Abstract
Aim:
This study tested for associations between SLCO1B1 polymorphisms and circulating estrogen levels in women with breast cancer treated with letrozole or exemestane.
Patients & methods:
Postmenopausal women with hormone-receptor positive breast cancer were genotyped for SLCO1B1*5 (rs4149056) and rs10841753. Pretreatment and on-treatment plasma estrogens and aromatase inhibitor (AI) concentrations were measured. Regression analyses were performed to test for pharmacogenetic associations with estrogens and drug concentrations.
Results:
SLCO1B1*5 was associated with elevated pretreatment estrone sulfate and an increased risk of detectable estrone concentrations after 3 months of AI treatment.
Conclusion:
These findings suggest SLCO1B1 polymorphisms may have an effect on estrogenic response to AI treatment, and therefore may adversely impact the anticancer effectiveness of these agents.
Keywords: : breast cancer, exemestane, letrozole, pharmacogenomics, SLCO1B1
Approximately 80% of breast cancers are hormone-dependent and estrogen receptor positive (ER+) [1]. The primary antihormone therapy options for early stage ER+ breast cancer are the selective estrogen receptor modulator tamoxifen and the aromatase inhibitors (AIs). AIs prevent the recurrence of ER+ breast cancer in postmenopausal women by inhibiting estrogen production and depleting systemic estrogens, which, in turn, deprives the tumor of its endogenous growth signal [2,3]. Several clinical trials have established AIs as first-line adjuvant therapy for postmenopausal women with ER+ breast cancer as they result in improved overall survival when compared with tamoxifen [4,5].
The three commonly used third-generation AIs, the steroidal AI exemestane and two nonsteroidal AIs anastrozole and letrozole, are similarly effective [6,7]. Despite the improved survival seen with these agents, an estimated 19.1% of women recur within 10-year when receiving AI treatment [8]. One proposed mechanism of AI resistance is insufficient systemic estrogen suppression, which is supported by the finding that the more potent third-generation AIs (exemestane, letrozole and anastrozole) demonstrate efficacy after failure of an early-generation AI [9]. Although all of the third-generation AIs have superior efficacy to previous generations, there is large variability in the magnitude of estrogen suppression in patients receiving treatment, with a subset of patients even experiencing an increase in estrogen concentrations [10].
Several studies have investigated the role of germline genetics in the variability in the effectiveness or toxicity from AI therapy [11]. OATP1B1 is a hepatic uptake transporter expressed on the sinusoidal membrane of hepatocytes, and it is encoded by the SLCO1B1 gene [12]. Known substrates include endogenous substances such as estrogens and exogenous substances including methotrexate, caspofungin and several HMG-CoA reductase inhibitors [12]. It has been hypothesized that OATP1B1 may also impact the pharmacokinetics of exemestane [13]. SLCO1B1 is polymorphic, with a common, low-activity SNP, SLCO1B1*5 (rs4149056). Past studies have suggested that patient carrying this SNP have higher systemic estrogen concentrations prior to AI treatment [14] and higher exemestane concentrations during treatment [13]. Another SLCO1B1 polymorphism, rs10841753, results in increased expression of the OATP1B1 transporter, resulting in decreased systemic estrogens prior to AI treatment [14]. Based on these prior findings, we hypothesized that functional polymorphisms in SLCO1B1 may be associated with estrogenic response to AI treatment. In our primary analysis, we tested whether SLCO1B1*5 was associated with increased risk of maintaining detectable circulating estrogens after 3 months of AI treatment. Secondary objectives included replicating the association for SLCO1B1*5 with higher pretreatment estrogen concentrations and steady-state AI concentrations, and conducting similar pharmacogenetic association testing for rs10841753, with the opposite expected direction of effect based on the prior evidence that this SNP has the opposite effect on OATP1B1 expression and pretreatment estrogen concentrations.
Patients & methods
Patient cohort
This is a secondary pharmacogenetic analysis of the Exemestane and Letrozole Pharmacogenetics study, a prospective, open-label, clinical trial conducted by the Consortium on Breast Cancer Pharmacogenomics (COBRA). Study design and inclusion criteria have previously been described in detail (ClinicalTrials.gov identifier: NCT00228956) [15]. Briefly, 503 postmenopausal women with stage 0–III hormone receptor-positive breast cancer were enrolled and initiated on an AI as adjuvant therapy. Patients were randomized 1:1 to receive oral exemestane 25 mg once daily or letrozole 2.5 mg once daily. Stratification was based on prior chemotherapy, tamoxifen and bisphosphonate therapy. Surgery, radiation and/or systemic chemotherapy were completed prior to enrollment. Recruitment took place from August 2005 through July 2009 at the University of Michigan Rogel Cancer Center, Sidney Kimmel Comprehensive Cancer Center and Indiana University Melvin and Bren Simon Cancer Center. All patients signed written informed consent, the clinical trial was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Institutional Review Boards at each site.
DNA samples & genotyping
Whole blood samples were collected at enrollment for isolation of germline DNA and genetic assessment. DNA extraction was performed using Qiamp DNA Blood Maxi Kits (Qiagen, CA, USA) as previously described [16]. Genotype determination for SLCO1B1*5 (rs4149056) and rs10841753 were conducted using Taqman® Allelic Discrimination assays according to manufacturer’s instructions (Applied Biosystems, CA, USA). Reactions were carried out using 10 ng of DNA with Genotyping Master Mix (Applied Biosystems) in a CFX96 real-time PCR detection system (BioRad, WI, USA) for 40 cycles. Totally, 10% of samples were randomly retested for quality control and results were 100% concordant.
Estrogen concentration sample collection & measurement
Prior to AI treatment initiation and after 3 months of AI treatment, whole blood samples were collected for measurement of estrone (E1), estrone sulfate (E1S) and estradiol (E2), as previously described [17]. Plasma concentrations were measured using gas chromatography–tandem mass spectrometry by inVentiv Health (NJ, USA). Methods for determining lower limits of quantification (LLOQs) have previously been described in detail (E2 = 1.25 pg/ml, E1 = 3.12 pg/ml, E1S = 3.13 pg/ml) [17].
AI concentration sample collection & measurement
Plasma concentrations of both AIs were measured at steady-state after 1 or 3 months of treatment. Patients were instructed to take their daily dose of AI 2 hours prior to blood sample collection to approximate steady-state maximum concentration [18]. Liquid chromatography–tandem mass spectrometry was used to quantify exemestane concentrations and high-performance liquid chromatography with fluorescence detection was used to quantify letrozole concentrations. Method development was described in detail by Desta et al. [16].
Statistical methods
Pharmacogenetic analyses were conducted assuming additive genetic effects, resulting in three genotype cohorts for each polymorphism (wild-type, heterozygous, variant homozygous). The effect of each SLCO1B1 genotype on baseline estrogen concentration was analyzed using linear regression, designating estrogen concentrations below the LLOQ as the LLOQ value for this analyses. The effect of SLCO1B1 genotype on the presence of detectable estrogens (concentration >LLOQ) after 3 months of therapy was analyzed using logistic regression. A nonparametric test was used to investigate the association between SLCO1B1 genotypes and steady-state exemestane and letrozole plasma concentrations. All significant univariate associations were tested in post-hoc analyses stratified by AI arm and were tested in multivariable models controlling for age, BMI, smoking status, prior tamoxifen therapy and prior hormone replacement therapy (HRT) and tested within each of the treatment arms. All analyses were conducted with a two-sided α = 0.05 in SAS v9.4. All datasets on which the conclusions of the report rely are available on request.
Results
Patient characteristics
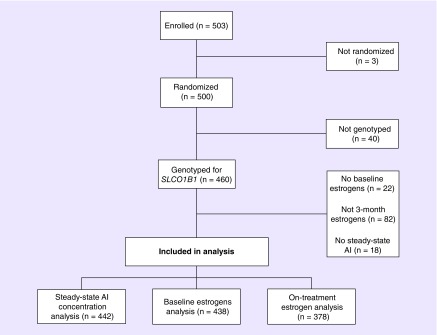
Demographic information of enrolled patients has been previously described in detail [19]. Subjects included in this analysis were similar to the overall cohort with a median age of 59 years, 88.3% were white and a mean BMI of 29.9 kg/m2 (Table 1). Genotype information was available for 460 of the 500 patients enrolled in the Exemestane and Letrozole Pharmacogenetics trial (Figure 1) and all SNPs were within expected distributions of Hardy–Weinberg equilibrium (p > 0.05).
Table 1. . Patient demographics.
| Demographics | Included in any analysis (n = 460) | Baseline estrogens analysis (n = 438) | On-treatment estrogen analysis (n = 378) |
|---|---|---|---|
| Age (years) | 59.24 (8.79) | 59.41 (8.67) | 59.80 (8.49) |
| Race: | |||
| – White | 406 (88.3%) | 387 (88.4%) | 334 (88.4%) |
| – Black | 42 (9.1%) | 40 (9.1%) | 34 (9.0%) |
| – Other | 12 (2.6%) | 11 (2.5%) | 10 (2.6%) |
| BMI (kg/m2) | 29.92 (6.47) | 29.97 (6.50) | 30.01 (6.51) |
| Prior treatment: | |||
| – Tamoxifen | 166 (36.1%) | 154 (35.2%) | 130 (34.4%) |
| – HRT | 226 (29.1%) | 218 (49.8%) | 193 (51.1%) |
| SLCO1B1*5 genotype: | |||
| – SLCO1B1*1/*1 | 334 (72.6%) | 323 (73.7%) | 282 (74.6%) |
| – SLCO1B1*1/*5 | 119 (25.9%) | 108 (24.7%) | 91 (24.1%) |
| – SLCO1B1*5/*5 | 7 (1.5%) | 7 (1.6%) | 5 (1.3%) |
| rs10841753 genotype: | |||
| – Wild-type (TT) | 309 (67.2%) | 296 (67.6%) | 253 (66.9%) |
| – Heterozygous (CT) | 139 (39.2%) | 130 (29.7%) | 115 (30.4%) |
| – Homozygous (CC) | 12 (2.6%) | 12 (2.7%) | 10 (2.7%) |
n (%) or mean (SD).
HRT: Hormone replacement therapy.
Figure 1. . Consort Diagram. This figure depicts patient flow from enrollment on the ELPh clinical trial into the three analyses.
Association of SLCO1B1 genotype with pretreatment estrogens
Pretreatment plasma estrogen and genotype information was available in 438 of the 500 patients enrolled. This analysis demonstrated that each SLCO1B1*5 variant allele was associated with a 51% (95% CI: 29–76%; p < 0.001) increase in pretreatment E1S concentrations (Table 2 & Figure 2). This association maintained significance (p < 0.0001) in a multivariable model controlling for relevant clinical covariates. The SLCO1B1*5 polymorphism was not associated with pretreatment E1 or E2 (p > 0.05).
Table 2. . Linear regression of SLCO1B1 polymorphisms on estrogen plasma concentrations prior to aromatase inhibitor initiation.
| SNP | Estrogen | Univariate analysis (N = 438) | Multivariable model† | ||||
|---|---|---|---|---|---|---|---|
| Concentration change per variant allele | 95% CI | p-value | Concentration change per variant allele | 95% CI | p-value | ||
| SLCO1B1*5 | E1 | 2% | -10–16 | 0.76 | |||
| E1S | 51% | 29–76 | <0.001 | 59% | 35–86 | <0.0001 | |
| E2 | 6% | -11–26 | 0.51 | ||||
| rs10841753 | E1 | -7% | -17–5 | 0.25 | |||
| E1S | -20% | -31 to -8 | 0.002 | -19% | -30 to -6 | 0.004 | |
| E2 | -5% | -19–11 | 0.48 | ||||
Corrected for age, BMI, smoking status, prior tamoxifen therapy and prior hormone replacement therapy.
E1: Estrone; E1S: Estrone sulfate; E2: Estradiol.
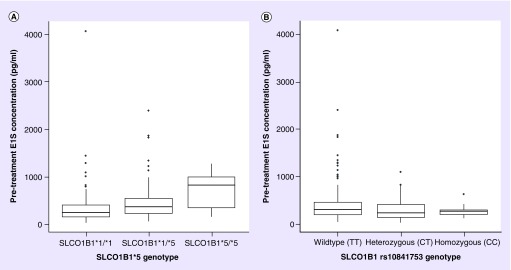
Figure 2. . Pretreatment plasma estrone sulfate concentrations (pg/ml) stratified by SLCO1B1 genotype.
Concentrations below the LLOQ were censored at that value. Patients carrying SLCO1B1*5 (left) allele had significantly greater median E1S concentration (SLCO1B1*1/*1: n = 323, median = 222.0 pg/ml [interquartile range (IQR): 134.5–381.5]; SLCO1B1*1/*5: n = 108, median = 341.5 pg/ml [IQR: 208.75–533.75]; and SLCO1B1*5/*5, n = 7, median = 800.0 pg/ml [IQR: 324–975.5]). The opposite effect was seen for carrying rs10841753 (right) although the association was not as strong (SLCO1B1 wild-type T/T: n = 296, median = 285.5 pg/ml [IQR: 163.5–431.25]; SLCO1B1 heterozygous C/T: n = 130, median = 203.5 pg/ml [IQR: 114.75-387.0]; and SLCO1B1 homozygous variant C/C: n = 12, median = 238.0 pg/ml [IQR: 174.25–272.0]). In the box and whisker plot, the middle line represents the median, the box represents the IQR and the whiskers extend to 1.5 × IQR.
E1S: Estrone sulfate.
In contrast, the variant (C) allele of rs10841753 was associated with a 20% ([95% CI: -31%, -8%], p = 0.002) decrease in plasma E1S concentrations prior to treatment. This association also maintained significance in a multivariable model (p = 0.004) (Table 2). This SNP was not associated with pretreatment E1 or E2.
Association of SLCO1B1 genotype with on-treatment estrogens
Totally, 378 patients had SLCO1B1 genotype information and estrogen plasma concentrations measured after 3 months of AI and were included in the primary analysis. Each SLCO1B1*5 allele was associated with an 84% increased risk of failing to achieve undetectable E1 after 3 months of AI treatment (odds ratio [OR]: 1.84; 95% CI: 1.08–2.14; p = 0.025) (Figure 3 & Table 3), which maintained significance in a multivariable model (p = 0.037). In a post-hoc stratified analysis, SLCO1B1*5 was not associated with risk of detectable E1 in either the letrozole (p = 0.18) or exemestane (p = 0.07) arm, but the effect in each arm was similar to that seen in the overall cohort (OR: 1.78 and 1.92, respectively; Supplementary Table 1). This SNP was not associated with risk of detectable E1S or E2 after 3 months of AI treatment.
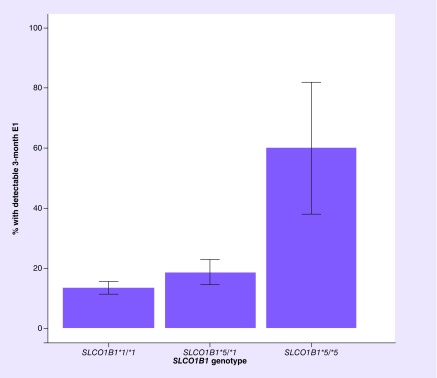
Figure 3. . Percent of patients with detectable estrone following 3 months of aromatase inhibitor treatment stratified by SLCO1B1*5 genotype.
Risk of detectable estrone (E1) after 3 months of treatment was higher in patients with SLCO1B1*5/*5 (n=5, 60%) compared with SLCO1B1*1/*5, n=91, 18.7%) or SLCO1B1*1/*1 (n=282, 13.5%) patients. Each bar represents the percent of patients with detectable E1 and error bars indicate the standard error.
Table 3. . Logistic regression of SLCO1B1 polymorphisms on detectable estrogen plasma concentrations following 3 months of aromatase inhibitor therapy.
| SNP | Estrogen | N > LLOQ (N = 378) | Univariate analysis | Multivariable model† | ||||
|---|---|---|---|---|---|---|---|---|
| Odds ratio | 95% CI | p-value | Odds ratio | 95% CI | p-value | |||
| SLCO1B1*5 | E1 | 58 (15%) | 1.84 | 1.08–3.14 | 0.025 | 1.82 | 1.04–3.21 | 0.037 |
| E1S | 242 (64%) | 1.41 | 0.88–2.24 | 0.15 | ||||
| E2 | 45 (12%) | 0.78 | 0.39–1.59 | 0.50 | ||||
| rs10841753 | E1 | 58 (15%) | 0.75 | 0.43–1.32 | 0.32 | |||
| E1S | 242 (64%) | 0.61 | 0.41–0.90 | 0.013 | 0.63 | 0.42–0.95 | 0.028 | |
| E2 | 45 (12%) | 0.75 | 0.40–1.40 | 0.36 | ||||
Corrected for age, BMI, smoking status, prior tamoxifen therapy and prior hormone replacement therapy.
E1: Estrone, E1S: Estrone sulfate, E2: Estradiol, LLOQ: Lower limit of quantification.
Based on these findings, we conducted a post-hoc linear regression analysis to determine if patients with higher pretreatment E1S were more likely to have detectable E1 after 3 months of AI treatment; however, these results were not statistically significant (regression coefficient = 1.03 [95% CI: 0.82–1.31]; p = 0.78).
Each rs10841753 variant allele was associated with a decreased risk of failing to achieve undetectable E1S concentrations after 3 months of AI therapy (OR: 0.61; 95% CI: 0.41–0.90; p = 0.013); however, there was no significant effect on E1 or E2. This analysis maintained significance in the multivariate analysis controlling for clinical covariates (Table 3). In the post-hoc stratified analysis, the association of rs10841753 with risk of detectable E1S plasma concentrations was confined to the exemestane arm (OR: 0.36; 95% CI: 0.19–0.68; p = 0.002) and was not seen in the letrozole arm (OR: 0.68; p = 0.19).
AI concentrations & SLCO1B1 expression
There was no association between SLCO1B1*5 or rs10841753 and steady-state plasma concentrations of letrozole or exemestane (all p > 0.05, Supplementary Table 2).
Discussion
The main objective of this secondary analysis was to test for associations between SLCO1B1 polymorphisms and plasma estrogen and AI concentrations during treatment of patients with ER+ breast cancer. Patients carrying SLCO1B1*5 had an increased risk of failing to achieve undetectable E1 following 3 months of AI treatment and patients carrying rs10841753 had decreased risk of failing to achieve undetectable E1S. Neither polymorphism was associated with steady-state AI drug concentrations.
We hypothesized that patients carrying SLCO1B1*5 would be at higher risk of detectable estrogens during treatment, based on previous studies that these patients have higher pretreatment estrogens. Consistent with this hypothesis, patients carrying SLCO1B1*5 were more likely to have detectable E1 concentrations after 3 months of AI treatment. We further hypothesized that this could potentially be due to metabolic conversion of E1S to E1 via estrone sulfatase [20] in patients taking AIs; however, our post-hoc analysis did not identify an association between pretreatment E1S and the risk of detectable E1 after 3 months of treatment. Given its inverse effect relative to SLCO1B1*5, we hypothesized that patients carrying rs10841753 would have decreased risk of detectable estrogens following 3 months of AI treatment. Although no association with E1 was found, patients carrying rs10841753 had decreased risk of detectable E1S, which is likely due to lower pretreatment E1S in these patients. We genotyped rs10841753 due to its previously reported association with pretreatment estrogens [14], but the associations for this intronic SNP are likely due to its linkage disequilibrium (r2 = 0.79, d’ = 1) with the known high-activity SLCO1B1*14 (rs11045819) polymorphism [21]. This was the first attempt, to our knowledge, to investigate pharmacogenetic associations for SNPs in SLCO1B1 with estrogenic response to AI [11]. Similar prior studies have focused on SNPs in the aromatase enzyme, CYP19A1, including intriguing findings for rs7176005 and rs6493437 [22]; however, these associations have not been validated [11,23].
Our finding that SLCO1B1*5 results in elevated pretreatment E1S levels is consistent with the findings of a previously published genome-wide association study of estrogen concentrations measured prior to treatment with anastrozole or exemestane in patients on the MA.27 clinical trial [14]. We further replicated their finding that rs10841753 is associated with lower pretreatment E1S [14]. High-circulating E2 has been associated with risk for breast cancer [24,25], but the clinical implications of E1 and E1S concentrations are unknown [26]. E1 is the most abundant estrogen in postmenopausal women [27], but the lack of association for SLCO1B1*5 or rs10841753 with breast cancer risk in large genome-wide association study suggests that these polymorphisms and E1 or E1S levels do not have any clinically consequence [28].
AIs prevent the recurrence of ER+ breast cancer by inhibiting estrogen production and depleting systemic estrogens, depriving the tumor of its requisite growth signal [27]. We found that patients carrying SLCO1B1*5 were at higher risk of having detectable E1 during AI treatment, implying that these patients may have worse outcomes when receiving AI treatment. However, there is no direct evidence that estrogen suppression below a certain threshold is necessary for treatment effectiveness. For example, although pharmacologic studies demonstrate that letrozole has more potent estrogen suppression and aromatization inhibition in vivo than exemestane or anastrozole, no differences in relapse rates or mortality have been observed in trials comparing these agents head-to-head [2,3]. Notably, a meta-analysis comparing letrozole and anastrozole reported a trend toward superior breast cancer related- and all cause-mortality for letrozole [29]. If the relationship between estrogen suppression and AI treatment efficacy were validated, plasma estrogen measurement during treatment may be clinically useful, particularly in patients carrying SLCO1B1*5 who may be at higher risk for treatment failure. However, at this time there is insufficient direct evidence of this association to recommend using this approach in practice.
No associations were found for these SLCO1B1 SNPs and steady-state concentrations of exemestane or letrozole. Our results are in contrast to those from a highly controlled pharmacokinetic analysis of 14 healthy volunteers that reported 284% higher exemestane exposure in SLCO1B1*5 carriers [13]. Our inability to replicate this finding using drug levels measured in a large patient cohort indicates that these SNPs are unlikely to exert a clinically meaningful effect on AI concentrations. Furthermore, there is no established association between exemestane or letrozole concentrations with magnitude of estrogen suppression, as we have previously reported from an analysis of this cohort [18], or with efficacy or toxicity of AI treatment [11,30,31]; therefore, the clinical relevance of discovering predictors of AI pharmacokinetics is limited.
This secondary pharmacogenetic analysis of functional SNPs in SLCO1B1 conducted within a prospectively accrued cohort of patients with ER+ breast cancer discovered intriguing associations with estrogenic response to AI treatment and replicated previous findings regarding pretreatment estrogens. However, this study has several limitations that should be considered. Most importantly, treatment outcomes data are unavailable in this cohort of patients who represented a broad cross-section of women taking AIs for treatment of both in situ and stage I–IIIB primary breast cancers. Therefore, the observed association with a surrogate endpoint needs to be replicated in independent patient cohorts with on-treatment estrogen measurements and/or long-term outcomes data. Additionally, although we used estrogen assays that were highly sensitive for their time, we were unable to quantify estrogen levels below the LLOQ, resulting in classification of many patients as ‘undetectable’ at 3 months and precluding quantitative analyses. Also, simultaneous investigation of multiple SNPs and medications increases the risk of false discovery. Another limitation is our inability to investigate the effect of OATP1B1 drug interactions, which would be expected to phenocopy functional SCLO1B1 polymorphisms, due to the relative lack of knowledge about which drugs induce or inhibit this transporter [32]. Last, the SLCO1B1*5 allele has a relatively low minor allele frequency (MAF = 0.13), leading to small numbers of patients with the variant genotype in pharmacogenetic analyses and somewhat limiting the clinical usefulness of genotyping for this SNP to guide clinical practice.
Conclusion
In conclusion, in this cohort of postmenopausal women with ER+ breast cancer, we found that the SLCO1B1*5 allele was associated with increased risk of failing to achieve undetectable E1 after 3 months of AI treatment. Future work is required to replicate this finding in independent cohorts and determine whether this SNP is associated with AI treatment outcomes. If validated, this SNP could be useful to predict which patients may be at increased risk of breast cancer relapse during AI treatment, in whom alternative treatment options or estrogen monitoring during treatment could be considered.
Summary points.
Aromatase inhibitors deplete systemic estrogens and are first-line adjuvant treatment in postmenopausal women with hormone-receptor positive breast cancer.
A subset of patients receiving AI treatment continue to have measurable systemic estrogens, which is one proposed mechanism of AI treatment failure.
OATP1B1 is a transporter involved in hepatic uptake and regulation of estrogens.
The SLCO1B1 polymorphism rs4149065 (SLCO1B1*5) decreases OATP1B1 transporter activity while rs10841753 increases OATP1B1 expression.
Associations of both polymorphisms with systemic estrogen conjugates have been reported, but no studies have assessed whether these polymorphisms affect estrogenic response to AI treatment.
In this analysis, SLCO1B1*5 was associated with elevated pretreatment estrone sulfate and an increased risk of maintaining detectable estrone despite 3 months of AI treatment.
SLCO1B1 rs10841753 was associated with depressed pretreatment estrone sulfate and an decreased risk of detectable estrone sulfate following 3 months of AI treatment.
No association was detected for SLCO1B1 polymorphisms and steady-state AI plasma concentrations in this secondary analysis of a large patient cohort.
These findings confirm that SLCO1B1 polymorphisms are associated with pretreatment estrogens and suggest they may be associated with estrogenic response to AI treatment.
Replication of this association and validation that this has a meaningful effect on AI treatment outcomes is necessary for translation into clinical practice.
Supplementary Material
Acknowledgments
The authors wish to posthumously recognize D Flockhart, who co-chaired the COnsortium on BReast cancer phArmacogomics (COBRA) and passed during preparation of this manuscript.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/pgs-2019-0020
Financial & competing interests disclosure
This research was supported by Pharmacogenetics Research Network Grant Number U-01 GM61373 David A Flockhart (DAF) and Clinical Pharmacology Training Grant Number 5T32-GM08425 (DAF) from the National Institute of General Medical Sciences, NIH from Grant Numbers M01-RR000042 (University of Michigan), M01-RR00750 (Indiana University) and M01-RR00052 (Johns Hopkins University) from the National Center for Research Resources (NCRR), a component of the NIH, the Breast Cancer Research Foundation (BCRF; N003173 to JM Rae and DF Hayes), the National Cancer Institute (5T32CA083654-12, PI Jeremy Taylor), the National Institute of General Medical Sciences (GM099143 to JM Rae) and the NIH through the University of Michigan’s Cancer Center Support Grant (P30 CA046592) by the use of the following Cancer Center Core: University of Michigan DNA Sequencing Core. In addition, these studies were supported by grants from Pfizer (DF Hayes), Novartis Pharma AG (DF Hayes), the Fashion Footwear Association of New York/QVC Presents Shoes on Sale (DF Hayes). Drugs were supplied by Novartis and Pfizer. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Howlader N, Altekruse SF, Li CI. et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. JNCI J. Natl Cancer Inst. 106(5), pii:dju055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geisler J, Haynes B, Anker G, Dowsett M, Lønning PE. Influence of letrozole and anastrozole on total body aromatization and plasma estrogen levels in postmenopausal breast cancer patients evaluated in a randomized, cross-over study. J. Clin. Oncol. 20(3), 751–757 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Geisler J, King N, Anker G. et al. In vivo inhibition of aromatization by exemestane, a novel irreversible aromatase inhibitor, in postmenopausal breast cancer patients. Clin. Cancer Res. 4(9), 2089–2093 (1998). [PubMed] [Google Scholar]
- 4.Cuzick J, Sestak I, Baum M. et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol. 11(12), 1135–1141 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Regan MM, Neven P, Giobbie-Hurder A. et al. Assessment of letrozole and tamoxifen alone and in sequence for postmenopausal women with steroid hormone receptor-positive breast cancer: the BIG 1-98 randomised clinical trial at 8·1 years median follow-up. Lancet Oncol. 12(12), 1101–1108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith I, Yardley D, Burris H. et al. Comparative efficacy and safety of adjuvant letrozole versus anastrozole in postmenopausal patients with hormone receptor-positive, node-positive early breast cancer: final results of the randomized Phase III femara versus anastrozole clinical evaluation (FACE) trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 35(10), 1041–1048 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Goss PE, Ingle JN, Pritchard KI. et al. Exemestane versus anastrozole in postmenopausal women with early breast cancer: NCIC CTG MA.27 – a randomized controlled Phase III trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 31(11), 1398–1404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Early Breast Cancer Trialists’ Collaborative Group. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet 386(10001), 1341–1352 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Miller WR, Larionov AA. Understanding the mechanisms of aromatase inhibitor resistance. Breast Cancer Res. 14(1), 201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingle JN, Buzdar AU, Schaid DJ. et al. Variation in anastrozole metabolism and pharmacodynamics in women with early breast cancer. Cancer Res. 70(8), 3278–3286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertz DL, Henry NL, Rae JM. Germline genetic predictors of aromatase inhibitor concentrations, estrogen suppression and drug efficacy and toxicity in breast cancer patients. Pharmacogenomics 18(5), 481–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol. 158(3), 693–705 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory BJ, Chen SM, Murphy MA, Atchley DH, Kamdem LK. Impact of the OATP1B1 c.521T>C single nucleotide polymorphism on the pharmacokinetics of exemestane in healthy postmenopausal female volunteers. J. Clin. Pharm. Ther. 42(5), 547–553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudenkov TM, Ingle JN, Buzdar AU. et al. SLCO1B1 polymorphisms and plasma estrone conjugates in postmenopausal women with ER+ breast cancer: genome-wide association studies of the estrone pathway. Breast Cancer Res. Treat. 164(1), 189–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry NL, Giles JT, Ang D. et al. Prospective characterization of musculoskeletal symptoms in early stage breast cancer patients treated with aromatase inhibitors. Breast Cancer Res. Treat. 111(2), 365–372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desta Z, Kreutz Y, Nguyen AT. et al. Plasma letrozole concentrations in postmenopausal women with breast cancer are associated with CYP2A6 genetic variants, body mass index, and age. Clin. Pharmacol. Ther. 90(5), 693–700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robarge JD, Desta Z, Nguyen AT. et al. Effects of exemestane and letrozole therapy on plasma concentrations of estrogens in a randomized trial of postmenopausal women with breast cancer. Breast Cancer Res. Treat. 161(3), 453–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz DL, Speth KA, Kidwell KM. et al. Variable aromatase inhibitor plasma concentrations do not correlate with circulating estrogen concentrations in postmenopausal breast cancer patients. Breast Cancer Res. Treat. 165(3), 659–668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry NL, Azzouz F, Desta Z. et al. Predictors of aromatase inhibitor discontinuation as a result of treatment-emergent symptoms in early-stage breast cancer. J. Clin. Oncol. 30(9), 936–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasqualini JR, Chetrite G, Blacker C. et al. Concentrations of estrone, estradiol, and estrone sulfate and evaluation of sulfatase and aromatase activities in pre- and postmenopausal breast cancer patients. J. Clin. Endocrinol. Metab. 81(4), 1460–1464 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Ramsey LB, Moncrieffe H, Smith CN. et al. Association of SLCO1B1*14 allele with poor response to methotrexate in juvenile idiopathic arthritis patients. ACR Open Rheumatol. 1, 58–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Ellsworth KA, Moon I. et al. Functional genetic polymorphisms in the aromatase gene CYP19 vary the response of breast cancer patients to neoadjuvant therapy with aromatase inhibitors. Cancer Res. 70(1), 319–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghimenti C, Mello-Grand M, Grosso E. et al. Regulation of aromatase expression in breast cancer treated with anastrozole neoadjuvant therapy. Exp. Ther. Med. 5(3), 902–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuhrman BJ, Schairer C, Gail MH. et al. Estrogen metabolism and risk of breast cancer in postmenopausal women. JNCI J. Natl Cancer Inst. 104(4), 326–339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Key T, Appleby P, Barnes I, Reeves G. Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J. Natl Cancer Inst. 94(8), 606–616 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Brown SB, Hankinson SE. Endogenous estrogens and the risk of breast, endometrial, and ovarian cancers. Steroids 99, 8–10 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Smiley DA, Khalil RA. Estrogenic compounds, estrogen receptors and vascular cell signaling in the aging blood vessels. Curr. Med. Chem. 16(15), 1863–1887 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Pankratz VS, Fredericksen Z. et al. Common variants associated with breast cancer in genome-wide association studies are modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 19(14), 2886–2897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lønning PE. The potency and clinical efficacy of aromatase inhibitors across the breast cancer continuum. Ann. Oncol. 22(3), 503–514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrie AE, Rose RV, Choi Y-H. et al. Letrozole concentration is associated with CYP2A6 variation but not with arthralgia in patients with breast cancer. Breast Cancer Res. Treat. 172(2), 371–379 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kadakia KC, Kidwell KM, Seewald NJ. et al. Prospective assessment of patient-reported outcomes and estradiol and drug concentrations in patients experiencing toxicity from adjuvant aromatase inhibitors. Breast Cancer Res. Treat. 164(2), 411–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shitara Y. Clinical importance of OATP1B1 and OATP1B3 in drug–drug interactions. Drug Metab. Pharmacokinet. 26(3), 220–227 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.