Abstract
KH-type splicing regulatory protein (KHSRP) is a multifunctional nucleic acid binding protein implicated in key aspects of cancer cell biology: inflammation and cell-fate determination. However, the role KHSRP plays in colorectal cancer (CRC) tumorigenesis remains largely unknown. Using a combination of in silico analysis of large data sets, ex vivo analysis of protein expression in patients, and mechanistic studies using in vitro models of CRC, we investigated the oncogenic role of KHSRP. We demonstrated KHSRP expression in the epithelial and stromal compartments of both primary and metastatic tumors. Elevated expression was found in tumor versus matched normal tissue, and these findings were validated in larger independent cohorts in silico. KHSRP expression was a prognostic indicator of worse overall survival (hazard ratio, 3.74; 95% CI, 1.43–22.97; P = 0.0138). Mechanistic data in CRC cell line models supported a role of KHSRP in driving epithelial cell proliferation in both a primary and metastatic setting, through control of the G1/S transition. In addition, KHSRP promoted a proangiogenic extracellular environment by regulating the secretion of oncogenic proteins involved in diverse cellular processes, such as migration and response to cellular stress. Our study provides novel mechanistic insight into the tumor-promoting effects of KHSRP in CRC.
Almost all the major oncogenic signaling pathways result in the reprogramming of translation. This impacts on both cancer initiation and progression by directing selective translation of specific tumor-promoting mRNAs.1 RNA-binding proteins (RBPs), such as KH-type splicing regulatory protein (KHSRP; alias KSRP or FUBP2), are capable of recognizing sequence-specific regulatory elements in the 3′-untranslated region, such as AU-rich elements that regulate transcript-specific translation, promoting mRNA splicing and stability.2 KHSRP regulates translation by mediating mRNA decay through direct binding and recruitment of proteins involved in RNA degradation, including the poly(A)-specific ribonuclease, exosome components, and decapping enzymes.3, 4 Alternatively, KHSRP can regulate translation independently of mRNA decay, by either promoting translational inhibition5 or indirectly promoting maturation of selected miRNA precursors.6 Adding a further layer of complexity, KHSRP also acts as a transcription factor, inducing the expression of the MYC oncogene.7
The KHSRP-mediated control of mRNA translation and stability has been extensively studied in the context of innate immunity.5, 8 Proinflammatory cytokines (eg, IL-6, IL-8, tumor necrosis factor-α, and IL-1β) and inflammatory mediators, such as inducible nitric oxide synthase, are directly targeted and negatively regulated by KHSRP.9, 10 However, the role of KHSRP is most likely context dependent, with the balance between KHSRP and other RBPs with similar or divergent regulatory effects an important consideration.11
Direct evidence for the involvement of KHSRP in cancer is accumulating. KHSRP has been implicated in the pathogenesis of small-cell lung cancer,12 osteosarcoma,13 and breast cancer.14 Several distinct mechanisms have been proposed (eg, regulation of cell differentiation in P19 mouse teratocarcinoma cells15; deregulation of oncosuppressive miRNAs, such as let-7a and miR-30c16; or control of transforming growth factor-β–dependent regulation of epithelial-to-mesenchymal transition17).
The role of KHSRP in colorectal cancer (CRC), however, remains underappreciated. Although other RBPs have been implicated in cellular transformation in the intestinal epithelium [namely, epithelial splicing regulatory protein 1,18 Apobec-1,19 Musashi (MSI)-1,20 and MSI-221], KHSRP remains largely underinvestigated in this indication. Interestingly, adenomatous polyposis coli (APC), a tumor suppressor frequently mutated in CRC, is itself an RBP capable of regulating the translation of mRNAs associated with cell adhesion, motility, and other cellular processes crucial for carcinogenesis,22 suggesting the importance of RBP-mediated translation control in the gut. Herein, we use a combination of bioinformatic, in vitro, and ex vivo approaches to dissect the role of KHSRP in both regulation of cell proliferation and inflammatory environment in CRC and provide evidence for a novel prognostic role of this RBP in intestinal tumorigenesis.
Materials and Methods
Ethics Approval and Consent to Participate
All human tissue used in this study was obtained with the informed written consent of the patient; ethical approval was granted by the Research and Ethics Committee of St. Vincent's University Hospital (Dublin, Ireland). The study was performed in accordance with the Declaration of Helsinki.
Bioinformatic Analysis of Publicly Available Data Sets
CRC data sets in the Oncomine database (http://www.oncomine.org, last accessed November 2014; registration required; Thermo Fisher Scientific, Ann Arbor, MI) were searched using the differential expression module for cancer versus normal tissue; output of the resulting database searches was downloaded for offline plotting, along with detailed gene expression results for two representative cohorts [GSE9348 and The Cancer Genome Atlas (TCGA)]. A similar analysis was conducted without restricting for cancer type. Differential expression analysis of RNA-sequencing data for 12 tumor-normal pairs in TCGA database was previously described and made available by the authors.23 Gene expression (RNA sequencing) and the frequency of genetic alterations for KHSRP in TCGA cohort were analyzed using the cBio portal (http://www.cbioportal.org, last accessed November 2016).24 All other data sets were downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo, last accessed November 13, 2018), using the indicated accession number.
Patient Samples and Tissue Microarrays
Fresh-frozen tumor and adjacent normal tissue samples were obtained from 16 patients and were used for protein isolation and Western blot analysis (Western blot cohort), as described thereafter. Tissue microarrays were accessed from a previously described cohort of formalin-fixed, paraffin-embedded archival resected tumor tissue samples.25 Patient details for both cohorts are reported in Table 1. For immunohistochemistry (IHC), tissue microarray sections were incubated with a 1:250 dilution of a validated (The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000088247-KHSRP/antibody, last accessed April 2019) rabbit polyclonal anti-KHSRP antibody (catalog number HPA034739; RRID:AB_10601582; Sigma-Aldrich, St. Louis, MO) and secondary antibody horseradish peroxidase (Dako Agilent, Santa Clara, CA). Endogenous peroxidase activity was blocked using 3% hydrogen peroxide, and non-specific binding was blocked with casein buffer. Diaminobenzidine was used to visualize staining, and sections were counterstained with hematoxylin, dehydrated, and mounted. Stained tissue microarray sections were acquired with a ScanScope XT high-throughput scanning system (Aperio Technologies, Leica Biosystems, Buffalo Grove, IL), and images were scored by two independent investigators (F.C. and J.J.P.) blinded to the clinical data. Both epithelial and stromal cells were assessed for staining intensity and percentage positivity; positive cell count was scored as a continuous variable (0% to 100%), whereas intensity was scored using a scale of 0 to 3 (correlating with negative, weak, medium, and strong staining, respectively). The mean value of each parameter from the two investigators was calculated, and parameters were averaged from all tissue cores related to the same patient; the two parameters were finally combined using the semiquantitative quickscore method,26 by multiplying the averaged intensity and positivity scores for epithelial and stromal cells separately. The quickscore values for tumor tissue were further divided by the quickscore values for normal tissue, to obtain a tumor/normal ratio score that was used to quantify the amplitude of the tumor-specific changes in protein expression. A similar strategy was used to obtain metastasis/normal scores. There was overall a good correlation between the scores obtained by the two independent assessors (weighted Cohen κ coefficient = 0.63).
Table 1.
Summary of Clinical Characteristics for the Patients Included in the Two Cohorts Analyzed by Either WB or TMA Analysis
| Variable | WB analysis cohort | TMA analysis cohort |
|---|---|---|
| Total patients, n | 16 | 62 |
| Tissue type | Fresh-frozen | FFPE |
| Age in years, mean (range) | 65.8 (50–82) | 63.5 (25–84) |
| Sex, n | ||
| Male | 9 | 34 |
| Female | 7 | 28 |
| Clinical stage, n | ||
| I | 0 | 1 |
| II | 3 | 2 |
| III | 4 | 22 |
| IV | 9 | 37 |
Data are given as number of patients, unless otherwise indicated.
FFPE, formalin fixed, paraffin embedded; TMA, tissue microarray; WB, Western blot.
Cell Culture and Transient and Stable Knockdown of KHSRP
SW480 and SW620 cells were obtained from ATCC (Manassas, VA) and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% amphotericin B (Fungizone; all from Invitrogen Life Technologies, Carlsbad, CA). Cell line identity was confirmed by analysis of short-term repeat loci, and cells were routinely tested for Mycoplasma infection. Cells were seeded in 24-well plates to be transfected with 30 pmol KHSRP siRNA (Silencer Select; Thermo Fisher Scientific) or nontargeting scramble siRNA using 2 μL Lipofectamine 2000 (Invitrogen Life Technologies) in serum- and antibiotic-free growth media for 6 hours, followed by incubation in standard growth media for a total of 48 hours after transfection. Alternatively, cells were transfected with 50 nmol/L KHSRP siRNA (SMARTpool: ON-TARGET plus) and corresponding Non-Targeting Pool (Dharmacon, Lafayette, CO). To generate stable KHSRP knockdown cell lines, advantage was taken of an ultracomplex pooled shRNA library targeting each annotated human protein-coding gene with 25 independent shRNAs on average (as well as 500 negative control shRNAs), which was previously described and used to perform genome-wide genetic interaction screens in mammalian cells.27 From this original library, three independently validated shRNA sequences targeting KHSRP were selected, along with one negative control scramble shRNA sequence; for each sequence, top and bottom oligonucleotides were synthesized (Integrated DNA Technologies, Coralville, IA). Two shRNA-expressing derivative cell lines were generated from SW480 and SW620 following a previously published protocol.28 The lentiviral plasmid used for shRNA expression was pMK1201, which is a modified version of pMK120028 based on the design of pINDUCER10 for tetracycline-induced expression of the shRNA coupled with a fluorescent reporter (turbo red fluorescent protein) and a puromycin resistance sequence for positive selection.29 To clone each individual shRNA into the lentiviral backbone, 2 μL of top and bottom oligonucleotides was annealed at 95°C for 5 minutes in a buffer containing 100 mmol/L potassium acetate, 30 mmol/L HEPES-KOH (pH = 7.4), and 2 mmol/L magnesium acetate. Annealed oligonucleotides (0.01 μmol/L) were ligated in 25 ng of the pMK1201 vector predigested with BstXI (New England Biolabs, Ipswich, MA) and gel purified; ligation was performed at room temperature for 2 hours using 2000 U of T4 DNA ligase (New England Biolabs). DH5α cells (Thermo Fisher Scientific) were transformed and plated onto ampicillin-containing Luria broth plates overnight at 37°C. Single colonies were picked and expanded for plasmid DNA miniprep (Qiagen, Germantown, MA). Correct insertion of shRNA sequences was confirmed by sequencing using the 5′ pSico-Eco-insert-seq.28 For lentivirus preparation, the second-generation virion packaging vector psPAX2 (plasmid number 12260; Addgene, Cambridge, MA) and the vesicular stomatitis virus G glycoprotein envelope plasmid pMD2.G (plasmid number 12259; Addgene) from Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) were used. The producer cell line was 293T, a highly transfectable derivative of human embryonic kidney cell 293 (ATCC), which was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% amphotericin B. 293T cells were transfected in 10-cm plates using the calcium phosphate method: 2.5 μg of scramble shRNA vector or a pooled mix of 0.83 μg each of the three KHSRP shRNA vectors was combined with 0.58 μg of pMD2.G and 1.92 μg of psPAX2 and transfected with 12.5 μmol/L chloroquine diphosphate and 12.5 mmol/L calcium chloride. Transfection medium was replaced after 16 hours with full Dulbecco’s modified Eagle’s medium, and virus-containing conditioned medium was harvested two times at 48 and 72 hours after transfection; pooled harvests from each condition were precleared by centrifugation, filtered through a 0.45-μm membrane, and used immediately to transduce SW480 and SW620. Cells were selected with 1 μg/mL puromycin for 2 weeks, expanded, and stock frozen. A fresh stock of cells was used each time for experiments.
Protein Isolation, Immunoblotting, Immunofluorescence, and Enzyme-Linked Immunosorbent Assay
Cells or tissue homogenate were lysed in radioimmunoprecipitation assay buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl, 1% Triton, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with a protease and phosphatase inhibitor cocktail (Roche Applied Science, Burgess Hill, UK) and 1 mmol/L phenylmethylsulfonyl fluoride (Sigma-Aldrich). Total protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Total proteins (10 to 20 μg) were separated onto 10% or 4% to 20% gradient SDS-PAGE gels and wet transferred to PVDF using the mini-PROTEAN TGX system (Bio-Rad, Hercules, CA). Membranes were preblocked with 5% low-fat dry milk in Tris-buffered saline and Tween 20 and incubated with a rabbit polyclonal anti-KHSRP antibody (Sigma-Aldrich; catalog number HPA034739; RRID:AB_10601582), a monoclonal anti–c-Myc antibody (D84C12; Cell Signaling, Danvers, MA), a monoclonal anti–ephrin receptor B2 (clone D2X2I; Cell Signaling), or a monoclonal anti-S100A11 (clone EPR11171B; Abcam, Cambridge, MA), and either anti-rabbit or anti-mouse horseradish peroxidase–conjugated secondary antibody (Bio-Rad). Protein loading normalization was performed with anti–β-actin or anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies (Cell Signaling) or Coomassie stain (Thermo Fisher Scientific). Proteins were visualized by chemiluminescence with enhanced chemiluminescence substrate (Thermo Fisher Scientific) and semiquantified using ImageJ software version 2.0.0-rc-69/1.52n (NIH, Bethesda, MD; http://imagej.nih.gov/ij). For immunofluorescence, cells were fixed on chamber slides with 4% formaldehyde, permeabilized with 0.5% Tween in phosphate-buffered saline (PBS), and stained using the same anti-KHSRP antibody and a Cy3–anti-rabbit, with DAPI and phalloidin counterstain (Dako Agilent); slides were imaged on a Leitz DM40 microscope (Leica Microsystems, Wetzlar, Germany) equipped with the AxioCam system and AxioVision 3.0.6 for image acquisition. Human IL-8 and vascular endothelial growth factor were quantified in cell culture supernatants using DuoSet ELISA (R&D Systems, Minneapolis, MN), following manufacturer's instructions.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from cells using the EZNA Total RNA Kit (Omega, VWR, Radnore, PA). One microgram of RNA was reverse transcribed into cDNA using the RT Omniscript cDNA kit (Qiagen), and 20 ng of cDNA template was amplified on a LightCycler-480 (Roche, Indianapolis, IL) using a FAST SYBR Green Master Mix (Invitrogen Life Technologies) under the following conditions: preincubation step at 95°C for 5 minutes was followed by 45 cycles of denaturation at 95°C for 10 seconds, annealing at 60°C for 10 seconds, and elongation at 72°C for 10 seconds. GAPDH was used as the housekeeping gene. The mRNA expression levels of all samples were normalized to the housekeeping gene, and the ΔΔCt method was used to calculate relative expression (treatment versus control). The following primers were used in real-time quantitative PCR: KHSRP, 5′-CCGCTTACTACGGACAGACC-3′ (forward) and 5′-ATTCATTGAGCCTGCTGCTGT-3′ (reverse); and GAPDH, 5′-CGGATTTGGTCGTATTGGGCGCCTG-3′ (forward) and 5′-CAGCATCGCCCCACTTGATTTTGGA-3′ (reverse). For validation of microarray candidate genes, single-strand cDNA synthesis and PCR amplification were performed in a one-step reaction using the Brilliant II QRT-PCR Master Mix (Dako Agilent) and TaqMan gene expression assays (Invitrogen Life Technologies). RNA (25 ng) was loaded in 96-well plate format and amplified with 0.9 mmol/L of primers and 0.25 mmol/L of minor groove binder probe. The reaction was performed in a CFX Connect instrument (Bio-Rad) with the following parameters: a reverse transcription step at 50°C for 30 minutes was followed by a preincubation step at 95°C for 10 minutes, 50 cycles of denaturation at 95°C for 15 seconds, and annealing/extension at 60°C for 1 minute. Relative quantification of gene expression was calculated with the comparative cycle threshold method, normalizing for GAPDH expression levels. TaqMan gene expression assays for FABP3 (GeneID: 2170; assay ID: Hs00997360_m1), SMG1 (GeneID: 23049; assay ID: Hs00979691_m1), MT1F (GeneID: 4494; assay ID: Hs00744661_sH), and GAPDH (GeneID: 2597; assay ID: Hs02786624_g1) contained minor groove binder probes with a FAM reporter dye.
Gene Expression Microarray
Total RNA was extracted from cells using the RNeasy Plus kit (Qiagen); the integrity and concentration of RNA were confirmed using the RNA 6000 Nano kit on a Bioanalyzer 2100 (Dako Agilent), with reported RNA integrity numbers > 9. Amplified cDNA for gene expression analysis was prepared with the Ovation PicoSL WTA System V2 (Nugen, San Carlos, CA); labeled cDNA targets were generated with the Encore Biotin Module (Nugen) and hybridized to a GeneChip Human Transcriptome Array 2.0 (Affymetrix, Thermo Fisher Scientific) following manufacturer's instructions. CEL files were analyzed with the Affymetrix Expression Console software version 1.4.1, using the Affymetrix Human Transcripome Array 2.0 library files and the HTA-2_0.na35.2.hg19 annotation files. A robust multi-array average workflow was performed, which used a quantile normalization procedure and a general background correction. The resulting normalized CHP files were then imported in the Affymetrix Transcriptome Analysis Console software version 3.0.0 to test for differential expression using a paired one-way repeated-measures (analysis of variance) approach and the default filter criteria (fold change > |2|, and P < 0.05). The full data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus, and they can be accessed with the Gene Expression Omnibus series accession number GSE112329. Overrepresentation analysis was used to identify canonical pathways and functional processes of biological importance within the list of differentially expressed genes; the analysis was performed using GeneTrail2 version 1.5 (https://genetrail2.bioinf.uni-sb.de, last accessed April 2016).30 Network analysis was performed using STRING version 10.0 (https://string-db.org, last accessed April 2016).31 STRING is a database of known and predicted protein-protein interactions, including both direct (physical) and indirect (functional) associations, inferred from experimental data and computational predictions. STRING computes a global score by combining the probabilities from the different types of evidence and correcting for the probability of randomly observing an interaction. Proteins in the network are then clustered on the basis of the distance matrix obtained from the global scores, using the K-means clustering algorithm.32
Cell-Based Assays
Cell proliferation was measured by neutral red staining, following an established protocol.33 Briefly, cells were seeded in 24-well plates (20,000 cells/well) 24 hours after transfection (for the transient knockdown) or 24 hours after doxycycline treatment (for the stable knockdown), and they were allowed to grow for 7 days. Cells were stained with 33 mg/L neutral red (Sigma-Aldrich) for 2 hours, then washed in PBS and imaged (for representative images of the plates); cells were destained using a solution of 50% ethanol and 1% glacial acetic acid in water, and absorbance was measured at 540 nm to quantify staining. Real-time cell proliferation was measured using an E-plate 16 and an RTCA DP Analyser (xCelligence System; Roche), which uses gold microelectrodes fused to the bottom surface of a microtiter plate well. The impedance of electron flow at the interface between electrodes and culture medium, caused by adherent cells, is reported using a unitless parameter called the cell index. Cells were seeded at 10,000 cells/well, and the recording of the cell index occurred every 15 minutes during the first 6 hours, and every hour for the rest of the period. Cells seeded at low density (5000 cells/well in 6-well plates) for colony-formation assay were grown for 7 days, then fixed in methanol/citric acid (4:1 v/v), stained in 2% crystal violet, and counted with ImageJ software. The plating efficiency was calculated by dividing the number of colonies by the original seeding density, and the surviving fraction was determined by comparing the plating efficiency of transfected versus control wells. The total colony area was calculated for each biological replicate by averaging the area of all colonies in replicate wells. Spheroids were generated by seeding 20,000 cells/well in 24-well plates on growth factor–reduced Matrigel (BD Biosciences, San Jose, CA) following the three-dimensional on-top protocol.34 After 7 days, spheroids were stained with neutral red (as outlined above), imaged (averaging three to four fields per well at ×10 magnification), and quantified via ImageJ software. Scratch-wound assays were performed in culture insert two-well μ-dishes (Ibidi, Martinsried, Germany) and quantified with ImageJ software (using MiToBo plugin), and representative images were prepared with WimScratch software version 4.0 (Ibidi). Invasion assays were performed using Matrigel-coated Boyden chambers (8-μm pore size; BioCoat; BD Biosciences) for 72 hours against 10% fetal bovine serum as chemoattractant. Invaded cells were fixed using 1% glutaraldehyde, stained with 0.1% crystal violet, and imaged with a light microscope at a ×10 magnification, averaging five random fields per insert. Crystal violet staining was quantified with ImageJ software. For tube-formation assays, human dermal microendothelial cells (a gift from Prof. Ursula Fearon, Trinity College Dublin, Dublin, Ireland) were seeded on Matrigel-coated 96-well plates (10,000 cells/well) in MCDB131 medium (Invitrogen Life Technologies) with 40% conditioned medium from SW620 cells. The tube analysis was performed manually after 6 hours by counting five sequential fields (×10 magnification) with a focus on the surface of the Matrigel, and a connecting branch between two discrete endothelial cells was counted as one tube. Representative images were prepared with WimTube (Ibidi).
Flow Cytometry
Cell cycle was analyzed by propidium iodide staining. Cells were resuspended at 1 to 2 × 106 cells/mL in PBS and fixed in 70% ethanol for 2 hours at −20°C, washed twice in PBS, and resuspended in staining solution [0.1% (v/v) Triton X-100, 10 μg/mL propidium iodide, and 100 μg/mL DNase-free RNase A in PBS] for 30 minutes at room temperature. Data were acquired on a CyAn ADP (Beckman Coulter, Brea, CA) using a 488-nm excitation laser, and data deconvolution was performed in FlowJo (Becton Dickinson, Franklin Lakes, NJ). To quantify the mitotic index, fixed cells were permeabilized with 0.25% Triton X-100 in PBS for 20 minutes on ice, blocked with 10% bovine serum albumin in PBS, and incubated with 1 μg of an Alexa-Fluor 488–conjugated anti–phosphorylated histone H3 (Ser10) antibody (catalog number 650804; BioLegend, San Diego, CA). After washing, cells were resuspended in PBS containing 50 μg/mL propidium iodide for 15 minutes on ice before analysis on a CyAn ADP using a 488-nm excitation laser. Mitotic cells with 4N DNA and elevated levels of phosphorylated histone H3 can be detected by plotting FL1-H (FITC) versus FL3-A (propidium iodide).
Shotgun Proteomics
Stably transfected SW620 cells were treated with vehicle or 500 ng/mL doxycycline for 4 days to induce shRNA-mediated knockdown of KHSRP, as outlined above. Cells were then incubated in serum-free medium for 16 hours, and triplicate conditioned media for each condition were harvested, buffer exchanged, and concentrated in a final volume of 300 μL PBS using Amicon centrifugal filter units with a 3-kDa cutoff membrane (MilliporeSigma, Burlington, MA). From each sample, 10 μg total proteins was denatured with 6 mol/L urea, disulfide bonds were reduced with 10 mmol/L dithiothreitol, and free thiols were then alkylated with 12.5 mmol/L iodoacetamide. The final volume was diluted threefold into 25 mmol/L ammonium bicarbonate, and trypsin digestion was performed with a 1:50 mass ratio of sequencing-grade trypsin (Promega, Madison, WI)/total protein for 16 hours at 37°C. Samples were acidified to approximately pH = 2 with formic acid, and peptides were desalted with C18 Desalting Tips (Rainin, Oakland, CA), lyophilized, and rehydrated in 0.1% formic acid. Peptide sequencing by liquid chromatography–tandem mass spectrometry (MS/MS) was performed on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoACQUITY (Waters, Milford, MA) ultraperformance liquid chromatography system and an EASY-Spray PepMap C18 column (Thermo Fisher Scientific; ES800; 3-m bead size; 75 m × 150 mm) for reversed-phase chromatography. Peptides were eluted over a linear gradient over the course of 60 minutes from 2% to 50% (v/v) acetonitrile in 0.1% formic acid. MS and MS/MS spectra were acquired in a data-dependent mode with up to six higher-energy collisional dissociation MS/MS spectra acquired for the most intense parent ions per MS. For data analysis, MS peak lists were generated with ProteoWizard MSConvert,35 and database searches were performed with Protein Prospector software version 5.16.1 (http://prospector.ucsf.edu/prospector/mshome.htm; University of California, San Francisco, San Francisco, CA)36 against the SwissProt human protein database (https://www.uniprot.org/proteomes/UP000005640, downloaded November 1, 2017). The database was concatenated with an equal number of fully randomized entries for estimation of the false-discovery rate. Database searching was performed with tolerances of 20 ppm for parent ions and 0.8 Da for fragment ions. For database searches, peptide sequences were matched to tryptic peptides with up to two missed cleavages. Carbamidomethylation of cysteine residues was used as a constant modification, and variable modifications included oxidation of methionine, N-terminal pyroglutamate from glutamine, N-terminal acetylation, and loss of N-terminal methionine. The following protein prospector score thresholds were selected to yield a maximum protein false-discovery rate of <1%: a minimum protein score of 15 and a minimum peptide score of 10 were used; maximum expectation values of 0.01 for protein and 0.05 for peptide matches were used. The list of proteins was further manually curated to include only proteins identified in at least two of three biological replicates, and with a minimum of two unique peptides identified in at least one biological replicate. The t-test analysis was performed on the normalized spectral abundance factor values for proteins, calculated as previously described37: normalized spectral abundance factor is the number of spectral counts (the total number of MS/MS spectra) identifying a protein, divided by the protein's length, divided by the sum of spectral counts/length for all proteins in the experiment. Gene Ontology pathway enrichment analysis was performed using the DAVID 6.8 database (https://david.ncifcrf.gov, last accessed January 2018).38 Label-free quantitation was used to compare relative abundance of selected proteins. The MaxQuant version 1.6.7.0 software package39 was used to obtain normalized peak areas for precursor ions from extracted ion chromatograms, using the MaxLFQ algorithm.40 Raw mass spectrometry data files and peak list files have been deposited with accession number MSV000082206 at ProteoSAFE (http://massive.ucsd.edu, last accessed June 2019).
Statistical Analysis and Data Availability
Statistical significance (P values) was determined with paired t-test or one-way analysis of variance for differences between experimental group mean values. Where data required nonparametric statistics, the Wilcoxon signed-rank test or the U-test was used for paired or unpaired experimental groups, respectively. The statistical test used in each experiment is specified in the relevant figure legends. Survival curves were calculated according to the Kaplan-Meier method and compared using a univariate Cox-Mantel log-rank test. Cox regression multivariate analysis was used to determine whether KHSRP expression was an independent predictor of survival (variables included age, sex, size, and stage), using SPSS version 20.0 (IBM, Armonk, NY). Two-tailed P < 0.05 was considered statistically significant. Data are represented as means ± SEM, unless stated otherwise. Analyses were performed using Prism 6 (GraphPad Software, La Jolla, CA) or R Studio version 1.1.463 (http://www.rstudio.com; RStudio, Inc., Boston, MA).
Gene expression microarray data from this study have been deposited in the Gene Expression Omnibus under accession number GSE112329. Raw data from the proteomic analysis have been deposited with accession number MSV000082206 at ProteoSAFE (http://massive.ucsd.edu, last accessed June 2019).
Results
KHSRP Is Overexpressed in Colorectal Cancer
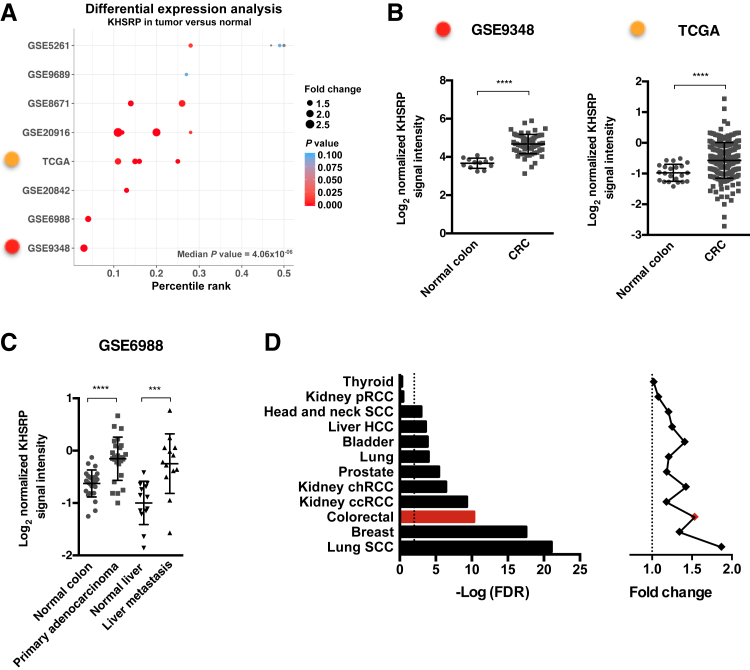
To profile KHSRP expression in CRC, a meta-analysis of CRC microarray data sets was performed from the Oncomine database. KHSRP expression was increased in tumor compared with normal tissue across 18 CRC patient cohorts from eight different data sets, with a median adjusted P = 4.06 × 10−6 (Figure 1A and Supplemental Table S1). KHSRP ranked in the top 5% of overexpressed genes in two of these analyses (from GSE9348 and GSE6988) and in the top 25% for eight other analyses. In the remaining eight analyses, KHSRP was not one of the most overexpressed genes (<25%). Only 4 of the 18 analyses (from GSE5261 and GSE9689) fell below the threshold of significance. Figure 1B shows individual data from the GSE9348 cohort41; in this data set, KHSRP expression was increased by twofold, on average, in 70 patients with early-stage (stage II) colorectal carcinoma compared with healthy controls (P = 4.6 × 10−11). In TCGA cohort42 (Figure 1B), KHSRP expression increased in 211 samples of colorectal adenocarcinoma (any stage) compared with normal tissue (pooled analysis, 1.36-fold change; P < 0.0001). In an additional analysis of the GSE6988 cohort,43 KHSRP expression increased in samples from either primary colon adenocarcinoma (n = 25) or liver metastases (n = 13) compared with matched normal colon mucosa and normal liver tissue, respectively (Figure 1C).
Figure 1.
In silico analysis of publicly available colorectal cancer (CRC) data sets reveals a consistent tumor-specific overexpression of KHSRP. A: Differential expression analysis of KHSRP in the Oncomine database returns 18 analyses (each comprising a different patient group compared with normal tissue) across eight different CRC data sets. The analyses are plotted by percentile ranking of KHSRP overexpression (as a percentage of all overexpressed genes in the data set); dots are sized in proportion to the fold change of KHSRP expression in tumor versus normal, and colored by P value (further details of the analyses are provided in Supplemental Table S1). B: Differential expression of KHSRP between tumor and normal tissue, showing individual data points, is reported for the two representative analyses indicated (with the color code referring to A). Mann-Whitney test was performed. C: Differential expression of KHSRP between paired matched tumor and normal tissue in primary adenocarcinoma and liver metastasis. Individual points are shown. Wilcoxon matched-pairs signed-rank test was performed. D: Differential expression analysis for KHSRP in RNA-sequencing data for the indicated 12 cancer types in The Cancer Genome Atlas (TCGA) data set. For each type, the fold change expression of KHSRP in tumor versus normal tissue and the associated false-discovery rate (FDR) adjusted P value are reported. Dashed lines indicate a fold change of 1. Data are expressed as means ± SD (B and C). ***P < 0.001, ****P < 0.0001. ccRCC, clear cell renal cell carcinoma; chRCC, chromophobe renal cell carcinoma; HCC, hepatocellular carcinoma; pRCC, papillary renal cell carcinoma; SCC, squamous cell carcinoma.
Increased KHSRP expression in tumor tissue was not limited to CRC but remained significant across a meta-analysis of 59 cancer types in the Oncomine database (median adjusted P = 0.004) (Supplemental Table S2). In addition to looking at expression of KHSRP across different cancer types, RNA-sequencing transcriptomic data available in TCGA database, from which differentially expressed genes were derived for 12 different cancer types against their respective normal tissue, were used23: KHSRP was significantly overexpressed to varying degrees in 10 cancer types (Figure 1D), with CRC being among the most highly significant (fold change overexpression = 1.53; false-discovery rate adjusted P = 5.02 × 10−11). The colon has also generally a high tissue-specific cancer expression of KHSRP across the entire spectrum of cancers represented in TCGA (Supplemental Figure S1). However, the frequency of genetic alterations (mutations, deletions, or amplifications) in KHSRP is <2% in the CRC TCGA data set (Supplemental Figure S2), which is low considering that the three most commonly mutated genes in CRC (APC, TP53, and KRAS) have mutation frequencies of 81%, 60%, and 43%, respectively.42 Collectively, these in silico data provided a framework to support additional investigations into the putative role of KHSRP in the pathogenesis of CRC.
KHSRP Is a Marker of Poor Prognosis in Colorectal Cancer
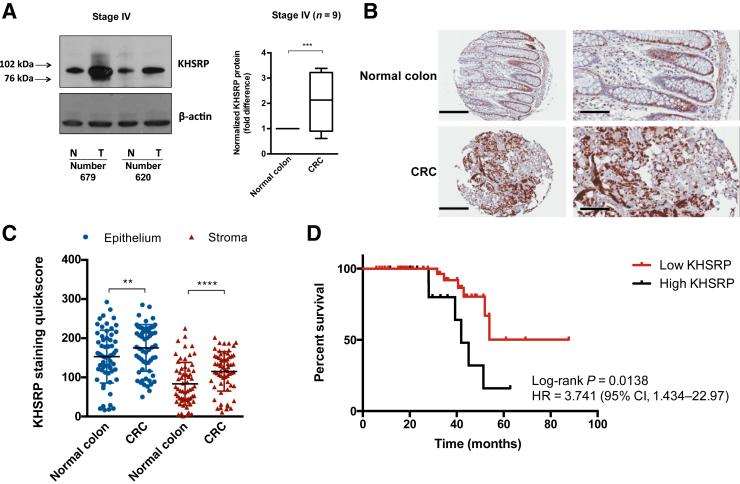
Next, KHSRP protein expression was investigated in primary tissue samples. Protein extracts were obtained from fresh-frozen colorectal tumor and normal adjacent tissue of 16 patients after surgical resection. Western blot analysis revealed a twofold increased expression (P = 0.0009) of KHSRP in stage IV CRC, compared with matched uninvolved (adjacent normal) tissue control (Figure 2A). Analysis of stage II to III patients showed a similar trend (Supplemental Figure S3A). The KHSRP antibody used for Western blot analysis detected a single band at approximately 82 kDa (Supplemental Figure S3B), the predicted size for the protein, confirming the specificity of this antibody, which has been previously orthogonally validated (Human Protein Atlas).
Figure 2.
Analysis of KHSRP expression in primary tissue samples reveals a tumor-specific overexpression with prognostic value. A: Western blot analysis of KHSRP in lysates from fresh-frozen tissue samples of patients with stage IV colorectal cancer (CRC), comparing tumor (T) with matched normal (N) tissue from each patient. A representative blot for two patients is shown along with semiquantitative densitometric analysis of all patients (the box-and-whisker plot depicts interquartile range, median, minimum, and maximum limits). A t-test was performed. B: Representative images from tissue microarray (TMA) sections of normal and CRC tissue stained for KHSRP. C: Quantitation of stromal and epithelial KHSRP staining in normal colon versus primary CRC tissue samples from the TMA. Individual points are shown. A Wilcoxon test was performed. D: T/N ratios in the epithelium were used to segregate patients, using 20% as a cutoff (T/N < 1.2 versus T/N > 1.2). Survival curves were calculated, according to the Kaplan-Meier method, and compared using the Cox-Mantel log-rank test. The hazard ratio (HR) and associated CI are shown. Data are expressed as means ± SD (C). n = 9 (A). **P < 0.01, ***P < 0.001, and ****P < 0.0001. Scale bars: 200 μm (B, left column); 100 μm (B, right column). Original magnification: ×10 (B, left column); ×20 (B, right column).
To expand on these initial results, and to compensate for the limited number of samples analyzed by Western blot analysis, IHC was performed to investigate KHSRP expression in tissue obtained from a cohort of 62 patients with late-stage (III to IV) CRC.25 KHSRP was expressed in both the epithelial and stromal compartments (Figure 2B). KHSRP expression was significantly higher in tumor versus matched normal tissue in both epithelium and stroma (Figure 2,B and C). This was mainly due to differences in percentage of positive cells, whereas the intensity values were similar overall (Supplemental Figure S3C). There were more KHSRP-positive cells in the epithelium than in the stroma (Figure 2C); however the increased expression in tumor compared with normal was more prominent in the stroma, as indicated by a higher tumor/normal ratio of KHSRP staining in the stroma (Supplemental Figure S3D). When tumor/normal ratios are plotted separately for each patient, most had increased KHSRP expression in the tumor compared with matched normal tissue (Supplemental Figure S3, E and F). High KHSRP expression (>20% increase in tumor versus normal tissue) was associated with decreased overall survival (Figure 2D) (hazard ratio, 3.74; 95% CI, 1.43–22.97; P = 0.0138) in a univariate analysis. Multivariate Cox regression analysis demonstrated that higher KHSRP expression in tumor epithelium was an independent predictor of poor survival, associated with a 2.5-fold increase in disease-associated death (P = 0.009). As most of the patients in this cohort were diagnosed with metastatic disease, relapse-free survival was not included in the analysis. There was also a weak, but significant, correlation of the KHSRP tumor/normal ratio with tumor size in both epithelium (Pearson r = 0.43; P = 0.0016) and stroma (Pearson r = 0.38; P = 0.0051).
For a limited number of cases (N = 10 patients), tissue was also available for the corresponding liver metastasis. In both the epithelium and the stroma, the KHSRP score was significantly higher in the metastatic tumor tissue compared with matched normal liver (Supplemental Figure S4A). Overall, the metastasis/normal ratio was positive for all except one patient, but the difference between metastasis and normal was vastly larger in the stroma than in the epithelium, with higher metastasis/normal ratios (Supplemental Figure S4B–D); this can be explained by the low expression of KHSRP in the normal liver stroma. Comparison of matched primary tumor and liver metastasis (n = 10 patients) showed that the expression of KHSRP is unchanged in the epithelial cells on migration; however, the metastatic stroma has a higher expression compared with the stroma in the primary tumor (Supplemental Figure S4, E and F).
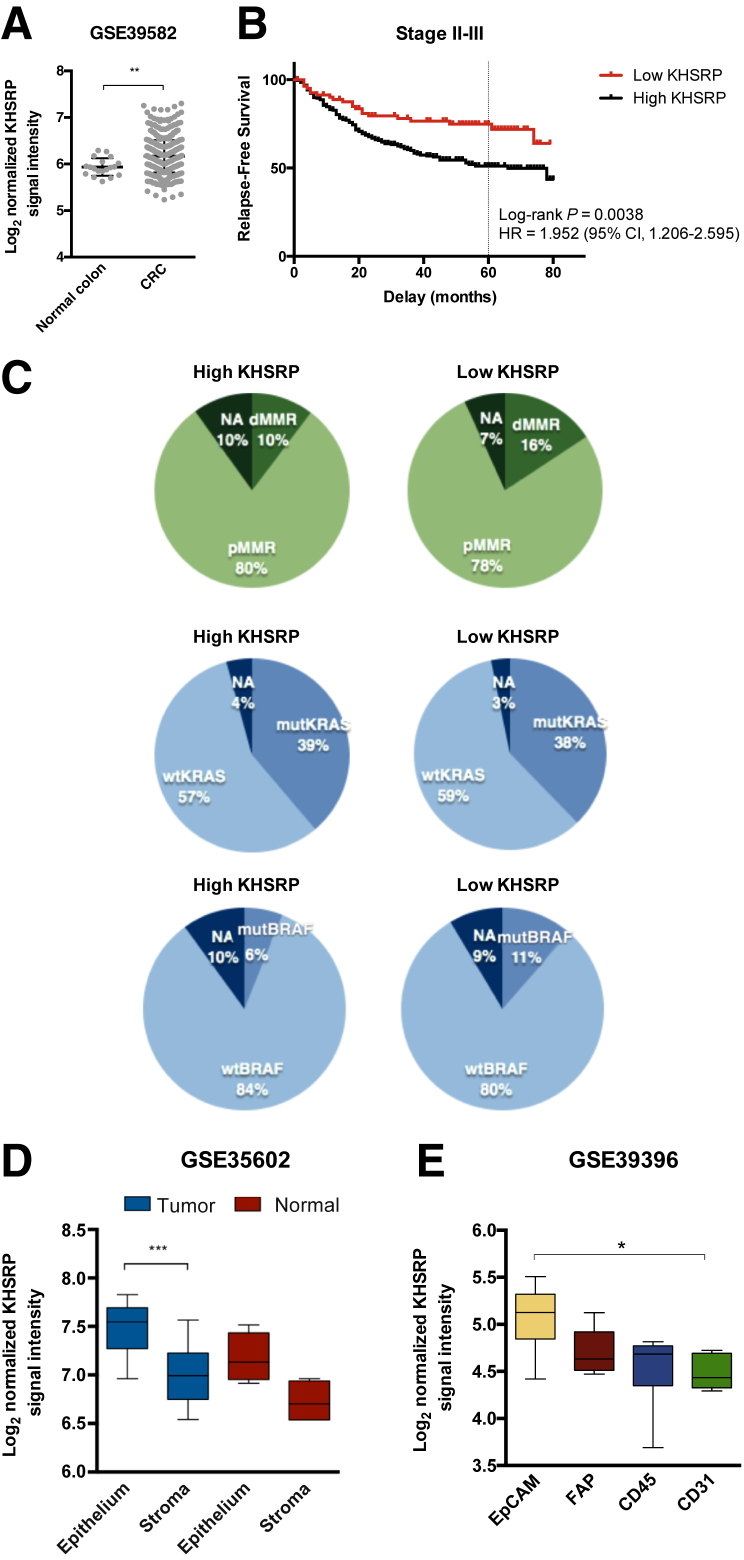
To validate the prognostic role of KHSRP in an independent cohort, a published data set of 566 patients with stage I to IV CRC in the French Tumor ID Cards program was used.44 KHSRP expression was increased in tumor compared with uninvolved (normal) tissue, in accordance with our previous data (Figure 3A). Survival analysis was restricted to stage II to III patients following recommendations from the original authors: high KHSRP expression was associated with a decreased 5-year relapse-free survival in this cohort (Figure 3B) (hazard ratio, 1.95; 95% CI, 1.21–2.60; P = 0.0038). KHSRP expression was not associated with other tumor features, such as loss of mismatch repair proteins or mutations in KRAS or BRAF (Figure 3C).
Figure 3.
In silico analysis of three independent cohorts validates the overexpression and prognostic significance of KHSRP. A: Log2 normalized signal intensity of KHSRP in a cohort of 566 colorectal cancer (CRC) patients and 19 normal controls, profiled by gene expression microarrays. Individual points are shown. A Mann-Whitney test was performed. B: Survival curves for patients segregated into high/low groups using the 25th percentile of the KHSRP probe signal intensity. The hazard ratio (HR) and associated CI for relapse-free survival at 5 years (dashed line) are shown. C: Distribution of patients with high/low KHSRP expression, according to clinical characteristics: mismatch repair deficient (dMMR; ie, microsatellite instable) or proficient (pMMR; ie, microsatellite stable), KRAS (codons 12 and 13) mutational status, and BRAF (p.V600E) mutational status. D: Expression of KHSRP in laser-capture microdissected epithelial and stromal cells from 13 CRC and normal colonic mucosa samples. A Kruskal-Wallis test was performed. E: Expression of KHSRP in a cohort of fluorescence-activated cell sorted cells from 14 CRC patients. CD45 indicates leukocytes, and CD31 indicates endothelial cells. A Kruskal-Wallis test was performed. D and E: The box-and-whisker plots depict interquartile range, median, minimum, and maximum limits. Data are expressed as means ± SD (A). *P < 0.05, **P < 0.01, and ***P < 0.01. EpCAM, epithelial cancer cell; FAP, cancer-associated fibroblast; mut-, mutant; NA, not available; wt-, wild-types.
To independently validate the expression of KHSRP in epithelial and stromal compartments from the IHC data, a transcriptomic data set of 13 CRC and normal colonic mucosa samples, in which epithelial cells and stromal cells had been microdissected by laser capture and profiled separately, was used.45 KHSRP was significantly overexpressed in the microdissected epithelial tumor compared with tumor stromal areas and, to a lesser extent, in the epithelium compared with the stroma of the normal colonic mucosa, in accordance with the IHC data (Figure 3D). Although there was a trend in increased KHSRP expression in tumor versus normal tissue for both the epithelium and the stroma, these comparisons were not statistically significant, probably because of the low number of samples. Finally, transcriptomic data from purified cell populations that were isolated from 14 dissociated human primary CRC samples were used.46 Fluorescence-activated cell sorting was used to enrich for epithelial cancer cells [epithelial cellular adhesion molecule (EpCAM+)], leukocytes (CD45+), endothelial cells (CD31+), and cancer-associated fibroblasts [fibroblast activation protein (FAP+)]. The expression of KHSRP was higher in the EpCAM+ epithelial cell population compared with the stromal cells, with the CD31+ endothelial cells showing the lowest KHSRP expression (Figure 3E).
KHSRP Is Involved in Promoting the Growth and Survival of CRC Cells
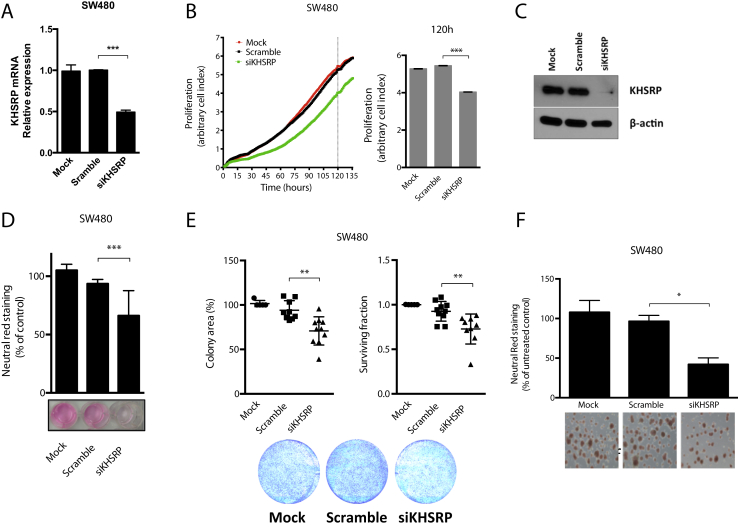
Next, an in vitro model of CRC was used to investigate the mechanistic role of KHSRP in epithelial cancer cells. The SW480 cell line, which was derived from a primary tumor of a moderately differentiated Duke B colon adenocarcinoma, was used.47 These cells constitutively express KHSRP with a diffuse intracellular pattern (Supplemental Figure S5). An initial screen with a pool of four different siRNA sequences targeting KHSRP effectively silenced mRNA expression by 50% (Figure 4A) and stably reduced proliferation when cells were monitored in real time for 6 days (Figure 4B). These initial results were then confirmed and extended using an independent siRNA that effectively knocked down KHSRP protein expression (Figure 4C): silencing of KHSRP reduced cell growth, as measured by neutral red uptake (Figure 4D); reduced clonogenic potential, measured by both number of colonies (surviving fraction) and total colony area (Figure 4E); and reduced spheroid formation in three-dimensional Matrigel cultures (Figure 4F). Similar results were obtained by silencing KHSRP in an independent cell line model, SW620, which was isolated from a lymph node after multiple metastases occurred in the same patient (Supplemental Figure S6). Furthermore, a conditional (doxycycline-dependent) shRNA-based stable knockdown model from both SW480 and SW620 cell lines was also developed, which further confirmed results obtained with the transient silencing model (Supplemental Figure S7).
Figure 4.
KHSRP is involved in growth and survival of an in vitro colorectal cancer (CRC) cell line model. A: Quantitative RT-PCR analysis of SW480 cells transfected with a pool of siRNAs targeting KHSRP (siKHSRP), a scramble control pool (Scramble), or transfection reagent control (Mock) for 48 hours. B: Cells were transfected, as in A, and continuously monitored for proliferation for the indicated time; full time-course growth traces are shown on the left. Dashed line indicates the time point of 5 days, for which quantification of cell index values (arbitrary measure of cell proliferation on the basis of impedance measurements) is reported on the right. C: Western blot analysis of KHSRP protein expression in SW480 cells transfected with an siRNA targeting KHSRP, a scramble control siRNA, or a mock (transfection reagent) control for 48 hours. D: Growth of cells transfected as in C, monitored after 7 days by neutral red assay; representative images from one experiment are shown, with staining quantification from triplicate assays. Growth is expressed as percentage of untreated control (cells with no transfection). E: Clonogenic potential for cells transfected as in C, measured by both colony area and surviving fraction. Representative images from one experiment are shown, with quantification from triplicate assays. F: Spheroids from cells transfected as in C, grown in Matrigel; representative images are shown with staining quantification from triplicate assays. Growth is expressed as percentage of untreated control (cells with no transfection). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (analysis of variance). Original magnification, ×10 (F)
KHSRP Regulates Cell Cycle and Affects the Tumor-Promoting Microenvironment
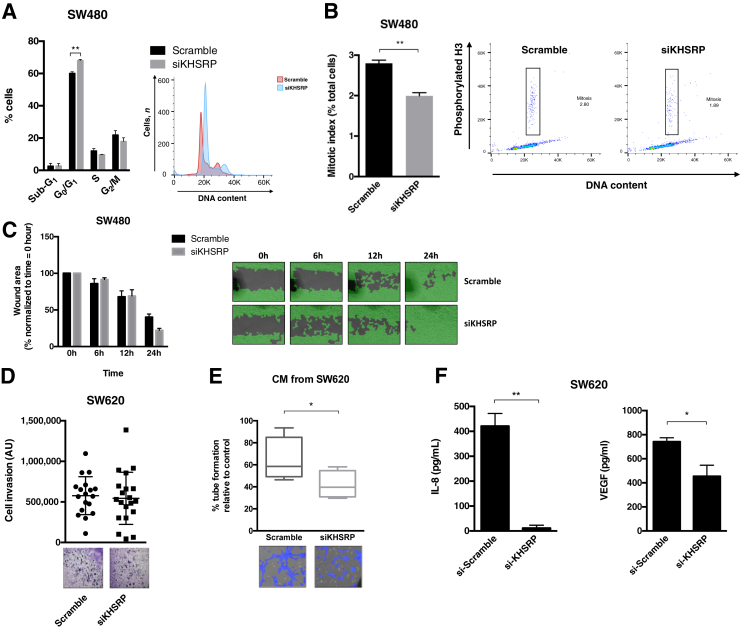
To investigate the mechanisms involved in the tumor-promoting effects of KHSRP, the transcriptomic profiles of siRNA-transfected KHSRP knockdown SW480 cells were analyzed, compared with scramble control transfection (Supplemental Figure S8, A and B). This preliminary analysis identified 135 genes differentially regulated on KHSRP knockdown (Supplemental Table S3), which are enriched for functional categories related to cell growth and survival (specifically, cell cycle and apoptosis), as well as cell signaling and known KHSRP-related functions (translation and protein metabolism), as shown by Gene Ontology term analysis (Supplemental Table S4). Three representative genes (FABP3, MTF1, and SMG1) were further validated by quantitative RT-PCR (Supplemental Figure S8C). The protein interactions inferred from the differentially expressed transcripts (Supplemental Figure S8D)31 included gene networks for cell cycle, transcription, and protein synthesis, further suggesting a role for KHSRP in cell cycle control. The effects of KHSRP knockdown on the cell cycle distribution of SW480 cells were next investigated. KHSRP siRNA caused an increase in the G0/G1 population (Figure 5A), concomitant with a decrease in the mitotic index, measured as the number of cells positively stained by phosphorylated histone H3 (Figure 5B), indicative of a reduced proliferation rate due to a delayed G1/S transition. These data, together with those presented in Figure 4, suggest an involvement of KHSRP in driving epithelial cells into mitosis, thereby increasing proliferation. Furthermore, stable knockdown of KHSRP resulted in reduced expression of c-Myc (Supplemental Figure S7).
Figure 5.
KHSRP is involved in regulating cell cycle and mitosis, as well as influencing the protumorigenic extracellular environment in vitro. A: Cell cycle analysis of SW480 cells transfected with siRNAs targeting KHSRP (siKHSRP) or Scramble negative control siRNA; a representative histogram of propidium iodide staining from one experiment is shown, along with a bar graph of cell distribution into cell cycle phases across three replicate experiments. An analysis of variance was performed. B: Analysis of mitotic cells by quantitation of phosphorylated histone H3 in SW480 cells transfected as in A; a representative histogram is shown with bar graph of three replicate experiments. The boxed areas indicate mitotic cells (positive for phospho histone H3 and with 2X DNA content). A Mann-Whitney test was performed. C: Wound-healing assay of SW480 cells transfected as in A; representative images from one experiment are shown, with quantitation of wound area from two replicate experiments. D: Invasion of SW620 cells transfected with siKHSRP or Scramble negative control siRNA, measured by the capacity to migrate through Matrigel; quantification of invaded cells from three replicate experiments is shown, along with representative images. E: Endothelial tube formation of human dermal microendothelial cells (HDECs) treated with conditioned media (CM) from SW620 cells transfected as in D. Tube formation is expressed as percentage of control (HDECs treated with the same media without previous conditioning in SW620 cells). A t-test was performed. F: Expression of secreted IL-8 and vascular endothelial growth factor (VEGF) in the CM of SW620 cells transfected as in D, measured by enzyme-linked immunosorbent assay. A t-test was performed. *P < 0.05, **P < 0.01. Original magnification, ×10 (C–F). AU, arbitrary unit.
KHSRP has been involved in regulating cell migration and invasion in different noncolorectal tumor models.48 However, silencing of KHSRP did not influence cell migration (as measured by a wound-healing assay) in SW480 cells (Figure 5C) and did not impact the ability of the metastatic SW620 cells to invade through a Matrigel layer (Figure 5D). Nevertheless, the cell culture supernatant from SW620 cells with silenced KHSRP was able to reduce endothelial cell growth in a tube-forming assay (Figure 5E), suggesting that KHSRP may regulate the secretion of specific extracellular signaling mediators promoting angiogenesis. In support of this, CRC cells with silenced KHSRP expression secreted reduced levels of IL-8 and vascular endothelial growth factor (Figure 5F).
KHSRP Regulates the Secretion of Multiple Proteins by Tumor Cells
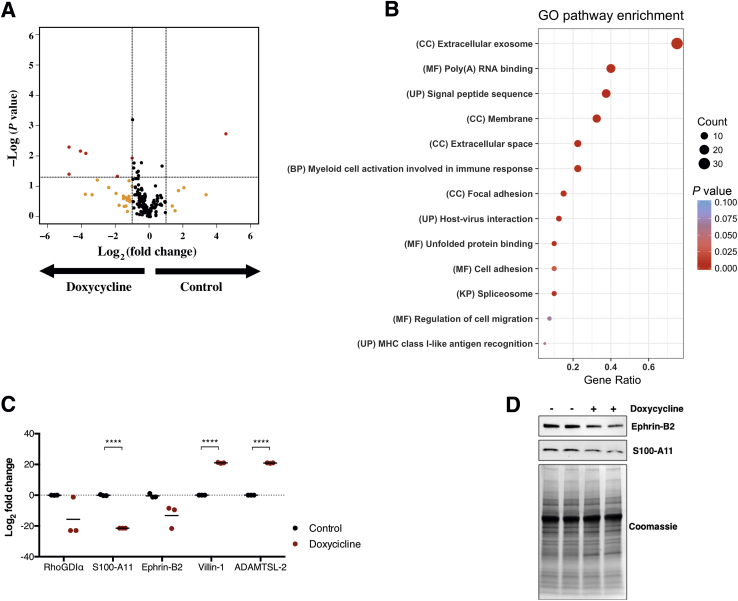
To further investigate the involvement of KHSRP in regulating the extracellular microenvironment, the conditional KHSRP knockdown cell line model (Supplemental Figure S7) was used to probe the secretome of the metastatic SW620 cell line on doxycycline-induced, shRNA-mediated knockdown of KHSRP. Shotgun proteomics analysis identified 191 proteins, of which 27 were unique for the control samples (ie, undetected in the doxycycline-treated samples) and 16 were unique for the doxycycline-treated samples (undetected in the control samples). Statistical analysis of spectral count data showed that 40 proteins were significantly underexpressed or overexpressed by >50% on doxycycline treatment; most of these differentially regulated proteins (33 of 40) were down-regulated (Figure 6A and Supplemental Table S5). Gene Ontology term analysis of the 40 highly differentially regulated proteins revealed an enrichment for known terms related to KHSRP function (eg, RNA binding and spliceosome) and expected terms related to the cellular location of the probed sample (eg, membrane and extracellular space); interestingly, there was also a significant enrichment for terms related to the protumorigenic extracellular microenvironment (eg, exosomes, focal adhesion, and cell migration), as well as terms broadly related to the immune response and specifically to myeloid cell activation (Figure 6B). Using label-free quantitation, specific examples of candidate proteins whose secretion is deregulated on KHSRP knockdown were validated (Figure 6C). Down-regulated proteins included the following: the Ras homolog family (Rho)–specific guanine nucleotide dissociation inhibitor–α, a master regulator of Rho GTPases that promotes cell migration49; the S100 calcium-binding protein A11, an endoplasmic reticulum–associated calcium-binding protein that regulates cell death50; and Ephrin-B2, a membrane-anchored protein that activates paracrine tyrosine kinase signaling, impacting proliferation, migration, and response to therapy.51 Conversely, up-regulated proteins included the following: the actin-binding protein Villin-1, a protein that, on cellular stress, induces intestinal epithelial cell death by necroptosis, as well as increased inflammation52; and a disintegrin and metalloproteinase with thrombospondin motifs–like protein 2, an extracellular matrix–residing protein with roles in matrix remodeling and modulation of metalloprotease activity.53 The KHSRP-dependent down-regulation of S100 calcium-binding protein A11 and Ephrin-B2 was further confirmed by Western blot analysis (Figure 6D).
Figure 6.
KHSRP regulates expression of key proteins that modulate the tumorigenic microenvironment. Differential expression of secreted proteins was analyzed by shotgun proteomics in conditioned media from SW620 cells treated with doxycycline for 4 days to induce shRNA-mediated KHSRP knockdown. A: Volcano plot showing differentially regulated proteins in the doxycycline-treated samples compared with control, colored in red (log2 fold change > |1|; P < 0.05). Proteins that are differentially regulated above the fold change threshold with a nonsignificant P value are colored in yellow. Dashed lines indicate the thresholds on both axes. B: Dot plot of results from a Gene Ontology (GO) analysis of the 40 highly differentially regulated proteins. GO terms are reported with their category and plotted by gene ratio (the number of genes in one GO term compared with the total). Dots are sized in proportion to the number of hits within a GO term and colored by P value. C: Label-free quantitation of relative abundance of the indicated five proteins in the doxycycline-treated versus control samples. D: Western blot analysis of Ephrin B2 and S100 calcium-binding protein A11 (S100-A11) in conditioned media from control or doxycycline-treated SW620 cells. A Coomassie stain is shown for protein loading control. ****P < 0.0001 (t-test). BP, biological process; CC, cellular component; KP, Kegg pathway; MF, molecular function; RhoGDIα, Ras homolog family–specific guanine nucleotide dissociation inhibitor–α; UP, uniprot annotation.
Discussion
The role of KHSRP in the pathogenesis and progression of CRC has not been extensively studied so far. This study provides orthogonal evidence supporting a protumorigenic role of KHSRP in CRC, on the basis of extensive mining of multiple publicly available data sets, analysis of patient samples, and in vitro cell line models. Our results are in agreement with recent reports in osteosarcoma,13 lung cancer,12, 54 papillary thyroid cancer,55 and esophageal squamous cell carcinoma.56
The IHC data provide evidence for KHSRP overexpression in the context of both primary tumor and metastasis. However, it was surprisingly found that KHSRP remained unchanged in the metastatic epithelial cells infiltrating the liver compared with that of the primary site, whereas the stromal expression was strongly up-regulated from a low basal expression in the normal hepatic stroma. This suggests that invading epithelial cells are capable of modifying the local microenvironment or, alternatively, that a local increase in KHSRP expression preexists and might contribute to the metastatic niche.57 In fact, KHSRP is involved in the transient expression of the chemokine C-X3-C motif chemokine ligand 1 in liver epithelial cells in response to interferon-γ stimulation,58 contributing to the control of cellular homeostasis during inflammation and macrophage infiltration. The proteomic data provide additional evidence in support of a putative role for KHSRP in regulating the tumor microenvironment, with implications for tumor progression and metastasis. To the best of our knowledge, this represents the first report of such an effect for KHSRP in shaping the extracellular environment of epithelial cells. Preliminarily, these data also suggest a possible role for KHSRP in regulating the immune response, both at the gene-expression level and at the secretome level (as reported herein by microarray and proteomic data). This suggests the intriguing idea that the reported negative prognostic role of KHSRP could be partly ascribed to a regulation of immune evasion mechanisms and provides a rationale for reconciling the oncogenic roles of KHSRP with its other known functions in shaping innate immunity during viral infection and inflammation. In fact, recent work from Bezzi et al,59 using genetically engineered mouse models of prostate cancer driven by the loss of specific tumor suppressor genes, has shown that the genetic landscape of epithelial tumor cells can directly influence the composition of immune cell infiltrates in primary tumors, through mechanisms based on distinct chemokine pools resulting from aberrant transcriptional and signaling programs. Furthermore, knockout mice studies have shown a requirement of KHSRP for the post-transcriptional control of type 1 and type 3 interferon genes.60, 61 Recent mechanistic evidence proved that KHSRP can directly associate and negatively regulate retinoic acid–inducible gene I receptor signaling during viral infection; this exerts a negative control on the activation of type I interferon innate immune responses during the recognition of pathogen-associated molecular patterns encoded by viral RNA.62 This could have potential consequences for the control of antitumor innate immune responses, as similar mechanisms are involved in the recognition of damage-associated molecular patterns and the immunogenicity of tumor cells.63 Consistently, KHSRP has been implicated in the innate immune response to oncogenic pathogens, such as Helicobacter pylori in the stomach64 and human papillomavirus in the uterus.65
One question that remains open is how overexpression of KHSRP is controlled, given that these data suggest it is not the product of a driver mutation event. A possible explanation could come from studying another RBP that is commonly overexpressed in CRC, MSI-1.66 Spears and Neufeld67 have demonstrated a reciprocal interaction between MSI-1 and APC through a double-negative feedback loop, whereby MSI-1 contributes to APC inactivation, an early initiating driver event in most sporadic CRC. This suggests the intriguing hypothesis that increased expression of RBPs (perhaps not limited to MSI-1) might coevolve as a passenger nonmutational event early in the adenoma-to-carcinoma sequence. Another hypothesis, related to the effect of KHSRP on the tumor microenvironment that these data begin to suggest, is that deregulation of RBPs could be selected for indirectly through negative regulation of immune surveillance, a relevant force shaping cancer evolution. In support of an early event hypothesis, the in silico data suggest that KHSRP expression is unchanged in hypermutated, microsatellite-instable CRC, a type of tumor characterized by a proportionally increased burden of passenger mutations.68 Nevertheless, specific mutations in KHSRP could still impact protein functions and show increased frequencies in some cancer types.48 Furthermore, although limited to a small fraction of the data set, the analysis from the Oncomine database provides information on colorectal mucinous carcinoma, a distinct form of CRC found in 10% to 15% of patients and characterized by distinct molecular features and mutation events69 in this subset of patients; KHSRP was not one of the most overexpressed genes, suggesting that it might be an event specifically related to adenocarcinoma.
There is also considerable interest in the suitability of different members of the translational control machinery as potential therapeutic targets in cancer, although the evidence so far is at the preclinical stage.70 Small-molecule inhibition of the RBP HuR in APCMin mice led to a reduction in small intestinal tumor formation and a concomitant reduction in c-Myc expression.71 Another recent report showed that inhibition of mammalian target of rapamycin complex 1 using rapamycin caused regression of established APC-deficient intestinal tumors, suggesting that inhibition of translation elongation using existing clinically approved drugs might benefit patients at high risk of developing CRC.72 The loss-of-function data in in vitro CRC models suggest that KHSRP could also have putative therapeutic implications and warrant further investigation into the suitability of such an approach.
In conclusion, this report sheds light onto the molecular role of KHSRP in CRC, providing for the first time comprehensive data in support of a protumorigenic role of this RBP through direct modulation of epithelial cell phenotype and indirect modulation of the tumor microenvironment.
Acknowledgments
We thank Drs. Giselle Knudsen [University of California, San Francisco (UCSF)], Sudipto Das (Royal College of Surgeons in Ireland), Janet McCormack [University College Dublin (UCD)], Barry Moran [Trinity College Dublin (TCD), Dublin, Ireland], and Dimitri Scholz (UCD) for technical assistance at various stages of this project; Dr. Martin Kampmann (UCSF) for providing advice and material for the generation of the conditional knockdown cell lines; Dr. Quing Xiong (Southwest University, Chongqing, China) for providing access to data; Dr. Tim Hacker and Sam Ivry (UCSF) for bioinformatics expertise; and Prof. Ursula Fearon (TCD) for providing human dermal microendothelial cells. Mass spectrometry was performed in collaboration with the UCSF Mass Spectrometry Facility (directed by Prof. Alma Burlingame).
F.C., E.J.R., J.O.S., and G.A.D. conceived and designed the study; F.C., K.O., M.T., L.B., F.O., R.P., S.N., and P.M. acquired data; F.C., K.O., J.J.P., K.K., S.F., K.S., J.O., and E.J.R. analyzed data; F.C., K.O., J.O.S., E.J.R., and C.S.C. wrote and revised the manuscript; E.J.R., K.S., G.A.D., D.F., and C.S.C. procured funding and support; M.T. and B.N. provided administrative, technical, or material support; E.J.R. and G.A.D. supervised the study.
Footnotes
Supported by the University College Dublin Foundation Newman Fellowship, funded by a donation from Merck Serono (F.C.); Science Foundation Ireland Industry Fellowship 14/IFB/2715 (F.C.); NIH grant P41CA196276 (C.S.C.); and Irish Health Research Board grant HRA _POR_2013_281 (E.J.R.).
Disclosures: None declared.
Current address of E.J.R., Department of Biological Sciences, School of Engineering and Science, University of Limerick, Limerick, Ireland; of R.P., Personalis Inc., Menlo Park, CA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.07.004.
Supplemental Data
RNA-sequencing expression data for KHSRP in The Cancer Genome Atlas (TCGA) cohort is plotted in increasing order, grouped by tumor type, with CRC highlighted in red (for details of the tumor type codes, refer to TCGA Data Portal; https://tcga-data.nci.nih.gov/docs/publications/tcga, last accessed November 13, 2018). ACC, adenoid cystic carcinoma; Adeno, adenocarcinoma; AML, acute myeloid leukemia; ccRCC, clear cell renal cell carcinoma; chRCC, chromophobe renal cell carcinoma; CS, carcinosarcoma; DLBC, diffuse large B-cell lymphoma; GBM, glioblastoma; HCC, hepatocellular carcinoma; pRCC, papillary renal cell carcinoma; SCC, squamous cell carcinoma.
The frequency of genetic alterations in KHSRP across four different colorectal cancer data sets, as reported in the cBio portal: Dana-Farber Cancer Institute (DFCI) 2016 (dbGap; https://www.ncbi.nlm.nih.gov/gap; accession number phs000722), Genentech (European Genone-phenome Archive; https://www.ebi.ac.uk/ega/home; accession number EGAS00001000288), Memorial Sloan Kettering Cancer Center (MSKCC; dbGaP accession number phs000790.v1.p1), and The Cancer Genome Atlas (TCGA; original publication42 and provisional data accessed November 2016; https://www.cancer.gov/tcga). CNA, copy number alteration.
A: Western blot analysis of KHSRP in lysates from fresh-frozen tissue samples of patients with stage II to III colorectal cancer (CRC), comparing tumor (T) with matched normal (N) tissue from each patient. A representative blot for two patients is shown along with semiquantitative densitometric analysis of all patients. P = 0.077 (t-test). B: Full-size image of the blot for KHSRP corresponding to A. C: Separate quantification of KHSRP staining intensity and percentage between normal and tumor tissue. Individual points are shown. D: Quantification of T/N ratio of KHSRP staining in epithelial and stromal compartments of a tissue microarray. Individual points are shown, and a dashed line indicates a ratio of 1. E and F: Waterfall plots of T/N ratios in the epithelium (E) or stroma (F) indicate patients with increased or decreased tumor-specific KHSRP expression. Data are expressed as means ± SD (C and D). N = 7 (A). *P < 0.05, ****P < 0.0001 (Wilcoxon).
A: Quantitation of stromal and epithelial KHSRP staining in normal liver versus liver metastasis (Met) tissue from colorectal cancer (CRC) patients from the tissue microarray. Individual points are shown. B and C: Quantification of metastasis/normal (M/N) ratio of KHSRP staining in epithelial and stromal compartments of TMA. D: Waterfall plots of M/N ratios for single patients are shown, along with a comprehensive plot of all patients. Individual points are shown, and a dashed line indicates a ratio of 1. E: Quantitation of stromal and epithelial KHSRP staining in primary CRC tumor versus liver metastasis tissue for 10 patients from the TMA. Individual points are shown. F: Comparison of tumor/normal (T/N) ratios in the primary tumor with the M/N ratios in the corresponding liver metastasis for both epithelial and stromal compartments of 10 patients in the TMA. Individual points are shown, and a dashed line indicates a ratio of 1. Data are expressed as means ± SD (A and D–F). n = 8 (D). *P < 0.05, **P < 0.01 (Wilcoxon).
Immunofluorescence images of SW480 (A) and SW620 (B) cells. Red: nuclei (DAPI); green: KHSRP; blue: F-actin (phalloidin). Original magnification, ×10.
A: Quantitative RT-PCR analysis of SW620 cells transfected with a pool of siRNAs targeting KHSRP (siKHSRP), a scramble control pool (Scramble), or transfection reagent control (Mock) for 48 hours. B: Cells were transfected as in A and continuously monitored for proliferation for the indicated time; full time-course growth traces are shown on the left. A dashed line indicates the time point of 5 days, for which quantification of cell index values (arbitrary measure of cell proliferation on the basis of impedance measurements) is reported on the right. C: Western blot analysis of KHSRP protein expression in SW620 cells transfected with an siRNA targeting KHSRP, a scramble control siRNA, or a mock (transfection reagent) control for 48 hours. D: Growth of cells transfected as in C, monitored after 7 days by neutral red assay; representative images from one experiment are shown, with staining quantification from triplicate assays. E: Clonogenic potential for cells transfected as in C, measured by both colony area and surviving fraction. Representative images from one experiment are shown, with quantification from triplicate assays. F: Spheroids from cells transfected as in C, grown in Matrigel; representative images are shown with staining quantification from triplicate assays. **P < 0.01, ****P < 0.0001 (analysis of variance) for all tests in the figure. Original magnification, ×10 (F).
SW620 and SW480 cells stably transfected with a conditionally expressible shRNA pool targeting KHSRP (M2) or a nontargeting control pool (Scramble). A: Phase-contrast and fluorescence images of SW620 cells treated with doxycycline or vehicle control for 4 days. Consistent expression of red fluorescent protein (RFP) indicates induced expression of the shRNA-containing cassette. B: Western blot (WB) analysis confirms effective knockdown of KHSRP protein expression. C: WB analysis of the same cells reveals reduction of c-Myc protein expression on knockdown of KHSRP. D: Growth of stably transfected SW620 cells in the presence and absence of doxycycline was quantified using the neutral red assay; colony formation was measured by both surviving fraction and colony area. E: WB analysis confirms effective knockdown of KHSRP protein expression in SW480 generated and treated as above. F: Growth of stably transfected SW480 cells in the presence and absence of doxycycline was quantified using the neutral red assay; colony formation was measured by both surviving fraction and colony area. ****P < 0.0001 (analysis of variance). Original magnification, ×10 (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Neg, negative.
Transcriptomic profile of SW480 cells transfected with KHSRP siRNA compared with scramble negative siRNA control. A: Volcano plot showing differentially regulated genes in green (down-regulated) and red (up-regulated) (log2 fold change > |1|; P < 0.05). Genes that are differentially regulated above the fold change threshold with a nonsignificant P value are orange. B: The distribution of the 135 significantly differentially regulated genes is depicted in the pie chart, using the same color scheme as in A. C: Quantitative RT-PCR analysis of the indicated genes in SW480 cells stably transfected with a conditionally expressible shRNA pool targeting KHSRP or a nontargeting control pool (Scramble), stimulated with doxycycline for 4 days to induce shRNA expression. D: Network map of predicted associations for the protein products of the 135 differentially regulated genes. Proteins are represented by nodes (smaller nodes indicate proteins with unknown structural information); edges represent the predicted functional associations, with the thickness of the line indicating the degree of confidence for the prediction of the interaction. Proteins are color coded arbitrarily by clustering, with the three main clusters annotated. **P < 0.01, ***P < 0.001 (t-test).
References
- 1.Truitt M.L., Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gherzi R., Lee K.-Y., Briata P., Wegmüller D., Moroni C., Karin M., Chen C.-Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Chou C.F., Mulky A., Maitra S., Lin W.J., Gherzi R., Kappes J., Chen C.Y. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol Cell Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhamija S., Kuehne N., Winzen R., Doerrie A., Dittrich-Breiholz O., Thakur B.K., Kracht M., Holtmann H. Interleukin-1 activates synthesis of interleukin-6 by interfering with a KH-type splicing regulatory protein (KSRP)-dependent translational silencing mechanism. J Biol Chem. 2011;286:33279–33288. doi: 10.1074/jbc.M111.264754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L., Zaysteva O., Nie Z., Mitchell N.C., Amanda Lee J.E., Ware T., Parsons L., Luwor R., Poortinga G., Hannan R.D., Levens D.L., Quinn L.M. Defining the essential function of FBP/KSRP proteins: Drosophila Psi interacts with the mediator complex to modulate MYC transcription and tissue growth. Nucleic Acids Res. 2016;44:7646–7658. doi: 10.1093/nar/gkw461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winzen R., Thakur B.K., Dittrich-Breiholz O., Shah M., Redich N., Dhamija S., Kracht M., Holtmann H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gherzi R., Chen C.-Y., Ramos A., Briata P. KSRP controls pleiotropic cellular functions. Semin Cell Dev Biol. 2014;34:2–8. doi: 10.1016/j.semcdb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Briata P., Chen C.-Y., Ramos A., Gherzi R. Functional and molecular insights into KSRP function in mRNA decay. Biochim Biophys Acta. 2013;1829:689–694. doi: 10.1016/j.bbagrm.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Suswam E.A., Nabors L.B., Huang Y., Yang X., King P.H. IL-1beta induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3' untranslated region: involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int J Cancer. 2005;113:911–919. doi: 10.1002/ijc.20675. [DOI] [PubMed] [Google Scholar]
- 12.Tong L., Luo Y., Wei T., Guo L., Wang H., Zhu W., Zhang J. KH-type splicing regulatory protein (KHSRP) contributes to tumorigenesis by promoting miR-26a maturation in small cell lung cancer. Mol Cell Biochem. 2016;422:61–74. doi: 10.1007/s11010-016-2806-y. [DOI] [PubMed] [Google Scholar]
- 13.Pruksakorn D., Teeyakasem P., Klangjorhor J., Chaiyawat P., Settakorn J., Diskul-Na-Ayudthaya P., Chokchaichamnankit D., Pothacharoen P., Srisomsap C. Overexpression of KH-type splicing regulatory protein regulates proliferation, migration, and implantation ability of osteosarcoma. Int J Oncol. 2016;49:903–912. doi: 10.3892/ijo.2016.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santarosa M., Del Col L., Viel A., Bivi N., D'Ambrosio C., Scaloni A., Tell G., Maestro R. BRCA1 modulates the expression of hnRNPA2B1 and KHSRP. Cell Cycle. 2010;9:4666–4673. doi: 10.4161/cc.9.23.14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovarelli M., Bucci G., Pasero M., Gherzi R., Briata P. KSRP silencing favors neural differentiation of P19 teratocarcinoma cells. Biochim Biophys Acta. 2013;1829:469–479. doi: 10.1016/j.bbagrm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Walerych D., Lisek K., Sommaggio R., Piazza S., Ciani Y., Dalla E., Rajkowska K., Gaweda-Walerych K., Ingallina E., Tonelli C., Morelli M.J., Amato A., Eterno V., Zambelli A., Rosato A., Amati B., Wiśniewski J.R., Del Sal G. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol. 2016;18:897–909. doi: 10.1038/ncb3380. [DOI] [PubMed] [Google Scholar]
- 17.Puppo M., Bucci G., Rossi M., Giovarelli M., Bordo D., Moshiri A., Gorlero F., Gherzi R., Briata P. miRNA-mediated KHSRP silencing rewires distinct post-transcriptional programs during TGF-β-induced epithelial-to-mesenchymal transition. Cell Rep. 2016;16:967–978. doi: 10.1016/j.celrep.2016.06.055. [DOI] [PubMed] [Google Scholar]
- 18.Fagoonee S., Picco G., Orso F., Arrigoni A., Longo D.L., Forni M., Scarfò I., Cassenti A., Piva R., Cassoni P., Silengo L., Tolosano E., Aime S., Taverna D., Pandolfi P.P., Brancaccio M., Medico E., Altruda F. The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget. 2017;8:10007–10024. doi: 10.18632/oncotarget.14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanc V., Henderson J.O., Newberry R.D., Xie Y., Cho S.-J., Newberry E.P., Kennedy S., Rubin D.C., Wang H.L., Luo J., Davidson N.O. Deletion of the AU-rich RNA binding protein Apobec-1 reduces intestinal tumor burden in Apc(min) mice. Cancer Res. 2007;67:8565–8573. doi: 10.1158/0008-5472.CAN-07-1593. [DOI] [PubMed] [Google Scholar]
- 20.Sureban S.M., May R., George R.J., Dieckgraefe B.K., McLeod H.L., Ramalingam S., Bishnupuri K.S., Natarajan G., Anant S., Houchen C.W. Knockdown of RNA binding protein Musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458.e2. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 21.Wang S., Li N., Yousefi M., Nakauka-Ddamba A., Li F., Parada K., Rao S., Minuesa G., Katz Y., Gregory B.D., Kharas M.G., Yu Z., Lengner C.J. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat Commun. 2015;6:6517. doi: 10.1038/ncomms7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preitner N., Quan J., Nowakowski D.W., Hancock M.L., Shi J., Tcherkezian J., Young-Pearse T.L., Flanagan J.G. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158:368–382. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng L., Bian X.-W., Li D.K., Xu C., Wang G.M., Xia Q.Y., Xiong Q. Large-scale RNA-seq transcriptome analysis of 4043 cancers and 548 normal tissue controls across 12 TCGA cancer types. Sci Rep. 2015;5:13413. doi: 10.1038/srep13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noonan S.A., Morrissey M.E., Martin P., Biniecka M., Ó'Meachair S., Maguire A., Tosetto M., Nolan B., Hyland J., Sheahan K., O'Donoghue D., Mulcahy H., David F., O'Sullivan J. Tumour vasculature immaturity, oxidative damage and systemic inflammation stratify survival of colorectal cancer patients on bevacizumab treatment. Oncotarget. 2018;9:10536–10548. doi: 10.18632/oncotarget.24276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Detre S., Saclani Jotti G., Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassik M.C., Kampmann M., Lebbink R.J., Wang S., Hein M.Y., Poser I., Weibezahn J., Horlbeck M.A., Chen S., Mann M., Hyman A.A., Leproust E.M., McManus M.T., Weissman J.S. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kampmann M., Bassik M.C., Weissman J.S. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat Protoc. 2014;9:1825–1847. doi: 10.1038/nprot.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., Schmitt E.M., Bernardi R.J., Fu X., Bland C.S., Cooper T.A., Schiff R., Rosen J.M., Westbrook T.F., Elledge S.J. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stöckel D., Kehl T., Trampert P., Schneider L., Backes C., Ludwig N., Gerasch A., Kaufmann M., Gessler M., Graf N., Meese E., Keller A., Lenhof H.-P. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics (Oxford, England) 2016;32:1502–1508. doi: 10.1093/bioinformatics/btv770. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sylvain Brohée J.V.H. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinform. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Repetto G., del Peso A., Zurita J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat Protoc. 2008;3:1125–1131. doi: 10.1038/nprot.2008.75. [DOI] [PubMed] [Google Scholar]
- 34.Lee G.Y., Kenny P.A., Lee E.H., Bissell M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers M.C., Maclean B., Burke R., Amodei D., Ruderman D.L., Neumann S. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalkley R.J., Baker P.R., Medzihradszky K.F., Lynn A.J., Burlingame A.L. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol Cell Proteomics. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 38.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 40.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong Y., Downey T., Eu K.W., Koh P.K., Cheah P.Y. A “metastasis-prone” signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ki D.H., Jeung H.-C., Park C.H., Kang S.H., Lee G.Y., Lee W.S., Kim N.K., Chung H.C., Rha S.Y. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 44.Marisa L., de Reyniès A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.-C., Schiappa R., Guenot D., Ayadi M., Kirzin S., Chazal M., Fléjou J.-F., Benchimol D., Berger A., Lagarde A., Pencreach E., Piard F., Elias D., Parc Y., Olschwang S., Milano G., Laurent-Puig P., Boige V. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishida N., Nagahara M., Sato T., Mimori K., Sudo T., Tanaka F., Shibata K., Ishii H., Sugihara K., Doki Y., Mori M. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res. 2012;18:3054–3070. doi: 10.1158/1078-0432.CCR-11-1078. [DOI] [PubMed] [Google Scholar]
- 46.Calon A., Espinet E., Palomo-Ponce S., Tauriello D.V.F., Iglesias M., Céspedes M.V., Sevillano M., Nadal C., Jung P., Zhang X.H.F., Byrom D., Riera A., Rossell D., Mangues R., Massagué J., Sancho E., Batlle E. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leibovitz A., Stinson J.C., McCombs W.B., McCoy C.E., Mazur K.C., Mabry N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 48.Briata P., Bordo D., Puppo M., Gorlero F., Rossi M., Perrone-Bizzozero N., Gherzi R. Diverse roles of the nucleic acid-binding protein KHSRP in cell differentiation and disease. Wiley Interdiscip Rev RNA. 2016;7:227. doi: 10.1002/wrna.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao L., Wang H., Li J., Liu Y., Ding Y. Overexpression of Rho GDP-dissociation inhibitor alpha is associated with tumor progression and poor prognosis of colorectal cancer. J Proteome Res. 2008;7:3994–4003. doi: 10.1021/pr800271b. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y., Kuramitsu Y., Baron B., Kitagawa T., Akada J., Tokuda K., Cui D., Nakamura K. PERK/CHOP contributes to the CGK733-induced vesicular calcium sequestration which is accompanied by non-apoptotic cell death. Oncotarget. 2015;6:25252–25265. doi: 10.18632/oncotarget.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasquale E.B. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy S., Esmaeilniakooshkghazi A., Patnaik S., Wang Y., George S.P., Ahrorov A., Hou J.K., Herron A.J., Sesaki H., Khurana S. Villin-1 and gelsolin regulate changes in actin dynamics that affect cell survival signaling pathways and intestinal inflammation. Gastroenterology. 2018;154:1405–1420.e2. doi: 10.1053/j.gastro.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mead T.J., Apte S.S. ADAMTS proteins in human disorders. Matrix Biol. 2018;71-72:225–239. doi: 10.1016/j.matbio.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bikkavilli R.K., Zerayesus S.A., Van Scoyk M., Wilson L., Wu P.-Y., Baskaran A., Tang K., Raheem S., Samuelson B.A., Reddy N.M., Reddy S.P., Cool C.D., Kosmider B., Avasarala S., Winn R.A. KSRP promotes post-transcriptional destabilization of Spry4 transcripts in non-small cell lung cancer. J Biol Chem. 2017;292:7423–7434. doi: 10.1074/jbc.M116.757906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gou Q., Gao L., Nie X., Pu W., Zhu J., Wang Y., Liu X., Tan S., Zhou J.K., Gong Y., He J., Wu K., Xie Y., Zhao W., Dai L., Liu L., Xiang R., Wei Y.Q., Zhang L., Peng Y. Long noncoding RNA AB074169 inhibits cell proliferation via modulation of KHSRP-mediated CDKN1a expression in papillary thyroid carcinoma. Cancer Res. 2018;78:4163–4174. doi: 10.1158/0008-5472.CAN-17-3766. [DOI] [PubMed] [Google Scholar]
- 56.Fujita Y., Masuda K., Hamada J., Shoda K., Naruto T., Hamada S., Miyakami Y., Kohmoto T., Watanabe M., Takahashi R., Tange S., Saito M., Kudo Y., Fujiwara H., Ichikawa D., Tangoku A., Otsuji E., Imoto I. KH-type splicing regulatory protein is involved in esophageal squamous cell carcinoma progression. Oncotarget. 2017;8:101130–101145. doi: 10.18632/oncotarget.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Descot A., Oskarsson T. The molecular composition of the metastatic niche. Exp Cell Res. 2013;319:1679–1686. doi: 10.1016/j.yexcr.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 58.Xia Z., Lu Y., Li X., Mao T., Chen X.-M., Zhou R. Upregulation of KSRP by miR-27b provides IFN-γ-induced post-transcriptional regulation of CX3CL1 in liver epithelial cells. Sci Rep. 2015;5:17590. doi: 10.1038/srep17590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bezzi M., Seitzer N., Ishikawa T., Reschke M., Chen M., Wang G., Mitchell C., Ng C., Katon J., Lunardi A., Signoretti S., Clohessy J.G., Zhang J., Pandolfi P.P. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med. 2018;24:165–175. doi: 10.1038/nm.4463. [DOI] [PubMed] [Google Scholar]
- 60.Lin W.-J., Zheng X., Lin C.-C., Tsao J., Zhu X., Cody J.J., Coleman J.M., Gherzi R., Luo M., Townes T.M., Parker J.N., Chen C.-Y. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol. 2011;31:3196–3207. doi: 10.1128/MCB.05073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidtke L., Schrick K., Saurin S., Käfer R., Gather F., Weinmann-Menke J., Kleinert H., Pautz A. The KH-type splicing regulatory protein (KSRP) regulates type III interferon expression post-transcriptionally. Biochem J. 2019;476:333–352. doi: 10.1042/BCJ20180522. [DOI] [PubMed] [Google Scholar]
- 62.Soonthornvacharin S., Rodriguez-Frandsen A., Zhou Y., Galvez F., Huffmaster N.J., Tripathi S., Balasubramaniam V.R.M.T., Inoue A., de Castro E., Moulton H., Stein D.A., Sánchez-Aparicio M.T., De Jesus P.D., Nguyen Q., König R., Krogan N.J., García-Sastre A., Yoh S.M., Chanda S.K. Systems-based analysis of RIG-I-dependent signalling identifies KHSRP as an inhibitor of RIG-I receptor activation. Nat Microbiol. 2017;2:17022. doi: 10.1038/nmicrobiol.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green D.R., Ferguson T., Zitvogel L., Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9:353–363. doi: 10.1038/nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li N., Cao M., Yi S., Cheng J., Wang L., Tao Y., Wu D., Peng J., Zhang M., Qi P., Zhao J. Effects of the RNA-binding protein, KSRP, on innate immune response against Helicobacter pylori infection in mice. Biochem Biophys Res Commun. 2018;495:1573–1579. doi: 10.1016/j.bbrc.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 65.Xu J., Liu H., Yang Y., Wang X., Liu P., Li Y., Meyers C., Banerjee N.S., Wang H.K., Cam M., Lu W., Chow L.T., Xie X., Zhu J., Zheng Z.M. Genome-wide profiling of cervical RNA-binding proteins identifies human papillomavirus regulation of RNASEH2A expression by viral E7 and E2F1. MBio. 2019;10:e02687-18. doi: 10.1128/mBio.02687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kudinov A.E., Karanicolas J., Golemis E.A., Boumber Y. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res. 2017;23:2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spears E., Neufeld K.L. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J Biol Chem. 2011;286:4946–4950. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martincorena I., Raine K.M., Gerstung M., Dawson K.J., Haase K., Van Loo P., Davies H., Stratton M.R., Campbell P.J. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hugen N., Brown G., Glynne-Jones R., de Wilt J.H., Nagtegaal I.D. Advances in the care of patients with mucinous colorectal cancer. Nat Rev Clin Oncol. 2016;13:361–369. doi: 10.1038/nrclinonc.2015.140. [DOI] [PubMed] [Google Scholar]
- 70.Silvera D., Formenti S.C., Schneider R.J. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 71.Lang M., Berry D., Passecker K., Mesteri I., Bhuju S., Ebner F., Sedlyarov V., Evstatiev R., Dammann K., Loy A., Kuzyk O., Kovarik P., Khare V., Beibel M., Roma G., Meisner-Kober N., Gasche C. HuR small-molecule inhibitor elicits differential effects in adenomatosis polyposis and colorectal carcinogenesis. Cancer Res. 2017;77:2424–2438. doi: 10.1158/0008-5472.CAN-15-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Faller W.J., Jackson T.J., Knight J.R.P., Ridgway R.A., Jamieson T., Karim S.A., Jones C., Radulescu S., Huels D.J., Myant K.B., Dudek K.M., Casey H.A., Scopelliti A., Cordero J.B., Vidal M., Pende M., Ryazanov A.G., Sonenberg N., Meyuhas O., Hall M.N., Bushell M., Willis A.E., Sansom O.J. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-sequencing expression data for KHSRP in The Cancer Genome Atlas (TCGA) cohort is plotted in increasing order, grouped by tumor type, with CRC highlighted in red (for details of the tumor type codes, refer to TCGA Data Portal; https://tcga-data.nci.nih.gov/docs/publications/tcga, last accessed November 13, 2018). ACC, adenoid cystic carcinoma; Adeno, adenocarcinoma; AML, acute myeloid leukemia; ccRCC, clear cell renal cell carcinoma; chRCC, chromophobe renal cell carcinoma; CS, carcinosarcoma; DLBC, diffuse large B-cell lymphoma; GBM, glioblastoma; HCC, hepatocellular carcinoma; pRCC, papillary renal cell carcinoma; SCC, squamous cell carcinoma.
The frequency of genetic alterations in KHSRP across four different colorectal cancer data sets, as reported in the cBio portal: Dana-Farber Cancer Institute (DFCI) 2016 (dbGap; https://www.ncbi.nlm.nih.gov/gap; accession number phs000722), Genentech (European Genone-phenome Archive; https://www.ebi.ac.uk/ega/home; accession number EGAS00001000288), Memorial Sloan Kettering Cancer Center (MSKCC; dbGaP accession number phs000790.v1.p1), and The Cancer Genome Atlas (TCGA; original publication42 and provisional data accessed November 2016; https://www.cancer.gov/tcga). CNA, copy number alteration.
A: Western blot analysis of KHSRP in lysates from fresh-frozen tissue samples of patients with stage II to III colorectal cancer (CRC), comparing tumor (T) with matched normal (N) tissue from each patient. A representative blot for two patients is shown along with semiquantitative densitometric analysis of all patients. P = 0.077 (t-test). B: Full-size image of the blot for KHSRP corresponding to A. C: Separate quantification of KHSRP staining intensity and percentage between normal and tumor tissue. Individual points are shown. D: Quantification of T/N ratio of KHSRP staining in epithelial and stromal compartments of a tissue microarray. Individual points are shown, and a dashed line indicates a ratio of 1. E and F: Waterfall plots of T/N ratios in the epithelium (E) or stroma (F) indicate patients with increased or decreased tumor-specific KHSRP expression. Data are expressed as means ± SD (C and D). N = 7 (A). *P < 0.05, ****P < 0.0001 (Wilcoxon).
A: Quantitation of stromal and epithelial KHSRP staining in normal liver versus liver metastasis (Met) tissue from colorectal cancer (CRC) patients from the tissue microarray. Individual points are shown. B and C: Quantification of metastasis/normal (M/N) ratio of KHSRP staining in epithelial and stromal compartments of TMA. D: Waterfall plots of M/N ratios for single patients are shown, along with a comprehensive plot of all patients. Individual points are shown, and a dashed line indicates a ratio of 1. E: Quantitation of stromal and epithelial KHSRP staining in primary CRC tumor versus liver metastasis tissue for 10 patients from the TMA. Individual points are shown. F: Comparison of tumor/normal (T/N) ratios in the primary tumor with the M/N ratios in the corresponding liver metastasis for both epithelial and stromal compartments of 10 patients in the TMA. Individual points are shown, and a dashed line indicates a ratio of 1. Data are expressed as means ± SD (A and D–F). n = 8 (D). *P < 0.05, **P < 0.01 (Wilcoxon).
Immunofluorescence images of SW480 (A) and SW620 (B) cells. Red: nuclei (DAPI); green: KHSRP; blue: F-actin (phalloidin). Original magnification, ×10.
A: Quantitative RT-PCR analysis of SW620 cells transfected with a pool of siRNAs targeting KHSRP (siKHSRP), a scramble control pool (Scramble), or transfection reagent control (Mock) for 48 hours. B: Cells were transfected as in A and continuously monitored for proliferation for the indicated time; full time-course growth traces are shown on the left. A dashed line indicates the time point of 5 days, for which quantification of cell index values (arbitrary measure of cell proliferation on the basis of impedance measurements) is reported on the right. C: Western blot analysis of KHSRP protein expression in SW620 cells transfected with an siRNA targeting KHSRP, a scramble control siRNA, or a mock (transfection reagent) control for 48 hours. D: Growth of cells transfected as in C, monitored after 7 days by neutral red assay; representative images from one experiment are shown, with staining quantification from triplicate assays. E: Clonogenic potential for cells transfected as in C, measured by both colony area and surviving fraction. Representative images from one experiment are shown, with quantification from triplicate assays. F: Spheroids from cells transfected as in C, grown in Matrigel; representative images are shown with staining quantification from triplicate assays. **P < 0.01, ****P < 0.0001 (analysis of variance) for all tests in the figure. Original magnification, ×10 (F).
SW620 and SW480 cells stably transfected with a conditionally expressible shRNA pool targeting KHSRP (M2) or a nontargeting control pool (Scramble). A: Phase-contrast and fluorescence images of SW620 cells treated with doxycycline or vehicle control for 4 days. Consistent expression of red fluorescent protein (RFP) indicates induced expression of the shRNA-containing cassette. B: Western blot (WB) analysis confirms effective knockdown of KHSRP protein expression. C: WB analysis of the same cells reveals reduction of c-Myc protein expression on knockdown of KHSRP. D: Growth of stably transfected SW620 cells in the presence and absence of doxycycline was quantified using the neutral red assay; colony formation was measured by both surviving fraction and colony area. E: WB analysis confirms effective knockdown of KHSRP protein expression in SW480 generated and treated as above. F: Growth of stably transfected SW480 cells in the presence and absence of doxycycline was quantified using the neutral red assay; colony formation was measured by both surviving fraction and colony area. ****P < 0.0001 (analysis of variance). Original magnification, ×10 (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Neg, negative.
Transcriptomic profile of SW480 cells transfected with KHSRP siRNA compared with scramble negative siRNA control. A: Volcano plot showing differentially regulated genes in green (down-regulated) and red (up-regulated) (log2 fold change > |1|; P < 0.05). Genes that are differentially regulated above the fold change threshold with a nonsignificant P value are orange. B: The distribution of the 135 significantly differentially regulated genes is depicted in the pie chart, using the same color scheme as in A. C: Quantitative RT-PCR analysis of the indicated genes in SW480 cells stably transfected with a conditionally expressible shRNA pool targeting KHSRP or a nontargeting control pool (Scramble), stimulated with doxycycline for 4 days to induce shRNA expression. D: Network map of predicted associations for the protein products of the 135 differentially regulated genes. Proteins are represented by nodes (smaller nodes indicate proteins with unknown structural information); edges represent the predicted functional associations, with the thickness of the line indicating the degree of confidence for the prediction of the interaction. Proteins are color coded arbitrarily by clustering, with the three main clusters annotated. **P < 0.01, ***P < 0.001 (t-test).