2295–2300. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Rep domain of Wheat dwarf virus, 6q1m