Abstract
Background
Riemerella anatipestifer is one of the most serious infectious disease-causing pathogens in the duck industry. Drug administration is an important method for prevention and treatment of infection in duck production, leading to widespread drug resistance in R. anatipestifer.
Methods
For a total of 162 isolates of R. anatipestifer, the MICs were determined for a quinolone antimicrobial agent, namely, nalidixic acid, and three fluoroquinolones, namely, ciprofloxacin, enrofloxacin and ofloxacin. The gyrA, parC, and parE gene fragments were amplified by PCR to identify the mutation sites in these strains. Site-directed mutants with mutations that were detected at a high frequency in vivo were constructed (hereafter referred to as site-directed in vivo mutants), and the MICs of these four drugs for these strains were determined.
Results
In total, 100, 97.8, 99.3 and 97.8% of the 137 R. anatipestifer strains isolated between 2013 and 2018 showed resistance to nalidixic acid, ciprofloxacin, enrofloxacin, and ofloxacin, respectively. The high-frequency mutation sites were detected in a total of 162 R. anatipestifer strains, such as Ser83Ile and Ser83Arg, which are two types of substitution mutations of amino acid 83 in GyrA; Val799Ala and Ile811Val in ParC; and Val357Ile, His358Tyr, and Arg541Lys in ParE. MIC analysis results for the site-directed in vivo mutants showed that the strains with only the Ser83Ile mutation in GyrA exhibited an 8–16-fold increase in MIC values, and all mutants showed resistance to ampicillin and ceftiofur.
Conclusions
The resistance of R. anatipestifer to quinolone agents is a serious problem. Amino acid 83 in GyrA is the major target mutation site for the fluoroquinolone resistance mechanism of R. anatipestifer.
Keywords: Riemerella anatipestifer, Fluoroquinolone resistance, Point mutant, gyrA gene, parC gene, parE gene
Background
Riemerella anatipestifer (R. anatipestifer) is a nonmotile Gram-negative rod-shaped bacterium that is usually isolated from ducks, geese, and turkeys. R. anatipestifer infection can cause pericarditis, peritonitis, fibrinous exudation, diarrhea and neurological symptoms in ducks, which lead to reduced growth rates and high mortality and consequently to great economic loss [1].
Fluoroquinolone antibiotics are one of the most potent broad-spectrum agents commonly used to treat a range of infections [2, 3]. Due to the widespread and indiscriminate use of fluoroquinolone antibiotics, the resistance of R. anatipestifer to fluoroquinolones is particularly serious [4, 5].
Many types of antibiotic resistance mechanisms of R. anatipestifer have been reported, including those against chloramphenicol [6]; florfenicol [7]; aminoglycosides [8]; macrolides, lincosamides and streptogramin B (collectively referred to as MLS) [9, 10]; and tetracycline [11]. However, few articles on the mechanism of fluoroquinolone resistance associated with R. anatipestifer have been published. Resistance to quinolones can occur via multiple mechanisms, including target-site mutations, multidrug resistance (MDR) efflux pumps, changes in membrane permeability, and plasmid-mediated quinolone resistance (PMQR) genes [12].
DNA gyrase and topoisomerase IV can both relax positively supercoiled DNA, albeit with different focuses. DNA gyrase is essential for DNA replication, transcription, and recombination [12]. Topoisomerase IV unlinks newly replicated DNA, thereby allowing chromosome segregation at cell division [13]. Studies on target mutations in quinolone-resistance-determining regions (QRDRs) show that these mutations are the most prevalent mechanism of quinolone resistance. Mutations involved in fluoroquinolone resistance have been extensively studied in other species in recent decades, such as Escherichia coli [14], Mycobacterium tuberculosis [15], and Shigella flexneri [16]; however, research on R. anatipestifer is limited. In addition, there may also be major mutations in non-QRDRs [17, 18]. Therefore, in addition to QRDRs, non-QRDRs are also worthy of attention.
Linda [19], Viktòria Làzàr [20] and Csaba Pà [21] et al. all showed that quinolone-resistant E. coli mutants exhibited cross-resistance to other types of antibiotics. In addition, Webber et al. [22] reported that DNA gyrase mutants of Salmonella typhimurium L821(Ser83Phe) and L825 (Asp87Gly) showed different susceptibilities to some antimicrobials, such as beta-lactams, aminoglycosides, and folate synthesis inhibitors.
In this study, we determined the minimum inhibitory concentration (MIC) values of four quinolone drugs for 162 R. anatipestifer isolates to investigate the prevalence of quinolone resistance in R. anatipestifer. Then, we detected mutations in the gyrA, parC and parE gene fragments of the 162 R. anatipestifer isolates and constructed R. anatipestifer mutants with high-frequency in vivo mutation sites to explore whether mutations at these sites contribute to the quinolone resistance mechanism of R. anatipestifer and to investigate whether these mutations can cause cross-resistance.
Methods
R. anatipestifer strains
A total of 162 R. anatipestifer strains were used in this study. Among these strains, 137 clinical isolates for fluoroquinolone resistance analysis were obtained from 48 duck farms from 2013 to 2018, 15 other clinical isolates were isolated from 14 duck farms from 1998 to 2012, one strain was obtained from the American Type Culture Collection (ATCC) and 9 strains were obtained from the Culture Collection of the University of Gothenburg (CCUG). Detailed strain information is shown in Additional file 1: Table S1.
All R. anatipestifer isolates were cultured on 5% blood tryptic soy agar (TSA; Oxoid, Basingstoke, UK). These inoculated plates were incubated at 37 °C in a 5% CO2 atmosphere for 24 h. Polymerase chain reaction (PCR) amplification of 16S rRNA was performed to identify R. anatipestifer as described previously [9].
Minimum inhibitory concentrations (MICs) of quinolones
MICs of nalidixic acid (Aladdin, USA), ciprofloxacin (Aladdin, USA), enrofloxacin (Aladdin, USA) and ofloxacin (Meilunbio, China) for 162 R. anatipestifer isolates were determined using the standard microscale broth dilution method, performed according to Clinical and Laboratory Standard Institute (CLSI) criteria [23], to analyze the antimicrobial susceptibility of randomly collected isolates between 1932 and 2018.
To explore whether the R. anatipestifer site-directed in vivo mutants showed cross-resistance phenotypes with other nonquinolone antibiotics, the MICs of several other nonquinolone antibiotics such as ampicillin (Aladdin, USA), ceftiofur (Meilunbio, China), amikacin (Meilunbio, China), florfenicol (Aladdin, USA), doxycycline (Meilunbio, China) and lincomycin (Aladdin, USA) were determined for these strains.
The concentration of these antimicrobial agents in 96-well plates ranged from 0.25 to 1024 μg/mL, except that of nalidixic acid, which ranged from 0.5 to 2048 μg/mL. E. coli ATCC 25922 was used for quality control. The experiments were repeated in triplicate.
PCR amplification of the gyrA, parC and parE genes
Thirty-seven complete genomes of R. anatipestifer strains were sequenced by our team in a previous study, and these sequences were uploaded to the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/genome) database. The accession numbers of these complete genomes are listed in Additional file 2: Table S2. We compared the gyrA, gyrB, parC, and parE genes of these thirty-seven complete genomes. The comparison results are shown in Additional file 3-6: Table S3-S6. A region with a high mutation probability that included a large number of mutations was selected as the target region for PCR amplification. The region includes more than just the QRDR. Five fragments were selected (GyrA amino acids 51–327 and 442–726; ParC amino acids 349–615 and 610–857; and ParE amino acids 312–590). There were no eligible areas in the GyrB. The primers were designed with ATCC 11845 as the template to amplify the above five sequences. In addition to the 37 whole-genome sequencing strains, the remaining 125 clinical isolates were prepared as DNA templates by heat and lysis. The above five regions of 125 clinical isolates were amplified to obtain the mutant sites in these regions. In addition, Sanger sequencing was also carried out for the above five regions of the 37 whole-genome sequencing strains to verify the accuracy of the sequences. PCR primers are listed in Additional file 7: Table S7.
Sequence alignment analysis
The sequenced fragments of the 125 clinical isolates and the 37 complete genomes of R. anatipestifer strains were compared with the genome of ATCC 11845 (as a reference strain) by BLAST [24]. The genomic sequence of ATCC 11845 was downloaded from NCBI. MEGA 7 [25] was used for alignment and to obtain mutation sites in all R. anatipestifer strains.
Construction of R. anatipestifer mutants in vivo
The regions where the high-frequency mutation sites were located were selected to design primers. The target fragments of the three genes of ATCC 11845 were amplified via PCR, and the amplified fragments were ligated to a NcoI/XhoI-digested pOES suicide vector to generate pOES-fragments (gyrA fragment-1, 514 bp; gyrA fragment-2, 944 bp; parC fragment-1, 454 bp; parC fragment-2, 433 bp; parE fragment, 1026 bp). Subsequently, pOES fragments were transformed into E. coli DH5α. The recombinant plasmids were extracted using the E.Z.N.A® Plasmid Mini Kit I (Omega Bio-Tek, USA). The QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene, CA) was used to construct the site-directed mutations in recombinant plasmids. Subsequently, these recombinant plasmids containing site-directed mutations were transformed into E. coli S17–1. The transconjugants were selected on LB plates with ampicillin (Amp, 100 μg/mL).
The donor strain E. coli S17–1 harboring the suicide vector pOES fragments and the recipient strain R. anatipestifer ATCC 11845 were grown on LB and sheep blood plates, respectively, at 37 °C overnight. Targeted mutants were constructed by introducing recombinant DNA into the cells by conjugation [26]. The transconjugants were selected on blood agar plates supplemented with cefoxitin (Cfx, 1 μg/mL) and kanamycin (Kana, 50 μg/mL). Transconjugants were shaken in GC broth (GCB) at 37 °C overnight to lose the suicide plasmid. Then, 200 μL of the appropriate dilution was plated on GCB agar supplemented with 13 mM p-Cl-Phe to select clones without the plasmid [27]. Clones were screened by PCR using primers cfx P1/P2 to choose Cfx-sensitive colonies. Then, the Cfx-sensitive colonies were sequenced with the corresponding primers. The primers and plasmids used in this study are shown in Table 1. Growth curves were plotted for the ATCC 11845 strain and R. anatipestifer mutants grown in TSB medium at 37 °C. OD600 was recorded at 2-h intervals.
Table 1.
Antibiotic resistance phenotypes of R. anatipestifer strains collected between 2013 and 2018
| Years | Antibiotics | MICs (μg/mL) and proportion of strains (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | ||
| 2013 n = 13 | NA | 7.69 | 61.5 | 30.8 | ||||||||
| CIP | 7.69 | 7.69 | 84.6 | |||||||||
| ENR | 7.69 | 7.69 | 7.69 | 53.8 | 23.1 | |||||||
| OFX | 7.69 | 46.2 | 23.1 | 23.1 | ||||||||
| 2014~2015 n = 6 | NA | 16.7 | 83.3 | |||||||||
| CIP | 16.7 | 16.7 | 50 | 16.7 | ||||||||
| ENR | 16.7 | 16.7 | 50 | 16.7 | ||||||||
| OFX | 16.7 | 16.7 | 33.3 | 16.7 | 16.7 | |||||||
| 2016 n = 18 | NA | 11.1 | 77.8 | 11.1 | ||||||||
| CIP | 5.6 | 55.6 | 38.9 | |||||||||
| ENR | 5.6 | 22.2 | 27.8 | 33.3 | 11.1 | |||||||
| OFX | 11.1 | 44.4 | 44.4 | |||||||||
| 2017 n = 60 | NA | 10 | 42 | 8 | ||||||||
| CIP | 3.3 | 5.0 | 1.7 | 21.7 | 61.7 | 6.7 | ||||||
| ENR | 8.3 | 1.7 | 8.3 | 28.3 | 36.7 | 16.7 | ||||||
| OFX | 3.3 | 5.0 | 11.7 | 28.3 | 20 | 18.3 | 13.3 | |||||
| 2018 n = 40 | NA | 17.5 | 57.5 | 25 | ||||||||
| CIP | 7.5 | 35 | 35 | 22.5 | ||||||||
| ENR | 2.5 | 50 | 10 | 12.5 | 25 | |||||||
| OFX | 2.5 | 47.5 | 12.5 | 22.5 | 15 | |||||||
aNA nalidixic acid; CIP ciprofloxacin; ENR enrofloxacin; OFX ofloxacin
Results
Minimum inhibitory concentrations (MICs) of nalidixic acid, ciprofloxacin, enrofloxacin, and ofloxacin
The MICs of four antibacterial drugs were determined for a total of 162 R. anatipestifer strains. The MIC values for all strains are shown in Additional file 1: Table S1.
Since the number of collected strains before 2013 was limited, we selected 137 R. anatipestifer strains from 48 farms from 2013 to 2018 to analyze the prevalence of quinolone resistance (Table 1). The MIC values of four quinolone drugs for the 137 R. anatipestifer strains were all greater than 90%. A total of 100% (137/137) of the R. anatipestifer strains were resistant to nalidixic acid. Meanwhile, R. anatipestifer strains that showed resistance to ciprofloxacin, enrofloxacin, or ofloxacin were determined to be 97.8% (134/137), 99.3% (136/137) or 97.8% (134/137) of the total, respectively.
Amino acid substitutions of GyrA, GyrB and ParC in quinolone-resistant R. anatipestifer
We sequenced five gene fragments of gyrA, parC and parE of 162 R. anatipestifer strains. The detailed amino acid substitutions in the amplified fragments are shown in Additional file 1: Table S1. In GyrA, amino acid position 83 contained two mutation types: Ser83Arg (26/162) and Ser83Ile (125/162). Moreover, the strains that were resistant to ciprofloxacin (149/162), enrofloxacin (151/162), and ofloxacin (149/162) all had mutations at position 83. In addition, the mutation frequencies of Cys465Arg (69/162) in GyrA; Val586Thr/Ala (67/162), Val799Ala (117/162), and Ile811Val (109/162) in ParC; and Val357Ile (131/162), His358Tyr (131/162), Arg541Lys (133/162), and Asp564Lys (72/162) in ParE were all higher than 40%; these sites were defined as high-frequency mutation sites. Asn202Glu (52/162) in GyrA and Ile390Thr (45/162), Gly752Val (42/162), Val768Ala (42/162), Glu827Asp (42/162), and Met833Ile (42/162) in ParC had mutation frequencies between 20 and 40%; these sites were defined as medium-frequency mutation sites. Twenty-eight amino acid mutations with mutation frequencies lower than 20% were observed (the detailed mutation sites are shown in Additional file 1: Table S1).
The correspondence between different mutation types and MIC values of different drugs are shown in Additional file 8: Table S8. The number of mutants containing Ser83Ile in GyrA; Val799Ala and Ile811Val in ParC; and Val357Ile, His358Tyr, and Arg541Lys in ParE was the highest (44/162), whereas mutants with Ser83Ile in GyrA and Val357Ile, His358Tyr and Arg541Lys in ParE were the second most frequent (21/162, 13.0%). Compared to the ATCC 11845 strain, the strains with these mutation types exhibited a 32- to 512-fold increase in the MICs of the fluoroquinolone drugs, but the MIC value of nalidixic acid did not change significantly.
Characterization of R. anatipestifer mutants
Eight mutant strains, which were selected based on sites with mutation frequencies greater than 40%, were constructed to explore the contribution of different mutation sites to the resistance of R. anatipestifer. The MIC values of four antibacterial drugs were determined for the mutants (Table 2). Regardless of the amino acid substitution, the MIC of nalidixic acid did not change significantly. The high-frequency mutations Val799Ala and Ile811Val in ParC and/or Val357Ile, His358Tyr and Arg541Lys in ParE, which were present without the amino acid 83 substitution in GyrA, had no significant effect on MIC values. Compared to the parent strain, only the mutant strains with the amino acid 83 substitution in GyrA, namely, the ATCC gyrA (Ser83Ile) and ATCC gyrA (Ser83Ile) + parE (Val357Ile, His358Tyr, Arg541Lys) strains, exhibited an 8- to 16-fold enhancement in resistance to fluoroquinolones. In contrast, the high-frequency mutation sites Cys465Arg in GyrA; Val799Ala and Ile811Val in ParC; and Val357Ile, His358Tyr and Arg541Lys in ParE had no effect on quinolone resistance.
Table 2.
MICs for site-directed R. anatipestifer mutants
| Mutants | MICs (μg/mL)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NA | CIP | ENR | OFX | AMP | EFT | AK | FFC | DO | MY | |
| ATCC 11845 | 256 | 0.25 | 0.25 | 0.25 | < 0.25 | 1 | 64 | 2 | < 0.25 | 0.25 |
| ATCC gyrA (Ser83Ile) | 512 | 2 | 4 | 4 | 64 | 128 | 128 | 2 | < 0.25 | 0.25 |
| ATCC gyrA (Cys465Arg) | 256 | 0.25 | < 0.25 | 0.25 | 16 | 32 | 32 | 2 | < 0.25 | < 0.25 |
| ATCC parC (Val586Ala) | 256 | 0.25 | < 0.25 | 0.5 | 64 | 128 | 64 | 2 | < 0.25 | < 0.25 |
| ATCC parC (Ile811Val) | 256 | < 0.25 | < 0.25 | 0.25 | 64 | 128 | 64 | 2 | < 0.25 | 0.25 |
| ATCC parC (Val799Ala, Ile811Val) | 512 | 0.25 | 0.25 | 0.25 | 64 | 128 | 64 | 2 | < 0.25 | 0.25 |
| ATCC parE (Val357Ile, His358Tyr) | 256 | 0.25 | 0.25 | 0.25 | 64 | 128 | 64 | 2 | < 0.25 | 0.25 |
| ATCC parE (Val357Ile, His358Tyr, Arg541Lys) | 512 | 0.25 | 0.25 | 0.25 | 32 | 128 | 64 | 2 | < 0.25 | 0.25 |
| ATCC gyrA (Ser83Ile) + parE (Val357Ile, His358Tyr, Arg541Lys) | 256 | 4 | 4 | 2 | 128 | 256 | 64 | 2 | < 0.25 | < 0.25 |
aNA nalidixic acid; CIP ciprofloxacin; ENR enrofloxacin; OFX ofloxacin; AMP ampicillin, EFT ceftiofur; AK amikacin; FFC florfenicol; DO doxycycline; MY lincomycin
To understand whether the site-directed mutants exhibit low sensitivity to other types of antibiotics, the MICs of six nonquinolone antibiotics were determined for these strains. The results showed that the MICs of ampicillin and ceftiofur were significantly increased for the mutants. Interestingly, regardless of which gene among gyrA, parC, and parE has amino acid substitutions, the MIC value of ampicillin and ceftiofur increased significantly.
To explore whether these site mutations would affect the adaptability of the strain, the growth curves of the site-directed mutant strains and the parent strain were plotted (Fig. 1). Compared to the parent strain, the mutants exhibited no significant difference in growth, except for the ATCC parC (Ile811Val) strain.
Fig. 1.
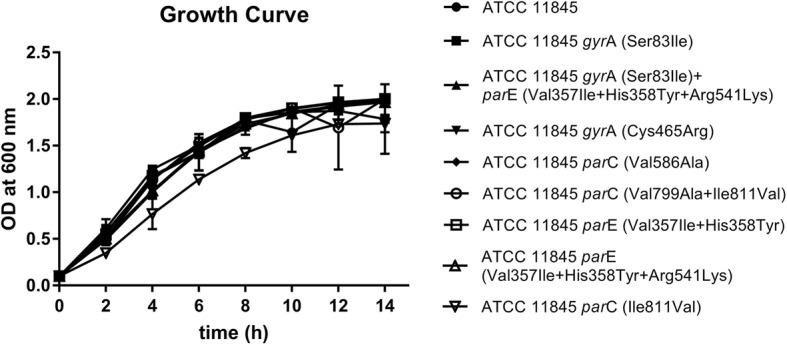
Growth curves for R. anatipestifer ATCC 11845 and R. anatipestifer site-directed mutants. R. anatipestifer ATCC 11845 and site-directed mutants were cultured (OD600 = 0.1) in 20 mL of TSB, and the growth curves were determined
Discussion
In the past six years, the resistance rate of R. anatipestifer to four quinolones has increased. The resistance rate to nalidixic acid, which was 60% before 2003 [28] and 87.4% between 1999 and 2009 [29], has risen to 100% between 2013 and 2018. The resistance rate to ciprofloxacin increased from 23.1 to 59.2% between 1998 and 2010 [29–31]. The resistance rate to enrofloxacin was approximately 60% between 1999 and 2010 [29, 30]. Meanwhile, Zhong et al. [31] and Zhang et al. [29] determined that the resistance rate of R. anatipestifer isolated from different years to ofloxacin was approximately 24%. Nevertheless, the resistance rate to ofloxacin increased to 51.5% between 2008 and 2010. The resistance rates of R. anatipestifer isolated between 2013 and 2018 to CIP, ENR and OFX were 97.8, 99.3 and 97.8%, respectively. Compared with the resistance rate determined in the past, it can be seen that the resistance of R. anatipestifer to quinolones has increased. The high prevalence of fluoroquinolone resistance in R. anatipestifer isolates from ducks may be due to the overuse and abuse of fluoroquinolones in duck disease treatment.
The MIC of nalidixic acid is interesting. The MIC values of nalidixic acid against ATCC 11845 and CCUG 18373, which were isolated in 1932 and 1955, respectively, were both 256 μg/mL, notwithstanding the fact that quinolones were not used in that period. Therefore, in addition to point mutations, there are other mechanisms that mediate resistance to nalidixic acid.
The emergence of mutations is consistent with the history of the development of quinolone drugs. In the 1980s, third-generation quinolone antibiotics were synthesized successively. The strains isolated from 1960 to 1980 are sensitive to FQs, while the RA-CH-2 and RCAD0392 strains isolated in the 1990s showed a resistant phenotype to FQs (Additional file 1: Table S1). The proportion of fluoroquinolone-resistant strains increased significantly. This phenomenon may be due to the increased use of FQs and/or the third-generation quinolone antibiotics being highly conducive to enriching R. anatipestifer resistant strains.
The presence of the amino acid 83 mutation increases the MIC value of fluoroquinolones. All fluoroquinolone-resistant isolates had a mutation at the amino acid 83 site in GyrA (Additional file 1: Table S1), and the site-directed mutants with an amino acid 83 mutation showed an 8- to 16-fold increase in MIC values. These results indicate that the amino acid 83 mutation in GyrA is critical to confer fluoroquinolone resistance to R. anatipestifer. This finding is consistent with previous research results [32, 33]. There are two mutant types at the amino acid 83 position in GyrA, Ser83Arg and Ser83Ile. Previous reports suggest that the mutation type Ser83Ile provides more resistance to FQs than Ser83Arg [29]. However, we found that several strains containing the Ser83Arg mutation type have FQ MICs ranging from 64 to 128 μg/mL. Both Ser83Ile and Ser83Arg can confer low-level or high-level resistance to strains, and Ser83Arg can also confer sensitivity to strains. The same type of mutation also has a variety of base substitution methods. The Ser83Ile group contains AGC-ATC and AGC-ATT mutations, while the Ser83Arg group contains AGC-AGA and AGC-CGC mutations. The same mutant types with different base substitutions had difference in resistance to FQs. For strains with Ser83Arg, some of which had the AGC-AGA base substitution type, the MICs of FQs were 4 μg/mL or less; the strains in which the base substitution type was AGC-CGC had MIC values of FQs greater than or equal to 8 μg/mL. The reason for this phenomenon remains to be explored.
It is strange that while more than 90% of R. anatipestifer strains have high-frequency mutations in parC and parE, these mutations have no significant effect on fluoroquinolone resistance. Perhaps these types of high-frequency mutations are highly adaptive to the evolution of the strain [12, 34]; however, the exact significance of these mutations remains to be explored. Interestingly, although these high-frequency mutations have no significant effect on fluoroquinolone resistance, they all increase the resistance of strains to beta-lactams. This result is consistent with previous reports. The reason for the change in the sensitivity to nonquinolones is probably due to altered patterns of supercoiling and hence global expression of stress response pathways [20, 22].
The RCAD0133 strain has 22 mutations, including all high-frequency mutations; however, this strain has a low level of resistance to fluoroquinolone drugs. These results suggest that not all mutations confer resistance to fluoroquinolones [35] and that the number of mutations is not related to the level of fluoroquinolone resistance.
The substitutions Asp87His in GyrA and Arg120Glu in ParC, which might be associated with fluoroquinolone resistance, were reported by Sun et al. [5]. Additionally, Zhang et al. [29] suggested that Gly81Asp in GyrA might play a sufficient role in enrofloxacin resistance. However, none of these three mutation sites were detected in this test. The Glu202Asn mutation in GyrA [29] and mutations at positions 357 and 358 in ParE, which may increase the resistance of RA to fluoroquinolones, were reported. These mutation sites are consistent with the detected mutations in this study, but the MIC results for the site-directed mutants showed that amino acids 357 and 358 had no significant effect on fluoroquinolone resistance. ATCC parC (Val799Ala, Ile811Val) grew significantly better than ATCC parC (Ile811Val) in the same media conditions (Fig. 1). This phenomenon may be due to the generation of compensatory mutations that increase the adaptability of the strain [36].
There are many strains that have the same mutation type but have different drug resistance phenotypes, such as RCAD0354 and RCAD0356, but these strains exhibit a 2- to 8-fold difference in MICs of FQs. This phenomenon indicates that there may be other mechanisms, such as other low-frequency mutations, undetected sites or efflux pumps.
Conclusions
The resistance rate of R. anatipestifer to four quinolones is considerably high in the duck industry. Sequencing results and the MIC of site-directed mutants showed that only the strains containing the amino acid 83 mutation exhibited resistance to FQs; the other high-frequency mutation sites did not significantly contribute to the MICs. This study highlights the importance of using fluoroquinolones reasonably and correctly to reduce the emergence of multidrug-resistant strains; moreover, it provides data for the molecular detection of fluoroquinolone-resistant R. anatipestifer strains.
Supplementary information
Additional file 1: Table S1. Information on the mutation sites in the gyrA, parC and parE genes of R. anatipestifer
Additional file 2: Table S2. Origin and accession numbers of 37 whole-genome sequences of R. anatipestifer
Additional file 3: Table S3. Amino acid sequence alignment of GyrA from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 4: Table S4. Amino acid sequence alignment of GyrB from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 5: Table S5. Amino acid sequence alignment of ParC from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 6: Table S6. Amino acid sequence alignment of ParE from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 7: Table S7. Primers and plasmids used in this study
Additional file 8: Table S8. Antibiotic phenotype and amino acid substitutions in GyrA, ParC and ParE of R. anatipestifer.
Acknowledgements
Not applicable.
Abbreviations
- ATCC
American Type Culture Collection
- CCUG
Culture Collection of the University of Gothenburg
- CLSI
Clinical and Laboratory Standards Institute
- MDR
multidrug resistance
- MIC
minimum inhibitory concentration
- NCBI
National Center for Biotechnology Information
- PCR
polymerase chain reaction
- PMQR
plasmid-mediated quinolone resistance
- QPDRs
quinolone-resistance-determining regions
- TSA
tryptone soy agar
- TSB
tryptone soy broth
Authors’ contributions
DZ, MZ and AC conceived and designed the project. MZ, JH, LZ, YL and ML determined the MICs for the R. anatipestifer isolates. MZ, XC, LP and YY performed PCR amplification of the gyrA, parC and parE genes. MZ, QY, YW, SZ and XZ constructed the R. anatipestifer mutants in vitro and characterized the mutants. JX, MW, RJ and SC performed sequence alignment analysis. MZ and DZ drafted and revised the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China under grant no. 2017YFD050080; China Agricultural Research System under grant no. CARS-42-17; Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System under grant no. CARS-SVDIP; International S&T Cooperation Program of Sichuan Province under grant no. 2017HH0026; and Science and Technology Innovation Program of Guizhou Academy of Agricultural Science under grant no. (2017)03. The funders had no role in study design and collection, analysis, and interpretation of data, or preparation of the manuscript.
Availability of data and materials
The data supporting the results of this paper are available in the NCBI WGS database. The accession numbers of R. anatipestifer genomes are included in Additional file 2: Table S2.
Ethics approval and consent to participate
The Animal Ethics Committee of the Sichuan Agricultural University waived the need for ethics approval and the need to obtain informed consent for the sample collection.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dekang Zhu and Mingyu Zheng contributed equally to this work.
Contributor Information
Dekang Zhu, Email: zdk24@sicau.edu.cn.
Mingyu Zheng, Email: zhengmyszz@163.com.
Jinge Xu, Email: xje0809@163.com.
Mingshu Wang, Email: mshwang@163.com.
Renyong Jia, Email: cqrc_jry@163.com.
Shun Chen, Email: shunchen@sicau.edu.cn.
Mafeng Liu, Email: liumafengra@163.com.
Xinxin Zhao, Email: xxinzhao@163.com.
Qiao Yang, Email: yangqiao721521@sina.com.
Ying Wu, Email: yingzi_no1@126.com.
Shaqiu Zhang, Email: shaqiu86@hotmail.com.
Yunya Liu, Email: yunnyaaliu@163.com.
Ling Zhang, Email: zl97451@126.com.
Yanling Yu, Email: yanling3525@163.com.
Anchun Cheng, Email: chenganchun@vip.163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-019-1659-4.
References
- 1.Ruiz JA, Sandhu TS. Riemerella anatipestifer infection. In: Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair VL, editors. Diseases of poultry, 13th edition. Hoboken: Wiley; 2013. p. 823–8.
- 2.Andersson M. I. Development of the quinolones. Journal of Antimicrobial Chemotherapy. 2003;51(90001):1–11. doi: 10.1093/jac/dkg212. [DOI] [PubMed] [Google Scholar]
- 3.Endtz HP, Ruijs GJ, van Klingeren B, Jansen WH, van der Reyden T, Mouton RP. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J Antimicrob Chemother. 1991;27(2):199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 4.Nhung NT, Chansiripornchai N, Carrique-Mas JJ. Antimicrobial resistance in bacterial poultry pathogens: a review. Front Vet Sci. 2017;4:126. doi: 10.3389/fvets.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun N, Liu J-H, Yang F, Lin D-C, Li G-H, Chen Z-L, Zeng Z-L, et al. Molecular characterization of the antimicrobial resistance of Riemerella anatipestifer isolated from ducks. Vet Microbiol. 2012;158(3–4):376–383. doi: 10.1016/j.vetmic.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y-P, Tsao M-Y, Lee S-H, Chou C-H, Tsai H-J. Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol. 2010;39(5):333–338. doi: 10.1080/03079457.2010.507761. [DOI] [PubMed] [Google Scholar]
- 7.Chen YP, Lee SH, Chou CH, Tsai HJ. Detection of florfenicol resistance genes in Riemerella anatipestifer isolated from ducks and geese. Vet Microbiol. 2012;154(3–4):325–331. doi: 10.1016/j.vetmic.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Yang F-F, Sun Y-N, Li J-X, Wang H, Zhao M-J, Su J, Zhang Z-J, Liu H-J, Jiang S-J. Detection of aminoglycoside resistance genes in Riemerella anatipestifer isolated from ducks. Vet Microbiol. 2012;158(3–4):451. doi: 10.1016/j.vetmic.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Luo H, Liu M, Wang L, Zhou W, Wang M, Cheng A, Jia R, Chen S, Sun K, Yang Q, et al. Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 2015;44(3):162–168. doi: 10.1080/03079457.2015.1019828. [DOI] [PubMed] [Google Scholar]
- 10.Luo HY, Liu MF, Wang MS, Zhao XX, Jia RY, Chen S, Sun KF, Yang Q, Wu Y, Chen XY, et al. A novel resistance gene, lnu(H), confers resistance to lincosamides in Riemerella anatipestifer CH-2. Int J Antimicrob Agents. 2017. [DOI] [PubMed]
- 11.Zhu D-K, Luo H-Y, Liu M-F, Zhao X-X, Jia R-Y, Chen S, Sun K-F, Yang Q, Wu Y, Chen X-Y. Various profiles of tet genes addition to tet (X) in Riemerella anatipestifer isolates from ducks in China. Front Microbiol. 2018;9:585. doi: 10.3389/fmicb.2018.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22(8):438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Khodursky AB, Cozzarelli NR. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J Biol Chem. 1998;273(42):27668–27677. doi: 10.1074/jbc.273.42.27668. [DOI] [PubMed] [Google Scholar]
- 14.Ren YN, Zhen PP, Jian LI, Guo YF, Wang LH, Ren JY, et al. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of QRDR,acrR,marR and soxR. China Anim Husbandry Vet Med. 2012;39:11–16. [Google Scholar]
- 15.Singhal R, Reynolds PR, Marola JL, Epperson LE, Arora J, Sarin R, Myneedu VP, Strong M, Salfinger M. Sequence analysis of fluoroquinolone resistance-associated genes gyrA and gyrB in clinical Mycobacterium tuberculosis isolates from patients suspected of having multidrug-resistant tuberculosis in New Delhi, India. J Clin Microbiol. 2016;54(9):2298–2305. doi: 10.1128/JCM.00670-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin T, Bi R, Fan W, Kang H, Ma P, Gu B. Novel mutations in quinolone resistance-determining regions of gyrA, gyrB, parC and parE in Shigella flexneri clinical isolates from eastern Chinese populations between 2001 and 2011. Eur J Clin Microbiol Infect Dis. 2016;35(12):2037–2045. doi: 10.1007/s10096-016-2761-2. [DOI] [PubMed] [Google Scholar]
- 17.Al-Emran HM, Heisig A, Dekker D, Adu-Sarkodie Y, Cruz Espinoza LM, Panzner U, von Kalckreuth V, Marks F, Park SE, Sarpong NJ. Detection of a novel gyrB mutation associated with fluoroquinolone-nonsusceptible Salmonella enterica serovar Typhimurium isolated from a bloodstream infection in Ghana. Clin Infect Dis. 2016;62(suppl_1):S47–S49. doi: 10.1093/cid/civ790. [DOI] [PubMed] [Google Scholar]
- 18.Nawaz M, Sung K, Kweon O, Khan S, Nawaz S, Steele R. Characterisation of novel mutations involved in quinolone resistance in Escherichia coli isolated from imported shrimp. Int J Antimicrob Agents. 2015;45(5):471–476. doi: 10.1016/j.ijantimicag.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Strand L, Jenkins A, Henriksen IH, Allum AG, Grude N, Kristiansen BE. High levels of multiresistance in quinolone resistant urinary tract isolates of Escherichia coli from Norway; a non clonal phenomen? BMC Res Notes. 2014;7(1):376. doi: 10.1186/1756-0500-7-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lázár V, Nagy I, Spohn R, Csörgő B, Györkei Á, Nyerges Á, Horváth B, Vörös A, Busa-Fekete R, Hrtyan M. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun. 2014;5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pál C, Papp B, Lázár V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015;23(7):401–407. doi: 10.1016/j.tim.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webber MA, Ricci V, Whitehead R, Patel M, Fookes M, Ivens A, Piddock LJ. Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. MBio. 2013;4(4):e00273–e00213. doi: 10.1128/mBio.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-seventh Informational Supplement. CLSI document M100-S27. Wayne: Clinical and Laboratory Standards Institute; 2017.
- 24.Altschul Stephen F., Gish Warren, Miller Webb, Myers Eugene W., Lipman David J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao H, Cheng X, Zhu D, Wang M, Jia R, Chen S, Chen X, Biville F, Liu M, Cheng A. TonB energy transduction systems of Riemerella anatipestifer are required for iron and hemin utilization. PLoS One. 2015;10(5):e0127506. doi: 10.1371/journal.pone.0127506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu M, Huang Y, Liu J, Biville F, Zhu D, Wang M, Jia R, Chen S, Zhao X, Yang Q, et al. Multiple genetic tools for editing the genome of Riemerella anatipestifer using a counterselectable marker. App Microbiol Biotechnol. 2018;102(17):7475–7488. doi: 10.1007/s00253-018-9181-4. [DOI] [PubMed] [Google Scholar]
- 28.Chang C-F, Lin W-H, Yeh T-M, Chiang T-S, Chang Y-F. Antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks and the efficacy of ceftiofur treatment. J Vet Diagn Investig. 2003;15(1):26–29. doi: 10.1177/104063870301500106. [DOI] [PubMed] [Google Scholar]
- 29.Zhang CZ. Antimicrobial susceptibility of Riemerella anatipestifer isolates from ducks and geese and the mutations of Gyrase associated with quinolone resistance. J Vet Med Taiwan. 2012;38(1):7–18. [Google Scholar]
- 30.Gyuris É, Wehmann E, Czeibert K, Magyar T. Antimicrobial susceptibility of Riemerella anatipestifer strains isolated from geese and ducks in Hungary. Acta Vet Hung. 2017;65(2):153–165. doi: 10.1556/004.2017.016. [DOI] [PubMed] [Google Scholar]
- 31.Zhong CY, Cheng AC, Wang MS, Zhu DK, Luo QH, De Zhong C, Li L, Duan Z. Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 2009;53(4):601–607. doi: 10.1637/8552-120408-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25(5):358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Yonezawa M, Takahata M, Banzawa N, Matsubara N, Watanabe Y, Narita H. Analysis of the NH2-terminal 83rd amino acid of Escherichia coli GyrA in quinolone-resistance. Microbiol Immunol. 1995;39(4):243–247. doi: 10.1111/j.1348-0421.1995.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 34.N W MJE. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007;13(1):5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 35.Santhosh KS, Deekshit VK, Venugopal MN, Karunasagar I, Karunasagar I. Multiple antimicrobial resistance and novel point mutation in Fluoroquinolone-resistant Escherichia coli isolates from Mangalore, India. Microb Drug Resist. 2017;23(8):994–1001. doi: 10.1089/mdr.2016.0142. [DOI] [PubMed] [Google Scholar]
- 36.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8(3):273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Information on the mutation sites in the gyrA, parC and parE genes of R. anatipestifer
Additional file 2: Table S2. Origin and accession numbers of 37 whole-genome sequences of R. anatipestifer
Additional file 3: Table S3. Amino acid sequence alignment of GyrA from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 4: Table S4. Amino acid sequence alignment of GyrB from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 5: Table S5. Amino acid sequence alignment of ParC from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 6: Table S6. Amino acid sequence alignment of ParE from the 37 whole-genome-sequenced R. anatipestifer strains
Additional file 7: Table S7. Primers and plasmids used in this study
Additional file 8: Table S8. Antibiotic phenotype and amino acid substitutions in GyrA, ParC and ParE of R. anatipestifer.
Data Availability Statement
The data supporting the results of this paper are available in the NCBI WGS database. The accession numbers of R. anatipestifer genomes are included in Additional file 2: Table S2.


