Abstract
Despite rapid evolution in the area of microbial natural products chemistry, there is currently no open access database containing all microbially produced natural product structures. Lack of availability of these data is preventing the implementation of new technologies in natural products science. Specifically, development of new computational strategies for compound characterization and identification are being hampered by the lack of a comprehensive database of known compounds against which to compare experimental data. The creation of an open access, community-maintained database of microbial natural product structures would enable the development of new technologies in natural products discovery and improve the interoperability of existing natural products data resources. However, these data are spread unevenly throughout the historical scientific literature, including both journal articles and international patents. These documents have no standard format, are often not digitized as machine readable text, and are not publicly available. Further, none of these documents have associated structure files (e.g., MOL, InChI, or SMILES), instead containing images of structures. This makes extraction and formatting of relevant natural products data a formidable challenge. Using a combination of manual curation and automated data mining approaches we have created a database of microbial natural products (The Natural Products Atlas, www.npatlas.org) that includes 24 594 compounds and contains referenced data for structure, compound names, source organisms, isolation references, total syntheses, and instances of structural reassignment. This database is accompanied by an interactive web portal that permits searching by structure, substructure, and physical properties. The Web site also provides mechanisms for visualizing natural products chemical space and dashboards for displaying author and discovery timeline data. These interactive tools offer a powerful knowledge base for natural products discovery with a central interface for structure and property-based searching and presents new viewpoints on structural diversity in natural products. The Natural Products Atlas has been developed under FAIR principles (Findable, Accessible, Interoperable, and Reusable) and is integrated with other emerging natural product databases, including the Minimum Information About a Biosynthetic Gene Cluster (MIBiG) repository, and the Global Natural Products Social Molecular Networking (GNPS) platform. It is designed as a community-supported resource to provide a central repository for known natural product structures from microorganisms and is the first comprehensive, open access resource of this type. It is expected that the Natural Products Atlas will enable the development of new natural products discovery modalities and accelerate the process of structural characterization for complex natural products libraries.
Short abstract
The Natural Products Atlas is a new online database of microbially derived natural product structures, designed as a comprehensive open access repository for the scientific community.
Introduction
The field of natural products is enjoying a period of rapid innovation and technological advancement as new tools are developed for unbiased characterization of natural product mixtures.1 These include chemical,2 biological,3 and bioinformatic4,5 approaches which are broadening our viewpoint on the diversity, distribution, and functions of natural products. However, despite the rapid evolution in this area, there is currently no open access database containing all microbially produced natural product structures. Lack of availability of these data is hampering the implementation of new ideas and approaches in this area. Many of these strategies, such as the prediction of mass spectrometry fragmentation patterns,6,7 rely on the availability of known compound databases against which to compare experimental data. In addition, the absence of a consensus structure data set precludes the integration of information from different natural products characterization platforms (e.g., MIBiG8 and GNPS9), limiting the interoperability of these individual resources.
The current landscape for microbial natural products databases is large, but fragmented. Existing databases are either commercial and do not make all structures accessible (e.g., Antibase, MarinLit, The Dictionary of Natural Products), or are free but have limited information on compound origin (e.g., Supernatural II10), are not easily downloadable (e.g., NPEdia), or are narrowly defined (e.g., StreptomeDB,11 AfroDB,12 and NuBBEDB13). In addition, most existing databases are not accurately referenced, with many data points being presented without details about their primary source. Absence of accurate referencing limits the value of these data, preventing researchers from easily evaluating the original source material for accuracy and validity. Many natural product databases are available to the community (Table 1), each of which offer different content and analysis tools. However, none of these provide comprehensive coverage of microbial natural product structures under FAIR principles.14 For this reason, we elected to create a new microbial natural products database, termed the Natural Products Atlas (www.npatlas.org).
Table 1. Examples of Existing Natural Product Structure Databases.
| database | description | URL | access | number of compounds | last revision | downloadable? |
|---|---|---|---|---|---|---|
| Dictionary of Natural Products | plant, microbial and marine natural products | 1a | commercial | 226 000 | 2019 | no |
| MarinLit | marine-derived natural products, including invertebrates, algae and microorganisms | 2b | commercial | ∼30 000 | July 2019 | no |
| Antibase | microbial natural products | 3c | commercial | 43 700 | May 2017 | no |
| StreptomeDB | natural products derived from bacteria of the genus Streptomyces | 4d | open access | 4040 | Aug. 2015 | yes |
| Supernatural II | chemical structures of primary and secondary metabolites and natural macromolecules | 5e | open access | 325 508 | April 2018 | no |
| NPEdia | natural products from plants and microorganisms, focused on structural features | 6f | open access | 49 610 | Jan, 2014 | no |
| AfroDB | natural products from African medicinal plants | 7g | open access | 1000 | Nov. 2013 | yes |
| NuBBEDB | natural products isolated from Brazil | 8h | open access | 2200 | Aug. 2017 | yes |
http://132.230.56.4/streptomedb2/ (Previously: http://www.pharmaceutical-bioinformatics.de/streptomedb2/).
Downloadable at https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0078085 (ZINC searchable: http://zinc.docking.org/catalogs/afronp/).
At a minimum, any natural product database should contain information on compound structures, names, producing organisms, and isolation reference. Extracting even these basic data from the primary literature for all microbial natural products is a challenging objective. Structures have been reported as early as the late 1800s (e.g., polyporic acid, 187715). Over the intervening period, both article layout (e.g., structure representation) and style (e.g., phraseology and terminology) have changed significantly, making it difficult to define rules for text mining across the full scientific literature. In addition, natural products science is an international discipline, and discoveries are reported in a broad range of languages beyond English, which limits options for text-based article prioritization. Finally, many of the early journal articles are either unavailable or are only available as images, making them unsuited for automated data extraction.
Natural product structures have been reported in a broad cross-section of journals, broadening the search scope substantially. Further, despite the excellent coverage provided by PubChem, ChEMBL, ChemSpider, and other resources, not all structures are currently available in open access structure repositories. When combined with issues of structure misassignment, structure reassignment, synonym creation, and taxonomic revision, it is perhaps clear why no such database is currently publicly available.
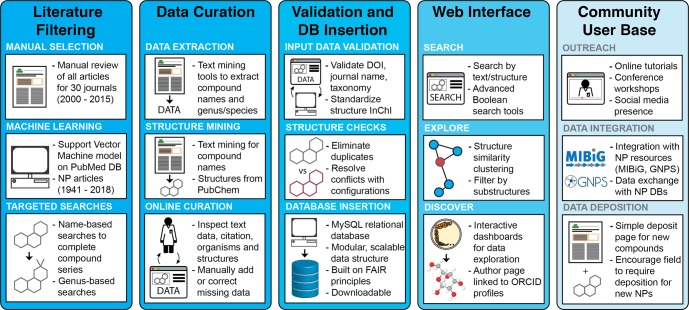
In order to address this issue, we have created a new automated curation platform designed to identify articles related to natural products discovery and extract the relevant information for final manual verification (Figure 1 and Supporting Information). From this curation effort, we have extracted 24 594 compounds from 10 481 journal articles spanning 306 journal titles. These results were inserted into a MySQL relational database, and an interactive online web portal was created to permit searching, filtering, and visualization of the data set. This online knowledge base contains a growing fraction of published microbial natural products and is designed to encourage additional data deposition and curation from the natural products community. To this end, we have incorporated online tools for new data deposition and the correction of existing data. It is hoped that this community-driven model will help to ensure that the data are of high quality, will improve the database in terms of overall coverage, and will assist with the long-term sustainability of this resource.
Figure 1.
Workflow for creation and curation of the Natural Products Atlas.
Results and Discussion
The Structure of the Natural Products Atlas
The Natural Products Atlas project aimed to create a comprehensive database of all microbially derived natural products and to make this database freely accessible to the research community. The long-term vision for the database is to include:
Both primary literature and patents
All microbial natural product structures, compound names, and synonyms
Physicochemical data (optical rotation, infrared, and ultraviolet absorptions and direct links to nuclear magnetic resonance and mass spectrometry databases)
Bioactivity data
All instances of total synthesis
All structural reassignments
Taxonomy for the original producing organism
Full list of all producing organisms
In addition, the database should comply with the requirements that:
All data are fully referenced
Entries include the original isolation paper for each compound (first instance where complete structure reported)
Curation and optimization should be community-driven
The data be open access and fully downloadable
In this initial release, we have accomplished many of these goals, while reserving others for future expansion. Specifically, we have focused on data from the primary scientific literature covering the period 1941–2018. Currently, these entries include both original isolation name and the original published taxonomy for the producing organism. With contributions from a large group of volunteer curators, we have incorporated ∼25 000 microbial natural product structures from this period. To be of the highest value, the Natural Products Atlas should be related to other natural products data resources. We have invested significant curation effort to generate accurate links between the Natural Products Atlas and both MIBiG (a biosynthetic gene cluster database) and GNPS (a database of mass spectrometry data for natural products). In addition, we have incorporated references to instances of total synthesis for Natural Products Atlas entries, as well as instances of structure reassignment through synthesis or reisolation.
Compound Selection
There is likely no single definition of “natural product” that would satisfy all researchers in the field. Nevertheless, we required a set of guiding principles for compound inclusion in order to construct the Natural Products Atlas. For this application, “natural product” was defined as any naturally occurring metabolite with a molecular weight less than 3000 Da produced by a microorganism. Primary metabolites, as defined by the KEGG database,16 were excluded. Shunt products from biosynthetic gene clusters were included if detected in wild-type organisms, but excluded if only found through genetic manipulation. Compounds derived from augmenting fermentation media with biosynthetic feedstocks were included if those feedstocks could plausibly have been found in nature (e.g., proteinogenic amino acids) but excluded if of synthetic origin (e.g., p-fluoro-phenylalanine). Finally, compounds produced by biotransformation (modification of a purified natural product by microbial fermentation with a different organism) were excluded.
Taxonomic Boundaries
The definition of microorganism was also complicated by a number of edge cases. We elected to include natural products from both lichens and macroscopic fungi (Basidiomycetes) as both are classified as fungi in most modern taxonomic ontologies. We included cyanobacteria, but excluded eukaryotic phytoplankton and other photosynthetic microalgae. Further, some taxonomic assignments have changed over time, meaning that the genus and species reported in the original literature have now been reassigned. Currently, the Natural Products Atlas contains taxonomic assignments as originally defined. A future goal is to align these taxonomic assignments with the Integrated Taxonomic Identification System (ITIS; www.itis.gov). This would provide automatic updates to species assignments and would improve interoperability with other taxonomy-based resources.
Natural Products Atlas Structure
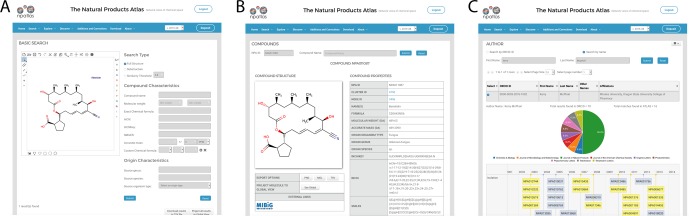
The Natural Products Atlas knowledge base is divided into three main sections: Search, Explore, and Discover (Figure 2). The search pages (Figure 2A) provide several different searching modalities and are designed to prioritize ease of use and flexibility. The basic search page permits rapid searching using a range of terms including structure, name, taxonomy, and so on. Substructure and similarity structure searching are both enabled, allowing researchers to dereplicate isolated compounds based on partial structures, or compare new compounds to the structural diversity of the known natural products world. The advanced search page allows more complex Boolean searches (e.g., molecular weight range x and date range y) and permits combinations of structure and text criteria. Results from these searches are displayed in paginated tables online or can be exported for offline use. For example, if a researcher is studying a given genus and wishes to create a compound reference library in a proprietary software package (e.g., LCMS software), they can search the genus name in the basic search page and then export the full results set as a single file.
Figure 2.
(A) Search interface (basic search). (B) Explore view (compound), (C) Discover view (author).
The Explore section (Figure 2B) provides different viewpoints on chemical similarity in natural products chemical space. Four levels of resolution are provided: compound, cluster, node, and global. This visualization is analogous to views of house, town, province, and globe in mapping applications like Google Maps. The compound page (Figure 3A) provides detailed information about each individual compound and includes export of various data types (compound MDL MOLfile, compound image, full TSV file for all compound data). This page also provides citations for the original isolation reference, instances of total synthesis, and instances of structural reassignment, each of which is accompanied by a DOI hyperlink to the journal article page.
Figure 3.
Four views in the Explore section. (A) Compound view, providing data for individual compounds. (B) Cluster view, illustrating compounds with close structural similarity. (C) Node view, illustrating clusters of compounds that are more distantly related. (D) Global view, presenting distribution of all chemical space in the Natural Products Atlas.
The cluster pages (Figure 3B) present groups of compounds in the Natural Products Atlas with close structural similarities. Structural similarity is defined by Morgan fingerprinting (radius = 2) and Dice similarity scoring (0.75 cutoff). This tool allows users to easily visualize the scope and diversity of similar structures in the data set. The next visualization layer, nodes, illustrates how clusters are related to one another at the level of compound class. These relationships are determined by taking the most interconnected member of each cluster and then scoring structural similarities between these cluster representatives using a less stringent similarity scoring method (Atom pairs fingerprinting and Dice similarity scoring (0.7 cutoff)). In the node networks (Figure 3C), node diameter is proportional to the number of compounds in each node. Finally, the global view (Figure 3D) presents all of the nodes in the Natural Products Atlas, arrayed as a spherical plot. The distribution of nodes in this spherical plot is an extension of van Krevelen diagrams,17 which represent molecules using C:H and C:O ratios from their molecular formulas. In the global plot, the C:H ratio defines the polar angle (the radial value in the xy-plane), the C:O ratio defines the azimuth value (the angle with the z-axis), and the C:N ratio sets the radius (distance from the origin). Because these three properties are typically similar for molecules of the same compound class, nodes cluster in the global view based on compound type. Importantly, this coordinate system is dependent on compound physical properties, so the positions of existing nodes will not change as new data are added to the Natural Products Atlas over time.
All the layers of the Explore section are interconnected, so that users can navigate between viewpoints from compound through to global (Figure 3). Node highlighting has been implemented to illustrate the position of compounds of interest in the plots. For example, if a user starts on a compound page (Figure 3A) and navigates from there to the corresponding cluster (Figure 3B), the position in the cluster network corresponding to the original compound is highlighted in red. This highlighting tracks through both node (Figure 3C) and global (Figure 3D) views, which helps users navigate these representations of the natural product chemical space.
In addition, search results can be projected onto the global view, illustrating the distribution of compound subsets in global chemical space. For example, functional group prevalence and distribution can be visualized by searching this group in the basic search structure section, and clicking the “project all results to global view” button on the results page. This generates a global visualization where every node containing this functional group is highlighted in red. A substructure search for compounds containing a pyridine motif returns 791 compounds, whose distributions in chemical space can be displayed by projection to the global view (Figure 4). Similar visualizations can be generated for any combination of search terms, making this a versatile tool for examining distributions of structural or taxonomic phenomena across microbial natural products chemical space.
Figure 4.
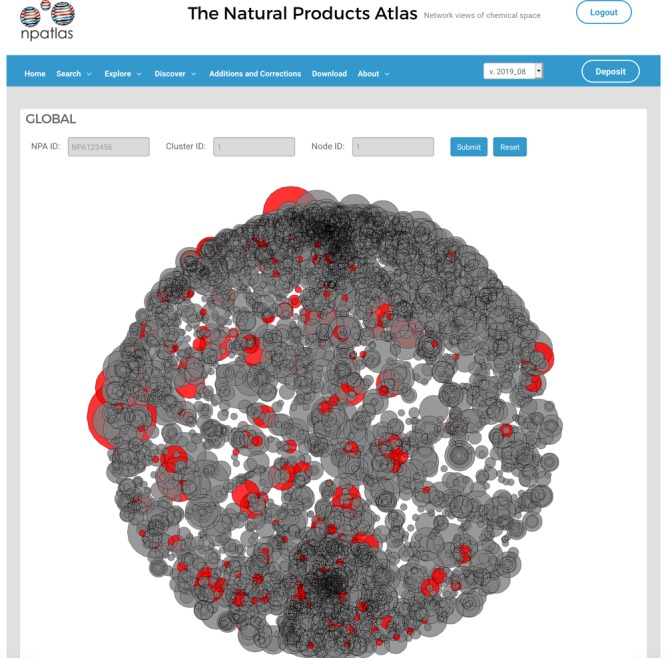
Global view, illustrating positions of all natural products containing a pyridine functional group as a substructure motif.
The Discover section (Figure 2C) is a suite of dashboards designed to present alternative viewpoints on natural products diversity. Currently the Natural Products Atlas contains three dashboards: Overview, Author, and Known Compound. The Overview dashboard provides general statistics about the contents and distribution of compounds and taxonomy in the database. The Author dashboard retrieves all linked publications for a given author from the ORCID database (https://orcid.org), and displays information about the compounds discovered in these publications, including discovery timelines, distribution of journals, and compound and citation links. The Known Compound dashboard presents an overview of pertinent data for compounds currently in the database, including timelines of discovery for cluster members and structures of related compounds.
It is envisioned that this section of the Web site will grow over time, as users request dashboards presenting alternative viewpoints on the data. For example, dashboards incorporating dereplication tools from mass spectrometry or nuclear magnetic resonance data are possible and could conceivably be of high value to the natural products community for rapidly identifying candidate structures from spectral data.
The Open Data Model
Open data principles are central to the design of the Natural Products Atlas. Committing to the open data model was critical to recruiting volunteer curators and getting buy-in from the natural products community. In line with these principles, the database is covered by a Creative Commons Attribution 4.0 International license and all data can be downloaded as a single flat file. Alternatively, results from searches can be downloaded as selected data files to permit simple data filtering, and data for individual compounds can be downloaded directly from the compound page in a standard format. The web interface is designed to be easy for other resources to link to. To facilitate the creation of links to individual pages, each compound page URL terminates in the Natural Products Atlas ID number (NPAID), making it straightforward to automate the generation of hyperlinks. Further, all pages on the Web site are open and do not require login credentials, with the exception of pages for data deposition.
Database Versioning
The Natural Products Atlas infrastructure includes support for database versioning. This is important because the addition of new compounds can impact both the contents and numbering of compound clusters and nodes. For example, addition of a new hybrid structure may create a link between two clusters that were previously separate. This could conceivably be very frustrating for researchers in the middle of a complex study using these data. To address this, the Web site presents the most recent data by default but permits users to specify any previous database version through a dropdown menu. It is also possible to download any of the previous versions of the database from the downloads section.
Database Construction
The Natural Products Atlas database was created by de novo searching of the primary literature using a two-stage process. Initially, we selected 30 journals known to contain articles reporting novel natural products discovery. A simple scoring system was used to compare title and abstract text against lists of positive and negative keywords. For example, “structure elucidation” and “natural product” were positive keywords, while “organometallic” and “essential oil” were negative score drivers. In addition, we prioritized any article containing one or more microbial genus names (excluding common pathogens) on the basis that most of these chemistry articles relate to natural products discovery. Articles that scored positively were formatted for manual review using an in-house curation software tool, and corrected to ensure that compound names, structures, source organisms and citation information were accurate for each article. Using this approach, the team reviewed ∼30 000 articles by hand to create an initial set of 12 924 compounds. Manual review of selected journal years suggested that this method captured ∼80% of all relevant articles.
This initial manual curation effort yielded a large training set of article titles and abstracts, divided into two groups; articles pertaining to natural products discovery, and those unrelated to natural products isolation. This training set is well suited for use in machine learning applications. Using a Support Vector Machine (SVM) model, we scanned titles and abstracts from 90 priority journals (1941–2018) to identify articles describing microbial natural products discovery. Compound names were extracted from titles and abstracts using an in-house text mining tool and the associated chemical structures extracted from public databases (PubChem, ChEMBL, ChemSpider) where available. Finally, CrossRef was used to confirm citation and DOI information, and the full data for each article was reviewed manually using an in-house online curation platform. Following this approach, we reviewed a further 14 700 articles, yielding a total of 13 236 compounds. To augment this data set, we performed manual searches based on common compound names, priority genera, and targeted authors. These searches filled in gaps in the data set (e.g., incomplete compound families) and captured compounds in journals not included in our original set of 90 titles. For more details about data curation, see Supporting Information.
Data Validation
Prior to insertion into the Natural Products Atlas, all data are validated to ensure data standardization and eliminate data duplication. This validation tool contains over 30 checks, including citation validation (e.g., is the journal name contained in the list of allowed journal names?), taxonomic validation (does this genus belong to the validated list of bacterial or fungal genera in the List of Prokaryotic names with Standing in Nomenclature (LPSN)18 or MycoBank?19) and structure validation (is this structure already present in the database?). Structure validation is particularly important, as the same structure can be present in the literature with different synonyms and different levels of configurational assignment. Without careful review, these structures would be entered as independent entries, erroneously increasing the number of entries in the database. For additional details about data validation, see Supporting Information.
Natural Product Atlas Identification Numbers
Articles that pass the validation step are inserted into the database, and each new compound is assigned a unique Natural Products Atlas Identification number (NPAID). These NPAIDs provide a fixed reference for each natural product that is independent of structure, taxonomy, or citation data. This is an important element of the Natural Products Atlas infrastructure, as it means that links to compounds can be maintained even if core information, such as structure or taxonomy, is updated or corrected. This improves interoperability with other resources, and provides a stable point of reference to each compound that will be preserved as the database expands.
Connectivity with Other Natural Products Databases
Numerous online repositories now exist that contain data related to microbial natural products. For example, in the area of biosynthesis, the Minimum Information about a Biosynthetic Gene cluster database (MIBiG) contains data on ∼1800 biosynthetic gene clusters and their associated natural products. This repository includes links to relevant manuscripts describing these gene clusters and includes tools to highlight other related gene clusters in the database. In the area of analytical chemistry, the Global Natural Products Social molecular networking database (GNPS) contains mass spectrometry and fragmentation data for a large number of natural products-based data sets and incorporates a growing suite of tools for comparing spectra between samples and the de novo prediction of fragmentation spectra.
Unfortunately, in many cases, the integration between independent natural products resources is poor. This is due to both variations in compound content between databases and to challenges with standardization of compound structures and trivial names. The Natural Products Atlas has collaborated with the developers of both MIBiG and GNPS to standardize structure representations between the three platforms, and to create bidirectional links to each database. Compounds which have entries in either platform display links to the respective databases on the compound page. In addition, the Natural Products Atlas data can be searched and filtered to include only compounds that have entries in one or both of these databases using terms in the Advanced Search section. It is envisioned that future releases of the Atlas will include links to a wider array of external databases including, for example, NMR spectra, biological activity data, or biogeographic information about compound collections.
The Value of the Natural Products Atlas
Rather than presenting a static list of natural product structures, the Natural Products Atlas is designed to offer users an interactive portal for natural products discovery. As data sets increase in size, methods for data filtering and visualization have a greater and greater impact on interpretation. The user interface for the Natural Products Atlas contains a suite of visualizations that we envisage will provide value to the natural products community by presenting compounds and search results from a variety of different perspectives. The expectation is that these tools will enable new modes of discovery in a range of subject areas, including natural products isolation, medicinal chemistry, and computational predictions for natural products spectral data.
For natural products isolation, the basic search page can be used to rapidly dereplicate compounds based on spectral characteristics. The data set can be filtered by accurate mass values (from mass spectrometry) or the presence of one or more functional groups (from NMR or MS/MS data). If the source organism is known, these results can be filtered on the basis of taxonomy, either by selecting only bacterial or fungal compounds, or by restricting results to a specific genus. If more than one genus is required, the Advanced Search page provides an expanded set of options for building more complex queries.
The structure-based search pane can be used in several different modalities. Structures can be directly compared to the existing data set using the Full Structure option. If chiral centers are defined, then results are restricted to direct matches. If chiral centers are left undefined, then all molecules with the same planar structure are returned as candidate matches. Substructure searching is available through the Substructure option. This option permits one or more substructures to be included in a single query, allowing users to identify candidate matching structures from partial NMR structural information. Finally, structurally related compounds can be identified using the Similarity Threshold option. This option is valuable for placing new discoveries in context with known natural product chemical space. For example, compounds that have few or no matches to known natural products even with a low similarity threshold cutoff might be of higher priority for biosynthetic investigation than compounds with large numbers of closely related analogues.
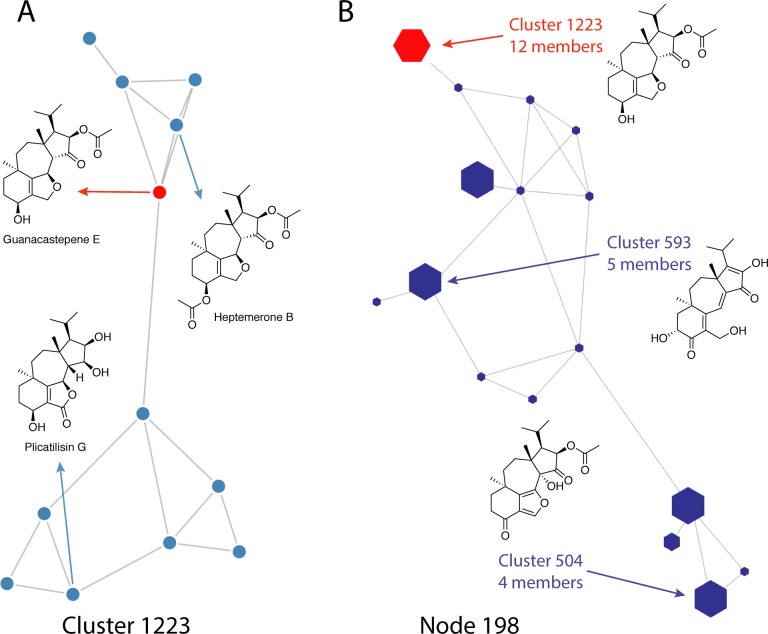
This concept of positioning in chemical space can be further explored using the tools in the Explore section. For example, if a known compound (e.g., guanacastepene E,20 NPAID 2040) is identified in a discovery project, the Cluster page can be used to determine other known natural products with close structural similarity in the same cluster (cluster 1223, Figure 5A). Switching to the Node view (Node 198, Figure 5B) reveals that this group of compounds is also related to many other structures, some closely (e.g., Cluster 593) and others more distantly (e.g., Cluster 504). Searching for Node 198 in the advanced search page affords a downloadable results table that includes relevant data for all the members of this node, including structures, taxonomy, and isolation references, permitting a more detailed evaluation of the relevant existing literature. This search modality is different to the results that would be obtained from a simple similarity search. The Node view includes “chaining” of clusters to highlight compounds with more distant structural relationships that are returned from similarity searches. This highlights common structural motifs and sites of variation within the full set of known microbial natural products, providing a more comprehensive view of related microbial natural product chemical space.
Figure 5.
(A) Cluster view for guanacastepene E (NPAID 2040, red circle) and related compounds. (B) Node view for guanacastepene E, illustrating distribution and connectivities of related clusters (purple hexagons) and example structures from each cluster.
The Natural Products Atlas also offers a number of valuable resources for medicinal chemists. At the simplest level, substructure searching can be used to identify compounds containing pharmacophore features of interest. Compounds can also be filtered on the basis of reports of total synthesis, highlighting molecules for which synthetic routes already exist or clusters of compounds for which no total synthesis has yet been reported. In situations where a specific compound has been identified as of interest for further development, the cluster and node views can be used to explore natural variations in structural features and, where published activity data exists, explore SAR features by compound class. Finally, data from the Natural Products Atlas can be used to identify areas of chemical space not occupied by a particular compound class. This information could be used to design non-natural diversity libraries around specific scaffolds or to create natural product mimics containing features and functional groups not encountered in these classes in the natural world.
Lastly, this database offers a valuable reference set for tools designed to predict spectral properties of natural products. For example, data sets containing predicted NMR chemical shifts or MS/MS fragmentation patterns could be used to improve annotation of unknown metabolites from complex mixtures. Indeed, the data from the Natural Products Atlas have already been incorporated into the network Annotation Propagation tool in GNPS with planned support for other in silico tools including DEREPLICATOR and DEREPLICATOR+.7 These tools predict compound identities from mass spectrometry fragmentation data by comparing experimental fragmentation patterns to those predicted from compound structure libraries. The inclusion of structures from the Natural Products Atlas increases the coverage of available chemical space, improving the percentage of known compounds with theoretical fragmentation spectra in the reference set. In an analogous future direction, detailed knowledge about the distribution and grouping of functional groups for different biosynthetic classes of natural products could prove valuable in predicting the functions of hypothetical proteins in biosynthetic gene clusters and for relating biosynthetic gene clusters and compound classes in large genome sequencing projects.
License Terms
The Natural Products Atlas is covered by a Creative Commons Attribution 4.0 International license (CC BY 4.0). This license means that users are free to both share and adapt the database, provided that they give credit to the Natural Products Atlas, provide a link to the CC BY 4.0 license and indicate if any changes were made. Full license terms can be found at https://creativecommons.org/licenses/by/4.0/.
Maintenance and Future Expansion
Community involvement is critical to the long-term viability of the Natural Products Atlas. Currently the database is growing at a rate of ∼1200 compounds per year, mostly through curation efforts by the authors of this manuscript. We hope that, as the profile of the Natural Products Atlas increases, users will be incentivized to deposit new compounds because of the annotation tools available in the platform. In order to facilitate user contributions, we have created a Deposit page where researchers can easily upload data from new articles. Deposition requires only the article DOI, compound names, SMILES structures, and the genus and species of the producing organism. We expect that most articles can be deposited in under 5 min, provided that authors have the structure files available. Similarly, we have created pages for reporting corrections and additions to existing entries. These pages each require fewer than five pieces of information and include both text and structure drawing tools as options for reporting structural corrections where required.
In addition to the incorporation of new compounds, ongoing effort is required to improve the coverage of the historical literature. To extend our coverage in this area, we continue to create targeted tools for highlighting missing compounds. For example, the names of compounds in the existing database can be used to identify missing members from compound series (e.g., salinipostins A–K) which can then be targeted for literature searching. To complement this effort, we are exploring options to integrate data from related databases from academia and industry to improve the coverage of compounds from previous decades.
In addition to the expansion of the database, we aim to increase both the types of data included in the Natural Products Atlas, and the range of tools available for data analysis. This effort is part of our ongoing development of the platform with input from a wide range of stakeholders. Possible future expansions include:
Extension of taxonomic classifications to include higher designations (phylum, order, etc.)
Addition of biosynthetic class assignments
Increased coverage of the patent literature
Increasing the number and variety of Discover dashboards
Increased coverage of structure reassignment and total synthesis data
Creation of a full application programming interface (API)
Both the MySQL database and the JavaScript front-end have been designed with scalability in mind. The infrastructure is therefore already in place to extend the content beyond microbial natural products. With appropriate resources and sufficient community involvement, the Natural Products Atlas would be well-positioned to incorporate natural products from marine invertebrates and ultimately plant natural products.
Conclusions
The Natural Products Atlas is the first fully open access knowledge base of microbial natural product structures build on FAIR principles. It contains a suite of interactive visualization tools to explore the chemical diversity of microbial natural products and is fully searchable and downloadable, permitting researchers to query and filter the data from a wide array of different perspectives. The base frameworks for both data curation and online visualization are scalable and could readily be extended to other classes of source organisms. We hope that the Natural Products Atlas will become a central point of reference for tools and resources that pertain to microbial natural product structures and that this will catalyze the integration of data sets that focus on a wide array of attributes of natural products.
Acknowledgments
We thank Evan Bolton (PubChem) and John Blunt and Murray Munro (MarinLit) for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00806.
A summary of the design and use of the data curation toolbox and the data validation methods used to create the Natural Products Atlas database (PDF)
Author Contributions
R.G.L. designed the project; J.V.S. created the online data curation system; R.G.L., J.V.S., G.J., A.S., D.B., and A.H. designed and created the database structure and the web interface; J.V.S., V.A., M.J.B., D.B., F.C.N., L.C., C.C., T.N.C., J.L.C.L., D.A.D., K.R.D., J.M.E., M.M.G., F.P.J.H., A.H., A.H., D.I., A.K., J.H.L., S.L., N.L., D.L., C.S.M., J.M., R.P.N., T.J.O., J.P., L.M.S., A.S., S.S., B.T., T.A.T., M.V., J.J.J.v.d.H., D.V., D.W., K.E.Z., and R.G.L. curated the data; J.J.J.v.d.H., B.T., M.H.M., J.V.S., and R.G.L. created the links to MIBiG; M.W., P.C.D., and J.V.S. created the links to GNPS; and R.G.L. wrote the manuscript. All authors have given approval to the final version of the manuscript.
This work was supported by funding from NSERC Discovery (RGL); National Institutes of Health grants U41-AT008718 (RGL), R01-GM125943 (LMS), F31-AT010098 (JME), F31-CA236237 (KEZ), T32-AT007533 (JLCL) and D43-TW010530 (TAT); National Science Foundation grants IOS-1557914 (MJB), IOS-1656475 (MJB), MCB-1817955 (LMS), and GRFP 2017247469 (AFS); the Biotechnology and Biological Sciences Research Council (BBSRC), UK BB/R022054/1 (KRD); the Carnegie Trust for the Universities of Scotland under a Collaborative Research Grant (KRD); The Netherlands eScience Center—NLeSC ASDI eScience grant, ASDI.2017.030 (JJJvdH, MHM); the São Paulo Research Foundation (FAPESP) postdoctoral fellowship 2014/12343–2 (FCN); a FAPESP Research Internship Abroad 2016/22573–0 (FCN); NSERC PGSD (DW).
The authors declare the following competing financial interest(s): RGL and MW are consultants for Sirenas. PCD is on the scientific advisory board for Sirenas. MHM is a member of the Scientific Advisory Board of Hexagon Bio and co-founder of Design Pharmaceuticals. MW is a founder of Ometa Labs.
Supplementary Material
References
- Wolfender J.-L.; Litaudon M.; Touboul D.; Queiroz E. F. Innovative Omics-Based Approaches for Prioritisation and Targeted Isolation of Natural Products - New Strategies for Drug Discovery. Nat. Prod. Rep. 2019, 36 (6), 855–868. 10.1039/C9NP00004F. [DOI] [PubMed] [Google Scholar]
- Caesar L. K.; Nogo S.; Naphen C. N.; Cech N. B. Simplify: A Mass Spectrometry Metabolomics Approach to Identify Additives and Synergists from Complex Mixtures. Anal. Chem. 2019, 91 (17), 11297–11305. 10.1021/acs.analchem.9b02377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan E. A.; Kwon G.; Clemenceau J. R.; Fisher K. W.; Vaden R. M.; Shaikh A. F.; Neilsen B. K.; Kelly D.; Potts M. B.; Sung Y.-J.; et al. A Genome-Wide Functional Signature Ontology Map and Applications to Natural Product Mechanism of Action Discovery. Cell Chem. Biol. 2019, 26, 1380. 10.1016/j.chembiol.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshko D.; Mohimani H.; Tracanna V.; Hajirasouliha I.; Medema M. H.; Korobeynikov A.; Pevzner P. A. Biosynthetic SPAdes: Reconstructing Biosynthetic Gene Clusters from Assembly Graphs. Genome Res. 2019, 29 (8), 1352–1362. 10.1101/gr.243477.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanjary M.; Steinke K.; Ziemert N. AutoMLST: An Automated Web Server for Generating Multi-Locus Species Trees Highlighting Natural Product Potential. Nucleic Acids Res. 2019, 47 (W1), W276–W282. 10.1093/nar/gkz282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K.; Fleischauer M.; Ludwig M.; Aksenov A. A.; Melnik A. V.; Meusel M.; Dorrestein P. C.; Rousu J.; Böcker S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16 (4), 299–302. 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- Mohimani H.; Gurevich A.; Shlemov A.; Mikheenko A.; Korobeynikov A.; Cao L.; Shcherbin E.; Nothias L.-F.; Dorrestein P. C.; Pevzner P. A. Dereplication of Microbial Metabolites through Database Search of Mass Spectra. Nat. Commun. 2018, 9 (1), 4035. 10.1038/s41467-018-06082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H.; Kottmann R.; Yilmaz P.; Cummings M.; Biggins J. B.; Blin K.; de Bruijn I.; Chooi Y. H.; Claesen J.; Coates R. C.; et al. Minimum Information about a Biosynthetic Gene Cluster. Nat. Chem. Biol. 2015, 11 (9), 625–631. 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34 (8), 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P.; Erehman J.; Gohlke B.-O.; Wilhelm T.; Preissner R.; Dunkel M. Super Natural II—a Database of Natural Products. Nucleic Acids Res. 2015, 43 (D1), D935–D939. 10.1093/nar/gku886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klementz D.; Döring K.; Lucas X.; Telukunta K. K.; Erxleben A.; Deubel D.; Erber A.; Santillana I.; Thomas O. S.; Bechthold A.; et al. StreptomeDB 2.0—an Extended Resource of Natural Products Produced by Streptomycetes. Nucleic Acids Res. 2016, 44 (D1), D509–D514. 10.1093/nar/gkv1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntie-Kang F.; Zofou D.; Babiaka S. B.; Meudom R.; Scharfe M.; Lifongo L. L.; Mbah J. A.; Mbaze L. M.; Sippl W.; Efange S. M. N. AfroDb: A Select Highly Potent and Diverse Natural Product Library from African Medicinal Plants. PLoS One 2013, 8 (10), e78085 10.1371/journal.pone.0078085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon A. C.; Valli M.; Dametto A. C.; Pinto M. E. F.; Freire R. T.; Castro-Gamboa I.; Andricopulo A. D.; Bolzani V. S. NuBBEDB: An Updated Database to Uncover Chemical and Biological Information from Brazilian Biodiversity. Sci. Rep. 2017, 7 (1), 7215. 10.1038/s41598-017-07451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. D.; Dumontier M.; Aalbersberg Ij. J.; Appleton G.; Axton M.; Baak A.; Blomberg N.; Boiten J.-W.; da Silva Santos L. B.; Bourne P. E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3 (1), 160018. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlschmidt C. Ueber Eine Neue in Der Natur Vorkommende Organische Säure. Justus Liebig’s Ann. der Chemie 1877, 187 (2–3), 177–197. 10.1002/jlac.18771870204. [DOI] [Google Scholar]
- Ogata H.; Goto S.; Sato K.; Fujibuchi W.; Bono H.; Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27 (1), 29–34. 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Krevelen D. Graphical Statistical Method for the Study of Structure and Reaction Processes of Coal. Fuel 1950, 29, 269–284. [Google Scholar]
- Parte A. C. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res. 2014, 42 (D1), D613–D616. 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V.; Vu D.; Amor A. B. H.; van de Wiele N.; Brouwer C.; Jabas B.; Szoke S.; Dridi A.; Triki M.; Daoud S. Ben; et al. MycoBank Gearing up for New Horizons. IMA Fungus 2013, 4 (2), 371–379. 10.5598/imafungus.2013.04.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S. F.; Bondi S. M.; Clardy J. The Guanacastepenes: A Highly Diverse Family of Secondary Metabolites Produced by an Endophytic Fungus. J. Am. Chem. Soc. 2001, 123 (40), 9900–9901. 10.1021/ja016176y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.