Abstract
Purpose
To assess the relative cost-effectiveness of cascade genetic testing in asymptomatic relatives of patients with dilated cardiomyopathy (DCM) compared with periodical clinical surveillance.
Methods
A decision-analytic model, combining a decision tree and a Markov model, was used to determine the lifetime costs and quality-adjusted life years (QALYs) for the two strategies. Deterministic and probabilistic sensitivity analyses were undertaken to assess the robustness of findings and to explore decision uncertainty.
Results
The incremental cost per additional QALY of cascade genetic testing prior to periodical clinical surveillance of first-degree relatives compared with periodical clinical surveillance alone was estimated at approximately AUD $6100. At established thresholds of cost-effectiveness, there is a 90% probability that cascade genetic testing is cost-effective. Extensive sensitivity analyses, including the addition of second-degree relatives, did not alter the conclusions drawn from the main analysis.
Conclusion
Using cascade genetic testing to guide clinical surveillance of asymptomatic relatives of patients with DCM is very likely to be cost-effective. As the DCM pathogenic variant detection rate rises and new evidence for personalized treatment of at-risk individuals becomes available, the cost-effectiveness of cascade testing will further increase.
Keywords: dilated cardiomyopathy, genomics, economic evaluation, cost-effectiveness
INTRODUCTION
Heart failure is a leading cause of morbidity and mortality, posing a significant health and economic burden on both patients and society. The prevalence of heart failure rises with age to about 12% in those over the age of 60 years, and about 5% of people above this age have impaired systolic function.1 Dilated cardiomyopathy (DCM) is a common cause of heart failure and is defined as a primary myocardial disorder characterized by left ventricular (LV) dilatation and contractile dysfunction in the absence of abnormal loading conditions or coronary disease sufficient to cause global systolic dysfunction.2 The prevalence of asymptomatic idiopathic DCM is likely to be ≥1 in 250 individuals3 with approximately 20% to 35% of DCM cases reported as familial, although with incomplete and age-dependent penetrance.4
Genetic testing is not routinely recommended for all patients diagnosed with DCM and their relatives.5,6 According to these guidelines, genetic testing is recommended for patients who have DCM with associated significant cardiac conduction disease and/or a family history of premature unexpected sudden death.5,6 Of the large number of known disease and candidate genes for DCM, certain core genes have robust evidence of causation (i.e., TNb, LMNA, MYH7, TNNT2, BAG3, RBM20, TNNC1, TNNI3, TPM1, SCN5A, PLN) and should be included as part of any planned genetic testing approaches.7,8 The diagnostic yield is estimated to be approximately 25–40% in families with autosomal dominant inheritance and 8–25% in isolated cases of DCM.9–13 In the absence of genetic testing, it is recommended that all close blood relatives of individuals with suspected familial DCM undergo a lifetime of periodical clinical surveillance.14 periodical clinical surveillance of relatives typically occurs at a frequency of 1–5 years and involves clinical screening with physical examination, electrocardiography, and echocardiography.
In clinical practice, however, genetic testing for DCM has its limitations. Although new sequencing technology unravels a large number of variants, a major challenge is to determine the variants that are likely to be disease-causing. For example, in some cardiomyopathy-related genes, rare or even high impact variants (i.e., those that significantly alter the encoded protein) that were previously thought to be potentially pathogenic have been subsequently observed in a reference population database, thus casting doubt regarding their true clinical association.15 Nevertheless, genetic testing for DCM may have important implications for the management of family members. If a genetic diagnosis is confirmed in the proband, that information can allow for the early identification of relatives with the pathogenic variant. Identifying those relatives means that they can be targeted for closer surveillance, which may facilitate earlier introduction of heart failure–related treatment. Also, identifying relatives without the pathogenic variant means they can be released from all future clinical surveillance and their offspring have no risk of carrying the gene, which has psychological benefits for the individual as well as economic implications.
The main objective of the present study is to assess the relative cost-effectiveness of the addition of genetic testing in individuals with DCM to inform risk assessment and guide clinical surveillance through cascade testing in asymptomatic relatives of patients with DCM compared with periodical clinical surveillance without genetic testing.
MATERIALS AND METHODS
This economic evaluation builds upon a Melbourne Genomics Health Alliance’s DCM study. In this study, 87 patients with idiopathic DCM or other nonhypertrophic cardiomyopathies (e.g., restrictive cardiomyopathies) underwent exome sequencing between April 2016 and September 2017. Criteria for offering exome sequencing were (1) diagnosis under the age of 40, or (2) had a family history (>2 members per family) of DCM and/or early (<35 years) sudden unexplained death. Patients with previously recorded hypertension were included if it was thought that mildly elevated blood pressure might be an amplifier of the phenotype rather than the primary cause of the DCM. Eligible patients were enrolled within Melbourne, Australia from four tertiary cardiovascular centers. Variants were rigorously classified based on principles outlined by the American College of Medical Genetics and Genomics (ACMG) standards for interpretation of sequence variants.16 Variant classifications were reviewed in a multidisciplinary team meeting attended by clinical geneticists, cardiologists, genetic counselors, medical scientists, and bioinformaticians. For the purposes of the cost-effectiveness modeling, and given the uncertainty and lack of evidence on how the condition is likely to progress in people with variants of unknown significance, a conservative dichotomous assumption was used to indicate that a pathological variant is either found or not found.
All dollar amounts are Australian Dollars.
This study seeks to model the benefits of genetic testing for DCM on family members based on adult clinical data collected from the Melbourne Genomics DCM study and evidence published in peer reviewed literature. An expected value analysis was applied on a hypothetical cohort of clinically unaffected first-degree relatives whose proband had a clinical diagnosis of DCM. The starting age was 18 years and the cohort was tracked annually until death. Similar to other studies in the cardiac genetic literature,17 it was assumed that cascade testing would be undertaken at the age of 18 to remove ethical concerns related to predictive genetic testing of children. The study received Human Research Ethics Committee approval (HREC/13/MH/326) and complied with the Declaration of Helsinki. Participants provided written informed consent.
Decision model
A model-based cost–utility analysis was undertaken to compare the cost-effectiveness of cascade genetic testing in asymptomatic relatives of a patient with DCM compared with periodical clinical surveillance based on the outcome of cost per additional quality-adjusted life year (QALY). A QALY expresses the quality and quantity of life in a common metric and allows for comparison across diseases and conditions and it is the recommended outcomes measure for economic evaluation in Australia18 and abroad.19 The analysis was undertaken from the perspective of the Australian health-care system. All costs are presented in 2018 Australian dollars (currently 0.63 euro, 0.72 US dollars). An annual discount rate of 5% was applied for both costs and outcomes per Australian Pharmaceutical Benefits guidelines.18 A lifetime horizon was adopted and when more than one source was available to inform model parameters, evidence was synthesized using Bayesian methods.
Model structure
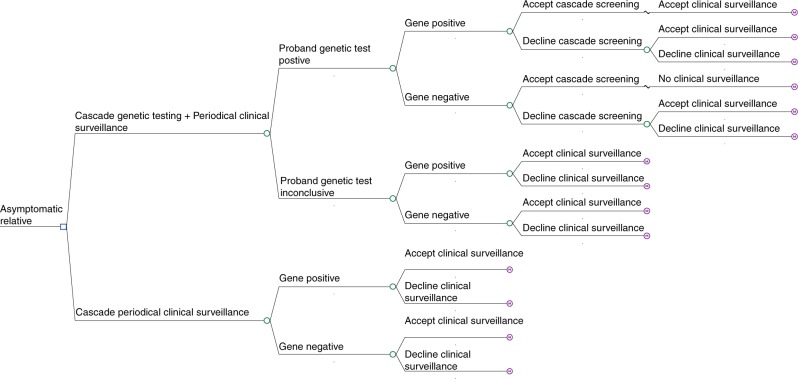
A decision-analytic model, combining a decision tree and a Markov model, was used to describe the options being compared and the possible pathways following them (Fig. 1). The decision tree replicates the cascade genetic and periodical clinical surveillance pathways faced by a relative of a DCM diagnosed proband. Cascade genetic testing is offered to those relatives whose proband returns a positive genetic diagnosis. Given the evidence suggesting that not all relatives take up genetic testing, the model allowed 40% of relatives to accept cascade testing.20,21 For these relatives, an identification of a pathogenic variant will mean a lifetime of periodical clinical surveillance. If a variant is not identified, relatives are at a population-level risk of developing the condition and, therefore, will be exempt from clinical surveillance. For relatives who have agreed to take up genetic testing it was assumed that they would then accept periodic surveillance in the presence of a positive test result. In the “no genetic testing” arm and the arms for which a genetic diagnosis has not been confirmed, relatives have the option to accept or decline periodical clinical surveillance. Evidence suggests that about 48% of first-degree relatives are expected to accept clinical surveillance in the presence of an unknown proband genetic diagnosis.20,21
Fig. 1.
Decision tree of genetic testing and periodical clinical surveillance versus periodical clinical surveillance alone.
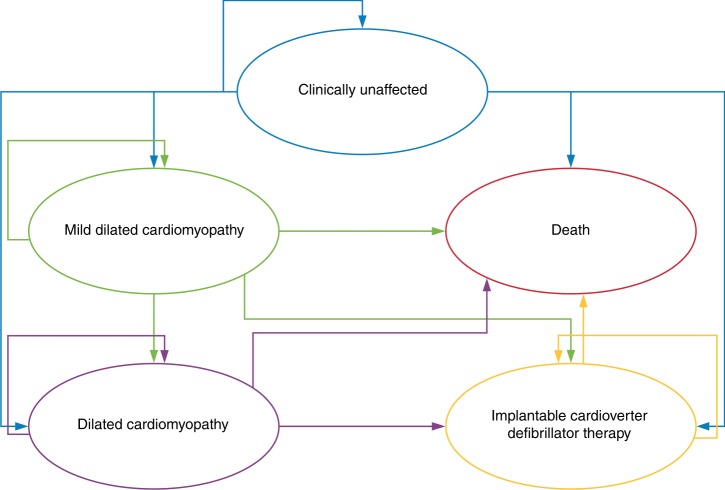
Subsequently, the natural history of relatives was simulated in annual cycles using a Markov model. Relatives with a pathogenic variant started in a clinically unaffected state and could either remain unaffected or move into one of three states: preclinical/mild DCM (MDCM), a state with features of early DCM with a normal sized or mildly dilated left ventricle;22 DCM; and death. MDCM and DCM were health states that encompassed the gradual decline of a patient’s health status associated with progression of cardiomyopathy until they received advanced treatments such as an implantable cardioverter defibrillator (ICD), or died from sudden cardiac death (SCD) or from other causes. The model structure of patients with a pathogenic variant clinical pathway is shown in Fig. 2. Patients without a pathogenic variant enter a Markov model that has two arms: alive or death from other causes. The model was developed in TreeAge Pro 2018 software.
Fig. 2.
Markov model of the natural history of a patient with a genetic diagnosis of dilated cardiomyopathy. The natural history of disease was simulated using a Markov model. Every year patients can remain in their current risk state, progress to mild dilated cardiomyopathy, dilated cardiomyopathy, or death. When patients are in the mild dilated cardiomyopathy or dilated cardiomyopathy health state they can progress to the implantable cardioverter defibrillator state or the death state (sudden cardiac death or other causes).
Probabilities
In the analysis it was assumed that familial DCM occurs in 35% of idiopathic DCM cases. This average estimate is based on clinical and family studies in DCM summarized in a review by Burkett et al.23 Recent genetic studies on patients with familial DCM have identified a monogenic cause in 17.4–50% of patients.10,24 In the Melbourne Genomics DCM study, as a result of a more stringent classification (in particular with the classification of truncating TTN variants), the need to establish gene–disease associations, and the inclusion of patients without a family history of DCM, the diagnostic rate of finding a monogenic cause was 13.7%. We therefore estimated that with the current state of evidence, a causative variant cannot be established in the remaining 21.3% (familial prevalence of 35% in DCM pathogenic variant yield 13.7%) of individuals with DCM.
Assuming an autosomal dominant transmission, first-degree relatives have a 50% chance of inheriting a pathogenic variant, and therefore, only 17.5% of our modeled cohort was expected to have a monogenic cause of DCM (i.e., 50% × 35%). In a cohort of first-degree relatives, about 7% (i.e., 13.7% × 50%) would be expected to have an identifiable variant. Health state transition probabilities were estimated based on various published data sources, representative of an at-risk individual of DCM (Table S1). The probability of receiving an ICD was based on a study by Gigli et al.22 and encompassed both the probability of receiving an ICD or cardiac resynchronization therapy (CRT). The annual risk of all-cause mortality was based on Australian life tables obtained from the Australian Bureau of Statistics.25
The only distinction between genetic testing and periodical clinical surveillance in terms of clinical outcomes was for the patients who were receiving periodical clinical surveillance relative to those who were not. It was expected that an individual who agrees to genetic testing and is gene positive would also agree to periodical clinical surveillance. A relative risk difference of 1.9 for SCD was applied between gene-positive family members receiving periodical clinical surveillance compared with their counterparts who were not once DCM symptoms have been developed.26
Outcomes
Age group–specific Australian population utility data were applied to clinically unaffected and gene-negative health states.27 A gene-negative diagnosis (no pathogenic variant identified) has been linked with a utility gain.17 We assumed that the utility gain is temporary (lasting a year) with individuals adapting and returning to standard population mean scores. DCM utility values were sourced from a study by Ingles et al.,28 in patients with cardiac genetic diseases (Table S2). The study also collected information on at-risk relatives. Participants completed the Short Form-36 (SF-36) measure and their responses were converted to SF-6D utility scores using Australian population norm utility weights.27 To estimate QALYs, utility scores were combined with estimates of the duration within the different health states.
Costs
Australian health-care perspective was adopted and costs were sought from the National Hospital Cost Data Collection Report Round 20 (2015–2016),29 the Australian Medicare Benefits Schedule30 and Pharmaceutical Benefits Schedule,31 and the Victorian Clinical Genetics Services price list (available from vcgs@vcgs.org.au). These are shown in Table S3. Where necessary, costs were inflated to 2018 dollars.
The cost of performing exome sequencing with analysis of up to 100 genes in a proband was $1200 and the initial and follow-up genetic counseling costs were $184 and $147 respectively (Table S3).32 These costs were spread across the number of relatives that the proband’s genetic test results would benefit. Based on data collected as part of the Melbourne Genomics DCM study, the number of first-degree relatives who could benefit from a proband’s genetic test results was five. The cost of cascade genetic testing incurred by a relative was $250 and a relative incurred the same genetic counseling costs as a proband.
Clinical surveillance involved consultation with a cardiologist, electrocardiography, and echocardiography and the costs are listed in Table S3. Surveillance occurred every 2 years and the sensitivity analysis accounted for clinical surveillance occurring with a frequency of 1 to 5 yearly.33 Event costs for ICD/CRT implant and SCD were also included using unit cost estimates from National Hospital Cost Data Collection Report Round 20 (2015–2016).29 The cost of the ICD/CRT implant was weighted based on the rates observed in the Gigli et al. study.22 The annual cost of the pharmacological management of DCM included the average costs of guideline-directed medical therapy.34 These include combination therapy with a cardioselective β-blocker and renin–angiotensin system inhibition with either angiotensin-converting enzyme (ACE) inhibitors, angiotensin receptor blockers or angiotensin receptor–neprilysin inhibitors and/or aldosterone antagonists with loop diuretics. The annual cost of managing MDCM included the average medication dose and costs of an ACE inhibitor.34 No medications were assumed for individuals at risk of DCM.
Cost-effectiveness analyses
Results were presented using incremental cost-effectiveness ratios (ICERs). Base-case analysis presented results for a cohort made up of only first-degree relatives. In a sensitivity analysis, the cost-effectiveness of cascade testing in first and second-degree relatives was assessed. For this analysis, the rates of genetic diagnosis and test acceptance rates changed based on the relationship of the relative but the clinical pathway probabilities of a genetic DCM patient remained the same. The composition of the modeled cohort including both first and second-degree relatives was assumed to be made up of 70% first-degree relatives to reflect on average the published evidence.20,21,35 Given the Melbourne Genomics DCM study identified that exome sequencing would benefit five first-degree relatives, it was assumed that the number of first and second-degree relatives who could benefit from a proband’s exome sequencing results was seven.
Sensitivity analyses were conducted to explore the robustness of the results to plausible variations in key assumptions and variations in the analytical methods used, and to consider the broader issue of the generalizability of the results. One-way sensitivity analyses explored how ranging key model inputs over plausible ranges impacted on model results using a tornado diagram. The probabilistic sensitivity analysis explored the effect of the uncertainty around the mean values of each model parameter on model results using a Monte Carlo simulation. Within each simulation, parameter values were randomly drawn from relevant probability distributions. Cost parameters were given a gamma distribution, while utilities and transition probabilities were given a beta distribution.36 The proportion of probands with a monogenic cause and the proportion of probands identified by exome sequencing were given a beta distribution. Where standard errors were required and were not reported in the sources used, it was assumed the standard error was equal to the mean.36 The lifetime costs and effects of an at-risk relative were simulated 10,000 times using Monte Carlo second-order simulations. To present the proportion of simulations that favored cascade genetic testing over clinical surveillance alone at each threshold of willingness to pay, cost-effectiveness acceptability curves (CEACs) were used.36 A CEAC summarizes the decision uncertainty in the cost-effectiveness results by showing the probability that each intervention is cost-effective across different thresholds of willingness to pay per additional QALY.
RESULTS
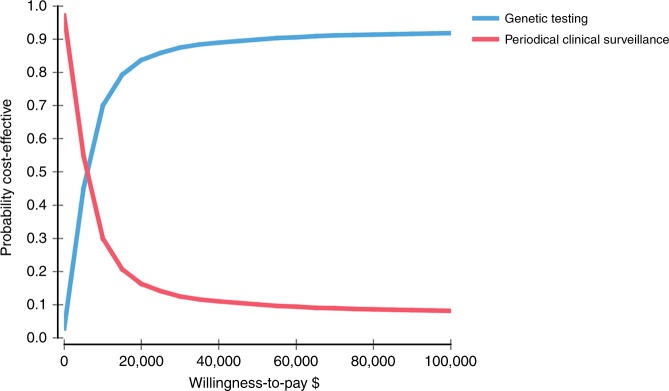
The estimated costs and QALYs for patients undergoing the different diagnosis strategies are presented in Table 1. The addition of cascade genetic testing to guide periodical clinical surveillance of first-degree relatives in probands with DCM resulted in a cost of $3600 per relative. This cost was estimated to be $300 larger per relative compared with periodical clinical surveillance alone ($3300). Cascade genetic testing as a guide for clinical surveillance also led to approximately 15 QALYs per first-degree relative. This was 0.04 QALYs more for genetic testing compared with the estimated benefit of periodical clinical surveillance alone. The incremental cost per QALY of cascade genetic testing compared with periodical clinical surveillance alone was estimated at approximately $6100 for first-degree relatives. This estimate falls well below the established $40,000–$70,000 cost-effective threshold observed in Australian health economic literature.37 Fig. 3 shows the results of the probabilistic sensitivity analysis. At a threshold of $50,000 per QALY, the probability that genetic testing with clinical surveillance is cost-effective compared with clinical surveillance alone is 90%.
Table 1.
Cost-effectiveness results
| Genetic testing | Periodical clinical surveillance | |
|---|---|---|
| Main analysis (first-degree relatives) | ||
| Expected cost per relativea | $3600 | $3300 |
| Expected QALYs per relativea | 14.96 | 14.92 |
| Expected ICER per relative | $6100 | |
| Sensitivity analysis (first and second-degree relatives) | ||
| Expected cost per relative | $3100 | $2900 |
| Expected QALYs per relative | 14.84 | 14.80 |
| Expected ICER per relative | $5100 | |
| Probabilistic sensitivity analysis (first-degree relatives) | ||
| Cost per relativea | $3600 ($1600) | $3400 ($1600) |
| QALY per relativea | 14.98 (2.00) | 14.94 (2.00) |
| ICER per relative | $6000 | |
ICER incremental cost-effectiveness ratio, QALY quality-adjusted life year.
a(Standard deviation). All dollar amounts are Australian Dollars.
Fig. 3.
Cost-effectiveness acceptability curve illustrating the probability of cost-effectiveness of genetic testing and periodical clinical surveillance versus periodical clinical surveillance alone for a given cost-effectiveness threshold value for first-degree relatives.
Cascade genetic testing of both first and second-degree relatives resulted in a reduction of costs for the different comparators ($3100 for genetic testing versus $2900 for clinical surveillance alone) and in a difference of $200 per relative. The same QALY gain of 0.04 QALYs was observed for cascade testing of first and second-degree compared with first-degree relatives only. The incremental cost per QALY was estimated at approximately $5100.
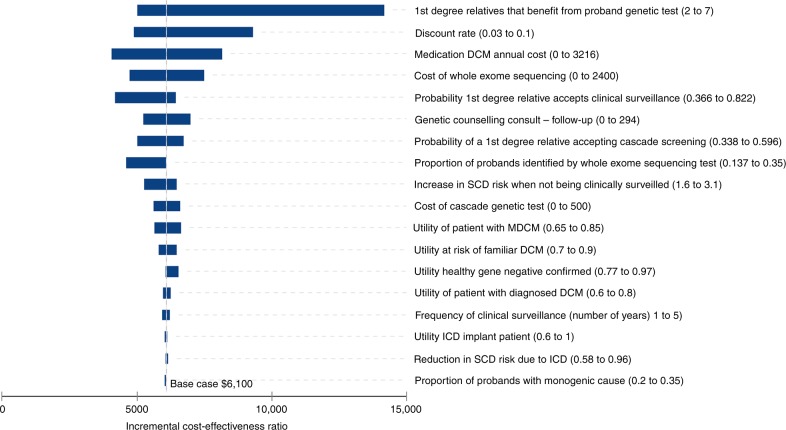
The results of one-way sensitivity analyses are presented in the tornado diagram of Fig. 4 and Table S4. The results suggest that the model may be sensitive to certain parameters but still supports base-case analysis. The results were sensitive to the number of relatives who benefit from proband exome sequencing as well as its cost and detection rate. The detection rate of probands identified by exome sequencing was tested to an upper bound limit of 35%. This represented a scenario in which exome sequencing was able to identify all probands carrying an underlying monogenic cause for DCM. When the detection rate rose, the cost-effectiveness of cascade testing increased.
Fig. 4.
Tornado plot of the results of the univariate sensitivity analysis. Vertical line is at $6100 per quality-adjusted life year and represents the base-case analysis. Refer to sensitivity analysis in supplementary table 4 for actual values. DCM dilated cardiomyopathy, ICD implantable cardioverter defibrillator, MDCM mild DCM, SCD sudden cardiac death.
DISCUSSION
This study assessed the cost-effectiveness of cascade genetic testing in asymptomatic relatives of a patient with DCM compared with periodical clinical surveillance. The strength of genetic testing is its potential to discharge those without a disease-causing variant from a lifetime of clinical surveillance. Our main results showed that cascade genetic testing following exome sequencing in the proband is very likely to be cost-effective compared with periodical clinical surveillance alone. Similarities in the cost of genetic testing and the management guidelines for the treatment of DCM,6,34 as well as extensive sensitivity analyses conducted as part of the economic evaluation, are likely to support the generalizability of our findings to other contexts, such as the United States and Europe.
Similar to our findings, other studies have also found that the addition of cascade genetic testing to routine clinical surveillance is cost-effective in the care of families with other forms of cardiomyopathy in Australia and the UK.17,38 In comparison with these studies, this analysis did not assume that all relatives would undergo predictive genetic testing if it was available, or that they would all attend for regular clinical surveillance in the absence of a genetic diagnosis. The inclusion of “real-world” acceptance rates for cascade genetic testing and periodical clinical surveillance was crucial in reflecting the closest costs and benefits of genetic testing for DCM. This may be a reason as to why cost-effectiveness estimates in this study are substantially larger, although still well below the established threshold of willingness to pay per additional QALY threshold in Australia. The higher costings could also be attributed to the lower monogenic origin of DCM compared with other genetic conditions such as hypertrophic cardiomyopathy (HCM). Previous modeling studies for HCM have implemented a diagnostic yield of 63% (refs. 17,38), although recent evidence suggests that diagnostic yield in HCM ranges between 32% and 42% (refs. 15,39). In this analysis a diagnostic yield of 13.7% for DCM was used. These studies also modeled less expensive and less advanced tests rather than the exome sequencing that was used in our analysis.
DCM genetic testing is recommended for a subset of patients who have DCM and significant cardiac conduction disease and/or a family history of premature unexpected sudden death.5 In contrast, genetic testing is recommended for any patient with an established diagnosis of HCM.40 These recommendations were published 8 years ago and do not take into account the recent improved analytical approaches with the use of next-generation sequencing strategies with associated lower costs. Indeed, our findings indicate that genetic testing in patients with DCM is very likely to be cost-effective and thus should be offered to more patients with DCM than is currently recommended.
A limitation of genetic testing for DCM in clinical practice is the diagnostic rate of exome sequencing. The value used within the model was based on the Melbourne Genomics DCM study. If a proband returns an uninformative genetic finding this means that unless their variant is identified in the future, relatives are resigned to a lifetime of clinical surveillance, a similar situation to if exome sequencing for the proband was not available at all. When varying the diagnostic rate in the sensitivity analysis across plausible ranges, genetic testing remained cost-effective. It is important to consider that the 13.7% diagnostic rate used in this study is based on data collected as part of the Melbourne Genomics DCM study. In this study, testing was only offered to those diagnosed under the age of 40 or those with a known family history of DCM or early SCD. If genetic testing for DCM was adopted as part of routine care, variation could be expected surrounding the diagnostic pick-up rate.
There are several model limitations to consider. It was assumed that a pathological variant was either found or not found. The effect of additional costs related to genetic testing, such as segregation costs, on model results was captured though the sensitivity analyses. Even when the costs were raised to $2400, genetic testing of first-degree relatives still remained highly cost-effective. Currently no published evidence is available to suggest that cardiac genetic testing results in improved clinical outcomes. For relatives, a gene-negative diagnosis means a relative no longer requires periodical clinical surveillance, however, the possible benefits of improved clinical outcomes when a pathogenic variant is identified remain uncertain. In the model, the likelihood of receiving an ICD or the rate of progression from MDCM to DCM did not vary between those who had a genetic diagnosis confirmed and those who did not. The model did however make a distinction between those who were receiving periodical clinical surveillance and those who were not. It was assumed that any individual who agreed to genetic testing and had a gene-positive diagnosis would agree to periodical clinical surveillance. Therefore, the prevention of SCD due to uptake of periodical clinical surveillance was an important focus in our model. A relative risk increase of 1.9 for SCD was based on a study by Grunig et al. that analyzed whether periodical clinical surveillance improved survival outcomes in those with a diagnosis of DCM.26 The relative risk increase was a very conservative estimate and is the only distinction in outcomes between genetic testing and periodical clinical surveillance. As new evidence in the personalized treatment of at-risk individuals becomes available, the cost-effectiveness of cascade testing is expected to increase.
The utility scores used to inform the effects of each health state were the same as those used in the study by Ingles et al.17 These values were based on Australian patients and their families using the SF-36.28 Unlike that study, our model only accounted for a utility improvement higher than general population norms in the first year following a pathogenic variant–negative diagnosis, before reverting back to normal age-dependent population levels. Uncertainty around the utility estimates was explored in deterministic and probabilistic sensitivity analyses, and all reinforced the cost-effectiveness of cascade genetic testing. Further research, using different patient-reported outcome measures, is needed to determine how DCM impacts on broader aspects of a person’s quality of life and wellbeing.
In conclusion, this article suggests that the addition of genetic testing in asymptomatic relatives of patients with DCM to guide periodical clinical surveillance is cost-effective in comparison with periodical clinical surveillance alone. As the DCM pathogenic variant detection rate rises and new evidence for personalized treatment of at-risk individuals becomes available, the cost-effectiveness of cascade testing will further increase. This has important implications for the evaluation of DCM and suggests that those with a family history of the condition should have improved access to specialized cardiac genetic services.
Supplementary information
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-019-0582-2) contains supplementary material, which is available to authorized users.
References
- 1.van Riet EE, Hoes AW, Wagenaar KP, Limburg A, Landman MA, Rutten FH. Epidemiology of heart failure: the prevalence of heart failure and ventricular dysfunction in older adults over time. A systematic review. Eur J Heart Fail. 2016;18:242–252. doi: 10.1002/ejhf.483. [DOI] [PubMed] [Google Scholar]
- 2.Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 4.Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 5.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 6.NHFA CSANZ Heart Failure Guidelines Working Group. Atherton JJ, Sindone A, et al. National Heart Foundation of Australia and Cardiac Society of Australia and New Zealand: guidelines for the prevention, detection, and management of heart failure in Australia 2018. Heart Lung Circ. 2018;27:1123–1208. doi: 10.1016/j.hlc.2018.06.1042. [DOI] [PubMed] [Google Scholar]
- 7.Hershberger RE, Givertz MM, Ho CY, et al. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2018;20:899–909. doi: 10.1038/s41436-018-0039-z. [DOI] [PubMed] [Google Scholar]
- 8.Bondue A, Arbustini E, Bianco A, et al. Complex roads from genotype to phenotype in dilated cardiomyopathy: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res. 2018;114:1287–1303. doi: 10.1093/cvr/cvy122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas J, Frese KS, Peil B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–1135a. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 10.Lakdawala NK, Funke BH, Baxter S, et al. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 12.Morales A, Hershberger RE. The rationale and timing of molecular genetic testing for dilated cardiomyopathy. Can J Cardiol. 2015;31:1309–1312. doi: 10.1016/j.cjca.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Spaendonck-Zwarts KY, van Rijsingen IA, van den Berg MP, et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years’ experience. Eur J Heart Fail. 2013;15:628–636. doi: 10.1093/eurjhf/hft013. [DOI] [PubMed] [Google Scholar]
- 14.Fatkin D, members of the CSANZ Cardiac Genetic Diseases Council Writing Group. Guidelines for the diagnosis and management of familial dilated cardiomyopathy. Heart Lung Circ. 2011;20:691–693. doi: 10.1016/j.hlc.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2016;19:192. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingles J, McGaughran J, Scuffham PA, Atherton J, Semsarian C. A cost-effectiveness model of genetic testing for the evaluation of families with hypertrophic cardiomyopathy. Heart. 2012;98:625–630. doi: 10.1136/heartjnl-2011-300368. [DOI] [PubMed] [Google Scholar]
- 18.Pharmaceutical Benefits Advisory Committee (PBAC). Guidelines for preparing submissions to the Pharmaceutical Benefits Advisory Committee. Australian Government, Department of Health and Ageing, Canberra: Australia, 2008.
- 19.National Institute for Health and Care Excellence (NICE). Guide to the methods of technology appraisal. NICE, London: UK, 2013. [PubMed]
- 20.Christiaans I, Birnie E, Bonsel GJ, Wilde AA, van Langen IM. Uptake of genetic counselling and predictive DNA testing in hypertrophic cardiomyopathy. Eur J Hum Genet. 2008;16:1201–1207. doi: 10.1038/ejhg.2008.92. [DOI] [PubMed] [Google Scholar]
- 21.Miller EM, Wang Y, Ware SM. Uptake of cardiac screening and genetic testing among hypertrophic and dilated cardiomyopathy families. J Genet Couns. 2013;22:258–267. doi: 10.1007/s10897-012-9544-4. [DOI] [PubMed] [Google Scholar]
- 22.Gigli M, Stolfo D, Merlo M, et al. Insights into mildly dilated cardiomyopathy: temporal evolution and long‐term prognosis. Eur J Heart Fail. 2017;19:531–539. doi: 10.1002/ejhf.608. [DOI] [PubMed] [Google Scholar]
- 23.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–981. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 24.Minoche AE, Horvat C, Johnson R, et al. Genome sequencing as a first-line genetic test in familial dilated cardiomyopathy. Genet Med. 2019;21:650–662. doi: 10.1038/s41436-018-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Australian Bureau of Statistics. Life tables, Australia. Canberra, Australia; 2015–2017. https://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/3302.0.55.0012015-2017?OpenDocument. Accessed July 2018.
- 26.Grunig E, Benz A, Mereles D, et al. Prognostic value of serial cardiac assessment and familial screening in patients with dilated cardiomyopathy. Eur J Heart Fail. 2003;5:55–62. doi: 10.1016/S1388-9842(02)00179-4. [DOI] [PubMed] [Google Scholar]
- 27.Norman R, Church J, van den Berg B, Goodall S. Australian health-related quality of life population norms derived from the SF-6D. Aust N Z J Public Health. 2013;37:17–23. doi: 10.1111/1753-6405.12005. [DOI] [PubMed] [Google Scholar]
- 28.Ingles J, Yeates L, Hunt L, et al. Health status of cardiac genetic disease patients and their at-risk relatives. Int J Cardiol. 2013;165:448–453. doi: 10.1016/j.ijcard.2011.08.083. [DOI] [PubMed] [Google Scholar]
- 29.Australian Government. National Hospital Cost Data Collection Cost Report Round 20 (2015-2016). Canberra, Australia 2015–2016. https://www.ihpa.gov.au/publications/national-hospital-cost-data-collection-public-hospitals-cost-report-round-20-0. Accessed August 2018.
- 30.Medicare Australia. Medicare Benefits Schedule (MBS) item statistics reports. 2018. http://medicarestatistics.humanservices.gov.au/statistics/mbs_item.jsp. Accessed July 2018.
- 31.Medicare Australia. Pharmaceutical Benefits Schedule item reports. 2018. http://medicarestatistics.humanservices.gov.au/statistics/pbs_item.jsp. Accessed July 2018.
- 32.Stark Z, Lunke S, Brett GR, et al. Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med. 2018;20:1554–1563. doi: 10.1038/gim.2018.37. [DOI] [PubMed] [Google Scholar]
- 33.The Cardiac Society of Australia and New Zealand. Guidelines for the diagnosis and management of familial dilated cardiomyopathy 2013. https://www.csanz.edu.au/wp-content/uploads/2014/12/Familial-Dilated-Cardiomyopathy_2013-November.pdf. Accessed July 2018.
- 34.Yancy CW, Jessup M, Bozkurt B, et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. J Am Coll Cardiol. 2017;70:776–803. doi: 10.1016/j.jacc.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Mahon NG, Murphy RT, MacRae CA, Caforio AL, Elliott PM, McKenna WJ. Echocardiographic evaluation in asymptomatic relatives of patients with dilated cardiomyopathy reveals preclinical disease. Ann Intern Med. 2005;143:108–115. doi: 10.7326/0003-4819-143-2-200507190-00009. [DOI] [PubMed] [Google Scholar]
- 36.Briggs A, Sculpher M, Claxton K. Decision modelling for health economic evaluation. Oxford, UK: Oxford University Press; 2006. [Google Scholar]
- 37.George B, Harris A, Mitchell A. Cost-effectiveness analysis and the consistency of decision making: evidence from pharmaceutical reimbursement in Australia (1991 to 1996) Pharmacoeconomics. 2001;19:1103–1109. doi: 10.2165/00019053-200119110-00004. [DOI] [PubMed] [Google Scholar]
- 38.Wordsworth S, Leal J, Blair E, et al. DNA testing for hypertrophic cardiomyopathy: a cost-effectiveness model. Eur Heart J. 2010;31:926–935. doi: 10.1093/eurheartj/ehq067. [DOI] [PubMed] [Google Scholar]
- 39.Bagnall RD, Ingles J, Dinger ME, et al. Whole genome sequencing improves outcomes of genetic testing in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2018;72:419–429. doi: 10.1016/j.jacc.2018.04.078. [DOI] [PubMed] [Google Scholar]
- 40.Authors/Task Force members. Elliott PM, Anastasakis A, et al. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2733–2779. doi: 10.1093/eurheartj/ehu283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.