Abstract
Rapid and transient expression of in vitro transcribed mRNA (IVT mRNA) in target cells is a current major challenge in genome engineering therapy. To improve mRNA delivery efficiency, a series of amphiphilic polyaspartamide derivatives were synthesized to contain various hydrophobic moieties with cationic diethylenetriamine (DET) moieties in the side chain and systematically compared as mRNA delivery vehicles (or mRNA-loaded polyplexes). The obtained results demonstrated that the side chain structures of polyaspartamide derivatives were critical for the mRNA delivery efficiency of polyplexes. Interestingly, when the mRNA delivery efficiencies (or the luciferase expression levels in cultured cells) were plotted against an octanol–water partition coefficient (log P) as an indicator of hydrophobicity, a log P threshold was clearly observed to obtain high levels of mRNA expression. Indeed, 3.5 orders of magnitude difference in the expression level is observed between −2.45 and −2.31 in log P. This threshold of log P for the mRNA transfection efficiency apparently correlated with those for the polyplex stability and cellular uptake efficiency. Among the polyaspartamide derivatives with log P > −2.31, a polyaspartamide derivative with 11 residues of 2-cyclohexylethyl (CHE) moieties and 15 residues of DET moieties in the side chains elicited the highest mRNA expression in cultured cells. The optimized polyplex further accomplished highly efficient, rapid, and transient IVT mRNA expression in mouse brain after intracerebroventricular and intrathecal injection. Ultimately, the polyplex allowed for the highly efficient target gene deletion via the expression of Streptococcus pyogenes Cas9 nuclease-coding IVT mRNA in the ependymal layer of ventricles in a reporter mouse model. These results demonstrate the utility of log P driven polymer design for in vivo IVT mRNA delivery.
Short abstract
Optimized hydrophobicity enhanced the amphiphilic polymer-based mRNA transfection into cells, allowing for gene deletion in the mouse brain by delivering Cas9-mRNA and chemically synthesized sgRNA.
1. Introduction
In vitro transcribed (IVT) mRNA drugs allow for rapid and transient protein expression in cells, which is quite advantageous for the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (CRISPR/Cas9)-mediated genome editing and therapeutic applications.1−3 The CRISPR/Cas9 technology can be potentially applied for treatment of central nervous system (CNS) disorders including Alzheimer’s diseases and Huntington disease.4,5 Particularly, rapid expression profiles of Cas9-coded IVT mRNA (Cas9-mRNA) in target cells more likely permit the efficient binding between Cas9 protein and cotransfected single guide RNA (sgRNA) for facilitated genome editing, associated with the minimal risk of nuclease-induced genome instability.6 In addition, IVT mRNA drugs have displayed an excellent safety profile in clinical trials as IVT mRNA is not inserted into the host genome, which is in sharp contrast to plasmid DNA (pDNA) and viral vectors.1,3 However, the high susceptibility of IVT mRNA to degradation by ribonucleases (RNases) in cerebrospinal fluid is an inherent drawback that results in substantially low availability of IVT mRNA drugs.
To overcome the bottleneck of IVT mRNA drugs, nonviral delivery technologies, such as nanoparticle carriers, have been rigorously developed for the protection and rapid internalization of IVT mRNA into target cells.7−10 Among these, polycations are one of the most widely developed materials because they can readily form nanosized polyion complexes, termed polyplexes, with IVT mRNA by simple mixing in aqueous media.11−13 Notably, recent combinatorial approaches based on large molecular libraries have demonstrated several key structures, e.g., “ionizable” cationic moieties, for accelerated endosomal escape of polyplexes.7,10,14 Besides the endosomal escape, polyplex stability is also critical for the transfection efficiency of fragile IVT mRNA especially under harsh in vivo conditions. Whereas higher polyplex stability is preferable for efficient cellular internalization of intact IVT mRNA, it more likely disturbs the prompt release of IVT mRNA from the polyplex in the target cell cytoplasm, thereby creating a need for optimization of polyplex stability. To this end, a viable approach is fine-tuning of the hydrophobicity of amphiphilic polycations to optimize the polyplex stability to be tolerable in extracellular milieu and dissociable in the cytoplasm.15−17 However, the optimal hydrophobicity of amphiphilic polycations has not been studied systematically for IVT mRNA-loaded polyplexes, and also critical physicochemical parameters (or indicators) in this regard have not been reported so far.
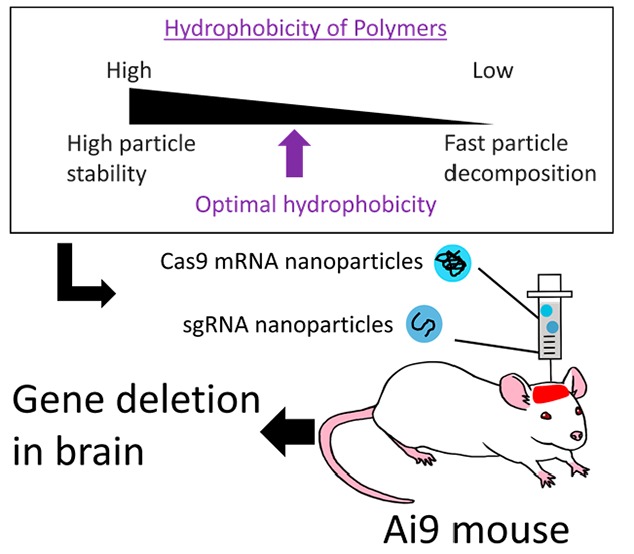
This study aims to design an amphiphilic polycation library to optimize the hydrophobic moieties for efficient, rapid, and transient expression of IVT mRNA in vitro and in vivo, directed toward the Cas9-mediated genome editing in the brain (Scheme 1). To this end, a series of amphiphilic polyaspartamide derivatives are prepared from the same parent poly(β-benzyl-l-aspartate) (PBLA) to systematically investigate the hydrophobic effects on physicochemical and biological properties of IVT mRNA-loaded polyplexes. Particularly, aliphatic amines with a slightly different structure are randomly introduced into the side chains of polyaspartamide derivatives along with cationic diethylenetriamine (DET) moieties (Figure 1). Of note, the DET moiety is selected as an IVT mRNA-condensing and endosome-escaping cationic unit that can induce the acidic pH-selective membrane disruption through the distinctive change in the protonation status from the membrane-inert monoprotonated state at physiological neutral pH (∼7.4) to the membrane-active diprotonated state at endosomal acidic pH (∼5.5).11,12,14 Importantly, the polyplex properties, including the transfection efficiency, are plotted against octanol–water partition coefficients (log P) of the amphiphilic polyaspartamide derivatives (PAsp(DET/R)s) as an indicator of hydrophobicity.18−20 In this way, a log P threshold for high mRNA transfection efficiency is clearly found as a critical parameter. Ultimately, the optimized polyplex is subjected to intracerebroventricular and intrathecal administration to deliver Cas9-mRNA into the brain. The obtained results demonstrate the utility of the log P driven polymer design and the feasibility of the optimized polyplexes for the in vivo local IVT mRNA delivery and gene editing.
Scheme 1. Schematic Illustration of the Present Study.
Firstly, a series of amphiphilic polyaspartamide derivatives are synthesized to optimize the polymer hydrophobicity for higher IVT mRNA delivery efficiency in cultured cells. Then, the optimized polyplexes are prepared with Cas9-mRNA and chemically synthesized sgRNA for the target gene deletion in the mouse ventricular area via intracerebroventricular or intrathecal administration.
Figure 1.
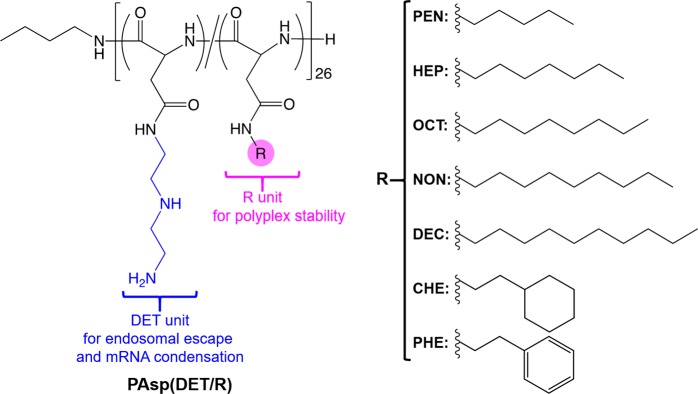
Chemical structures of amphiphilic polyaspartamide derivatives with DET moieties (PAsp(DET/R)). PEN, pentyl; HEP, heptyl; OCT, octyl; NON, nonyl; DEC, decyl; CHE, 2-cyclohexylethyl; and PHE, 2-phenylethyl. PAsp(DET/R)s have the mixed sequence of α and β isomers,21 yet only α isomer is shown here for simplicity.
2. Results and Discussion
2.1. Synthesis of Amphiphilic Polyaspartamide Derivatives
A series of PAsp(DET/R)s were synthesized by the simultaneous aminolysis reactions of PBLA with DET and various aliphatic amines (Scheme S1). The parent polymer, PBLA, was first synthesized by the ring-opening polymerization of BLA-NCA initiated by n-butylamine. The Mw/Mn and degree of polymerization (DP) of PBLA were determined to be 1.18 and 26 from the size exclusion chromatography (SEC) chart (Figure S1A) and 1H NMR spectrum (Figure S1B), respectively. Then, the mixture of DET and aliphatic amines (molar ratio = 1:2) was reacted with PBLA for 1 h.11 The aliphatic amines were chosen to have the slightly different number of carbon atoms (i.e., 5, 7, 8, 9, and 10) in the linear structure, 8 with cyclohexyl ring (CHE), and phenyl ring (PHE), for fine-tuning of the hydrophobicity of PAsp(DET/R). All PAsp(DET/R)s were confirmed to have comparable introduction rates of aliphatic amines, i.e., ∼40%, in the side chains from the 1H NMR spectra (Figures S2–S4), as summarized in Table S1. In a similar manner, PAsp(DET/CHE)s having different introduction rates of CHE moieties, i.e., ∼10% and ∼30%, were prepared to examine the effect of introduction rates of hydrophobic moieties (Table S2). Further, PAsp(DET/CHE)s, as well as PAsp(DET)s, having different DPs of 63 and 121, were prepared to evaluate the effect of DP. In PAsp(DET/CHE)s with DP = 63 (Mw/Mn = 1.19) and 121 (Mw/Mn = 1.34), the introduction rates of CHE moieties were determined to be ∼50% and ∼40%, respectively, from their 1H NMR spectra (data not shown). The hydrophobicity of PAsp(DET/R) series was determined by measuring a log P value. To this end, PAsp(DET/R)s were labeled with Alexa647 and dissolved in a biphasic mixture of 1-octanol and HEPES buffer (10 mM, pH 7.3). Then, the fluorescence intensities of Alexa647 distributed in each phase were measured to calculate a log P value (Tables S1 and S2). The log P values of PAsp(DET/R)s were ranging between −1.9 and −2.6. The obtained values showed an apparent linear correlation with the log P values calculated for the corresponding hydrophobic moiety by ChemDraw Professional Version 17: pentane (log P = 2.58), heptane (log P = 3.42), octane (log P = 3.84), nonane (log P = 4.25), decane (log P = 4.67), cyclohexylethane (log P = 3.25), and phenylethane (log P = 2.94). This finding suggests that the log P value of PAsp(DET/R)s can be roughly estimated from that of aliphatic amines.
2.2. Preparation and Characterizations of IVT mRNA-Loaded Polyplexes
Polyplex samples were prepared by mixing PAsp(DET/R)s with gaussia luciferase (GLuc)-coded mRNA (GLuc-mRNA) with the base number of 783 at various N/P ratios in 10 mM HEPES buffer (pH 7.3). Each polyplex sample was termed R-polyplex, where R represents the hydrophobic moiety in PAsp(DET/R), as summarized in Table S3. The mRNA loading into polyplexes was confirmed by agarose gel electrophoresis (Figure S5). The free mRNA band disappeared at N/P ≥ 2 in all PAsp(DET/R)s, indicating that all mRNA molecules were associated with PAsp(DET/R)s at N/P ≥ 2. Considering that DET moieties have an amine protonation degree of approximately 50%,12 the N/P of 2 corresponds to a charge-neutralization point between protonated amines in PAsp(DET/R) and phosphates in IVT mRNA. Thus, the result obtained from gel electrophoresis suggests the charge-stoichiometric polyplex formation between PAsp(DET/R) and GLuc-mRNA. Also, it is suggested that the introduction of hydrophobic R moieties did not affect the charge-stoichiometric complexation behavior. Hydrodynamic diameter (DH), polydispersity index (PDI), and ζ potential of polyplex samples prepared at N/P = 3 and 5 were measured with a Zetasizer (Table S4). While the DH values of polyplexes prepared at N/P = 3 were around 130 nm, the polyplexes at N/P = 5 exhibited smaller DH values of around 100 nm with a relatively narrow PDI of around 0.2. Considering that the ζ potentials of the polyplexes were considerably increased with the change in N/P from 3 to 5, the higher surface potential more likely provided the polyplexes with higher colloidal stability (or electrostatic repulsive force), permitting the formation of smaller and narrowly distributed polyplexes at the higher N/P. It should be noted that the polyplex samples prepared at the same N/P exhibited the similar ranges of DH and ζ potential regardless of log P values. These data indicate that all PAsp(DET/R)s successfully formed IVT mRNA-loaded polyplexes in 10 mM HEPES buffer (pH 7.3).
2.3. Evaluation of Protein Expression Efficiency of Polyplexes in Cultured Cells
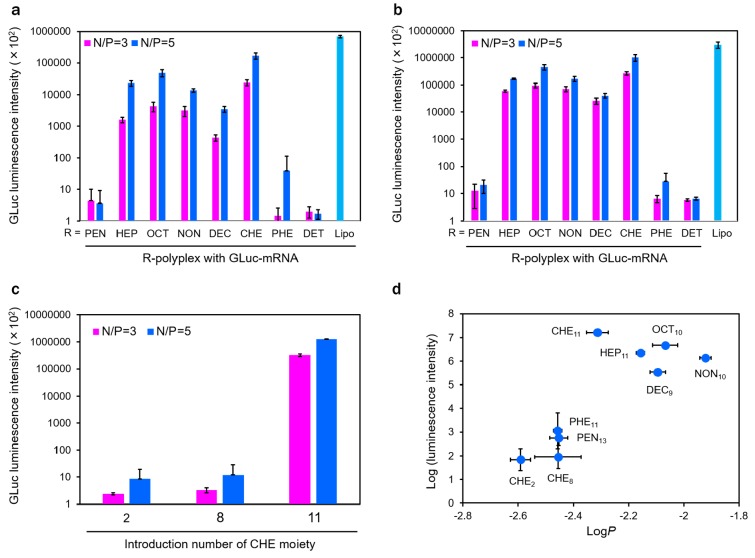
The IVT mRNA delivery efficiencies of polyplexes were determined in cultured mouse myoblast (C2C12) and neuroblastoma (Neuro-2a) cells. Each PAsp(DET/R) was mixed with GLuc-mRNA at N/P = 3 and 5 to form polyplex samples. The polyplex samples were transfected into the cells at 50 ng mRNA/well. The cells were incubated for 24 h, and GLuc expression levels were measured with a luminescence plate-reader. The obtained GLuc expression profiles were apparently similar between C2C12 cells and Neuro-2a cells (Figure 2A,B). In comparison among PAsp(DET/R)s with linear alkyl chains (or linear series), HEP-, OCT-, NON-, and DEC-polyplexes showed appreciably higher luminescence intensities than PEN- and DET-polyplexes, indicating that the alkyl chains with more than 5 carbons should be required for the significant IVT mRNA transfection into the cultured cells. Particularly, PAsp(DET/OCT) with 8 carbons in the hydrophobic moiety displayed the highest transfection efficiency in the linear series. This result suggests that the alkyl chain of 8 carbons might impart the optimal balance in IVT mRNA-loaded polyplex stability. A further comparison among OCT-, CHE-, and PHE-polyplexes with 8 carbons in the hydrophobic moieties (or 8-carbon series) reveals that the luminescence intensity of CHE-polyplex was higher than those of the other polyplexes and comparable to that of Lipofectamine 2000 (Lipo) as a reference. Interestingly, PHE-polyplex showed an almost background level of luminescence intensity, which is sharply contrasted with that of CHE-polyplex. Note that the similar IVT mRNA expression profile was obtained for the transfection of firefly luciferase (FLuc)-coded IVT mRNA (FLuc-mRNA) with the base number of ∼1900 into cultured C2C12 cells (Figure S6), indicating that the obtained transfection profiles were not limited for a specific IVT mRNA. These results demonstrate that a subtle change in hydrophobicity of PAsp(DET/R) dramatically improved the delivery efficiency of IVT mRNA-loaded polyplexes. In addition, the polyplexes prepared at N/P = 5 exhibited higher luminescence intensities than at N/P = 3, possibly due to the higher ζ potential values, which might facilitate the adsorptive endocytosis of polyplexes through the binding to anionic glycocalyx on the cell membranes and/or more efficient endosomal escape triggered by larger amounts of PAsp(DET/R) comprising the endosome-disrupting DET moieties.
Figure 2.
Luminescence intensities of cultured cells transfected with GLuc-mRNA. C2C12 cells (A) and Neuro-2a cells (B) were transfected by varying IVT mRNA-loaded polyplex samples prepared from PAsp(DET/R)s (DP = 26) at N/P ratios of 3 and 5. Lipofectamine 2000 (Lipo) was utilized as a reference. (C) C2C12 cells were transfected by PAsp(DET/CHE) with varying introduction rates of CHE moiety (DP = 26). The cells were incubated with each polyplex (50 ng mRNA/well) for 24 h. All results are expressed as mean ± SD (n = 4). (D) Correlation between GLuc-mRNA expression and log P. The expression levels of GLuc-mRNA were determined from the luminescence intensities (C2C12 cells, N/P = 5) in parts A and C.
The effect of substitution degrees of hydrophobic moiety in the polyaspartamide side chains on IVT mRNA transfection was also investigated for the most effective PAsp(DET/CHE). PAsp(DET/CHE)s with three different substitution degrees of CHE moiety were used for the polyplex formation with GLuc-mRNA. Note that all the polyplexes exhibited the similar DH, PDI, and ζ potential at N/P = 3 and 5 (Table S5). The CHE-polyplexes with ∼10% and ∼30% of CHE moieties (CHE2/26- and CHE8/26-polyplexes, respectively) induced 5 orders of magnitude lower GLuc-mRNA expression than that with ∼40% CHE moieties (CHE-polyplex) (Figure 2C). This result indicates that a certain amount of CHE moiety was required for the enhanced IVT mRNA transfection by CHE-polyplex. The effect of DP of PAsp(DET/CHE) on IVT mRNA delivery was further examined in comparison with PAsp(DET) control. Note that PAsp(DET/CHE)s with DPs of 63 and 121 formed IVT mRNA-loaded polyplexes with comparable DH and ζ potentials to that with DP of 26 (Table S5). Whereas the higher DP of 121 was needed for PAsp(DET) to elicit the significant luminescence intensity, all the CHE-polyplexes (CHE-, CHE32/63-, and CHE47/121-polyplexes) induced appreciably higher GLuc expression compared with the most effective DET0/121-polyplex (Figure S7). Thus, the DP of PAsp(DET/CHE) did not affect the high level of GLuc-mRNA expression at least in the range between 26 and 121.
A dose-dependent manner of GLuc-mRNA expression by CHE-polyplex (DP = 26, N/P = 5) was further examined in cultured C2C12 cells. The CHE-polyplex showed a distinct dose-dependent increase in luminescence intensity between 0.5 and 10 ng mRNA/well, and the increment reached a plateau at 10 ng mRNA/well (Figure S8). The apparent saturation of GLuc expression level might be due to the limitation of available free ribosomes, which is reported to be 105–106 molecules/cell in the cytoplasm.22,23 A time-dependent manner of GLuc-mRNA expression by CHE-, OCT-, and DET-polyplexes (DP = 26, N/P = 5) was also measured every 2 h in cultured C2C12 cells (Figure S9). The CHE-polyplex elicited a 10-fold higher luminescence intensity for all measurement times, compared with the OCT-polyplex, indicating that the higher mRNA expression by CHE-polyplex was not limited for a specific measurement time. Meanwhile, the viability of C2C12 cells was measured after treatment with CHE-polyplex (DP = 26, N/P = 5) for 24 h (Figure S10). No reduction in the cell viability was induced at 10–200 ng mRNA/well, indicating the negligible cytotoxicity of CHE-polyplex at these concentrations.
For a comprehensive interpretation, the correlation between the hydrophobicity of PAsp(DET/R) and the IVT mRNA expression efficiency of polyplex was investigated. Herein, the GLuc-mRNA expression efficiencies in cultured C2C12 cells were plotted against log P (Figure 2D). There is an apparent threshold of log P between −2.3 and −2.4 for the GLuc-mRNA expression efficiency. The PAsp(DET/R)s bearing log P > – 2.4, i.e., R = CHE11, HEP11, DEC9, OCT10, and NON10, elicited the appreciably higher mRNA transfection efficiency, compared with those bearing log P < – 2.4, i.e., R = PEN13, PHE11, CHE2, and CHE8. This result clearly indicates that the hydrophobicity of amphiphilic polycations should be optimized for the polyplex-mediated IVT mRNA delivery. Particularly, PAsp(DET/CHE) with a log P of −2.31 realized the optimal balance in hydrophobicity for the enhanced mRNA delivery under the tested condition.
2.4. Cellular Uptake and Intracellular Trafficking of mRNA-Loaded Polyplexes
To elucidate the underlying mechanism for the appreciably high IVT mRNA expression by CHE-polyplexes, the cellular uptake of IVT mRNA was examined by a flow cytometer using Cy5-labeled GLuc-mRNA (Cy5-mRNA). C2C12 cells were incubated with naked Cy5-mRNA or Cy5-mRNA-loaded polyplexes (DP = 26, N/P = 5) for 4 h before flow cytometric analysis (Figure S11). In the linear series, HEP-, OCT-, NON-, and DEC-polyplexes showed much higher mean fluorescence intensity derived from Cy5-mRNA, compared with DET- and PEN-polyplexes. Also, in the 8-carbon series, the mean fluorescence intensities obtained from the cells treated with OCT- and CHE-polyplexes were obviously higher than that with PHE-polyplex. These results indicate that the appreciably lower IVT mRNA expression by PEN-, PHE-, and DET-polyplexes should be ascribed to the inefficient cellular uptake of IVT mRNA payloads. Interestingly, the cellular uptake efficiencies plotted against log P clearly show their linear correlation (R2 = 0.951) except for CHE-polyplex (Figure S12), where the polyplexes with higher log P enabled more efficient cellular uptake of Cy5-mRNA. This finding suggests that the hydrophobicity of PAsp(DET/R)s determined the cellular uptake efficiency of polyplexes. The reason why the CHE-polyplex did not fit the line remains elusive. One possible explanation is that the cyclohexyl ring structure in the CHE moiety might be more effectively intercalated into the hydrophobic membrane domain due to the high affinity to saturated acyl chains in lipids for facilitated adsorptive endocytosis of CHE-polyplex.24,25
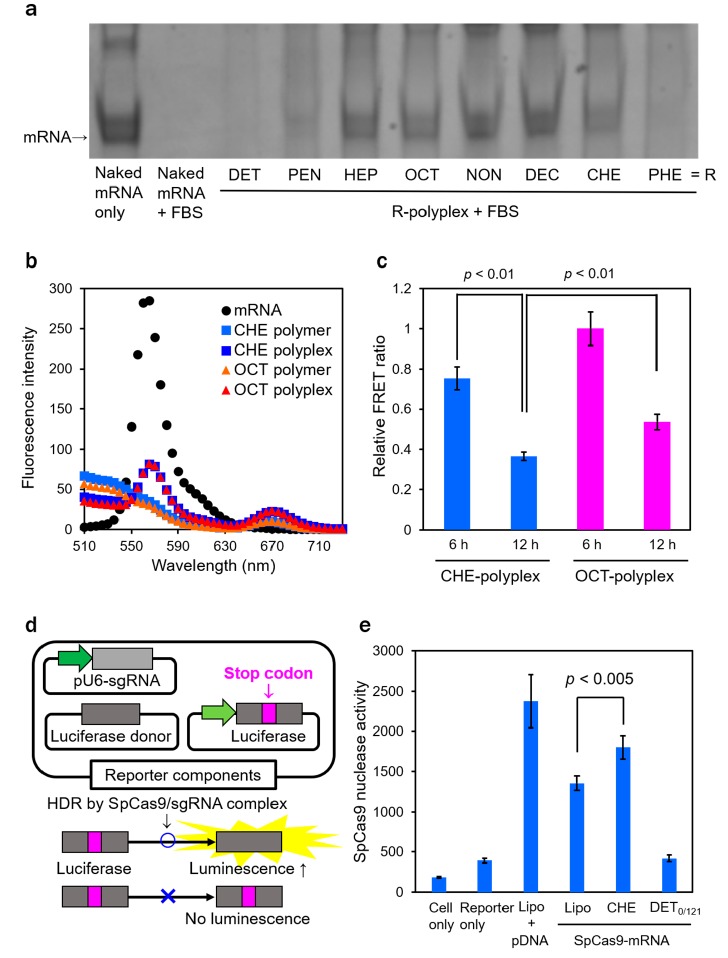
The inefficient cellular uptake of DET-, PEN-, and PHE-polyplexes might be due to lower polyplex stabilities, resulting in higher susceptibility to enzymatic degradation. Thus, the polyplex (or IVT mRNA payload) stabilities were evaluated through incubation with 10% FBS for 1 h at 37 °C (Figure 3A). Whereas the intact IVT mRNAs were clearly observed in HEP-, OCT-, NON-, DEC-, and CHE-polyplexes, DET-, PEN-, and PHE-polyplexes displayed smaller amounts of intact IVT mRNA on the gel. This result demonstrates that the polyplexes showing the inefficient cellular uptake of IVT mRNA were intolerable in the FBS solution. Again, the amounts of remaining intact IVT mRNA were plotted against log P (Figure S13). A positive correlation was clearly observed between the amounts of remaining intact IVT mRNA and log P, indicating that lower hydrophobicity in PAsp(DET/R)s resulted in the polyplex formation with lower stability. Lower stabilities of polyplexes with lower log P were also confirmed from the decrease in scattered light intensity (SLI) of polyplex samples in 150 mM NaCl solution (Figure S14). Larger decreases in relative SLI were observed for the polyplexes with lower log P values, suggesting that the physiological salt condition considerably destabilized the IVT mRNA-loaded polyplexes with the lower log P, probably due to attenuated electrostatic interactions between IVT mRNA and PAsp(DET/R)s. In contrast, PAsp(DET/R)s with higher log P were more tolerable to the physiological salt condition presumably due to their stronger hydrophobic interaction.
Figure 3.
(A) Acrylamide gel electrophoresis of IVT mRNA after incubation of polyplexes (N/P = 5) in 10% FBS for 1 h at 37 °C. After incubation, polyplex samples were purified with a RNeasy mini kit for mRNA release. (B) Fluorescence emission profiles of naked Cy3-mRNA, Alexa647-PAsp(DET/CHE), Alexa647-PAsp(DET/OCT), and their polyplexes (20 ng/μL mRNA, N/P = 5). (C) Relative FRET ratios (Alexa647 intensity/Cy3 intensity) derived from C2C12 cells treated with polyplexes with Cy3-mRNA and Alexa647-PAsp(DET/R) (N/P = 5, 500 ng mRNA/well). The Cy3 and Alexa647 intensities were determined by flow cytometer, and the FRET ratios were normalized to that obtained from OCT-polyplex in 6 h of incubation. All results are expressed as mean ± SE (n = 3). (D) Schematic illustration of a luciferase-reporter pDNA HDR assay. The pDNA encoding luciferase with an in-frame stop codon produces an inactive luciferase protein that cannot catalyze luciferin. sgRNA-associated SpCas9 can repair the pDNA to the full-length luciferase sequence. The repaired pDNA produces an active luciferase protein that can catalyze the luciferin substrate for emitting the luminescence. (E) SpCas9 nuclease activity determined by luminescence intensities in cultured HeLa cells. The cells were initially transfected with the reporter components by Lipo and incubated for 24 h. The cells were then transfected with CHE-polyplexes (DP = 26, N/P = 5), DET0/121-polyplex (DP = 121, N/P = 5), or Lipo prepared from SpCas9-mRNA for 24 h. As a positive control, the cells were transfected with SpCas9 pDNA by Lipo concurrently with the reporter components and incubated for a total of 48 h. Results are expressed as mean ± SD (n = 4).
Interestingly, the similar levels of cellular uptake efficiencies were observed for HEP-, OCT-, NON-, DEC-, and CHE-polyplexes, suggesting that the much higher IVT mRNA expression efficiency of CHE-polyplexes should be ascribed to the other transfection step following the cellular internalization. In detail, the CHE-polyplex exhibited 10-fold higher luciferase expression in the time-dependent profiles, compared to OCT-polyplex (Figure S9). Nevertheless, the CHE-polyplex showed lower cellular uptake efficiency than OCT-polyplex until 6 h of incubation (Figure S15). Thus, the endosomal escape abilities of CHE- and OCT-polyplexes were observed as a next transfection step by confocal laser scanning microscopic (CLSM) observation (Figure S16). The CLSM images display the similar subcellular distribution of Cy5-mRNA between CHE- and OCT-polyplexes. This is probably due to the fact that both polyplexes comprised similar amounts of DET moieties as an endosome-disrupting unit. To gain more insight into the higher mRNA expression by CHE-polyplex, the IVT mRNA releasability of CHE-polyplex inside cells was evaluated in comparison with OCT-polyplex by flow cytometry combined with a FRET technique. First, the FRET intensity of each polyplex prepared from Cy3-mRNA and Alexa647-PAsp(DET/R) (DP = 26, N/P = 5) was compared in a buffer solution (Figure 3B). Both CHE- and OCT-polyplexes exhibited similar FRET intensities (defined as fluorescence intensity (FI) at 670 nm/FI at 565 nm) of 0.28. These FRET intensities were apparently higher than the background signal (∼0.035) calculated from naked Cy3-mRNA (FI at 565 nm: 285) and free PAsp(DET/R)s (FI at 670 nm: ∼10), indicating the effective condensation of IVT mRNA by both PAsp(DET/OCT) and PAsp(DET/CHE). Then, we assumed that the IVT mRNA delivered by CHE-polyplex should be more rapidly decondensed inside the cells, compared with OCT-polyplex, to allow for the more rapid and higher mRNA expression. To verify this assumption, the FRET intensities of the Cy3-mRNA/Alexa647-PAsp(DET/R) pair in the cells treated with CHE- and OCT-polyplexes were quantitatively analyzed after 6 and 12 h of incubation by flow cytometer (Figure 3C). The FRET intensities derived from CHE-polyplex at 6 and 12 h were significantly lower than those of OCT-polyplex at 6 and 12 h, respectively. This result confirms that PAsp(DET/CHE) was more rapidly detached from IVT mRNA within the cells compared with PAsp(DET/OCT). Thus, the rapid decondensation of IVT mRNA in CHE-polyplex in the cells more likely allowed for the more rapid and higher IVT mRNA expression.
2.5. In Vitro SpCas9-mRNA Delivery by CHE-Polyplex
The CRISPR/Cas9 technology can site-specifically modify the genome in a wide variety of cell types3,26 and may change the modality of treatment for complex heritable and somatic disorders.27 Herein, the nuclease activity of SpCas9 was investigated by a luciferase-reporter pDNA homology-directed repair (HDR) assay (Figure 3D).28 The measurement of luciferase luminescence intensity allows us to estimate the SpCas9-mRNA activity. Human cervical cancer, HeLa cells, were first transfected with reporter components, i.e., sgRNAs, StopFluc reporter, and luciferase donor, using Lipofectamine 3000 (Lipo). Of note, HeLa cells were used here because the significant gene editing activity by the reporter components was successfully demonstrated in our previous study.28 As the optimized formulation, CHE-polyplex was tested for the delivery of SpCas9-mRNA with the base number of ∼4500 in comparison with a commercially available transfection reagent (Lipo). After 24 h of incubation with the reporter components, the cells were additionally incubated with SpCas9-mRNA-loaded samples for another 24 h. The Lipo loaded with pDNA encoding SpCas9 (Lipo/SpCas9 pDNA) was used as a positive control. Luminescence intensities of nontreated cells and the cells treated with the reporter components were 190 ± 10 and 390 ± 30, respectively (Figure 3E). In contrast, CHE-polyplex showed significantly higher SpCas9 nuclease activity (1800 ± 150) than Lipo (1350 ± 90) and DET0/121-polyplex (420 ± 40), demonstrating that PAsp(DET/CHE) had a superior transfection activity of SpCas9-mRNA in this HDR assay. Note that the luminescence intensity of the cells treated with the SpCas9 pDNA-loaded Lipo as a positive control was 2380 ± 330. This higher intensity might be due to long-lasting expression of SpCas9 and the longer contact time of pDNA-loaded Lipo with the cells (48 h).
2.6. In Vivo mRNA Delivery to Brain
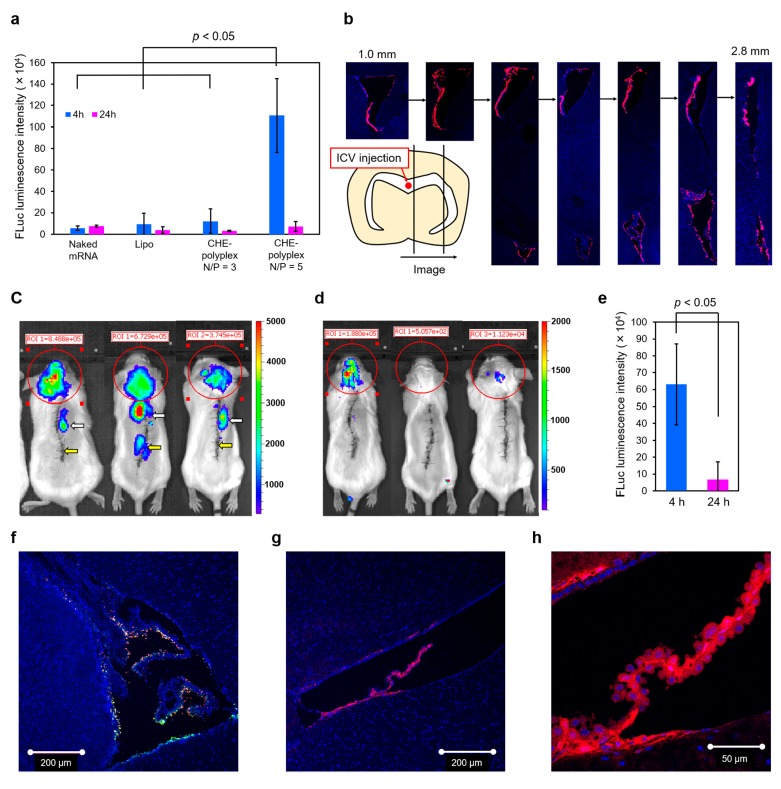
We further investigated the in vivo IVT mRNA delivery efficiency of CHE-polyplex in the mouse brain. This experiment supposes that CRISPR/Cas9 technology can be potentially applied for the treatment of Alzheimer’s disease4 and Huntington disease.5 FLuc-mRNA-loaded CHE-polyplexes (N/P = 3 and 5) and Lipo were intracerebroventricularly administered into the mice. The protein expression efficiency of FLuc-mRNA in the brain was quantitatively evaluated by IVIS measurement 4 and 24 h after administration (Figure 4A and Figure S17). The CHE-polyplex (N/P = 5) showed significantly higher luminescence intensities at 4 h, compared with CHE-polyplex (N/P = 3) and Lipo. Then, the luminescence intensities of the CHE-polyplex (N/P = 5) rapidly decreased after 24 h, indicating that the CHE-polyplex efficiently elicited the protein production in an early time period after injection even under the in vivo condition. The higher FLuc expression by CHE-polyplex (N/P = 5) is probably due to its higher cellular uptake, compared to the CHE-polyplex (N/P = 3). The higher ζ potential of the CHE-polyplex (N/P = 5) might induce the rapid internalization into the brain cells, which is in the small volume (∼40 μL) of cerebrospinal fluid (CSF).29 The distribution of CHE-polyplexes in the brain ventricular area after intracerebroventricular injection was further observed for Cy5-mRNA-loaded CHE-polyplex (N/P = 5) and naked Cy5-mRNA (Figure 4B). The brain was excised in 4 h postadministration and sliced as a sagittal section. DAPI-stained nuclei and Cy5-mRNAs were shown in blue and red, respectively, in the CLSM images. The Cy5 fluorescence derived from CHE-polyplex was distributed mainly in the ependymal layer of third and lateral ventricles.30,31 The fluorescence was found in the brain section of even ∼2.8 mm from the injection site. As a control, the red fluorescence of naked Cy5-mRNA was mainly observed in the hippocampus and found in the brain section of ∼1.4 mm from the injection site (Figure S18A,B). CSF is produced from choroid plexus in the third, fourth, and lateral ventricle at a rate of ∼350 nL/min.29 CSF circulates in the lateral ventricle and eventually moves into the third ventricle.32 The wide range of distribution of Cy5 fluorescence (or Cy5-mRNA) by CHE-polyplex in the lateral ventricles indicates that CHE-polyplex had a significant durability even in the cerebrospinal flow.
Figure 4.
(A) Quantitative evaluation of FLuc-mRNA expression level after intracerebroventricular administration into mice. Polyplexes (DP = 26) and Lipo were administered at 4.5 μg FLuc-mRNA, and the luminescence intensities in the brain were measured by an IVIS instrument 4 and 24 h after injection. Naked mRNA was also administered as a control. Results are expressed as mean ± SD (n = 3). (B) Histological evaluation of the brain treated with CHE-polyplex (DP = 26, N/P = 5) loaded with Cy5-mRNA 4 h after injection. The brain was serially sectioned from the center to the side as illustrated. DAPI-stained nuclei and Cy5-mRNAs were shown in blue and red, respectively. (C, D) Bioluminescence images of mice intrathecally treated with FLuc-mRNA-loaded CHE-polyplex (DP = 26, N/P = 5). The FLuc expression was recorded at 4 h (C) and 24 h (D) postadministration into mice (3.0 μg FLuc-mRNA/mouse). White and yellow arrows indicate the sample and catheter injection sites, respectively. (E) Quantitative evaluation of FLuc-mRNA expression level after the intrathecal administration into mice. Results are expressed as mean ± SD (n = 3). (F) CLSM image of brain distribution of Cy5-mRNA/Cy3-sgRNA-loaded CHE-polyplexes (DP = 26, N/P = 5) 4 h after intracerebroventricular injection (1.5 μg Cy5-mRNA and 1.5 μg Cy3-sgRNA/mouse). Cy5-mRNA, Cy3-sgRNA, and DAPI-stained nuclei are shown in red, green, and blue, respectively. (G, H) tdTomato fluorescence images of Ai9 mice intracerebroventricularly treated with SpCas9-mRNA/sgRNA-loaded CHE-polyplexes (DP = 26, N/P = 5). The tdTomato expression was recorded at 2 days postadministration into the mice (1.5 μg SpCas9-mRNA and 1.5 μg sgRNA/mouse). DAPI-stained nuclei and tdTomato are shown in blue and red, respectively. The ventricles were observed by using 10× (G) and 40× (H) objectives.
We further examined a potential availability of CHE-polyplex in the intrathecal administration because most administration protocols of therapeutic nucleic acids in clinical trials for treatment of CNS disorder employ intrathecal injections.33 FLuc-mRNA-loaded CHE-polyplex (N/P = 5) was intrathecally administered into the subarachnoid space at Th10. The luciferase protein expression in the mouse was quantitatively evaluated by IVIS measurement at 4 and 24 h postadministration (Figure 4C,D). The luciferase expression was observed in the brain probably because the CHE-polyplex stably diffused into the CSF of subarachnoid space at 4 h. We also observed luciferase expression at the injection site on the spinal canal. The luminescence intensities of CHE-polyplex at 4 h rapidly decreased to 1/10 at 24 h (Figure 4E), similar to the intracerebroventricular administration. Overall, these results demonstrate that CHE-polyplex has a strong potential for the local brain delivery of therapeutic IVT mRNA.
Ultimately, the gene editing activity of CHE-polyplex was validated in the Ai9 mouse model, where a STOP cassette prevents the transcription of fluorescence protein, tdTomato.34 The gene deletion via a double-stranded break in the Ai9 DNA sequence turns on the expression of the fluorescence protein. First, CHE-polyplexes were separately prepared from Cy5-mRNA and Cy3-labeled chemically synthesized sgRNA at N/P = 5, and were intracerebroventricularly administered into BALB/c mice.35 The distribution of two different polyplexes in the brain was observed because they need to be taken up by the same cells for the gene deletion. At 4 h postadministration, the brain was excised and sliced as a sagittal section, followed by nuclear staining with DAPI. The CLSM images of the tissue section displayed that the fluorescences of Cy5-mRNA and Cy3-sgRNA were apparently overlapped in the ependymal layer of ventricles and choroid plexus (Figure 4F and Figure S19), indicating that the two different polyplexes were internalized similarly into the brain cells. Next, CHE-polyplexes were separately prepared from SpCas9-mRNA and sgRNA at N/P = 5 and intracerebroventricularly administered into Ai9 mice. tdTomato fluorescence was observed in the brain tissue section 2 days after the administration. The tdTomato fluorescence was also mainly detected in the ependymal layer of ventricles and choroid plexus (Figure 4G), consistent with the distribution of Cy5-mRNA- and Cy3-sgRNA-loaded polyplexes. Clearly, the fluorescence was observed in both nucleus and cytosol areas (Figure 4H). These data demonstrate that CHE-polyplex efficiently delivered SpCas9-mRNA and sgRNA into the brain cells to edit the genomic DNA through intracerebroventricular administration. It should be further noted that the present polymer design can be extended for the brain-targeted systemic delivery of IVT mRNA by installing specific ligand molecules, such as glucose, peptides, and proteins, to cross the blood–brain barrier via the transcellular pathway.36−38
2.7. Safety Statement
No unexpected or unusually high safety hazards were encountered.
3. Conclusion
In summary, a series of amphiphilic polyaspartamide derivatives were synthesized to optimize the polyplex-mediated IVT mRNA delivery. The obtained results reveal that the polyaspartamide side chain structures and their contents dramatically altered the IVT mRNA expression efficiency. Importantly, the in vitro mRNA expression profiles were well-documented by a parameter of hydrophobicity, log P. The 3.5 orders of magnitude higher mRNA expression efficiencies were obtained from the higher log P group (with log P > −2.3), compared with the lower log P group (with log P < −2.4). In the higher log P group, the polyaspartamide derivative bearing the lower log P, i.e., PAsp(DET/CHE11)26, accomplished the highest IVT mRNA expression efficiency, probably due to the optimized balance in hydrophobicity for the significant polyplex stability in extracellular milieu and the prompt release of mRNA payloads inside the cells. Ultimately, the CHE-polyplex achieved the gene deletion in the brain through intracerebroventricular injection into the Ai9 mice. Altogether, the present study, for the first time, provides a useful index for the optimization of hydrophobicity of polycations for the enhanced IVT mRNA delivery. Also, the optimized derivative, PAsp(DET/CHE11)26, was demonstrated to have a high potential for the IVT mRNA-mediated genome engineering.
Acknowledgments
This work was financially supported by Center of Innovation (COI) program from Japan Science and Technology Agency (JST) and Grants-in-Aid for Scientific Research (KAKENHI Grant: 18K18378 for H.J.K.) from Ministry of Education Culture, Sports, Science and Technology (MEXT). This work was also partially supported by Basic Science and Platform Technology Program for Innovative Biological Medicine for K.M. and Research and Development of Core Technologies for Gene and Cell Therapy (JP18ae0201009) for H.J.K. from Japan Agency for Medical Research and Development (AMED), and a grant JSPS Core-to-Core Program, A. Advanced Research Networks. H.J.K. thanks Dr. S. Uchida and Dr. N. Yoshinaga at The University of Tokyo for their help with IVT mRNA preparation. Dr. Y. Lee at Department of Chemistry, Seoul National University, provided a valuable discussion regarding hydrophobicity.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00843.
Materials and methods, compositions, log P, DP, introduction rates, nomenclature of polyplexes, DH, PDI, ζ potential, synthetic procedure, 1H NMR spectra, agarose gel electrophoresis, luciferase assay in various conditions, cellular viability, cellular uptake profiles, cellular uptake efficiencies plotted against log P, naked mRNA amounts plotted log P, scattering light intensity at 0 mM and 150 NaCl, time-dependent cellular uptake, CLSM images, bioluminescence images, and histological evaluation (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Sahin U.; Karikó K.; Türeci Ö. mRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discovery 2014, 13, 759–780. 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Vallazza B.; Petri S.; Poleganov M. A.; Eberle F.; Kuhn A. N.; Sahin U. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. WIREs Interdiscip. Rev. RNA 2015, 6, 471–499. 10.1002/wrna.1288. [DOI] [PubMed] [Google Scholar]
- Wang H.-X.; Li M.; Lee C. M.; Chakraborty S.; Kim H.-W.; Bao G.; Leong K. W. CIRSPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem. Rev. 2017, 117, 9874–9906. 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- György B.; Lööv C.; Zaborowski M. P.; Takeda S.; Kleinstiver B. P.; Commins C.; Kastanenka K.; Mu D.; Volak A.; Giedraitis V.; Lannfelt L.; Maguire C. A.; Joung J. K.; Hyman B. T.; Breakefield X. O.; Ingelsson M. CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol. Ther.--Nucleic Acids 2018, 11, 429–440. 10.1016/j.omtn.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Chang R.; Yang H.; Zhao T.; Hong Y.; Kong H. E.; Sun X.; Qin Z.; Jin P.; Li S.; Li X.-J. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 2017, 127, 2719–2724. 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. B.; Platt R. J.; Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015, 21, 121–131. 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski P. S.; Capasso Palmiero U.; Huang Y.; Rudra A.; Langer R.; Anderson D. G. Ionizable amino-polyesters synthesized via ring opening polymerization of tertiary amino-alcohols for tissue selective mRNA delivery. Adv. Mater. 2018, 30, 1801151. 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Wang W.; He Y.; Li Y.; Yan E. Z.; Zhang K.; Irvine D. J.; Hammond P. T. Structurally programmed assembly of translation initiation nanoplex for superior mRNA delivery. ACS Nano 2017, 11, 2531–2544. 10.1021/acsnano.6b08447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. A.; Xu Y.; Tao W.; Ubellacker J. M.; Lim M.; Aum D.; Lee G. Y.; Zhou K.; Zope H.; Yu M.; Cao W.; Oswald J. T.; Dinarvand M.; Mahmoudi M.; Langer R.; Kantoff P. W.; Farokhzad O. C.; Zetter B. R.; Shi J. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018, 2, 850–864. 10.1038/s41551-018-0284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. B.; Zhang S.; Kos P.; Xiong H.; Zhou K.; Perelman S. S.; Zhu H.; Siegwart D. J. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Angew. Chem., Int. Ed. 2017, 56, 1059–1063. 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K.; Oba M.; Nakanishi M.; Fukushima S.; Yamasaki Y.; Koyama H.; Nishiyama N.; Kataoka K. Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J. Am. Chem. Soc. 2008, 130, 16287–16294. 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]
- Uchida H.; Itaka K.; Nomoto T.; Ishii T.; Suma T.; Ikegami M.; Miyata K.; Oba M.; Nishiyama N.; Kataoka K. Modulated protonation of side chain aminoethylene repeats in N-substituted polyaspartamides promotes mRNA transfection. J. Am. Chem. Soc. 2014, 136, 12396–12405. 10.1021/ja506194z. [DOI] [PubMed] [Google Scholar]
- Cabral H.; Miyata K.; Osada K.; Kataoka K. Block copolymer micelles in nanomedicine applications. Chem. Rev. 2018, 118, 6844–6892. 10.1021/acs.chemrev.8b00199. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Nishiyama N.; Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem. Soc. Rev. 2012, 41, 2562–2574. 10.1039/C1CS15258K. [DOI] [PubMed] [Google Scholar]
- McKinlay C. J.; Benner N. L.; Haabeth O. A.; Waymouth R. M.; Wender P. A. Enhanced mRNA delivery into lymphocytes enabled by lipid-varied libraries of charge-altering releasable transporters. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E5859–E5866. 10.1073/pnas.1805358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton O. S.; Kauffman K. J.; McClellan R. L.; Kaczmarek J. C.; Zeng M. D.; Andresen J. L.; Rhym L. H.; Heartlein M. W.; DeRosa F.; Anderson D. G. Customizable lipid nanoparticle materials for the delivery of siRNAs and mRNAs. Angew. Chem., Int. Ed. 2018, 57, 13582–13586. 10.1002/anie.201809056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H.; Nakatani T.; Furihata T.; Tange K.; Nakai Y.; Yoshioka H.; Harashima H.; Akita H. In vivo introduction of mRNA encapsulated in lipid nanoparticles to brain neuronal cells and astrocytes via intracerebroventricular administration. Mol. Pharmaceutics 2018, 15, 2060–2067. 10.1021/acs.molpharmaceut.7b01084. [DOI] [PubMed] [Google Scholar]
- Dharmaratne N. U.; Jouaneh T. M.; Kiesewetter M. K.; Mathers R. T. Quantitative measurements of polymer hydrophobicity based on functional group identity and oligomer length. Macromolecules 2018, 51, 8461–8468. 10.1021/acs.macromol.8b01747. [DOI] [Google Scholar]
- Li Y.; Zhao T.; Wang C.; Lin Z.; Huang G.; Sumer B. D.; Gao J. Molecular basis of cooperativity in pH-triggered supramolecular self-assembly. Nat. Commun. 2016, 7, 13214. 10.1038/ncomms13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa P.; Wever D. A.; Picchioni F.; Broekhuis A. A. Polymeric surfactants: synthesis, properties, and links to application. Chem. Rev. 2015, 115, 8504–8563. 10.1021/cr500129h. [DOI] [PubMed] [Google Scholar]
- Itaka K.; Ishii T.; Hasegawa Y.; Kataoka K. Biodegradable polyamino acid-based polycations as safe and effective gene carrier minimizing cumulative toxicity. Biomaterials 2010, 31, 3707–3714. 10.1016/j.biomaterials.2009.11.072. [DOI] [PubMed] [Google Scholar]
- Shah P.; Ding Y.; Niemczyk M.; Kudla G.; Plotkin J. B. Rate-limiting steps in yeast protein translation. Cell 2013, 153, 1589–1601. 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide A.; David A. The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis 2018, 7, 34. 10.1038/s41389-018-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnoud J.; Rossi G.; Marrink S. J.; Monticelli L. Hydrophobic compounds reshape membrane domains. PLoS Comput. Biol. 2014, 10, e1003873 10.1371/journal.pcbi.1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo H.; Ogita A.; Kanayama N.; Kawagishi M.; Sato E.; Yamamoto N.; Arai K.; Izawa M. A. Effects of the chemical specificity of benzoic acid and its analogs on osmotic fragility in erythrocytes of Sprague-Dawley rats in vitro. Eur. J. Pharmacol. 2013, 702, 142–148. 10.1016/j.ejphar.2013.01.029. [DOI] [PubMed] [Google Scholar]
- Doudna J. A.; Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Fellmann C.; Gowen B. G.; Lin P.-C; Doudna J. A.; Corn J. E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discovery 2017, 16, 89–100. 10.1038/nrd.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y.; Kawano F.; Nakajima T.; Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015, 33, 755–760. 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- Simon M. J.; Iliff J. J. Regulation of cerebrospinal fluid (CSF) flow in neurodegenerative, neurovascular and neuroinflammatory disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2016, 1862, 442–451. 10.1016/j.bbadis.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D. A.; Alvarez-Buylla A. The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harbor Perspect. Biol. 2016, 8, a018820 10.1101/cshperspect.a018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones-Hinojosa A.; Chaichana K. The human subventricular zone: a source of new cells and a potential source of brain tumours. Exp. Neurol. 2007, 205, 313–324. 10.1016/j.expneurol.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Fiorelli R.; Azim K.; Fischer B.; Raineteau O. Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development 2015, 142, 2109–2120. 10.1242/dev.119966. [DOI] [PubMed] [Google Scholar]
- Evers M. M.; Toonen L. J.; van Roon-Mom W. M. Antisense oligonucleotides in therapy for neurogenerative disorders. Adv. Drug Delivery Rev. 2015, 87, 90–103. 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Staahl B. T.; Benekareddy M.; Coulon-Bainier C.; Banfal A. A.; Floor S. N.; Sabo J. K.; Urnes C.; Munares G. A.; Ghosh A.; Doudna J. A. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017, 35, 431–434. 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A.; Bak R. O.; Clark J. T.; Kennedy A. B.; Ryan D. E.; Roy S.; Steinfeld I.; Lunstad B. D.; Kaiser R. J.; Wilkens A. B.; Bacchetta R.; Tsalenko A.; Dellinger D.; Bruhn L.; Porteus M. H. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. T.; Webster P.; Gale A.; Davis M. E. Transcytosis and brain uptake of transferrin-containing nanoparticles by tuning avidity to transferrin receptor. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 8662. 10.1073/pnas.1307152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku Y.; Kuwahara H.; Fukusato Y.; Mizoguchi A.; Ishii T.; Nitta K.; Matsumoto Y.; Toh K.; Miyata K.; Uchida S.; Nishina K.; Osada K.; Itaka K.; Nishiyama N.; Mizusawa H.; Yamasoba T.; Yokota Y.; Kataoka K. Glycaemic control boosts glucosylated nanocarrier crossing the BBB into the brain. Nat. Commun. 2017, 8, 1001. 10.1038/s41467-017-00952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Zhang J.; Meng F.; Zhong Z. Apolipoprotein E peptide-directed chimeric polymersomes mediate an ultrahigh-efficiency targeted protein therapy for glioblastoma. ACS Nano 2018, 12, 11070–11079. 10.1021/acsnano.8b05265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.