Abstract
Environmental DNA (eDNA) applications are transforming the standard of characterizing aquatic biodiversity via the presence, location and abundance of DNA collected from environmental samples. As eDNA studies use DNA fragments as a proxy for the presence of organisms, the ecological properties of the complex and dynamic environments from which eDNA is sampled need to be considered for accurate biological interpretation. In this review, we discuss the role that differing environments play on the major processes that eDNA undergoes between organism and collection, including shedding, decay and transport. We focus on a mechanistic understanding of these processes and highlight how decay and transport models are being developed towards more accurate and robust predictions of the fate of eDNA. We conclude with five recommendations for eDNA researchers and practitioners, to advance current best practices, as well as to support a future model of eDNA spatio-temporal persistence.
Keywords: environmental DNA, eDNA decay, taphonomy, eDNA transport, biodiversity, aquatic biodiversity
1. Introduction
The application of environmental DNA (eDNA) has experienced immense growth in recent years and is now used widely in community ecology, palaeo-environmental research, biomonitoring, conservation biology and invasion ecology [1–3]. Owing to their non-invasiveness, ease of field sampling and ability to detect cryptic species, eDNA-based tools are an attractive alternative to extensive direct sampling of ecosystems [2,3]. Despite this increasingly wide interest, uncertainties persist surrounding the physical processes that influence eDNA persistence and its fate within the environment. Because these techniques use fragments of DNA recovered from environmental samples to infer species presence, uncertainties in the relationship between the source organism(s) and the physical DNA molecules in the environment can significantly limit inferences made from eDNA-based tools and preclude their widespread application [3]. While eDNA refers to DNA recovered from any environmental sample (e.g. water [2], sediments [4] or air [5]), this review focuses on shed macro-organismal DNA collected from aquatic environments. We discuss other environments only where they are likely to impact collection from aquatic environments (e.g. within the benthic substrate).
One critical step towards overcoming these uncertainties is a better understanding of the mechanisms that influence eDNA presence and concentration through time and space. Conceptually, eDNA is shed from the host organism, then decreases in concentration as it is both transported from the origin location and undergoes decay until it can no longer be detected [3,6–8]. While eDNA studies are diverse in their objectives and scales of inquiry, most eDNA studies seek information on where and when a source organism was present (and perhaps even how many). For presence–absence surveys, these mechanisms influence how long eDNA can be detected, and how far away the source location may be. For quantitative eDNA surveys, the concentration of collected eDNA is further related to abundance of the target organisms [1].
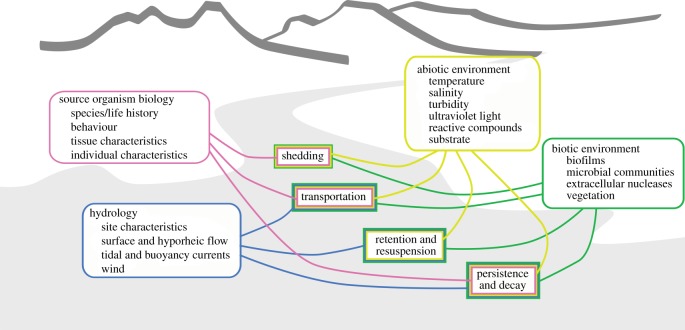
The relationship between the source organism and the concentration of eDNA at the point when it is measured (termed the ‘ecology of eDNA’ [3]) is complex, with multiple competing environmental processes influencing the relationship (figure 1). A growing body of literature shows that these processes are environment-specific (e.g. [3–6]), increasing the uncertainties when relating eDNA to the spatial and temporal distribution of source organisms [3]. The origin, transport and decay of eDNA influence the physical state of eDNA, and the physical state determines the mechanisms and consequences of the interactions between eDNA and the natural environment. As the use of eDNA continues to become a more widespread tool for research, conservation and industrial challenges [1,9], considering these mechanisms is critical for executing well-designed surveys and for robust interpretation of eDNA patterns.
Figure 1.
Mechanisms influencing the concentration and fate of eDNA in aquatic systems and environmental factors known or suspected to alter their rate and extent of action.
This review synthesizes recent research about the decay of biological material, from tissue to molecule, highlighting the nature of eDNA interactions with the environment, dependence on specific biological and environmental factors, and resulting implications for the interpretation of eDNA patterns. We focus on a mechanistic understanding of processes that affect eDNA within lentic, lotic and marine environments and discuss how those mechanisms are or are not captured in different transport and decay models currently used to predict the behaviour of eDNA. Where eDNA-specific information is underdeveloped, we have synthesized information from multiple disciplines to make predictions on how eDNA is likely to behave in the environment. Our review highlights the critical need for increased research towards an understanding of eDNA as physical particles within an environmental context, in order to understand and describe the relationship between the biological organism and its eDNA. To this effect, we provide five recommendations for future research that can fill these gaps in knowledge.
2. Origin of eDNA
For eDNA to accurately quantify species abundance, the amount of eDNA shed must exhibit causal correlations with the biomass or number of organisms present in the environment, ideally with estimates surrounding the uncertainties contributing to reductions in such correlations [10]. Understanding the origins of eDNA, i.e. the mechanisms behind and variability in the shedding of biological material from source organisms, is integral to developing such estimates.
Most eDNA probably originates from released urine and faecal matter [11,12], shed epithelial cells from external mucous layers [13] and tissue from decomposing organisms [14]. Shedding rates can be variable within species even when accounting for biomass [13,15–18], obscuring the relationship between collected eDNA and organism abundance. Stress has been linked to up to 100-fold increases in tissue shedding rates [13,16,19]. Age [19], diet [20], water temperature [16] and community structure [13] are among the numerous biotic and abiotic variables that cause significant variation in volume and rate of tissue shedding (see review in [21]). Consequently, this source of uncertainty needs to be considered in downstream analysis and ecological inference. For example, a change in abundance of eDNA for a given species may represent either changes in shedding rates (e.g. owing to stress) or changes in organism biomass or abundance.
3. Degradation and decay
The accurate and biologically relevant use of eDNA as a proxy for the presence, abundance and location of target organisms requires an understanding of the rates of eDNA decay (the reduction in detectable quantity of eDNA) and degradation (physical changes within the molecule/particle) in different environments. Although processes such as retention within substrate may contribute to eDNA removal from aquatic systems, a primary route of eDNA decay is the physical degradation of the tissue and particles comprising eDNA. Shed biological tissue will generally begin to degrade immediately from multicellular tissue fragments, to whole cells, separate organelles (e.g. mitochondria) and eventually to free (extracellular) DNA, which are then further degraded either by exogenous enzymes or by spontaneous chemical reactions. The extent of this degradation affects how eDNA can be captured [22] and what types of analyses can be performed [23].
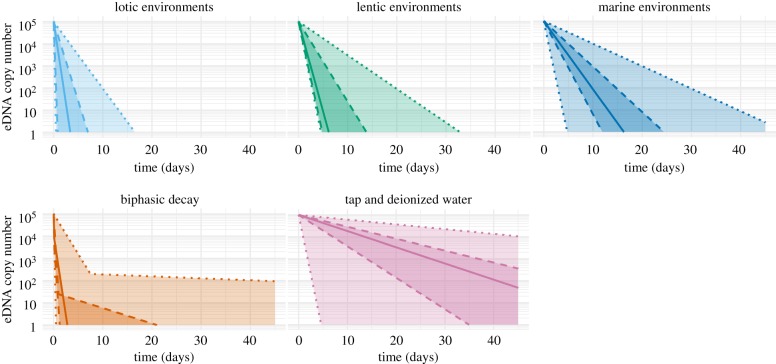
Although eDNA decay involves multiple processes, they are often combined into a single decay rate estimate. First-order estimates of eDNA decay rates vary considerably, from a half-life 0.7 h in a multi-species assay [24] to 71.1 h in Antarctic icefish [25] (figure 2, see [27]). Complex models of DNA decay where decay rates decrease over time have been shown to better explain aquatic eDNA decay than monophasic models [26,28,29]. Conceptually, this may correspond to the multiphasic mechanisms involved in tissue and eDNA decay [29], as different cellular compartments may have different liabilities, and individual degradation mechanisms may impact eDNA decay rates. If eDNA decay follows a multiphasic decay pattern, a monophasic model may underestimate the residency time of low concentration eDNA (if parametrized using data from the initial rapid decay phase) or significantly overestimate the initial eDNA concentration [29].
Figure 2.
Theoretical eDNA persistence over 45 days from an initial starting quantity of 105 copies, using decay constants derived from experimental studies, thereby illustrating the importance of considering ecosystem and model choice when predicting decay. Biphasic decay constants from both lentic and lotic ecosystems were combined, but bisphasic decay is likely to occur in marine and laboratory conditions as well. Solid lines indicate median decay constants (ordered by either T1/2 (monophasic models) or T99.99 (biphasic models)), dashed lines indicate quartile decay constants and dotted lines indicate extremes. Data from [26–28].
While numerous environmental characteristics influencing eDNA decay rates have been identified (e.g. water temperature [30–36], water turbidity [1,37], acidity [24,34,35] and salinity [38]), these characteristics have been applied to eDNA decay as a whole and are not linked to specific mechanisms of decay. An explicit focus on the taphonomy (post-mortem degradation processes) of eDNA is therefore critical to determining the relationship between eDNA detection and the current spatio-temporal state of the organisms being studied. The following sections review some of the major mechanisms by which tissue and DNA degrade and the implications for eDNA detectability.
(a). Cellular degradation
Many cells (e.g. intestinal or mucosal epithelial tissues) will begin to degrade via apoptosis before being shed [39]. Nuclear DNA is tightly packed in early-stage apoptosis before being hydrolysed into 180 bp fragments [40]. Changes in the mitochondrial membrane cause increased permeability allowing some mitochondrial nucleoids to be released into the cytoplasm of the cell [41], and most mitochondrial DNA is randomly fragmented during late-stage apoptosis [41,42]. Apoptosis generally concludes within 3 h to more than a day [43,44], but some cell enzymes may become inactive before apoptosis completes, triggering a switch to necrosis and potentially allowing longer fragments of DNA to survive [45].
Necrosis describes any uncontrolled cell death process, typically characterized by the swelling and bursting of cells as they lose osmotic control and release cell contents into the environment [39]. The outer mitochondrial membrane probably remains intact longer than other cellular structures [46], which may provide a protective effect to mtDNA. In both cadaver studies and eDNA studies, mtDNA has significantly lower decay rates than nuclear DNA, even with similar copy numbers [29,47,48]. Within the environment, free DNA (either nuclear or mitochondrial) is typically degraded by bacteria and extracellular nucleases [49–51] into smaller, unpredictable DNA fragments [26,29,52]. eDNA persistence can therefore be directly linked to microbial activity, trophic state [26,28,53] and the concentration of extracellular nucleases [54]. eDNA decay rates are also highly associated with temperature [33], which may be partially owing to increased concentrations of bacteria as well as increased rates of enzymatic and metabolic processes.
(b). Spontaneous degradation
DNA will undergo spontaneous (non-enzymatically catalysed) decomposition, leading to the fragmented and chemically modified DNA strands characteristic of ancient DNA (aDNA). The molecular mechanisms of spontaneous DNA decay have been reviewed extensively [55,56], as have their effect on genetic analysis [57]. Given a neutral buffer and moderate temperature, DNA is a highly stable molecule. At 25°C, cleavage of the phosphodiester bond (which links nucleotides together into a DNA chain) has an estimated half-life of 31 000 000 years; depurination of deoxyguanosine and deoxyadenosine have estimated half-lives of 70 years and 180 years, respectively; and deoxycytidine deamination has an estimated half-life of 120 years [58]. Environmental conditions further impact rates of molecular DNA modifications.
Temperature influences the rate of spontaneous decomposition, to the extent that thermal history is more important than age of material when successfully amplifying aDNA [59]. For example, deoxycytidine deamination (where cytosine bases are converted to uracil bases) has an estimated half-life (t1/2) of 120 years at 25°C and pH 7.5, but only 24 years at 37°C and more than 2600 years at 5°C [58]. The presence of deoxycytidine deamination in aDNA is used to discriminate between true aDNA and contemporary (contaminating) DNA (e.g. [60,61]). Thus, similar methods could be employed in eDNA to discriminate against DNA older than the temporal scope of the research [62]. However, using deoxycytidine deamination as a marker of ‘old’ DNA may only be a useful technique in warmer waters or for projects with long temporal scopes [58].
Elements within the environment may also interact to influence spontaneous DNA decay. For example, ultraviolet (UV) light can directly degrade DNA [55] but can also react with natural organic matter to form reactive oxygen species that cause oxidative damage and additional stress to eDNA [63]. Evidence for the impact of UV light on eDNA decay is currently mixed [34,64–66], and the effects of UV in correlation with natural organic matter uncertain. In another example, certain antibiotics (e.g. aminoglycosides) found in wastewater [67] can readily form metallo-complexes that increase the rate of phosphodiester bond hydrolysis by up to 5.2 × 107 times, rapidly degrading DNA [68,69].
While research on aDNA has a relatively deep legacy of research on the signature of spontaneous DNA degradation, there has also been a significant focus in a narrow range of environments that are favourable to DNA preservation [57]. Studies of DNA degradation in forensic taphonomy are perhaps more related to eDNA research, as these encompass a much wider range of environments [45]. Overall, environment-specific studies of eDNA decay mechanisms are critically needed to build informative models of eDNA persistence.
4. Transport of eDNA
Aquatic eDNA is collected in complex and variable hydrological systems, and therefore eDNA transport is a key consideration for inferring species presence and spatial distribution. This includes horizontal and vertical transport (in the suspended aquatic realm), retention in the benthic substrate and probability of resuspension into the water column [8,70] (electronic supplementary material, figure S1); the rates of which may be confounded with the residency of eDNA within its environment prior to decay. While the transport of eDNA is a new area of research [26,71], hydrological models that integrate physiochemical properties of organic matter with hydrological parameters of the system may give informative estimates of eDNA transport.
(a). Transport factors: eDNA as a particle
eDNA has been found in particles with diameters ranging from less than 0.2 µm to greater than 180 µm. Most eDNA is found in fragment sizes reflective of intact tissue or free mitochondria [13,16,22], although smaller particles begin to dominate within hours of the source organism being removed [16]. As colloidal particles, eDNA transport is non-conservative, meaning that it is impacted by factors other than advection (flow) and diffusion [72,73]. Because of the relatively large variation in particle size, eDNA is unlikely to be fully described by single parameter estimates of transport or diffusion [72]. However, some developed transport model characteristics may help predict factors influencing eDNA transport in aquatic environments [71].
Most eDNA studies use filtration methods that will preferentially capture eDNA particles that are categorized as fine particulate organic matter (i.e. with diameters between 1 mm and 0.5 µm) [13,74,75]. While eDNA has some fundamental differences from fine particulate organic matter as it is traditionally measured (in particular that eDNA tends to be over-dispersed or ‘clumpy’ as tissue contain multiple copies of DNA [22,76]), eDNA transport seems to follow similar transport dynamics as fine particulate organic matter [71,77].
(b). Transport factors: geochemical adsorption
Both DNA and cell surfaces reversibly adsorb to minerals such as those found in the streambeds [78,79]. Adsorption to sediments can protect DNA from degradation by nucleases as well as chemical damage [80,81] and can likewise protect cells and tissue fragment from shearing forces and microbial degradation [79]. This protective factor may explain increased persistence in sediments or stream bed substrates compared to water columns [51,82,83], as it is suspected that adsorbed DNA can be resuspended into the water column after aquatic eDNA has degraded [26].
Biological molecules bind to sediments via weak charge-based bonds such as electrostatic interactions [81]. As such, the binding capacity of eDNA in soil is heavily dependent on both the sediment type, proteins and other molecules exposed on cell surfaces, and configuration of the DNA molecule [79,81]. Inorganic clays adsorb more DNA than organic sediments [78,84], and large, supercoiled DNA strands (such as nuclear DNA packaged in apoptosis) have a lower binding affinity than smaller, less tightly packed fragments of DNA [85], potentially contributing to more rapid degradation of nuclear DNA or length-dependent eDNA degradation [86]. This binding affinity is further influenced by environmental properties, such as pH, dissolved salts, moisture, temperature and the presence of cell fragments [49,84,87–90]. Bacterial biofilms also increase adhesion and may promote the persistence of eDNA by using exogenous DNA as structural elements [91]. Resuspension is therefore a largely stochastic process [26] and decay times in sediment can vary from days and weeks to thousands of years [4,92,93].
(c). Transport in freshwater lakes and ponds
Horizontal dispersion of eDNA is typically limited in lentic ecosystems [35,94], with several studies finding a decrease in eDNA concentrations beyond approximately 100 m of the organisms [20,94–96]. Vertical transport in lentic ecosystems is primarily influenced by gravitational settling, as thermal stratification limits transport from water flow [97], and both faecal particles and associated eDNA are more concentrated in the lower layers of the water column [12,14,82]. Sampling from upper water layers has therefore been suggested to determine viable populations from eDNA [14]. The settling rate is determined by the size and density of the particle [98], although water turbulence or biotic breakdown may slow the sinking rate by breaking up particles [12]. As eDNA exists in non-uniform particle sizes and types [72], settling rates are difficult to estimate. In thermally stratified lakes, each layer may have its own microenvironment that may uniquely affect eDNA recovery and degradation. Tests with plasmid DNA have shown that degradation occurred in the upper stratified layer (the epilimnion) within 170 h, whereas no degradation occurred in the same time period in the lowest, coolest layer of water (the hypolimnion) [54].
Notably, large vertical and horizontal transports may be expected during lake overturns, when temperature-driven stratification is lost and whole lake mixing occurs [99]. In areas with significant seasonal temperature variation, lake overturns usually occur in the spring and autumn [97,99,100]. In other systems (e.g. areas with little seasonal variation, warm waters or in shallow lakes compared to their width), lake overturn can occur on a daily to weekly basis [97]. These events are expected to have significant but unpredictable impacts on eDNA transport. Consequently, collecting eDNA samples during lake overturns is discouraged [14].
(d). Transport in freshwater rivers and streams
While eDNA may be more homogenized in flowing water owing to the increased turbulence [71], downstream transport in lotic ecosystems is a concern and not yet well described. Repeated cycles of adsorption and resuspension to the benthic substrate result in eDNA being transported at a delayed rate compared to water velocity [8,72]. When taking repeated samples from the same site, site-specific factors have an impact on the explanatory potential of eDNA studies [101], but identifying and generalizing these factors is still difficult.
Most commonly, the decline in eDNA concentrations following downstream transport is described as a first-order exponential decline [8,26,71,77] using a model developed for fine particulate organic matter transport [102]. Under this model, the average transport distance of eDNA (Sp, defined as the distance in which 63.2% of particles will have been deposited [102]) has been measured or estimated to be between hundreds of metres to greater than 100 km [7,71,77,103,104]. As Sp is inherently non-generalizable between streams, the depositional velocity (vdep) is more informative. vdep is a particle property that describes the relationship between the transport distance (Sp) and the average flow velocity (u) and stream depth (h), captured by [8,71,102]. Initial measurements of the vdep of eDNA range from 0.146 to 0.535 mm s−1, similar to those of fine particulate organic matter [8,71]. This implies that the transport distance of the same particle would be eight times farther in a river twice as fast and four times as wide as a smaller headwater stream, stressing the importance of including hydrological factors in eDNA analysis [26,71]. Additional geomorphological factors may improve the accuracy or precision of such models (e.g. stream slope, average stream-scale form and longitudinal roughness as explanatory variables [73]).
Although DNA decay rates could easily be incorporated into transport models, retention appears to be more important than degradation for limiting transport in lotic systems [26,77,105]. As eDNA is transported downstream, both turbulence and gravitational settling result in eDNA particles being deposited into the sediment in the channel bed. These particles may persist in the substrate, be transported through the underflow or be resuspended into the water column owing to hyporheic (streambed) effects [8,72,82]. As sediment-bound DNA decays at a different rate than suspended DNA (discussed below), resuspension of eDNA has been suggested as a possible explanation for the unexpected persistence of eDNA [101]. The retention and resuspension dynamics of eDNA is a stochastic process [8] but can be influenced by channel bed characteristics, sediment type and the presence of biofilms that can capture or degrade eDNA [8,26,77]. Currently, the extent of these influences is not well described but probably increases uncertainty in factors contributing to eDNA concentration.
(e). Transport in marine ecosystems
In marine ecosystems, ocean currents driven by the Earth's rotation, water density, wind and tides combine to build a complex fabric of interacting currents, each influencing particle transport. One common method for investigating dispersal of passive particles in ocean systems is to use an oceanographic circulation model to track virtual particles in modelled velocity fields; these can include upwelling, along-shore currents, meso-scale eddies, wind-driven surface currents and tidal forcing (e.g. [106,107]). In a model of surface currents on the west coast of British Columbia, Canada, particle transport was predicted to have a median of 1 km and upper estimate of 4 km per day, depending on the source location [106]. When combined with reported decay rates, the distance of suspended eDNA transport before 50% decay can be as high as 1 km (median) or 12 km (upper estimate), although these estimates do not capture concentration changes owing to particle dilution and sinking.
Although these initial distance estimates suggest that eDNA should be highly mixed at the approximately 5 km scale, empirical studies of marine communities indicate remarkably high spatial turnover and site fidelity to the source communities. For example, Port et al. [108] found that eDNA was able to distinguish community assemblages from habitats separated by less than 60 m. Furthermore, within an intertidal zone where resident water is replaced twice per day, Kelly et al. [38] found that eDNA communities were not determined by the influx of tidal waters from elsewhere, but determined by local source organisms within ca 6 h. These findings suggest that local production and persistence of eDNA are more important than transport of eDNA from elsewhere [9]. This may be explained in part by dilution and sinking of eDNA, which can each contribute to diluted concentrations below detection limits [9,109], but experiments of distance decay from fixed sources considering different DNA sources and water flow regimes (calm versus mixed versus directional) would help to elucidate mechanisms, as well as important sources of variability of eDNA spatial fidelity.
(f). The effect of rare events
While rare or episodic events (e.g. storms and floods) are atypical, extreme weather is increasing in frequency [110] and can have a substantial effect on the transport and fate of organic materials [98]. Sediment resuspension is proportional to wind speed by a power of six, such that a 50% increase in wind speed leads to nearly 12 times more sediment resuspended [98]. Flooding, overland flooding and rain events can impact the discharge, dilution and source of eDNA in a system. For example, following a significant rain event, Staley et al. [111] observed a decrease in the relative abundance of human and freshwater fish eDNA sequences in creek and beach samples. Increases in the diversity and number of non-human mammal and bird eDNA sequences were also documented, but a reversal of this trend persisted in stormwater outfall samples. Given the potential influence of rare events, consideration should be given to the hydrological history of a site when designing or interpreting eDNA studies.
5. Synthesis and future directions
eDNA-based tools have been promising the potential for fast, sensitive and non-invasive detection of biodiversity, but as our review demonstrates, there are still several associated sources of error that generate uncertainty. As the spatial and temporal signal of eDNA is dependent heavily on the environment, a better understanding of the environmental processes that impact eDNA fate in aquatic systems are needed to better connect the eDNA collected to their organismal origins. To this end, we have developed five recommendations for future eDNA research.
-
(i)
Recommendation 1: integration of hydrological models to eDNA transport. The need for better transport estimates is becoming increasingly apparent, particularly in lotic ecosystems (e.g. streams), where variation in stream size and flow have a profound effect on eDNA transport, and in marine systems, where transport currents are now being modelled with increasingly greater precision [26,73]. Given this well-developed toolbox, we recommend the integration of hydrological modelling in eDNA sampling regimes (e.g. estimating average transport distances) become standard.
-
(ii)
Recommendation 2: controlled experiments in naturalized systems. While mesocosm experiments have provided notable insights into the behaviour and fate of eDNA, there are many important biotic and abiotic factors that are not captured in these systems, potentially contributing to inapplicable results (as illustrated in figure 2). Observational field studies of eDNA, while valuable, may not be well suited to describe the specific mechanistic impacts these factors have. We recommend an emphasis on an increased use of replicated, controlled experiments in naturalized systems (i.e. controlled, experimental systems that include numerous features of a natural ecosystem) when studying processes that affect eDNA and estimates of uncertainty, designed with an understanding of the potential mechanisms that impact these processes.
-
(iii)
Recommendation 3: ecosystem-specific parametrization of eDNA. Given the significant differences in transport and attenuation mechanisms between lentic, lotic and marine ecosystems, we recommend that eDNA parametrization and conclusions drawn from eDNA studies should be considered as ecosystem-specific. While this may have significant implications for eDNA applications in new systems and the need for increased basic research, we believe the marked difference in eDNA dynamics between environments necessitates this division.
-
(iv)
Recommendation 4: standardized collection and publication of environmental variables. We encourage eDNA practitioners to collect and include environmental data when collecting eDNA samples so that environmentally driven variation can eventually be assessed in a comparative approach (table 1).
-
(v)
Recommendation 5: focused development of process-based models. eDNA exists as a chemically active particle in highly variable and complex environments, and a mechanistic understanding of these interactions is crucial towards developing a full model predicting the relationships between eDNA and the organisms being studied. We recommend a focus towards elucidating the relative contribution of individual decay and transport processes in environment-specific contexts, and on describing and parametrizing eDNA decay within the context of specific mechanisms that contribute to patterns of bias and noise in varying environments. To achieve this goal, we encourage eDNA researchers to embrace cross-disciplinary collaborations, especially incorporating methodologies from disciplines such as forensic taphonomy and geochemistry to understand the mechanisms responsible for the decay and attenuation of eDNA.
Table 1.
Recommended environmental data to collect alongside eDNA samples.
| priority | variable | reference |
|---|---|---|
| recommended | date (season) | [112] |
| water temperature | [30–36] | |
| flow velocity and/or discharge | [7,71] | |
| water body width and depth | [7,35,71] | |
| salinity | [38] | |
| suggested | pH | [24,34,35] |
| turbidity | [1,37] | |
| microbial growth (as chlorophyll a or organic matter) | [18,26] | |
| substrate type | [8,105] | |
| nutrient levels | [105] | |
| geomorphological features (stream slope, average stream-scale form, longitudinal roughness, etc.) | [73] |
Despite the complexity in both situation and process that eDNA is exposed to, eDNA studies often show high fidelity to capture and visuals surveys [71,113]. While the understanding of the physical processes within these data is still relatively new, there is a wide foundation of observations, techniques and models from disciplines including hydrogeology, cell biology, chemistry, palaeontology, forensic taphonomy and ecology on which eDNA research can be built. As the adoption of eDNA techniques continues to grow, incorporation of the processes that impact eDNA can allow for a better prediction of the fate of eDNA and a more-finely honed tool for understanding global aquatic biodiversity.
Supplementary Material
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
This project was supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC). We are grateful for resources from the Bamfield Marine Sciences Centre while writing this manuscript.
References
- 1.Deiner K, et al. 2017. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. ( 10.1111/mec.14350) [DOI] [PubMed] [Google Scholar]
- 2.Ficetola GF, Miaud C, Pompanon FO, Taberlet P. 2008. Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. ( 10.1098/rsbl.2008.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes MA, Turner CR. 2016. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. ( 10.1007/s10592-015-0775-4) [DOI] [Google Scholar]
- 4.Willerslev E, et al. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791–795. ( 10.1126/science.1084114) [DOI] [PubMed] [Google Scholar]
- 5.Longhi S, Cristofori A, Gatto P, Cristofolini F, Grando MS, Gottardini E. 2009. Biomolecular identification of allergenic pollen: a new perspective for aerobiological monitoring? Ann. Allergy Asthma Immunol. 103, 508–514. ( 10.1016/S1081-1206(10)60268-2) [DOI] [PubMed] [Google Scholar]
- 6.Jerde CL, Olds BP, Shogren AJ, Andruszkiewicz EA, Mahon AR, Bolster D, Tank JL. 2016. Influence of stream bottom substrate on retention and transport of vertebrate environmental DNA. Environ. Sci. Technol. 50, 8770–8779. ( 10.1021/acs.est.6b01761) [DOI] [PubMed] [Google Scholar]
- 7.Jane SF, Wilcox TM, Mckelvey KS, Young MK, Schwartz MK, Lowe WH, Letcher BH, Whiteley AR. 2015. Distance, flow and PCR inhibition: EDNA dynamics in two headwater streams. Mol. Ecol. Resour. 15, 216–227. ( 10.1111/1755-0998.12285) [DOI] [PubMed] [Google Scholar]
- 8.Shogren AJ, Tank JL, Andruszkiewicz E, Olds B, Mahon AR, Jerde CL, Bolster D. 2017. Controls on eDNA movement in streams: transport, retention, and resuspension. Sci. Rep. 7, 5065 ( 10.1038/s41598-017-05223-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen BK, Bekkevold D, Clausen LW, Nielsen EE. 2018. The sceptical optimist: challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish. 19, 751–768. ( 10.1111/faf.12286) [DOI] [Google Scholar]
- 10.Iversen LL, Kielgast J, Sand-Jensen K. 2015. Monitoring of animal abundance by environmental DNA—an increasingly obscure perspective: a reply to Klymus et al., 2015. Biol. Conserv. 192, 479–480. ( 10.1016/j.biocon.2015.09.024) [DOI] [Google Scholar]
- 11.Williams JM, Duckworth CA, Burkitt MD, Watson AJM, Campbell BJ, Pritchard DM. 2015. Epithelial cell shedding and barrier function. Vet. Pathol. 52, 445–455. ( 10.1177/0300985814559404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wotton RS, Malmqvist B. 2001. Feces in aquatic ecosystems: feeding animals transform organic matter into fecal pellets, which sink or are transported horizontally by currents; these fluxes relocate organic matter in aquatic ecosystems. Bioscience 51, 537–544. ( 10.1641/0006-3568(2001)051[0537:FIAE]2.0.CO;2) [DOI] [Google Scholar]
- 13.Sassoubre LM, Yamahara KM, Gardner LD, Block BA, Boehm AB. 2016. Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ. Sci. Technol. 50, 10 456–10 464. ( 10.1021/acs.est.6b03114) [DOI] [PubMed] [Google Scholar]
- 14.Kamoroff C, Goldberg CS. 2018. An issue of life or death: using eDNA to detect viable individuals in wilderness restoration. Freshw. Sci. 37, 685–696. ( 10.1086/699203) [DOI] [Google Scholar]
- 15.Klymus KE, Richter CA, Chapman DC, Paukert C. 2015. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 183, 77–84. ( 10.1016/j.biocon.2014.11.020) [DOI] [Google Scholar]
- 16.Jo T, Murakami H, Yamamoto S, Masuda R, Minamoto T. 2019. Effect of water temperature and fish biomass on environmental DNA shedding, degradation, and size distribution. Ecol. Evol. 9, 1135–1146. ( 10.1002/ece3.4802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamoto T, Fukuda M, Katsuhara KR, Fujiwara A, Hidaka S, Yamamoto S, Takahashi K, Masuda R. 2017. Environmental DNA reflects spatial and temporal jellyfish distribution. PLoS ONE 12, e0173073 ( 10.1371/journal.pone.0173073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sansom BJ, Sassoubre LM. 2017. Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 51, 14 244–14 253. ( 10.1021/acs.est.7b05199) [DOI] [PubMed] [Google Scholar]
- 19.Maruyama A, Nakamura K, Yamanaka H, Kondoh M, Minamoto T. 2014. The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 9, e114639 ( 10.1371/journal.pone.0114639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosal R, Eichmiller JJ, Witthuhn BA, Sorensen PW. 2018. Attracting common carp to a bait site with food reveals strong positive relationships between fish density, feeding activity, environmental DNA, and sex pheromone release that could be used in invasive fish management. Ecol. Evol. 8, 6714–6727. ( 10.1002/ece3.4169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart KA. 2019. Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodivers. Conserv. 28, 983–1001. ( 10.1007/s10531-019-01709-8) [DOI] [Google Scholar]
- 22.Turner CR, Barnes MA, Xu CCY, Jones SE, Jerde CL, Lodge DM. 2014. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 5, 676–684. ( 10.1128/MCB.00849-10) [DOI] [Google Scholar]
- 23.Wilcox TM, Zarn KE, Piggott MP, Young MK, McKelvey KS, Schwartz MK. 2018. Capture enrichment of aquatic environmental DNA: a first proof of concept. Mol. Ecol. Resour. 18, 1392–1401. ( 10.1111/1755-0998.12928) [DOI] [PubMed] [Google Scholar]
- 24.Seymour M, et al. 2018. Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun. Biol. 1, 4 ( 10.1038/s42003-017-0005-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowart DA, Murphy KR, Cheng C-HC. 2018. Metagenomic sequencing of environmental DNA reveals marine faunal assemblages from the west Antarctic Peninsula. Mar. Genomics 37, 148–160. ( 10.1016/j.margen.2017.11.003) [DOI] [PubMed] [Google Scholar]
- 26.Shogren AJ, Tank JL, Egan SP, August O, Rosi EJ, Hanrahan BR, Renshaw MA, Gantz CA, Bolster D. 2018. Water flow and biofilm cover influence environmental DNA detection in recirculating streams. Environ. Sci. Technol. 52, 8530–8537. ( 10.1021/acs.est.8b01822) [DOI] [PubMed] [Google Scholar]
- 27.Collins RA, Wangensteen OS, O'Gorman EJ, Mariani S, Sims DW, Genner MJ. 2018. Persistence of environmental DNA in marine systems. Commun. Biol. 1, 185 ( 10.1038/s42003-018-0192-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichmiller JJ, Best SE, Sorensen PW. 2016. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 50, 1859–1867. ( 10.1021/acs.est.5b05672) [DOI] [PubMed] [Google Scholar]
- 29.Bylemans J, Furlan EM, Gleeson DM, Hardy CM, Duncan RP. 2018. Does size matter? An experimental evaluation of the relative abundance and decay rates of aquatic environmental DNA. Environ. Sci. Technol. 52, 6408–6416. ( 10.1021/acs.est.8b01071) [DOI] [PubMed] [Google Scholar]
- 30.Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM. 2014. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. ( 10.1021/es404734p) [DOI] [PubMed] [Google Scholar]
- 31.Lacoursière-Roussel A, Côté G, Leclerc V, Bernatchez L. 2016. Quantifying relative fish abundance with eDNA: a promising tool for fisheries management. J. Appl. Ecol. 53, 1148–1157. ( 10.1111/1365-2664.12598) [DOI] [Google Scholar]
- 32.Eichmiller JJ, Miller LM, Sorensen PW. 2016. Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish. Mol. Ecol. Resour. 16, 56–68. ( 10.1111/1755-0998.12421) [DOI] [PubMed] [Google Scholar]
- 33.Tsuji S, Ushio M, Sakurai S, Minamoto T, Yamanaka H. 2017. Water temperature-dependent degradation of environmental DNA and its relation to bacterial abundance. PLoS ONE 12, e0176608 ( 10.1371/journal.pone.0176608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strickler KM, Fremier AK, Goldberg CS. 2015. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 183, 85–92. ( 10.1016/j.biocon.2014.11.038) [DOI] [Google Scholar]
- 35.Goldberg CS, Strickler KM, Fremier AK. 2018. Degradation and dispersion limit environmental DNA detection of rare amphibians in wetlands: increasing efficacy of sampling designs. Sci. Total Environ. 633, 695–703. ( 10.1016/j.scitotenv.2018.02.295) [DOI] [PubMed] [Google Scholar]
- 36.Buxton AS, Groombridge JJ, Zakaria NB, Griffiths RA. 2017. Seasonal variation in environmental DNA in relation to population size and environmental factors. Sci. Rep. 7, 46294 ( 10.1038/srep46294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoeckle BC, Beggel S, Cerwenka AF, Motivans E, Kuehn R, Geist J. 2017. A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems. PLoS ONE 12, e0189119 ( 10.1371/journal.pone.0189119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly RP, Gallego R, Jacobs-Palmer E. 2018. The effect of tides on nearshore environmental DNA. PeerJ 6, e4521 ( 10.7717/peerj.4521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. 2009. Cell death. N. Engl. J. Med. 361, 1570–1583. ( 10.1056/NEJMra0901217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toné S, Sugimoto K, Tanda K, Suda T, Uehira K, Kanouchi H, Samejima K, Minatogawa Y, Earnshaw WC. 2007. Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp. Cell Res. 313, 3635–3644. ( 10.1016/j.yexcr.2007.06.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley JS, et al. 2018. Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J. 37, e99238 ( 10.15252/embj.201899238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tepper CG, Studzinski GP. 1993. Resistance of mitochondrial DNA to degradation characterizes the apoptotic but not the necrotic mode of human leukemia cell death. J. Cell. Biochem. 52, 325–361. ( 10.1002/jcb.240520311) [DOI] [PubMed] [Google Scholar]
- 43.Lemasters JJ, et al. 1998. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta Bioenerg. 1366, 177–196. ( 10.1016/S0005-2728(98)00112-1) [DOI] [PubMed] [Google Scholar]
- 44.Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516. ( 10.1080/01926230701320337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alaeddini R, Walsh SJ, Abbas A. 2010. Forensic implications of genetic analyses from degraded DNA: a review. Forensic Sci. Int. Genet. 4, 148–157. ( 10.1016/j.fsigen.2009.09.007) [DOI] [PubMed] [Google Scholar]
- 46.Henwood A. 1992. Exceptional preservation of dipteran flight muscle and the taphonomy of insects in amber. Palaios 7, 203 ( 10.2307/3514931) [DOI] [Google Scholar]
- 47.Foran DR. 2006. Relative degradation of nuclear and mitochondrial DNA: an experimental approach. J. Forensic Sci. 51, 766–770. ( 10.1111/j.1556-4029.2006.00176.x) [DOI] [PubMed] [Google Scholar]
- 48.Emmons AL, DeBruyn JM, Mundorff AZ, Cobaugh KL, Cabana GS. 2017. The persistence of human DNA in soil following surface decomposition. Sci. Justice 57, 341–348. ( 10.1016/j.scijus.2017.05.002) [DOI] [PubMed] [Google Scholar]
- 49.Levy-Booth DJ, et al. 2007. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 39, 2977–2991. ( 10.1016/j.soilbio.2007.06.020) [DOI] [Google Scholar]
- 50.Paul JH, Jeffrey WH, Deflaun MF. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corinaldesi C, Beolchini F, Dell'Anno A. 2008. Damage and degradation rates of extracellular DNA in marine sediments: implications for the preservation of gene sequences. Mol. Ecol. 17, 3939–3951. ( 10.1111/j.1365-294X.2008.03880.x) [DOI] [PubMed] [Google Scholar]
- 52.Yang W. 2011. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93. ( 10.1017/S0033583510000181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salter I. 2018. Seasonal variability in the persistence of dissolved environmental DNA (eDNA) in a marine system: the role of microbial nutrient limitation. PLoS ONE 13, e0192409 ( 10.1371/journal.pone.0192409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsui K, Honjo M, Kawabata Z. 2001. Estimation of the fate of dissolved DNA in thermally stratified lake water from the stability of exogenous plasmid DNA. Aquat. Microb. Ecol. 26, 95–102. ( 10.3354/ame026095) [DOI] [Google Scholar]
- 55.Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362, 709–715. ( 10.1038/362709a0) [DOI] [PubMed] [Google Scholar]
- 56.Gates KS. 2009. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 22, 1747–1760. ( 10.1021/tx900242k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rawlence NJ, Lowe DJ, Wood JR, Young JM, Churchman GJ, Huang YT, Cooper A. 2014. Using palaeoenvironmental DNA to reconstruct past environments: progress and prospects. J. Quat. Sci. 29, 610–626. ( 10.1002/jqs.2740) [DOI] [Google Scholar]
- 58.Schroeder GK, Wolfenden R. 2007. Rates of spontaneous disintegration of DNA and the rate enhancements produced by DNA glycosylases and deaminases. Biochemistry 46, 13 638–13 647. ( 10.1021/bi701480f) [DOI] [PubMed] [Google Scholar]
- 59.Smith CI, Chamberlain AT, Riley MS, Stringer C, Collins MJ. 2003. The thermal history of human fossils and the likelihood of successful DNA amplification. J. Hum. Evol. 45, 203–217. ( 10.1016/S0047-2484(03)00106-4) [DOI] [PubMed] [Google Scholar]
- 60.Orlando L, et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78. ( 10.1038/nature12323) [DOI] [PubMed] [Google Scholar]
- 61.Briggs AW, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14 616–14 621. ( 10.1073/pnas.0704665104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cristescu ME, Hebert PDN. 2018. Uses and misuses of environmental DNA in biodiversity science and conservation. Annu. Rev. Ecol. Evol. Syst. 49, 209–230. ( 10.1146/annurev-ecolsys-110617-062306) [DOI] [Google Scholar]
- 63.Leech DM, Snyder MT, Wetzel RG. 2009. Natural organic matter and sunlight accelerate the degradation of 17ß-estradiol in water. Sci. Total Environ. 407, 2087–2092. ( 10.1016/j.scitotenv.2008.11.018) [DOI] [PubMed] [Google Scholar]
- 64.Andruszkiewicz EA, Sassoubre LM, Boehm AB. 2017. Persistence of marine fish environmental DNA and the influence of sunlight. PLoS ONE 12, e0185043 ( 10.1371/journal.pone.0185043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilliod DS, Goldberg CS, Arkle RS, Waits LP. 2014. Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol. Ecol. Resour. 14, 109–116. ( 10.1111/1755-0998.12159) [DOI] [PubMed] [Google Scholar]
- 66.Mächler E, Osathanunkul M, Altermatt F. 2018. Shedding light on eDNA: neither natural levels of UV radiation nor the presence of a filter feeder affect eDNA-based detection of aquatic organisms. PLoS ONE 13, e0195529 ( 10.1371/journal.pone.0195529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tahrani L, Van Loco J, Ben Mansour H, Reyns T.. 2016. Occurrence of antibiotics in pharmaceutical industrial wastewater, wastewater treatment plant and sea waters in Tunisia. J. Water Health 14, 208–213. ( 10.2166/wh.2015.224) [DOI] [PubMed] [Google Scholar]
- 68.Sreedhara A, Freed JD, Cowan JA. 2000. Efficient inorganic deoxyribonucleases. Greater than 50-million-fold rate enhancement in enzyme-like DNA cleavage. J. Am. Chem. Soc. 122, 8814–8824. ( 10.1021/ja994411v) [DOI] [Google Scholar]
- 69.Sreedhara A, Cowan JA. 2001. Catalytic hydrolysis of DNA by metal ions and complexes. J. Biol. Inorg. Chem. 6, 337–347. ( 10.1007/s007750100209) [DOI] [PubMed] [Google Scholar]
- 70.McNair JN, Newbold JD. 2012. Turbulent particle transport in streams: can exponential settling be reconciled with fluid mechanics? J. Theor. Biol. 300, 62–80. ( 10.1016/j.jtbi.2012.01.016) [DOI] [PubMed] [Google Scholar]
- 71.Pont D, et al. 2018. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci. Rep. 8, 10361 ( 10.1038/s41598-018-28424-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shogren AJ, Tank JL, Andruszkiewicz EA, Olds B, Jerde C, Bolster D. 2016. Modelling the transport of environmental DNA through a porous substrate using continuous flow-through column experiments. J. R. Soc. Interface 13, 20160290 ( 10.1098/rsif.2016.0290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fremier AK, Strickler KM, Parzych J, Powers S, Goldberg CS. 2019. Stream transport and retention of environmental DNA pulse releases in relation to hydrogeomorphic scaling factors. Environ. Sci. Technol. 53, 6640–6649. ( 10.1021/acs.est.8b06829) [DOI] [PubMed] [Google Scholar]
- 74.Allan JD, Castillo MM. 2007. Stream Ecology, 2nd edn Dordrecht, Netherlands: Springer. [Google Scholar]
- 75.Evans NT, Lamberti GA. 2018. Freshwater fisheries assessment using environmental DNA: a primer on the method, its potential, and shortcomings as a conservation tool. Fish. Res. 197, 60–66. ( 10.1016/j.fishres.2017.09.013) [DOI] [Google Scholar]
- 76.Chambert T, Pilliod DS, Goldberg CS, Doi H, Takahara T. 2018. An analytical framework for estimating aquatic species density from environmental DNA. Ecol. Evol. 8, 3468–3477. ( 10.1002/ece3.3764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilcox TM, McKelvey KS, Young MK, Sepulveda AJ, Shepard BB, Jane SF, Whiteley AR, Lowe WH, Schwartz MK. 2016. Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol. Conserv. 194, 209–216. ( 10.1016/j.biocon.2015.12.023) [DOI] [Google Scholar]
- 78.Cai P, Huang Q, Zhang X, Chen H. 2006. Adsorption of DNA on clay minerals and various colloidal particles from an alfisol. Soil Biol. Biochem. 38, 471–476. ( 10.1016/j.soilbio.2005.05.019) [DOI] [Google Scholar]
- 79.Bradford SA, Morales VL, Zhang W, Harvey RW, Packman AI, Mohanram A, Welty C. 2013. Transport and fate of microbial pathogens in agricultural settings. Crit. Rev. Environ. Sci. Technol. 43, 775–893. ( 10.1080/10643389.2012.710449) [DOI] [Google Scholar]
- 80.Romanowski G, Lorenz MG, Wackernagel W. 1991. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl. Environ. Microbiol. 57, 1057–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou Y, Wu P, Zhu N. 2014. The protective effect of clay minerals against damage to adsorbed DNA induced by cadmium and mercury. Chemosphere 95, 206–212. ( 10.1016/j.chemosphere.2013.08.069) [DOI] [PubMed] [Google Scholar]
- 82.Turner CR, Uy KL, Everhart RC. 2015. Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol. Conserv. 183, 93–102. ( 10.1016/j.biocon.2014.11.017) [DOI] [Google Scholar]
- 83.Yoccoz NG, et al. 2012. DNA from soil mirrors plant taxonomic and growth form diversity. Mol. Ecol. 21, 3647–3655. ( 10.1111/j.1365-294X.2012.05545.x) [DOI] [PubMed] [Google Scholar]
- 84.Sirois SH, Buckley DH. 2019. Factors governing extracellular DNA degradation dynamics in soil. Environ. Microbiol. Rep. 11, 173–184. ( 10.1111/1758-2229.12725) [DOI] [PubMed] [Google Scholar]
- 85.Pietramellara G, Franchi M, Gallori E, Nannipieri P. 2001. Effect of molecular characteristics of DNA on its adsorption and binding on homoionic montmorillonite and kaolinite. Biol. Fertil. Soils 33, 402–409. ( 10.1007/s003740100341) [DOI] [Google Scholar]
- 86.Jo T, Murakami H, Masuda R, Sakata MK, Yamamoto S, Minamoto T. 2017. Rapid degradation of longer DNA fragments enables the improved estimation of distribution and biomass using environmental DNA. Mol. Ecol. Resour. 17, e25–e33. ( 10.1111/1755-0998.12685) [DOI] [PubMed] [Google Scholar]
- 87.Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6, 37–53. ( 10.1051/ebr:2007031) [DOI] [PubMed] [Google Scholar]
- 88.Cai P, Zhu J, Huang Q, Fang L, Liang W, Chen W. 2009. Role of bacteria in the adsorption and binding of DNA on soil colloids and minerals. Colloids Surf. B Biointerfaces 69, 26–30. ( 10.1016/j.colsurfb.2008.10.008) [DOI] [PubMed] [Google Scholar]
- 89.Morrissey EM, McHugh TA, Preteska L, Hayer M, Dijkstra P, Hungate BA, Schwartz E. 2015. Dynamics of extracellular DNA decomposition and bacterial community composition in soil. Soil Biol. Biochem. 86, 42–49. ( 10.1016/j.soilbio.2015.03.020) [DOI] [Google Scholar]
- 90.Pietramellara G, Ascher J, Ceccherini MT, Nannipieri P, Wenderoth D. 2007. Adsorption of pure and dirty bacterial DNA on clay minerals and their transformation frequency. Biol. Fertil. Soils 43, 731–739. ( 10.1007/s00374-006-0156-8) [DOI] [Google Scholar]
- 91.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int. J. Artif. Organs 34, 824–831. ( 10.5301/ijao.5000051) [DOI] [PubMed] [Google Scholar]
- 92.Wei N, Nakajima F, Tobino T. 2018. A microcosm study of surface sediment environmental DNA: decay observation, abundance estimation, and fragment length comparison. Environ. Sci. Technol. 52, 12 428–12 435. ( 10.1021/acs.est.8b04956) [DOI] [PubMed] [Google Scholar]
- 93.Buxton AS, Groombridge JJ, Griffiths RA. 2017. Is the detection of aquatic environmental DNA influenced by substrate type? PLoS ONE 12, e0183371 ( 10.1371/journal.pone.0183371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dunker KJ, Sepulveda AJ, Massengill RL, Olsen JB, Russ OL, Wenburg JK, Antonovich A. 2016. Potential of environmental DNA to evaluate northern pike (Esox lucius) eradication efforts: an experimental test and case study. PLoS ONE 11, e0162277 ( 10.1371/journal.pone.0162277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eichmiller JJ, Bajer PG, Sorensen PW. 2014. The relationship between the distribution of common carp and their environmental DNA in a small lake. PLoS ONE 9, e112611 ( 10.1371/journal.pone.0112611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moyer GR, Díaz-Ferguson E, Hill JE, Shea C. 2014. Assessing environmental DNA detection in controlled lentic systems. PLoS ONE 9, e103767 ( 10.1371/journal.pone.0103767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.MacIntyre S, Melack JM. 1995. Vertical and horizontal transport in lakes: linking littoral, benthic, and pelagic habitats. J. North Am. Benthol. Soc. 14, 599–615. ( 10.2307/1467544) [DOI] [Google Scholar]
- 98.Lick W. 2008. Sediment and contaminant transport in surface waters. Boca Raton, FL: CRC Press. [Google Scholar]
- 99.Boehrer B, Schultze M. 2008. Stratification of lakes. Rev. Geophys. 46, RG2005 ( 10.1029/2006RG000210) [DOI] [Google Scholar]
- 100.Hodges BR, Imberger J, Laval B, Appt J. 2000. Modeling the hydrodynamics of stratified lakes. In Hydroinformatics 2000 Conf., pp. 23–27 Iowa City, IA: University of Iowa College of Engineering. [Google Scholar]
- 101.Tillotson MD, Kelly RP, Duda JJ, Hoy M, Kralj J, Quinn TP. 2018. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 220, 1–11. ( 10.1016/j.biocon.2018.01.030) [DOI] [Google Scholar]
- 102.Webster JR, Benfield EF, Golladay SW, Hill BH, Hornick LE, Kazmierczak RF, Perry WB. 1987. Experimental studies of physical factors affecting seston transport in streams. Limnol. Oceanogr. 32, 848–863. ( 10.4319/lo.1987.32.4.0848) [DOI] [Google Scholar]
- 103.Deiner K, Fronhofer EA, Mächler E, Walser J-C, Altermatt F. 2016. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 7, 12544 ( 10.1038/ncomms12544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song JW, Small MJ, Casman EA. 2017. Making sense of the noise: the effect of hydrology on silver carp eDNA detection in the Chicago area waterway system. Sci. Total Environ . 605–606, 713–720. ( 10.1016/j.scitotenv.2017.06.255) [DOI] [PubMed] [Google Scholar]
- 105.Shogren AJ, Tank JL, Egan SP, Bolster D, Riis T. 2019. Riverine distribution of mussel environmental DNA reflects a balance among density, transport, and removal processes. Freshw. Biol. 64, 1467–1479. ( 10.1111/fwb.13319) [DOI] [Google Scholar]
- 106.Sunday JM, Popovic I, Palen WJ, Foreman MGG, Hart MW. 2014. Ocean circulation model predicts high genetic structure observed in a long-lived pelagic developer. Mol. Ecol. 23, 5036–5047. ( 10.1111/mec.12924) [DOI] [PubMed] [Google Scholar]
- 107.Cowen RK, Paris CB, Srinivasan A. 2006. Scaling of connectivity in marine populations. Science 311, 522–527. ( 10.1126/science.1122039) [DOI] [PubMed] [Google Scholar]
- 108.Port JA, O'Donnell JL, Romero-Maraccini OC, Leary PR, Litvin SY, Nickols KJ, Yamahara KM, Kelly RP. 2016. Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol. Ecol. 25, 527–541. ( 10.1111/mec.13481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Foote AD, et al. 2012. Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7, e41781 ( 10.1371/journal.pone.0041781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoegh-Guldberg O, et al. 2018. Impacts of 1.5°C of global warming on natural and human systems. In Global warming of 1.5°C. An IPCC special report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, Incheon, South Korea (eds Masson-Delmotte V, et al.), pp. 175–311. Geneva, Switzerland: IPCC. [Google Scholar]
- 111.Staley ZR, Chuong JD, Hill SJ, Grabuski J, Shokralla S, Hajibabaei M, Edge TA. 2018. Fecal source tracking and eDNA profiling in an urban creek following an extreme rain event. Sci. Rep. 8, 14390 ( 10.1038/s41598-018-32680-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Souza LS, Godwin JC, Renshaw MA, Larson E.. 2016. Environmental DNA (eDNA) detection probability is influenced by seasonal activity of organisms. PLoS ONE 11, e0165273 ( 10.1371/journal.pone.0165273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jeunen G-J, Knapp M, Spencer HG, Lamare MD, Taylor HR, Stat M, Bunce M, Gemmell NJ. 2019. Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 19, 426–438. ( 10.1111/1755-0998.12982) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.