Abstract
Purpose:
Microsatellite instability (MSI) and high tumor mutation burden (TMB-High) are promising pan-tumor biomarkers used to select patients for treatment with immune checkpoint blockade; however, real-time sequencing of unresectable or metastatic solid tumors is often challenging. We report a non-invasive approach for detection of MSI and TMB-High in the circulation of patients.
Experimental Design:
We developed an approach that utilized a hybrid-capture based 98 kb pan-cancer gene panel, including targeted microsatellite regions. A multifactorial error correction method and a novel peak finding algorithm were established to identify rare MSI frameshift alleles in cell-free DNA (cfDNA).
Results:
Through analysis of cfDNA derived from a combination of healthy donors and metastatic cancer patients, the error correction and peak finding approaches produced a specificity of >99% (n=163) and sensitivities of 78% (n=23) and 67% (n=15), respectively, for MSI and TMB-High. For patients treated with PD-1 blockade, we demonstrated that MSI and TMB-High in pre-treatment plasma predicted progression-free survival (hazard ratios 0.21 and 0.23, p=0.001 and 0.003, respectively). Additionally, we analyzed cfDNA from longitudinally collected plasma samples obtained during therapy to identify patients who achieved durable response to PD-1 blockade.
Conclusions:
These analyses demonstrate the feasibility of non-invasive pan-cancer screening and monitoring of patients who exhibit MSI or TMB-High and have a high likelihood of responding to immune checkpoint blockade.
STATEMENT OF TRANSLATIONAL RELEVANCE
Microsatellite instability and mismatch repair deficiency represent the first pan-cancer biomarker indication approved for treatment of patients with the immune checkpoint inhibitor, pembrolizumab. However, tumor biopsy or resection tissue is not easily obtained for genetic testing, and therefore, more accessible alternatives must be explored. Detection of circulating tumor DNA derived from plasma provides a viable alternative due to its non-invasive nature, ability to capture tumor heterogeneity, and affords the possibility of monitoring patient response to therapy. Here we describe the development of a liquid biopsy method to identify tumors in patients with MSI and high tumor mutation burden, and demonstrate the efficacy of the approach for determination of response to immune checkpoint blockade.
INTRODUCTION
Microsatellite instability (MSI) and mismatch repair (MMR) deficiency have recently been demonstrated to predict response to immune checkpoint blockade(1,2). The checkpoint inhibitor pembrolizumab is indicated for the treatment of patients with any unresectable or metastatic solid tumors identified as having either of these biomarkers(1,2). The accumulation of somatic mutations in cancers has the potential to result in the expression of neoantigens, which may elicit T-cell–dependent immune responses against tumors(3–5). MMR is a mechanism by which post-replicative mismatches in daughter DNA strands are repaired and replaced with the correct DNA sequence. MMR deficiency results in both MSI and high tumor mutation burden (TMB-High), which increases the likelihood that acquired somatic mutations may be transcribed and translated into proteins that are recognized as immunogenic neoantigens. Historically, testing for MSI has been restricted to screening for Hereditary Non-Polyposis Colorectal Cancer (HNPCC), which is often characterized by early age onset colorectal cancer and endometrial cancer, as well as other extracolonic tumors(6,7). HNPCC, commonly referred to as Lynch Syndrome, is caused by mutations in the DNA mismatch repair genes (MLH1, MSH2, MSH6 and PMS2)(8–15), as well as the more recently described, EPCAM(16). In addition to familial conditions, MSI can occur sporadically in cancer, and both hereditary and sporadic MSI patients respond to immune checkpoint blockade(1,2). A recent study, conducted across 39 tumor types and 11,139 patients to determine the landscape of MSI prevalence, concluded that 3.8% of these cancers across 27 tumor types displayed MSI, including 31.4% of uterine/endometrial carcinoma, 19.7% of colon adenocarcinoma, and 19.1% of stomach adenocarcinoma(17,18).
MSI can be detected by measuring the length of altered microsatellite sequences typically due to deletions of repetitive units that changes the lengths of these sequences in tumor DNA as compared with matched-normal DNA. Current methods for MSI testing, using tissue biopsies and resection specimens, include PCR-based amplification followed by capillary electrophoresis(19), and more recently, next-generation sequencing (NGS) based approaches(17,20–24), which are used to quantify microsatellite allele lengths. Both methodologies have sensitivity limitations for tissue applications due to polymerase-induced errors (stutter bands) and inaccurate estimation of homopolymer lengths in the PCR-based and NGS-based approaches, respectively.
Since MSI has become a valuable marker that predicts a robust response to checkpoint blockade, we were interested in utilizing circulating tumor DNA (ctDNA) to asses MSI status in patients with metastatic disease. Such an approach would be desirable because it is often not possible to readily obtain biopsy or resection tissue for genetic testing due to insufficient samples (biopsy size and tumor cellularity), exhaustion of specimens after pathologic analyses, logistical considerations for obtaining tumor and normal samples after initial diagnosis, or safety concerns related to additional tissue biopsy interventions(26). Plasma-based approaches offer the unique opportunity to obtain a rapid and real-time view of the primary tumor and metastatic lesions along with associated response to therapy. ctDNA can be used to monitor and assess residual disease in response to clinical intervention, such as surgery or chemotherapy(27–35), which can directly impact patient care. A novel method was recently described for determination of MSI in liquid biopsies using pre-PCR elimination of wild-type DNA homopolymers(25), but simply reports MSI status and was not designed to interrogate multiple genetic alterations as will be required in the future for tumor profiling applications. To determine the clinical impact of identifying tumors that harbor MSI and high tumor mutation burden, we developed and applied a 98 kb 58-gene targeted panel to non-invasively assess cancer patients with advanced disease treated with PD-1 blockade.
MATERIALS AND METHODS
Patients and Sample Collection
Formalin fixed paraffin embedded (FFPE) tumor and matched normal buffy coat specimens (n=61) from individuals with cancer were obtained after surgical resection through commercial biorepositories from BioIVT (Hicksville, NY, USA), Indivumed (Hamburg, Germany), and iSpecimen (Lexington, MA, USA). Plasma samples from healthy individuals (n=163) were procured through BioIVT during routine screening with negative results and no prior history of cancer. Human cells from previously characterized MSI cell lines were obtained from ATCC (Manassas, VA, USA) (n=5; LS180, LS411N, SNU-C2B, RKO, and SNU-C2A). Baseline and serial plasma samples from cancer patients with progressive metastatic carcinoma were obtained while patients were enrolled in a phase 2 clinical trial to evaluate immune checkpoint blockade with pembrolizumab(1,2). Radiographic and serum protein biomarker data for CEA and CA19–9 were collected as a part of routine clinical care. All samples were obtained under Institutional Review Board approved protocols with informed consent for research. Orthogonal testing of FFPE tissue was performed for MSI status using the Promega MSI analysis system (Madison, WI, USA) as recommended by the manufacturer.
Sample Preparation and Next-Generation Sequencing
FFPE Tumor and Normal Analyses
Sample processing from tissue or buffy coat, library preparation, hybrid capture, and sequencing were performed as previously described at Personal Genome Diagnostics (Baltimore, MD)(36,46). Briefly, DNA was extracted from FFPE tissue and matched normal buffy coat cells using the Qiagen FFPE Tissue Kit and DNA Blood Mini Kit, respectively (Qiagen, Hilden, Germany). Genomic DNA was sheared using a Covaris sonicator (Woburn, MA, USA), and subsequently used to generate a genomic library using the New England Biolabs (Ipswich, MA, USA) end-repair, A-tailing, and adapter ligation modules. Finally, genomic libraries were amplified and captured using the Agilent SureSelect XT in-solution hybrid capture system with a custom panel targeting the pre-defined regions of interest across 125 genes (Supplementary Table 1). Captured libraries were sequenced on the Illumina HiSeq 2000/2500 (Illumina, San Diego, CA, USA) with 100 bp paired end reads.
Plasma Analyses
Sample processing from plasma, library preparation, hybrid capture, and sequencing were performed as previously described at Personal Genome Diagnostics (Baltimore, MD)(36). cfDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Libraries were prepared with 5–250 ng of cfDNA using the NEBNext DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA). Targeted hybrid capture was performed using Agilent SureSelect XT in-solution hybrid capture system with a custom panel targeting the pre-defined regions of interest across 58 genes (Supplementary Table 4) according to the manufacturer protocol (Agilent Technologies, Santa Clara, CA, USA). Captured libraries were sequenced on the Illumina HiSeq 2000/2500 (Illumina, San Diego, CA, USA) with 100 bp paired end reads. For limit of detection analyses, the 1% MAF level was chosen based on the expected level of distinct sequencing coverage across the mononucleotide tracts, assuming a binomial distribution, requiring a minimum of three distinct observations.
Microsatellite Instability Analyses by Next-Generation Sequencing
Sequence data were aligned to the human reference genome assembly (hg19) using BWA-MEM(47). Reads mapping to microsatellites were excised using Samtools(48) and analyzed for insertion and deletion events (indels). In most cases, alignment and variant calling did not generate accurate indel calls in repeated regions due to low quality bases surrounding the microsatellites. Therefore, a secondary local realignment and indel quantitation was performed. Reads were considered for an expanded indel analysis if (i) the mononucleotide repeat was contained to more than eight bases inside of the start and end of the read, (ii) the indel length was ≤ 12 bases from the reference length, (iii) there were no single base changes found within the repeat region, (iv) the read had a mapping score of 60, and (v) ≤ 20 bases of the read were soft clipped for alignment. After read specific mononucleotide length analysis, error correction was performed for the mononucleotide indel to allow for an accurate quantitation among duplicated fragments using molecular barcoding. Indels were error corrected by using the ordered and combined read 1 and read 2 alignment positions with the molecular barcode. An indel with a given barcode was considered for downstream analysis if it had at least 2 observations and >50% of indels with the given barcode had consistent mononucleotide lengths. The error corrected mononucleotide length distribution based on indel size was subjected to a peak finding algorithm where local maxima were required to be greater than the error corrected distinct fragment counts of the adjacent lengths ± 2 bp. Identified peaks were further filtered to only include those which had > 3 error corrected distinct fragments at ≥ 1% of the absolute coverage. The shortest identified mononucleotide allele length was compared to the hg19 reference length. If the allele length was ≥ 3 bp shorter than the reference length, the given mononucleotide loci was classified as exhibiting instability. The sum of the error corrected distinct coverage of the shortest allele length for all tracts classified as MSI was divided by the sum of the total error corrected distinct coverage for all tracts classified as MSI to generate an MSI mutant allele fraction. This approach was applied across all mononucleotide loci. In the targeted 58 gene plasma panel, BAT25 (chr4:55598211–55598236 hg19), BAT26 (chr2:47641559–47641586 hg19), MONO27 (chr2:39536689–39536716 hg19), NR21 (chr14:23652346–23652367 hg19), and NR24 (chr2:95849361–95849384 hg19) mononucleotide loci were used for the determination of MSI status. In the targeted 125 gene targeted tissue panel, an additional 65 microsatellite regions were used for MSI classification (Supplementary Figure 5).
Tumor Mutation Burden Analyses by Next-Generation Sequencing
Next generation sequencing data were processed and variants were identified using the VariantDx custom software as previously described(36). A final set of candidate somatic mutations were selected for tumor mutation burden analyses based on: (i) variants enriched due to sequencing or alignment error were removed (≤5 observations or <0.30% mutant allele fraction), (ii) nonsynonymous and synonymous variants were included, but variants arising in non-coding regions were removed, (iii) hotspot variants annotated in COSMIC (version 72) were not included to reduce bias toward driver alterations (requiring a given genomic alteration to be mutated in at least 50 tumors with the exact nucleotide change), (iv) common germline SNPs found in dbSNP (version 138) were removed as well as variants deemed private germline variants based on the variant allele frequency, and (v) variants associated with clonal hematopoietic expansion were filtered as previously described and not included in the candidate variant set(36, 49).
In Silico TCGA Analyses
In order to evaluate the accuracy of the 98 kb targeted panel for prediction of TMB, a comparison to whole-exome sequencing data derived from The Cancer Genome Atlas (TCGA)(37) was performed by considering synonymous and nonsynonymous alterations, excluding known hotspot mutations which may not be representative of TMB in the tumor.
Microsatellite Instability and Tumor Mutation Burden Cutoff Selection from Plasma
For microsatellite instability analyses, samples were classified as MSI-H if ≥20% of loci were determined to be MSI, based on previous reports of these targeted loci when evaluating DNA derived from tumor tissue(19). For tumor mutation burden analyses, the TMB-High cutoff was determined based on in silico analyses of the 58 gene plasma panel compared to the TCGA whole exome analyses (r=0.91, p<0.0001; Pearson correlation) (Figure 2A). In this training cohort, we determined that a cutoff of five mutations (50.8 mutations/Mbp sequenced) in the targeted plasma panel could be used to identify tumors with exceptionally high TMB related to MMR deficiency (>36 mutations/Mbp of the whole exome) with >95% accuracy.
Figure 2. Plasma-Based Detection of High Tumor Mutation Burden.
(A) Using whole exome sequencing data derived from The Cancer Genome Atlas (TCGA), a significant positive correlation between the tumor mutation burden (TMB) evaluated in the 98 kb targeted regions compared to the whole exome analyses was observed (r=0.91, p<0.0001; Pearson correlation). (B) Comparison of the accuracy for determination of the TMB derived from the targeted panel in plasma at baseline compared to whole-exome analyses of matched archival tissue samples in 20 patients yielded a positive trend (r=0.38, p=0.095; Pearson correlation). (C) The overall TMB status at baseline was assigned as TMB-High or TMB-Low using a cutoff of 50.8 mutations/Mbp sequenced. In total, 13 patients were categorized as TMB-High and 16 patients as TMB-Low, with a median load of 152.4 mutations/Mbp sequenced and 20.3 mutations/Mbp sequenced, respectively. Additionally, 163 healthy donor cases were evaluated, all of which were determined to be TMB-Low, with a median load of 0 mutations/Mbp sequenced across the panel. Kaplan-Meier curves for progression free survival (D) and overall survival (E) among this same cohort of patients were determined using TMB status from pre-treatment plasma specimens with a cutoff of 50.8 mutations/Mbp sequenced. In TMB-High patients (n=13), median progression free survival and median overall survival were not reached. In TMB-Low patients (n=16), median progression free survival and median overall survival were 2.8 and 7.6 months, respectively.
Statistical Analyses
Due to small sample size, Firth’s Penalized Likelihood was used to evaluate significant differences between Kaplan-Meier curves for progression free survival and overall survival with the classifiers baseline MSI status and baseline TMB status. Pearson correlations were used to evaluate the significance of the association between TMB in the 58 gene targeted panel compared to whole-exome analyses, progression free and overall survival compared to residual protein biomarker levels, and progression free and overall survival compared to residual MSI and TMB allele levels. A student t-test was used to evaluate significant differences between the mean TMB level in TMB-High and TMB-Low patients.
RESULTS
Development of an assay to identify MSI in cell-free DNA
To identify MSI in tumor-derived cfDNA from the plasma as well as in tissue specimens we developed a highly sensitive error-correction approach incorporating the commonly-used mononucleotide tracts BAT25, BAT26, MONO27, NR21, and NR24 (Methods). To address the technical challenges associated with low level allele length polymorphisms obtained from NGS, we combined an error correction approach for accurate determination of insertions and deletions (indels) present in the cfDNA fragments, together with a digital peak finding (DPF) method for quantification of MSI-High (MSI-H) and microsatellite stable (MSS) alleles. Redundant sequencing of each cfDNA fragment was performed, and reads were aligned to the five microsatellite loci contained in the human reference genome (hg19). cfDNA sequences were then analyzed for indels through a secondary local alignment at these five microsatellite loci to more accurately determine the indel length. To perform the error correction, duplicated reads associated with each cfDNA molecule were consolidated, only recognizing indels present throughout barcoded DNA fragment replicates obtained through redundant sequencing. Finally, the DPF approach was applied across the error corrected distribution of indels to identify high confidence alleles which exhibit microsatellite instability (Figure 1A).
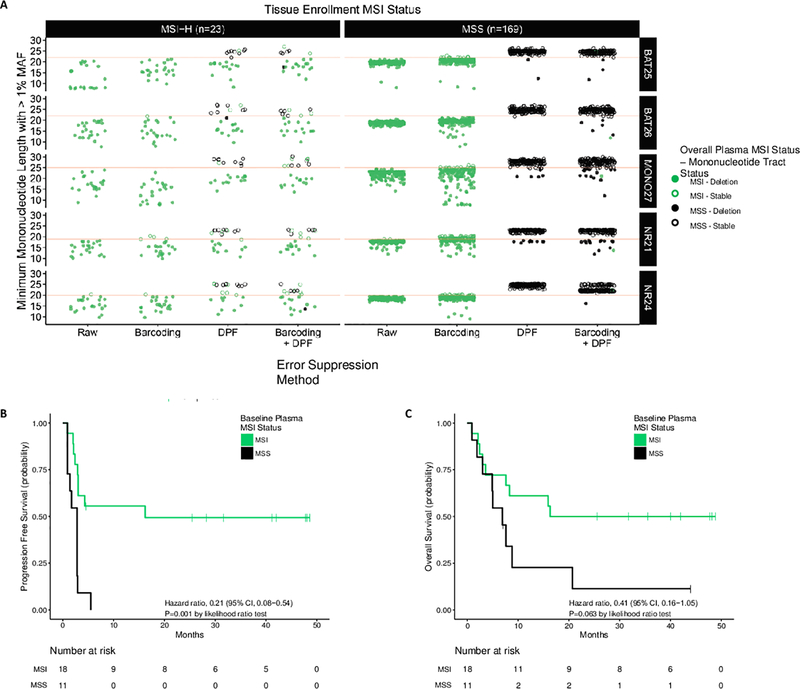
Figure 1. Plasma-Based Detection of Microsatellite Instability.
(A) Across the BAT25, BAT26, MONO27, NR21, and NR24 mononucleotide loci in 23 clinical MSI-H patients, 6 clinical MSS patients, and 163 healthy donor plasma specimens (169 total MSS cases), the error corrected mononucleotide count distribution was assessed with a DPF algorithm to identify mononucleotide alleles and determine MSI status. Prior to combined barcoding and DPF (Raw) and with barcoding alone, the majority of clinical MSS and healthy donor samples exhibit alleles below the cutoff for MSI and MSS classification (red line) making MSI-H and MSS cases indistinguishable. With the DPF algorithm alone and with combined barcoding and DPF, the majority of samples were correctly classified (15/23 MSI; 169/169 MSS and 18/23 MSI; 168/169 MSS respectively). Kaplan-Meier curves for progression free survival (B) and overall survival (C) among patients with progressive metastatic carcinoma were determined using MSI status from pre-treatment plasma specimens. In MSI patients (n=18*), median progression free survival and median overall survival were 16.2 and 16.3 months, respectively. In MSS patients (n=11*), median progression free survival and median overall survival were 2.8 and 6.9 months, respectively. *Five patients with a tissue enrollment status of MSI-H were classified as MSS using pre-treatment baseline cfDNA obtained from plasma.
To demonstrate the feasibility of this approach, we first evaluated the performance of the method for detection of MSI in formalin fixed, paraffin embedded (FFPE) tumor tissue specimens obtained from 31 MSI-H and 30 MSS tumors previously characterized with the PCR-based Promega MSI analysis system. In addition to these five mononucleotide markers, we sequenced 125 selected cancer genes which harbor clinically actionable genetic alterations consisting of sequence mutations (single base substitutions and indels), copy number alterations, and gene rearrangements in cancer (Supplementary Table 1). Analysis of these five mononucleotide loci, together with 65 additional microsatellite regions contained within the 125 gene panel, resulted in 100% sensitivity (31/31) and 100% specificity (30/30) for determination of MSI status using the patient-matched tumor and normal samples (Supplementary Tables 2 and 3). Across this cohort, microsatellite instability was observed as a tract length shortening compared to the matched normal tissue. Based on these data, only mononucleotide tract length shortening was considered for the subsequent analyses.
Next, we evaluated the signal-to-noise ratio in homopolymer regions from next-generation sequencing data obtained using cfDNA extracted from the plasma of healthy individuals and cancer patients. Together with the five mononucleotide loci, we developed a 98 kb, 58 gene panel for sequence mutation (single base substitutions and indels) analyses of clinically actionable genetic alterations in cancer(36) (Supplementary Table 4). To demonstrate the specificity of this approach for direct detection of MSI, we first obtained plasma from healthy donors (n=163), all of whom would be expected to be tumor-free and MSS. These analyses resulted in 2,600-fold distinct coverage across the 98 kb targeted panel and resulted in a per-patient specificity of 99.4% (162/163) for determination of MSI status (Figure 1A, Supplementary Tables 5 and 6). The single false positive result was obtained from a sample with 974-fold distinct coverage, lower than any late-stage cancer patient evaluated, indicating that the specificity of 99.4% is likely a lower bound for the intended use population. This is consistent with reduced cfDNA yields and lower coverage in healthy donor populations compared to cancer patients with metastatic disease(36).
Because ctDNA may be present at mutant allele fractions (MAFs) less than 5% even in patients with advanced cancer, we characterized the ability of DPF for sensitive and reproducible detection of MSI at low MAFs. Five previously characterized MSI cell line samples obtained from ATCC (LS180, LS411N, SNU-C2B, RKO, and SNU-C2A) were sheared to a fragment profile simulating cfDNA and diluted with normal DNA to yield a total of 25 ng evaluated at 1% MAF. Additionally, three of these cell lines (LS180, LS411N, and SNU-C2B) were evaluated at 1% MAF in triplicate within and across library preparation and sequencing runs (Supplementary Table 5). Based on the MAF observed in the parental cell line, the cases detected as MSI were confirmed to contain MSI alleles at MAFs of 1.2%-4.6%, with a median MSI allele MAF of 1.8%. Through our analysis, MSI was detected in 90% (18/20) of samples and demonstrated 93.3% (14/15) reproducibility within and across runs (Supplementary Table 6). For one case which was not detected as MSI, one MSI allele was identified, and for the other case, no MSI alleles were detected.
Assessment of MSI in cfDNA in patients treated with PD-1 blockade
To evaluate the analytical and clinical performance of this approach for determination of MSI in cfDNA from patients with late-stage cancers, we obtained baseline and serial plasma from patients with metastatic cancers, including 19 colorectal, 3 ampullary, 3 small intestine, 2 endometrial, 1 gastric, and 1 thyroid, with or without MMR deficiency, while enrolled in a clinical trial to evaluate response to immune checkpoint blockade with the PD-1 blocking antibody, pembrolizumab(1,2) (Supplementary Table 7). In total, 23 MSI-H cases and 6 MSS cases, determined through archival tissue-based analyses, were assessed at a baseline plasma timepoint, and 16 of the patients were evaluated across at least one additional plasma timepoint, including after approximately 2 weeks, 10 weeks, 20 weeks, and >100 weeks.
Patients with MSI tumors as determined by archival tissue analyses had improved progression-free survival (hazard ratio, 0.26; p=0.014, likelihood ratio test) and overall survival (hazard ratio, 0.27; p=0.02, likelihood ratio test) (Supplementary Figures 1A and 1B and Supplementary Table 8). In cfDNA, we detected MSI in 78.3% (18/23) of the MSI-H patients, and correctly identified 100% (6/6) of the MSS patients (Supplementary Table 6). Of the five cases that were MSI in the tumor tissue and MSS in the cfDNA, three were colorectal tumors (two patients exhibited progressive disease and the third was not evaluable) and two were small intestinal tumors (one patient exhibited a partial response and one exhibited progressive disease). Of these cases, two had no detectable ctDNA, two had low levels of ctDNA (average MAF of 0.4%, 1.1%), and one had an average sequence mutation MAF of 24.7%.
We evaluated pre-treatment MSI status in ctDNA to predict response and clinical outcome to treatment with PD-1 blockade. We assessed progression-free and overall survival to predict clinical outcome. Similar to tissue-based analyses, direct detection of MSI in baseline cfDNA could be used to predict progression free survival to immune checkpoint blockade (hazard ratio, 0.21; p=0.001, likelihood ratio test) (Figure 1B), but was not statistically significant for overall survival (hazard ratio, 0.41; p=0.063, likelihood ratio test) (Figure 1C). When considering only cases for which adequate ctDNA was detected (median sequence mutation MAF ≥0.5%), there were 25/29 cases evaluable, with 17/19 MSI-H cases detected and 6/6 MSS cases detected. For this subset of patients, direct detection of MSI in baseline cfDNA predicted progression free and overall survival to immune checkpoint blockade (hazard ratio, 0.15; p=0.001, likelihood ratio test and hazard ratio, 0.26; p=0.01, likelihood ratio test, respectively) (Supplementary Figure 2A–C).
Estimating tumor mutation burden in ctDNA
In addition to MSI status, we also evaluated the ability of our cfDNA panel to predict TMB across a range of tumor types, using whole exome sequencing data from 8,493 samples from The Cancer Genome Atlas (TCGA)(37). We considered synonymous and nonsynonymous alterations identified by TCGA and excluded known driver hotspot mutations as these have been selected during tumorigenesis and may not be representative of TMB in the tumor. These analyses demonstrated a positive correlation between predicted TMB from our targeted 58 gene plasma panel compared to the TCGA whole exome analyses (r=0.91, p<0.0001; Pearson correlation) (Figure 2A). We determined that a cutoff of five mutations in the targeted plasma panel corresponding to ~51 mutations/Mbp sequenced could be used to identify tumors with exceptionally high TMB related to MMR deficiency (>36 mutations/Mbp of the whole exome) at >95% accuracy.
Patients with TMB-High tumors (≥10 mutations/Mbp of the whole exome) as determined by analyses of archival tissue from 20 tumor samples (12 colorectal, 3 ampullary, 2 small intestine, 1 endometrial, 1 gastric, and 1 thryroid) had improved progression-free survival (hazard ratio, 0.24; p=0.021, likelihood ratio test) and overall survival (hazard ratio, 0.28; p=0.043, likelihood ratio test) (Supplementary Figures 1C and 1D). We also evaluated the accuracy of TMB derived from the targeted panel in 20 baseline plasma samples from these cases compared to whole-exome analyses of tumor and matched normal tissue in the same patients(1,2), and a similar trend was observed (r=0.38, p=0.095; Pearson correlation) (Figure 2B). This correlation was lower than that observed through in silico TCGA analyses, potentially due to the biological variability associated with low ctDNA levels and tumor heterogeneity. These patients were classified as either TMB-High or TMB-Low using a cutoff of 51 mutations/Mbp sequenced (selected through in silico TCGA analyses), which captured ten of the fifteen tumors categorized as TMB-High by archival tissue and provided a statistically significant difference in the TMB classification (p<0.0001, t-test) (Figure 2C). This algorithm was also applied to the same 163 healthy donor plasma samples and 100% (163/163) were determined to be TMB-Low (Figure 2C). When considering TMB classification as a predictor of clinical outcome for these patients enrolled in a clinical trial to evaluate response to immune checkpoint blockade with the PD-1 blocking antibody, pembrolizumab, baseline plasma TMB-High status was associated with favorable progression free survival (hazard ratio, 0.23; p=0.003 likelihood ratio test) and overall survival (hazard ratio, 0.26; p=0.008, likelihood ratio test) (Figures 2D and 2E). When we considered only cases for which adequate ctDNA was detected (median detected sequence mutation allele fraction ≥0.5%), there were 17/20 cases evaluable with archival tissue data available, with 9/12 TMB-High cases detected and 5/5 TMB-Low cases detected. When compared to progression free and overall survival, direct detection of TMB in baseline cfDNA (n=25) could be used to predict response to immune checkpoint blockade (hazard ratio, 0.27; p=0.013, likelihood ratio test and hazard ratio, 0.23; p=0.006, likelihood ratio test, respectively) with this ctDNA requirement (Supplementary Figure 2D–F). Interestingly, all five MSI-H patients exhibiting a complete response, determined through archival tissue analyses, were classified as TMB-High through plasma-based analyses, and six of seven MSI-H patients with progressive disease, determined through archival tissue analyses, were classified as TMB-Low through plasma-based analyses (Supplementary Tables 7 and 8).
Assessment of molecular remission and biomarker dynamics in patients treated with PD-1 blockade
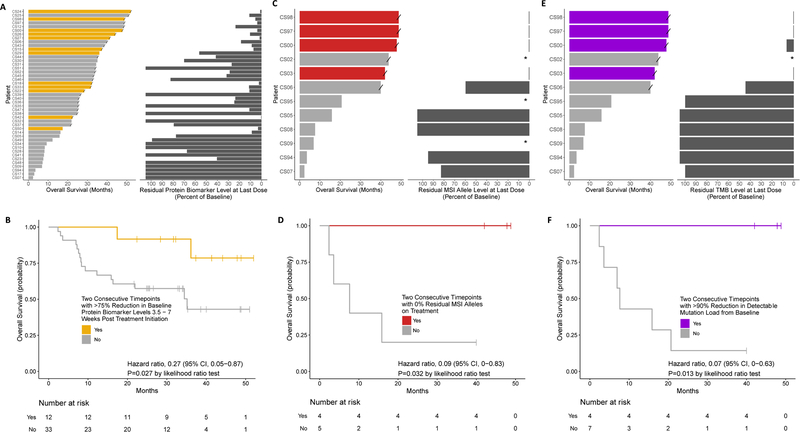
In addition to baseline plasma analyses, we hypothesized that molecular remission, as measured by changes in ctDNA levels during treatment, would also be predictive of long-term durable response to immune checkpoint blockade. We first evaluated the utility of monitoring serum tumor protein biomarkers (CA125, CEA, CA19–9, or PSA) for determination of response at 3.5–7 weeks post treatment initiation (Supplementary Table 7). We evaluated 45 patients with metastatic cancers with MMR deficiency and elevated baseline serum tumor protein biomarker levels while enrolled in a clinical trial to evaluate response to immune checkpoint blockade with the PD-1 blocking antibody, pembrolizumab. We landmarked the first time point between 3.5 and 7 weeks, and found that multiple consecutive timepoints with a >75% reduction in the baseline protein biomarker level resulted in improved overall and progression free survival (hazard ratio, 0.27; p=0.027 and hazard ratio, 0.38; p=0.052, likelihood ratio test, respectively) (Figures 3A and 3B and Supplementary Figures 3A and 3B). For 12 patients enrolled in this clinical study, when evaluating the on-treatment serial plasma samples for residual ctDNA levels, there was a significant inverse correlation between the overall and progression free survival when compared to the residual MSI allele levels at last dose (r=-0.91, p=0.0006 and r=-0.98, p<0.0001, respectively; Pearson correlation) (Figures 3C and Supplementary Figure 3C), however, only a limited subset of the timepoints were available for these analyses (Methods). We were able to correctly identify four of the six MSI patients who would achieve a long term durable clinical response requiring multiple consecutive on-treatment timepoints with 0% residual alleles displaying MSI, all four of which displayed a complete response (hazard ratio, 0.09; p=0.032, likelihood ratio test for overall survival) (Figure 3D and Supplementary Figure 3D). A similar trend was observed when considering patients with a >90% decrease in overall TMB across two timepoints when compared to baseline (hazard ratio, 0.07; p=0.013, likelihood ratio test for overall survival) (Figures 3E and 3F and Supplementary Figures 3E and 3F).
Figure 3. Serial Plasma-Based Overall Survival Analysis for Patients Treated with Immune Checkpoint Blockade.
(A) Evaluation of overall survival with the protein biomarker level at last dose (CA125, CEA, CA19–9, or PSA). A significant inverse correlation was observed between the overall survival in months when compared to the residual protein biomarker (r=-0.67, p=<0.001; Pearson correlation). (B) Kaplan-Meier curves for overall survival among patients with tissue enrollment status of MSI and detectable protein biomarker levels (n=45). For patients with two consecutive timepoints with >75% reduction in protein biomarker levels, landmarked 3.5–7 weeks post treatment initiation (n=12), median overall survival was not reached. For patients with ≤75% reduction in protein biomarker levels (n=33), median overall survival was 35.1 months. (C) Evaluation of overall survival compared to residual MSI allele levels at last dose. A significant inverse correlation was observed between the overall survival when compared to the residual MSI allele levels (r=-0.91, p=<0.001; Pearson correlation). (D) Kaplan-Meier curves for overall survival among patients with tissue enrollment status of MSI and detectable MSI status at baseline (n=9). For patients with two consecutive timepoints displaying no residual MSI alleles (n=4) median overall survival was not reached. For patients with multiple timepoints containing residual MSI alleles (n=5) median overall survival was 7.64 months. (E) Evaluation of overall survival compared to residual TMB levels at last dose. A significant inverse correlation was observed between the overall survival in months when compared to the residual TMB levels (r=-0.95, p=<0.001; Pearson correlation). (F) Kaplan-Meier curves for overall survival among patients with tissue enrollment status of MSI and detectable TMB levels at baseline (n=11). For patients with >90% reduction in TMB levels (n=4), median overall survival was not reached. For patients with ≤90% reduction in TMB levels (n=7), median overall survival was 7.64 months. “/” indicates a censored datapoint; “*” indicates cases where baseline protein biomarker, MSI or TMB was not detected and were not included in the subsequent analyses; In cases where residual protein biomarker, MSI or TMB levels increased when compared to baseline, values of greater than 100% are indicated.
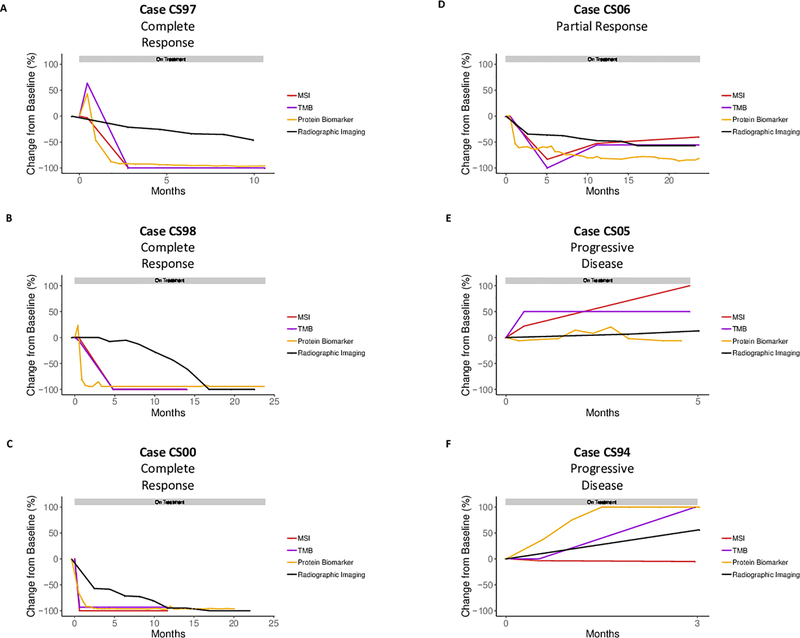
Additionally, for three patients (CS97, CS98, and CS00) with a complete response, one patient with a partial response (CS06), and two patients (CS05, and CS94) without a response to immune checkpoint blockade, circulating protein biomarkers (CEA, ng/mL or CA19–9, units/mL) and residual alleles exhibiting MSI and TMB were evaluated over time during treatment (Figure 4). In each of the patients exhibiting a complete response, there was a concurrent decrease in the circulating protein biomarker levels, the residual MSI alleles, and TMB levels, which correlated with reduced overall tumor volume as assessed by radiographic imaging. Patient CS97 demonstrated a partial radiographic response at 10.6 months, however, achieved a 100% reduction in residual MSI and TMB levels at 2.8 months. CS97 then went on to a complete radiographic response at 20.2 months (Supplementary Table 7). Patient CS98 appeared to develop new liver lesions at 20 weeks suggestive of progressive disease (Supplementary Figure 4). However, following an initial spike, protein biomarkers and residual MSI and TMB levels demonstrated a biochemical tumor response at 1.3 and 4.8 months. A liver biopsy demonstrated only inflammatory changes in the location where new lesions were noted, suggesting checkpoint therapy induced inflammation. Radiographic imaging finally demonstrated resolution of any hepatic lesions and a 100% reduction in tumor volume at 16.8 months. A similar pattern was observed for patient CS00 where significant reduction in protein biomarker and residual MSI and TMB levels occurred at 1.5 and 0.6 months, respectively, however, radiographic imaging did not demonstrate a 100% reduction in tumor volume until 17 months. These data suggest that the residual MSI allele burden and TMB levels are indicative of overall tumor response to immune checkpoint blockade.
Figure 4. Monitoring of Patients During Immune Checkpoint Blockade.
For three patients with a complete response to immune checkpoint blockade (CS97 (A), CS98 (B), and CS00 (C)), one patient with partial response (CS06 (D)), and two patients with progressive disease (CS05 (E) and CS94 (F)), residual alleles exhibiting MSI, TMB levels, circulating protein biomarkers (CEA, ng/mL and CA19–9, units/mL), and radiographic imaging were evaluated over time during treatment. In each case exhibiting a complete response, residual MSI and TMB alleles were reduced to 0% mutant allele fraction (MAF) between 0.61 and 4.81 months after first dose. For each patient, the grey horizontal bar represents time on treatment.
CONCLUSIONS/DISCUSSION
The checkpoint inhibitor pembrolizumab is now indicated for the treatment of adult and pediatric patients with unresectable or metastatic solid tumors identified as having MSI or MMR deficiency(1,2). However, it is often not possible to readily obtain biopsy or resection tissue for genetic testing due to insufficient material, exhaustion of the limited material available after prior therapeutic stratification, logistical considerations for tumor and normal sample acquisition after initial diagnosis, or safety concerns related to additional tissue biopsy interventions(26). We have described the development of an analytical method for simultaneous detection of MSI and TMB-High directly from cfDNA and demonstrated proof of concept for the clinical utility afforded through these analyses for the prediction of response to immune checkpoint blockade.
We present the first comprehensive tumor profiling approach for evaluation of MSI status from plasma utilizing next-generation sequencing. Specifically, MSISensor(20), MANTIS(22), and mSINGS(24) involve extraction of sequencing reads associated with microsatellite loci to create a distribution and compare to a matched normal or a panel of normal samples. MIRMMR(21) utilizes methylation and sequence mutational data from genes in the MMR pathway, from which a regression model is trained, and MSIseq(23) utilizes a single nucleotide variant and indel classifier to determine MSI status. The methods described herein for determination of MSI status are the first to employ error correction of the sequencing reads associated with microsatellite loci through molecular barcoding, together with local maxima detection of low-level microsatellite alleles associated with cfDNA.
Recently, Kim et al., have described the use of a 73 gene next-generation sequencing panel to correlate ctDNA mutational load scores with the mutational load calculated from tumor exome sequencing from 23 metastatic gastric cancer patients treated with pembrolizumab as salvage treatment(38). A second, larger scale study led by Gandara et al. was performed to evaluate the clinical utility of plasma TMB with a 1.1 Mb panel using samples collected prospectively from the POPLAR and OAK randomized clinical trials for second-line or higher patients with NSCLC(39). Interestingly, in our study all five MSI-H patients exhibiting a complete response were classified as TMB-High through plasma-based analyses, and six of seven MSI-H patients with progressive disease, determined through archival tissue analyses, were classified as TMB-Low through plasma-based analyses. These data suggest that a baseline TMB measurement in plasma may be more accurate than archival tissue, as was the case in this study, since it provides a real-time analysis and corrects for the sampling error that is inherent to tissue sequencing.
In addition to the baseline evaluation of cfDNA for response prediction, ctDNA monitoring represents an approach to obtain a real time analysis of tumor response to immune checkpoint blockade. Assessment of the efficacy of response to immune checkpoint inhibition has proven challenging utilizing imaging-based methodologies, particularly in the context of pseudoprogression, whereby an initial increase in tumor volume is observed, potentially due to immune cell infiltration, followed by tumor shrinkage(40,41). Therefore, cfDNA-based approaches for comprehensive genome profiling may be useful for the rapid determination of patients that ultimately may benefit from immune checkpoint blockade. This hypothesis has been previously demonstrated for melanoma patients treated with CTLA-4 blockade(42), as well as immune checkpoint blockade in NSCLC(34,43–45). Our data further support this hypothesis, and given the concordance with circulating protein biomarker data, suggest that the residual MSI allele burden and TMB prognostic signature could be applied to other tumor types where standardized protein biomarkers do not exist and may be an earlier predictor of response than radiographic imaging.
While every effort has been made to minimize potential confounding variables in the analyses described, the current study is limited to a small population of cancer patients and prospective clinical trials will need to be conducted across a broader range of tumor types to confirm these findings in a pan-cancer setting. Furthermore, the sensitivity for accurate detection of MSI and TMB-High are highly dependent upon ctDNA levels, and as such, there will be a proportion of patients with low levels of ctDNA for which these analyses will not be informative. TMB status and MSI status are highly correlated, and therefore, in the context of this study population, cannot differentiate the predictive value of each for determination of response to immune checkpoint blockade. Finally, for serial monitoring applications, while protein biomarker data was collected at similar time intervals for patients exhibiting a clinical response or lack of clinical response, sample availability across standardized timepoints was limited for evaluation of plasma MSI allele levels and TMB load. Nevertheless, these methods described herein provide feasibility for a viable diagnostic approach for screening and monitoring of patients who exhibit MSI or TMB-High and may respond to immune checkpoint blockade.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge D. Tsui for her data analysis contributions and critical review of the manuscript.
FINANCIAL SUPPORT
This research was supported in part by NIH Grants 1R43CA217544, CA121113, CA180950, NCI Contract HHSN261201600005C, Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (Grant Number SU2C-AACR-DT22–17), Stand Up to Cancer–Dutch Cancer Society International Translational Cancer Research Dream Team Grant (Grant Number SU2C-AACR-DT1415), and the Commonwealth Foundation. Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. B.R.B. is supported by NIH Grants 5P30GM114737 and P20GM103466.
Footnotes
CONFLICTS OF INTEREST
A.G., L.A.K., M.Z., D.M., E.L.V., F.R., D.R., S.A., and S.J. are employees of Personal Genome Diagnostics. J.R.W. has received consulting fees from Personal Genome Diagnostics. M.S. is a former employee of Personal Genome Diagnostics and current employee of Bristol-Myers Squibb. V.E.V. is a founder of Personal Genome Diagnostics, a member of its Scientific Advisory Board and Board of Directors, and owns Personal Genome Diagnostics stock, which is subject to certain restrictions under university policy. V.E.V. is a member of the Scientific Advisory Board of Ignyta. The terms of these arrangements are managed by Johns Hopkins University in accordance with its conflict of interest policies. Luis A. Diaz (L.A.D.) is a member of the board of directors of Personal Genome Diagnostics (PGDx) and Jounce Therapeutics. L.A.D. holds equity in PapGene, PGDx and Nephore. He is a paid consultant for PGDx and Phoremost. He is an uncompensated consultant for Merck. L.A.D. is an inventor of multiple licensed patents related to technology for circulating tumor DNA analyses and mismatch repair deficiency for diagnosis and therapy (WO2016077553A1) from Johns Hopkins University. Some of these licenses and relationships are associated with equity or royalty payments to L.A.D. The terms of all these arrangements are being managed by Johns Hopkins and Memorial Sloan Kettering in accordance with their conflict of interest policies. In addition, in the past 5 years, L.A.D. has participated as a paid consultant for Merck and for one-time engagements with Caris, Lyndra, Genocea Biosciences, Illumina and Cell Design Labs.
REFERENCES
- 1.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357(6349):409–13 doi 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science 2013;339(6127):1546–58 doi 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348(6230):69–74 doi 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 5.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer research 2008;68(3):889–92 doi 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 6.Warthin A Heredity with reference to carcinoma: As shown by the study of the cases examined in the pathological laboratory of the university of michigan, 1895–1913. Archives of internal medicine 1913;XII(5):546–55 doi 10.1001/archinte.1913.00070050063006. [DOI] [Google Scholar]
- 7.Lynch HT, Shaw MW, Magnuson CW, Larsen AL, Krush AJ. Hereditary factors in cancer. Study of two large midwestern kindreds. Archives of internal medicine 1966;117(2):206–12. [PubMed] [Google Scholar]
- 8.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 1994;371(6492):75–80 doi 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 9.Hendriks YM, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology 2004;127(1):17–25. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science 1994;263(5153):1625–9. [DOI] [PubMed] [Google Scholar]
- 11.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 1994;368(6468):258–61 doi 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 12.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 1993;75(6):1215–25. [DOI] [PubMed] [Google Scholar]
- 13.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993;75(5):1027–38. [DOI] [PubMed] [Google Scholar]
- 14.Lindblom A, Tannergard P, Werelius B, Nordenskjold M. Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nature genetics 1993;5(3):279–82 doi 10.1038/ng1193-279. [DOI] [PubMed] [Google Scholar]
- 15.Peltomaki P, Aaltonen LA, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, et al. Genetic mapping of a locus predisposing to human colorectal cancer. Science 1993;260(5109):810–2. [DOI] [PubMed] [Google Scholar]
- 16.Kempers MJ, Kuiper RP, Ockeloen CW, Chappuis PO, Hutter P, Rahner N, et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. The Lancet Oncology 2011;12(1):49–55 doi 10.1016/S1470-2045(10)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nature medicine 2016;22(11):1342–50 doi 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 18.Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen H-Z, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precision Oncology 2017(1):1–15 doi 10.1200/po.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. The Journal of molecular diagnostics : JMD 2006;8(3):305–11 doi 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30(7):1015–6 doi 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foltz SM, Liang WW, Xie M, Ding L. MIRMMR: binary classification of microsatellite instability using methylation and mutations. Bioinformatics 2017;33(23):3799–801 doi 10.1093/bioinformatics/btx507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget 2017;8(5):7452–63 doi 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang MN, McPherson JR, Cutcutache I, Teh BT, Tan P, Rozen SG. MSIseq: Software for Assessing Microsatellite Instability from Catalogs of Somatic Mutations. Scientific reports 2015;5:13321 doi 10.1038/srep13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clinical chemistry 2014;60(9):1192–9 doi 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 25.Ladas I, Yu F, Leong KW, Fitarelli-Kiehl M, Song C, Ashtaputre R, et al. Enhanced detection of microsatellite instability using pre-PCR elimination of wild-type DNA homo-polymers in tissue and liquid biopsies. Nucleic acids research 2018. doi 10.1093/nar/gky251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai EB, Pomykala K, Ruchalski K, Genshaft S, Abtin F, Gutierrez A, et al. Feasibility and Safety of Intrathoracic Biopsy and Repeat Biopsy for Evaluation of Programmed Cell Death Ligand-1 Expression for Immunotherapy in Non-Small Cell Lung Cancer. Radiology 2018;287(1):326–32 doi 10.1148/radiol.2017170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nature genetics 2013;45(1):12–7 doi 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Science translational medicine 2010;2(20):20ra14 doi 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science translational medicine 2012;4(136):136ra68 doi 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 30.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes, chromosomes & cancer 2010;49(11):1062–9 doi 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nature medicine 2008;14(9):985–90 doi 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson SJ, Rosenfeld N, Caldas C. Circulating tumor DNA to monitor metastatic breast cancer. The New England journal of medicine 2013;369(1):93–4 doi 10.1056/NEJMc1306040. [DOI] [PubMed] [Google Scholar]
- 33.Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nature communications 2015;6:7686 doi 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer research 2018. doi 10.1158/0008-5472.CAN-18-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phallen J, Leal A, Woodward BD, Forde PM, Naidoo J, Marrone KA, et al. Early Noninvasive Detection of Response to Targeted Therapy in Non-Small Cell Lung Cancer. Cancer research 2018. doi 10.1158/0008-5472.CAN-18-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Science translational medicine 2017;9(403) doi 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell systems 2018;6(3):271–81 e7 doi 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nature medicine 2018;24(9):1449–58 doi 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 39.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nature medicine 2018;24(9):1441–8 doi 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 40.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. The New England journal of medicine 2015;373(1):23–34 doi 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. The New England journal of medicine 2015;373(2):123–35 doi 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipson EJ, Velculescu VE, Pritchard TS, Sausen M, Pardoll DM, Topalian SL, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. Journal for immunotherapy of cancer 2014;2(1):42 doi 10.1186/s40425-014-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Annals of oncology : official journal of the European Society for Medical Oncology 2017;28(8):1996–2001 doi 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 44.Iijima Y, Hirotsu Y, Amemiya K, Ooka Y, Mochizuki H, Oyama T, et al. Very early response of circulating tumour-derived DNA in plasma predicts efficacy of nivolumab treatment in patients with non-small cell lung cancer. European journal of cancer 2017;86:349–57 doi 10.1016/j.ejca.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24(8):1872–80 doi 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Science translational medicine 2015;7(283):283ra53 doi 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60 doi 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–9 doi 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine 2014;20(12):1472–8 doi 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.