Abstract
Lithium (Li) is a medication long-used to treat bipolar disorder. It is currently under investigation for multiple nervous system disorders, including Alzheimer’s disease (AD). While perturbation of RNA levels by Li has been previously reported, its effects on the whole transcriptome has been given little attention. We, therefore, sought to determine comprehensive effects of Li treatment on RNA levels. We cultured and differentiated human neuroblastoma (SK-N-SH) cells to neuronal cells with all-trans retinoic acid (ATRA). We exposed cultures for one week to lithium chloride or distilled water, extracted total RNA, depleted ribosomal RNA and performed whole-transcriptome RT-sequencing. We analyzed results by RNA length and type. We further analyzed expression and protein interaction networks between selected Li-altered protein-coding RNAs and common AD-associated gene products. Lithium changed expression of RNAs in both non-specific (inverse to sequence length) and specific (according to RNA type) fashions. The non-coding small nucleolar RNAs (snoRNAs) were subject to the greatest length-adjusted Li influence. When RNA length effects were taken into account, microRNAs as a group were significantly less likely to have had levels altered by Li treatment. Notably, several Li-influenced protein-coding RNAs were co-expressed or produced proteins that interacted with several common AD-associated genes and proteins. Lithium’s modification of RNA levels depends on both RNA length and type. Li activity on snoRNA levels may pertain to bipolar disorders while Li modification of protein coding RNAs may be relevant to AD.
Subject terms: Neurodegeneration, Translational research
Introduction
Lithium (Li) has been used to effectively treat bipolar disorder for more than 60 years1. Li may affect cellular signaling processes and promote long-term neuroplasticity2,3. Li also seems to have neurotrophic properties affecting cell survival and apoptosis mechanisms4, and trace levels may to some extent prevent dementia5, suicide6, and homicide7. The breadth of effects for Li suggest that the metal may be a trace nutrient8. In particular, Li levels in drinking water correlated with reduced age-adjusted Alzheimer’s disease (AD) mortality9 and incidence10. Several investigations of using Li to treat AD or specific symptoms associated with AD are ongoing11–14. However, Li toxicity produces Parkinson’s disease (PD) - and AD-like outcomes. Li is currently under active investigation for a broad array of psychiatric and neurological disorders (including AD, Niemann-Pick disease, frontotemporal dementia, and ALS), as well as gastrointestinal disease, neoplasia, and other non-neurological conditions. As of June 11, 2019, the US National Institutes of Health (NIH) lists 141 planned, recruiting, or active clinical trials for Li effects on psychiatric and nervous system disorders, along with an additional 104 for conditions that do not involve the nervous system (http://www.clinicaltrials.gov).
The mechanisms underlying lithium therapeutic activity are still not clear15,16. Three principal hypotheses are its action on i) cyclic AMP17,18, ii) inositol depletion19, and iii) the inhibition of protein kinases, including glycogen synthase kinase, with subsequent activation of the wnt neurodevelopmental pathway20,21. Li also increases levels of the anti-apoptotic protein Bcl2 in the frontal cortex of rat brain22. Li displays general neuroprotective effects, including against excitotoxic lesions23 as well as protection against β-amyloid induced cell death24. Li increases N-acetyl-aspartate levels in human brain (a measure of neuronal viability) during therapeutic treatment, as measured by magnetic resonance spectroscopy25. This increase appears to be related to increases in grey matter volume26. Li, similarly to antidepressants, increases hippocampal neurogenesis27. Nevertheless, multiple clinical effects of Li are not entirely explained by prevailing theories28. Use of neuronally-differentiated pluripotent stem cells from bipolar patients who were responsive to Li vs non-responders revealed that neuronal hyperactivity responded to Li in responders but not in non-responders29,30. It should be noted that these cells, even though taken from living patients, represent early developmental stages of central nervous system (CNS) cells. Changes in adult neuronal cells from Li-responsive patients may be related to decreased phosphorylation of collapsin response mediator protein-2 (CRMP2) and increased dendritic spine density31.
Since so many pathways are apparently involved in Li treatment of bipolar disorder, and the pathways associated with possible Li treatment of other disorders, such as AD, are yet unknown, a reasonable question is how Li alters the transcriptome. Gene expression changes induced by Li treatment exist in both neuronal cultures and animals32–35. However, those reports were array-based assays, not whole transcriptome surveys. If a specific RNA were not a priori included on an array, Li effects on it would not be measured. Thus, we performed whole transcriptome sequencing of Li-treated vs. untreated cells. Neuronal cell lines have the advantage of standardization of biochemical characteristics, ease of adaptation to multiple experimental protocols, and simulation of adult CNS cells’ chemical and physiologic characteristics. We found that Li changes expression of multiple small RNA species, particularly non-coding small nucleolar RNAs (snoRNA), in all-trans retinoic acid (ATRA) differentiated human neuroblastoma neuronal (SK-N-SH) culture, but that small RNA species do not all respond to Li stimulation equally.
The snoRNAs may function in multiple neurobiological disorders and symptoms, including catatonia36. snoRNA levels differed between schizophrenic and control subjects in the anterior cingulate cortex37,38. snoRNA may also play a role in some features of autism39,40. Finally, specific snoRNAs undergo progressive changes with age41, and snoRNA levels vary with progressing Braak stages in AD42. We, therefore, investigated potential connections among Li-perturbed genes and AD-associated genes and proteins. Our results generated interaction networks that can inform future mechanistic research, relevant to AD, bipolar and other neuropsychiatric disorders.
Results
Cell culture response
No gross differences in cell morphology or survival were noted.
Small RNAs, particularly small nucleolar RNAs are over-represented among Li-influenced genes
We treated neuronally differentiated human cell cultures with Li and measured relative change vs. vehicle (distilled water)-treated cells in RNA levels, detected by whole-transcriptome sequencing. We found that log lengths of whole transcriptome sequencing had a bimodal distribution. Parameterized Gaussian mixture model43,44, cluster centers corresponded to 87 nucleotides (range 22 to 213) and 3047 nucleotides (range 233 to 116,854) in length (Fig. 1A, Table 1). Li significantly altered the expression of 207 RNA species of 15,394 sequenced (Supplementary Table 1) at a false discovery rate (FDR) ≤ 0.2, making Li influence an uncommon event. We did not consider magnitude of influence in our analysis, although the lowest absolute magnitude of change for RNA sequences with FDR ≤ 0.2 was −20% (Supplementary Table 2). Comparing the distribution of altered vs. non-altered sequences indicated that shorter RNA sequences predominated in the Li-influenced group (Fig. 1B), while relative distribution of non-influenced sequence lengths was more similar to the distribution of all sequence lengths (Fig. 1C). Of the 207 Li-influenced RNAs, 103 (49.8%) were in the “short” cluster vs 1031 of 15187 (6.8%) non-influenced RNAs.
Figure 1.

Size distribution of transcriptome, and relative distributions of Li-influenced vs. non-influenced transcriptome, and mean log-lengths of Li-influenced/-non-influenced RNAs. Both histograms and kernel density estimations44 of the distributions are shown. (A) Sequence frequencies by logarithms of sequence lengths. Log-lengths have a bimodal distribution with parameterized Gaussian mixture model clusters indicated as “Short” and “Long” and specific parameters in Table 1. (B–C) Sequences were separated by whether or not expression levels were altered by Li treatment. Relative frequencies within each group (Li-influenced vs. non-influenced) were calculated by dividing counts by each group’s respective total number of sequences and plotted vs log(length). (B) Frequency of Li-altered RNAs by length. Line is corresponding kernel density estimation. (C) Frequency of non-influenced RNAs by length. Line is corresponding kernel density estimation.
Table 1.
Gaussian mixture model clustering of transcriptome sequence lengths.
| Cluster | Center (log)a | Minimum (log) | Maximum (log) |
|---|---|---|---|
| Short | 87 (1.938) | 22 (1.342) | 213 (2.328) |
| Long | 3047 (3.484) | 233 (2.367) | 116,854 (5.068) |
aLog figures are mean of logs. All analysis was done on logs of lengths.
RNA species influenced response to Li
To investigate whether the difference simply reflected RNA transcript length or was also influenced by RNA species/type, we used logistic regression to model presence/absence of Li alteration by type of RNA and sequence length. The optimal model derived was “Li influenced ~ log(Length) + RNA Type”. We discovered an odds ratio (OR) of 0.729 +0.226/−0.171 (p ≤ 0.05) for log RNA length vs. Li influence, irrespective of RNA type, i.e., as RNA length decreased, likelihood of Li influence increased.
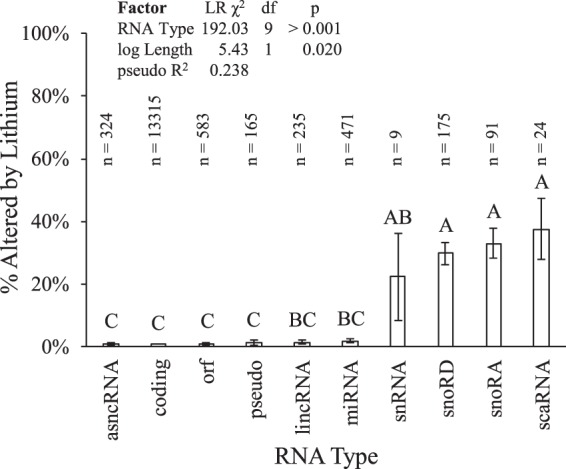
Of greater interest, we found that Li treatment had significantly different effects among RNA types/species, independent of sequence length. Two species (Small NF90 (ILF3) associated RNAs/snaRNA and vault RNA/vtRNA) were each represented in our sample by a single sequence. We excluded both RNA types from further analysis. When compared to overall levels of perturbation across all RNA types (effect coding), levels of 4 types were significantly more likely (OR > 1, p ≤ 0.05) to be altered by Li (Table 2), independently of RNA sequence length; specifically, small Cajal body-specific RNAs (scaRNA), 38% of scaRNA sequences; small nucleolar RNAs, H/ACA box (snoRA), 33% of snoRA sequences; small nucleolar RNAs, C/D box (snoRD), 30% of snoRD sequences; and small nuclear RNAs (snRNA), 22% of snRNA sequences. Levels of 4 types were significantly less likely (OR < 1, p ≤ 0.05) to be altered by Li treatment, specifically microRNA (miRNA), 1.7% of miRNA sequences; uncharacterized open reading frames (orf), 0.9% of orf sequences; protein coding RNA (coding), 0.7% of coding sequences; and antisense non-coding RNA (asncRNA), 0.6% of asncRNA sequences. To distinguish specific differences among the RNA species, we performed pairwise comparisons of ORs by RNA types, adjusted by FDR45 and found three overlapping groups (Fig. 2).
Table 2.
RNA types influenced by Li treatment.
| Speciesa | Total | Li Effect | percent ± SE | Logistic Modelingbc | Trendc. | |||
|---|---|---|---|---|---|---|---|---|
| altered | not | ORd | p | groupe | ||||
| snaRNAd | 1 | 0 | 1 | 0.00% ± 0.00% | na | na | na | na |
| asncRNA | 324 | 2 | 322 | 0.62% ± 0.44% | 0.21 +0.45/−0.17 | 0.023 | C | Down |
| coding | 13315 | 93 | 13222 | 0.70% ± 0.07% | 0.28 +0.24/−0.13 | <0.001 | C | Up |
| orf | 583 | 5 | 578 | 0.86% ± 0.38% | 0.29 +0.40/−0.19 | 0.009 | C | Down |
| pseudo | 165 | 2 | 163 | 1.21% ± 0.85% | 0.43 +0.92/−0.35 | 0.215 | C | Up |
| lincRNA | 235 | 3 | 232 | 1.28% ± 0.73% | 0.45 +0.79/−0.33 | 0.172 | BC | Up |
| miRNA | 471 | 8 | 463 | 1.70% ± 0.60% | 0.22 +0.27/−0.13 | <0.001 | BC | None |
| snRNA | 9 | 2 | 7 | 22.22% ± 13.86% | 3.47 +10.71/−2.89 | 0.110 | AB | Up* |
| snoRD | 175 | 52 | 123 | 29.71% ± 3.45% | 5.32 +4.76/−2.46 | <0.001 | A | Up* |
| snoRA | 91 | 30 | 61 | 32.97% ± 4.93% | 7.29 +6.03/−3.26 | <0.001 | A | Up* |
| scaRNA | 24 | 9 | 15 | 37.50% ± 9.88% | 9.95 +12.83/−5.76 | <0.001 | A | Up* |
| vtRNAd | 1 | 1 | 0 | 100.00% ± 0.00% | na | na | na | na |
| Sum | 15394 | 207 | 15187 | 1.36% | na | na | na | na |
aasncRNA: antisense noncoding RNA; coding: protein coding mRNA; lincRNA: long intergenic noncoding RNA; miRNA: micro-RNA; orf: uncharacterized RNA (open reading frame); pseudo: pseudogene RNA; scaRNA: small Cajal body specific RNA; snaRNA: small NF90 (ILF3) associated RNA; snoRA: small nucleolar RNAs, H/ACA box; snoRD: small nucleolar RNAs, C/D boxsnRNA: small nuclear RNA; vtRNA: vault RNA.
bModel coded to test the hypothesis of whether or coefficient differs from the mean of groups. It is appropriate for multiple pairwise comparisons.
cDerived from multinomial logistic modeling of Type + log(Length) vs. whether Li treatment significantly reduced, elevated, or had no effect on FC transcript levels. “*” indicates difference was significant at p < 0.05.
dOR is for effect of RNA type from the model (Li Effect) ~ log(Length) + (RNA Type). ± is 95% confidence intervals. OR marked “*” were significantly different from zero.
eMarginal means statistical group, FDR corrected. RNA species sharing letter did not significantly differ in odds of being altered by Li treatment, independent of RNA sequence length effect.
dExcluded from logistic model.
Figure 2.
Frequency/likelihood of Li alteration of RNA levels by type and relative distribution of small non-coding RNA species between Li-influenced vs. non-influenced “short” cluster RNAs. Estimated probabilities of alteration of RNA levels by RNA type, taking effect of transcript length into account. Estimated model log odds ratios were compared and pairwise comparison p values adjusted by FDR45. RNA types sharing a letter did not differ at p ≤ 0.05. Pseudo R2 is Efron’s.
We also investigated whether a given RNA species would be more likely to be upregulated or downregulated by Li treatment. We found that snRNA, snoRD, snoRA, and scaRNA were all significantly (p < 0.05) likely to have elevated levels after Li treatment (vs. reduced levels), while other RNA species had no significant effect (Table 3).
Table 3.
Li-influenced changes in RNA levels of genes implicated in AD.
| Gene | Product | Change | Ontologies | AD assoc | Co-exp. | PPI |
|---|---|---|---|---|---|---|
| BMP4 | bone morphogenetic protein 4 | +51% |
Regulation Cell comp.org Cell Proc Developmental Metabolic Processes |
83–85,109 | absent | present |
| PTMA | prothymosin α | +48% | 110 | present | present | |
| SOX5 | SRY box 5 | −45% |
Regulation Developmental Metabolic Processes |
111 | absent | present |
| RAB3A | ras-related protein 3A | −44% | 112,113 | present | present | |
| NRXN3 | neurexin-3 α | −43% | 88–90 | present | absent | |
| GLIS3 | GLIS family zinc finger 3 | −42% |
Developmental Multicellular Processes |
114,115 | present | present |
| HS6ST2 | heparan sulfate 6-O-sulfotransferase 2 | +41% | 116 | present | absent | |
| NMB | neuromedin B | +41% | 117–120 | present | absent | |
| HGF | hepatocyte growth factor | +38% | 121–124 | absent | present | |
| GPRC5B | G-protein coupled receptor family C group 5 member B | −34% |
Regulation Cell comp.org Immune Metabolic Proc Multicellular Processes Response to Stimulus |
125 | present | present |
| YAP1 | yes-associated protein 1 | −32% |
Regulation Metabolic Processes |
126 | present | present |
| SMAD6 | SMAD family member 6 | −29% |
Regulation Metabolic Processes Response to Stimulus Signaling |
127 | absent | present |
| GREM2 | gremlin 2 | −28% | 128 | present | absent | |
| IRS1 | insulin receptor substrate 1 | −28% | Regulation | 129 | present | present |
| IGFBP2 | insulin-like growth factor binding protein 2 | +28% | 130–133 | absent | present | |
| CUX2 | cut-like homeobox 2 | −27% |
Regulation Localization Metabolic Processes |
134 | absent | present |
| ZWINT | ZW10 interacting kinetochore protein | +27% | 135 | present | present | |
| PPARG | peroxisome proliferator-activated receptor γ | −26% |
Regulation Developmental Localization Metabolic Processes Response to Stimulus |
86,87 | present | present |
Li altered levels of specific protein-coding mRNAs and snoRNAs potentially associated with AD
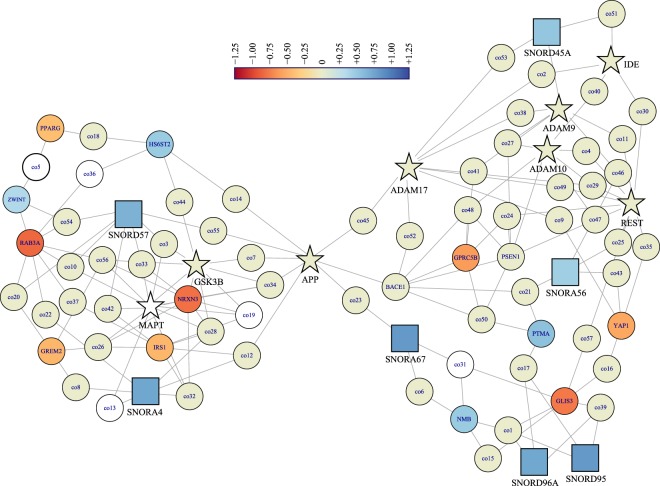
Although the majority of Li influence concentrated on small, noncoding RNAs, several protein-coding mRNAs influenced by Li in this study play a role in AD (Table 3). The genes are involved in several Gene Ontology biological functions, predominately biological regulation (GO:0065007) and metabolic processes (GO:0008152). We created two networks (human hippocampus gene co-expression and human hippocampus gene product interaction) using these genes plus a selection of “core” AD-related genes (Table 4). Networks showed several connections between the Li-influenced genes and AD-related genes. However, all such connections were second-degree, i.e., any AD-related gene did not co-express directly with a Li-influenced gene. Instead, a third gene co-expressed with both the AD-related gene and the Li-influenced gene (Fig. 3). On the other hand, when we built the protein-protein network, peroxisome proliferator-activated receptor γ (PPARγ), and SRY-related HMG-box 5 (SOX5) transcription factor directly interacted with the amyloid-β (Aβ) precursor protein (APP), along with SNORD14C (Fig. 4). Paradoxically, none of the snoRNAs associated with Braak staging (Table 5) appeared in either network. Thus, while Li may not directly modify expression of the better-known AD genes, a multi-target effect converging on APP could explain why micro-dosing may be effective, since APP could be the recipient of multiple outcome chains of Li activity.
Table 4.
“Core” AD genes compared to Li-influenced coding and snoRNA sequences.
| Genea | Productb | Functions |
|---|---|---|
| APP | amyloid β precursor protein | parental protein of neurotoxic/amyloidogenic Aβ peptide and neurotrophic sAPPα |
| BACE1 | β-secretase 1 | rate-limiting enzyme in production of Aβ from APP |
| ADAM9 | ADAM metallopeptidase domain 9 | α-secretase, non-amyloidogenic cleaving enzyme for APP |
| ADAM10 | ADAM metallopeptidase domain 10 | α-secretase, non-amyloidogenic cleaving enzyme for APP |
| ADAM17 | ADAM metallopeptidase domain 17 | α-secretase, non-amyloidogenic cleaving enzyme for APP |
| PSEN1 | presenilin 1 | critical constituent of γ-secretase complex, which completes APP cleavage processing |
| MME | membrane metalloendopeptidase | clearance enzyme for Aβ |
| IDE | insulin degrading enzyme | clearance enzyme for Aβ |
| MAPT | microtubule-associated protein τ | primary protein constituent of intraneuronal tangles typical of AD |
| GSK3B | glycogen synthase kinase 3β | primary kinase contributing to pro-tangle phosphorylation of microtubule-associated protein τ |
| REST | RE1-silencing transcription factor | transcription repression, varies significantly with age |
aNo genes in this table were significantly influenced by Li treatment in our data.
bProduct name as given in NCBI Gene database.
Figure 3.
Human hippocampus genetic co-expression network of snoRNA, selected Li-influenced protein coding genes and “core” AD genes. Gene symbols and Li-induced fold-change for snoRNAs, “core” AD genes (Table 5), and selected Li-influenced genes (Table 3) were analyzed for co-expression in human hippocampus by NetworkAnalyst. “Core” AD genes are indicated by star nodes. snoRNAs have square nodes. Color indicates log2 fold-change induced by Li treatment, according to legend. White nodes did not appear in our dataset. Additional genes inserted by NetworkAnalyst are in Supplementary Table 3.
Figure 4.
Human hippocampus gene product’s interaction network of snoRNA, selected Li-influenced protein coding genes and “core” AD genes. Gene symbols and Li-induced fold-change for snoRNAs, “core” AD genes (Table 5), and selected Li-influenced genes (Table 3) were analyzed for product interaction in human hippocampus by NetworkAnalyst. This map appears to reveal a multi-target convergence on APP. “Core” AD genes are indicated by star nodes. snoRNAs have square nodes. Color indicates log2 fold-change induced by Li treatment, according to legend. White nodes did not appear in our dataset. Additional genes inserted by NetworkAnalyst are in Supplementary Table 4.
Table 5.
Li-influenced snoRNA sequencesa associated with Braak staging42.
| Name | Li-induced Change |
|---|---|
| SCARNA22 | +41.7% |
| SCARNA3 | +92.9% |
| SCARNA6 | +52.0% |
| SNORA37 | +69.6% |
| SNORD104 | +83.9% |
| SNORD46 | +48.2% |
| SNORD60 | +42.1% |
| SNORD94 | +63.2% |
Discussion
We demonstrated two effects of Li treatment on the whole RNA transcriptome of a human neuronal cell line. First, Li treatment is more likely to perturb levels of a given RNA inversely to its sequence length, regardless of RNA type. Second, and more importantly, specific RNA type exerts a significant influence on whether or not Li treatment perturbs its levels. That is, snoRNAs of three types (snoRD, snoRA, and scaRNA) are all significantly more likely to be perturbed while miRNA is significantly less likely to be perturbed. It is particularly noteworthy that miRNAs, although often of the same approximate length as snoRNAs, were significantly less likely to have levels perturbed by Li treatment.
SnoRNAs act as a guide in processing nuclear and ribosomal RNAs46,47. The functions of snoRNAs may extend to regulation of alternative splicing, gene silencing, chromatin modification and may even exhibit miRNA-like function48–54. Understanding the role of snoRNAs in behavior and disease conditions is also emerging. Prader-Willi syndrome is a genetic disorder characterized by intellectual disabilities, hyperphagia, obesity, sleep disorders and behavioral problems; the mechanism involves a paternal deletion of chromosomal region 15q11-q13 harboring snoRNAs SNORD115 (elevated in this study by +84%, FDR = 0.05) and SNORD116 (elevated in this study by +74%, FDR = 0.01). Mice with a deletion of the Snord116 repeat cluster (Snord116del) show a diurnal disruption in feeding behaviors. Brain transcripts associated with Snord116del show upregulation of diurnally regulated genes, particularly Mtor (mammalian target of rapamycin), Crebp (Creb-binding protein), and Igf2r (insulin growth factor receptor) as well as clock genes, with accompanying greater body lean-ness and increased lipid oxidation during daylight hours55. Clock genes appear to be disrupted in bipolar disorder as well56,57, although the relationship is complex58; there is evidence implicating circadian rhythm changes in the response to Li59. SNORD115 also regulates maturation of serotonin receptor 2 C (5HT2C) by alternative splicing60,61.
It is notable that snoRNAs are active in chromatin modification. Genome wide association studies (GWAS) implicate histone methylation pathways as the most likely targets of snoRNA chromatin modification62. In addition, alternative splicing is likely to be important in bipolar disorder63.
Li influences snoRNA processing in a yeast model64, which may be enlightening to understand potential mechanism in mammalian cells. Specifically, Li appears to inhibit the bisphosphate nucleotidase Hal2p. This results in accumulation of adenosine 3′,5′-bisphosphate (pAp), which is normally broken down by Hal2p, and of pre-snoRNAs. Accumulation of pAp inhibits activities of RNases necessary for processing of snoRNAs. The human homologue for Hal2p is 3′(2′), 5′-bisphosphate nucleotidase 1, or PAP phosphatase (BPNT1). Li inhibits BPNT1 activity in crude human brain extracts65. Paradoxically, BPNT1 protein levels are reduced in the frontal cortex of bipolar patients65. Thus, the specific anti-bipolar activity of Li is unlikely to be a single mechanism.
Although Li influence on miRNA was very limited, some of the specific miRNAs with significantly altered expression play roles in neuropsychiatric disorders. For example, miR-10b was elevated 44% by Li treatment. Elevated expression of miR-10b, in cerebrospinal fluid (CSF), can serve as a diagnostic feature for Parkinson’s disease (PD)66. Paradoxically, miR-10b levels are reduced in PD brain, and greater levels of miR-10b associate with later age of motor onset. However, miR-10b is elevated in Huntington’s disease (HD) brains, and reduced levels of miR-10b associated with later age of motor onset in HD67,68. miR-24-1 was elevated 42% by Li treatment. This miRNA was downregulated in CSF samples of PD subjects69. It was also significantly reduced in CSF of AD vs. Control subjects70. miR-24 serves to reduce levels of Aβ through regulating expression of nicastrin71. Li regulation of glutamate metabotropic receptor 7 (GRM7) – a gene significantly associated in a GWAS study on bipolar patients – has been shown to act via miR-13472,73.
When we looked at potential networked effects for Li treatment vs. AD, we first note that none of the “core” AD genes we examined were altered by Li treatment in our experiment. However, when we examined both co-expression and gene product interaction, connections emerged that may be worthy of further investigation, particularly the interaction between SNORD14C and APP. Even though our Li treatment did not alter the levels of the core AD gene products, we did not test effects under more “pathogenic-like” conditions, such as oxidative stress, and it may be the differences between differences (non-stressed vs. stressed cells) that could further elucidate important pathways. Several of the connections we found in our network analysis, such as GSK3B, MAPT, ADAM17, and PSEN1 are also involved in other dementias than AD. However we hesitate to speculate on applicability of Li to multiple dementia disorders, since none of these were directly influenced by Li treatment in our study. Given that we restricted our work to a single differentiated cell line, we anticipate that much more can be learned by using other models, such as human primary brain cultures or induced pluripotent stem cell (iPSC) neuronal cultures.
Extending the role of Li treatment past bipolar disorder has revealed some challenges for its use. It is well known that, in higher doses, Li can be neurotoxic. It can suppress microtubule-associated protein tau and induce PD-like tremor and iron accumulation in brain74. Such iron accumulation is accompanied by reduction of levels of APP, which acts as an iron efflux protein75. Iron is only one of several metals that play a role in AD, in part due to the function of APP as an iron efflux protein75 that also responds to magnesium and lead76,77. Effective use of a metal as a drug may need to take into account interplay of multiple metals’ levels in the cells and environment.
Dosing “in the upper therapeutic range” (for bipolar disorder) can associate both with PD and AD-like symptoms and FDG-PET readouts, all of which were relieved by reduction of Li dose78. A larger-scale (>50,000 cases) retrospective study of Taiwanese patients administered Li reported mixed results5. On the other hand. low/micro-dose treatments improved agitation and stabilized cognitive impairment in AD79,80. When microdose Li was applied to animal AD models, it prevented memory loss and AD-like pathology81 as well as improved spatial memory and reduced Aβ42-induced neuroinflammation82.
Specific mechanisms of Li activity on AD may be explained by examining some of the Li-influenced coding gene products we mention herein. For example, BMP4 reduces hippocampal cell proliferation in animal AD models83, and BMP4 is elevated in AD84. Furthermore, APP regulates BMP/SMAD signaling in glial differentiation85. Another gene strongly implicated in AD is PPARγ, whose AD-related activities include possible inhibition of β-site amyloid cleaving enzyme 1 (BACE1)86 and SNP-based interaction with apolipoprotein E to alter AD risk87, among many other possible connections. Neurexin 3 (NRXN3) contains a polymorphism (rs17757879) that protects against AD in male subjects88. Furthermore, toxic Aβ oligomers interact with neurexins and reduce NRXN-mediated excitatory presynaptic organization89. Finally, NRXN3 is processed by the same α- and γ- secretases that produce the neurotrophic products of the “non-amyloidogenic” APP processing pathway90, including sAPPα. Other gene products we identified have their own potential associations with AD, as well, such as multiple snoRNAs that vary according to Braak stage42 and were also altered by Li-treatment in our study.
Our work herein highlights specific RNA species-specific responses to Li treatment, which would fuel mechanistic studies. While countless in silico pathways could have been found by examination of multiple databases, any claims connecting Li, snoRNAs, and disease states on that basis would be speculative. What we have experimentally discovered is that Li treatment has (at least) two distinct effects on RNA levels. The first is an RNA species-independent inverse relationship between RNA length and Li influence on RNA levels. The second is different effect levels by RNA type, irrespective of RNA length. The presence of miRNA as the least-influenced RNA type is particularly interesting, since it is in stark contrast against the majority of short RNA species. A different approach to investigating miRNA vs. Li treatment further underscored such “resistance” of miRNA to Li. When Li treatment response phenotypes in bipolar patients was screened by a large (1,693 sequence) genome-wide association study of pre-miRNA genes plus 20 kb flanking sequence, only one pre-miRNA gene had a nominally significant (p ≤ 0.05) sequence variation association, but when corrected for multiple comparisons, no polymorphisms in pre-miRNA plus flanking sequence showed any association with Li treatment phenotypes91. That study compared symptomatic Li response in patients to genetic sequences rather than response in cell culture transcriptome levels. This is not necessarily in conflict with known associations between miRNAs and bipolar disorder risk, since risk, progression, and treatment response may each be governed by distinct pathways.
Such differences may suggest potentially useful avenues for investigating short RNA species in the multiple disorders currently and potentially treated by Li. What mechanisms could make miRNA less susceptible to Li treatment than RNA types of similar length? Are there genetic variations in miRNA response to Li, and do these correspond to Li treatment resistance or facilitation in patients? Analogous converse questions could be asked regarding snoRNAs and Li. We must point out that our study was on “steady state” RNA levels after one week of Li treatment.
While effects of Li on RNA levels were previously reported32–35, our study differs from these prior works in one critical and significant way. The preceding studies were limited by using microarray profiling. While the scope of microarrays has become quite impressive, they still each have a fixed number (albeit potentially thousands) of pre-defined targets. Our survey was high-throughput sequencing with no a-priori presumptions of the presence (or absence) of any specific RNA transcript.
We finally need to note that, while ATRA-induced SK-N-SH cultures are often accepted as “neuronal”, they are not adult human neurons. Further uses of non-presumptive whole-transcriptome sequencing follow-up to Li treatment should be done in other neuronal cell lines and in systems such as neuronal and mixed neuron/glial induced pluripotent stem cell culture, at the very least. Unfortunately, human neuronal cell models are always approximations of the actual human brain, as are any animal models. ATRA-induced SK-N-SH cultures have important elements of reproducibility and availability, which allows for seminal discoveries to be confirmed and later tested in the more costly and difficult models.
Methods
Oversight and approval
All procedures were approved and overseen by the Institutional Biosafety Committee, Office of Research Compliance, Indiana University, and Indianapolis, Indiana, USA.
Cell culture and treatment
Human neuroblastoma (SK-N-SH) cells were obtained from ATCC and cultured as previously described92, then differentiated by ATRA93. Initial propagation was in Eagle’s modified minimum essential medium (MEM, ThermoFisher, Waltham MA) supplemented with 10% fetal bovine serum (FBS). For neuronal differentiation, a stock solution of 0.01 M ATRA (Sigma Prod. No. R2625; >98% HPLC purified), was prepared in absolute ethanol and stored in light protected vials at −20 °C and diluted with tissue culture medium right before use. Subsequent dilutions were made in growth medium with a final ethanol concentration of 0.1% (v/v) which did not affect the described system. During differentiation, cells were switched to MEM media with 1% FBS supplemented with 10 µM ATRA for two weeks. Cultures of treated SK-N-SH cells were subsequently treated with 1 mM Li chloride (LiCl) or vehicle (distilled water) for an additional week (n = 6 in each group). This dose is within a range of reported Li treatment of neuroblastoma cells characterized as “low” concentration94,95. Toxicity of Li treatment of neuroblastoma cells was reported to not occur until concentrations exceeded 10 mM96.
RNA extraction and sequencing
Total RNA was extracted using the RNeasy mini kit (Qiagen Company, Germantown, MD) and ribosomal RNA depleted with Ribo-Zero (Illumina, San Diego CA). Whole transcriptome sequencing was done via SOLiD RNA-Seq kit (ThermoFisher). Briefly, total RNA was fragmented with RNase III, hybridized and ligated to SOLiDTM adapters, then subject to reverse transcription with SOLiDTM RT primer mix. cDNA was then amplified and sequenced on a 5500xl Genetic Analyzer (ThermoFisher). Identified transcripts were classified into RNA types using the typology and databases at, specifically pseudogene.org97, HGNC98, and GeneCards99.
Statistical analysis
We analyzed quantitative transcriptome sequencing by edgeR100, which explicitly calculated fold differences in specific RNA levels between Li-treated and untreated cells and determined significance (p values and FDR) for each difference. We considered those RNAs that had level differences associated with FDR ≤ 0.2 to be “Li-influenced”, regardless of level of influence. We used the mclust43 package to explore possible clustered distribution of sequence lengths. Clustering with mclust used a univariate, unequal variances model to determine cluster borders and cluster centers were not predetermined.
RNA sequences were typologically classified by referring to three databases, specifically pseudogene.org97, HGNC98, and GeneCards99. The presence/absence of Li influence (significant change in RNA level in Li-treated vs. control cells) was determined by edgeR and coded as 1 or 0, 1 corresponding to Li-induced net RNA level change (either increase or decrease), by individual sequence. Magnitude of change was not considered in this study. Examination of the data showed that only 1.3% of sequences’ expression were influenced by Li treatment. Such a low frequency can introduce unacceptable bias into conventional logistic regression, which can be corrected by penalized log-likelihood estimation, such as via the logistf R package101. However, when we compared conventional and penalized logistic models for bias, we found that conventional methods produced no greater bias than did conventional regression for our data. Final analysis was, therefore, done with conventional logistic regression. A full model of “Li influenced ~ log(Length in bp) + RNA type + log(Length) × RNA type was constructed and assessed by second-order Akaike information criterion (AICc) using exhaustive combinations of predictors102 via the MuMIn R package102. Models were run with Effect contrast coding to explicitly compare Li influence of RNA species vs. the overall mean of Li influence frequency103. The effect coded model was further used for pairwise comparisons of RNA species by estimated marginal means104. Finally, the direction of Li-induced alteration (elevation, no change, or reduction of level) was modeled by multinomial logistic regression. Network analysis on selected coding gene products was performed vs. human hippocampus co-expression105 or protein-protein interaction106 dataset by NetworkAnalyst107, using the “minimum network” algorithm. Gene ontologies were analyzed by the “Gene Ontology System”108. For these utilities, default parameters were otherwise used.
Ethical approval and informed consent
All procedures were approved and overseen by the Institutional Biosafety Committee (IBC), Office of Research Compliance, Indiana University, and Indianapolis, Indiana, USA. No animals or human subjects or samples were used in this work.
Supplementary information
Acknowledgements
We thank Baindu Bayon and Clara Suh for their assistance in tissue cultures. DKL has received grant supports from NIH (R01AG051086, R21AG4687100 and P30AG010133). HJE has received grant support from NIH (U10 AA008401-30, U01MH109532-04 and P60 AA007611-32 [ARC]). The PGBD study was supported by 1U01MH092758-01 to JK.
Author contributions
B.M. analyzed data, figure construction, manuscript writing and references. Y.B. conducted the Lithium experiment and RNA preparation, and manuscript draft. Y.L. involved in whole-transcriptome RT-sequencing. N.C. assisted in cell culture experiments. H.J.E. reviewed data and manuscript editing. J.K. and J.I.N. contributed to manuscript writing and editing. D.K.L. planned, contributed reagents, materials and manuscript writing as well as final submission (corresponding).
Data availability
Authors agree to make materials, data and associated protocols promptly available to readers without undue qualifications in material transfer agreements. Raw RNA sequence data is in process of deposition in GEO, accession # pending.
Competing interests
Dr. Nurnberger is an investigator for Janssen.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bryan Maloney and Yokesh Balaraman.
Supplementary information
is available for this paper at 10.1038/s41598-019-54076-3.
References
- 1.Licht RW. Lithium: still a major option in the management of bipolar disorder. CNS neuroscience & therapeutics. 2012;18:219–226. doi: 10.1111/j.1755-5949.2011.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenox RH, Hahn CG. Overview of the mechanism of action of lithium in the brain: fifty-year update. The Journal of clinical psychiatry. 2000;61(Suppl 9):5–15. [PubMed] [Google Scholar]
- 3.Manji HK, Lenox RH. Lithium: a molecular transducer of mood-stabilization in the treatment of bipolar disorder. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 1998;19:161–166. doi: 10.1016/S0893-133X(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 4.Quiroz JA, Machado-Vieira R, Zarate CA, Jr., Manji HK. Novel insights into lithium’s mechanism of action: neurotrophic and neuroprotective effects. Neuropsychobiology. 2010;62:50–60. doi: 10.1159/000314310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng C, et al. Association Between Lithium Use and Risk of Alzheimer’s Disease. J. Clin. Psychiatry. 2017;78:e139–e145. doi: 10.4088/JCP.15m10304. [DOI] [PubMed] [Google Scholar]
- 6.Shiotsuki I, et al. Trace lithium is inversely associated with male suicide after adjustment of climatic factors. J. Affect. Disord. 2016;189:282–286. doi: 10.1016/j.jad.2015.09.070. [DOI] [PubMed] [Google Scholar]
- 7.Giotakos O, et al. A negative association between lithium in drinking water and the incidences of homicides, in Greece. Biol. Trace Elem. Res. 2015;164:165–168. doi: 10.1007/s12011-014-0210-6. [DOI] [PubMed] [Google Scholar]
- 8.Terao T. Is lithium potentially a trace element? World journal of psychiatry. 2015;5:1–3. doi: 10.5498/wjp.v5.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fajardo VA, Fajardo VA, LeBlanc PJ, MacPherson REK. Examining the Relationship between Trace Lithium in Drinking Water and the Rising Rates of Age-Adjusted Alzheimer’s Disease Mortality in Texas. J. Alzheimers Dis. 2018;61:425–434. doi: 10.3233/jad-170744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessing LV, et al. Association of Lithium in Drinking Water With the Incidence of Dementia. JAMA psychiatry. 2017;74:1005–1010. doi: 10.1001/jamapsychiatry.2017.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsunaga S, et al. Lithium as a Treatment for Alzheimer’s Disease: A Systematic Review and Meta-Analysis. J. Alzheimers Dis. 2015;48:403–410. doi: 10.3233/jad-150437. [DOI] [PubMed] [Google Scholar]
- 12.Morris G, Berk M. The Putative Use of Lithium in Alzheimer’s Disease. Curr Alzheimer Res. 2016;13:853–861. doi: 10.2174/1567205013666160219113112. [DOI] [PubMed] [Google Scholar]
- 13.Forlenza OV, De-Paula VJ, Diniz BS. Neuroprotective effects of lithium: implications for the treatment of Alzheimer’s disease and related neurodegenerative disorders. ACS Chem. Neurosci. 2014;5:443–450. doi: 10.1021/cn5000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devanand DP, et al. Lithium Treatment for Agitation in Alzheimer’s disease (Lit-AD): Clinical rationale and study design. Contemp. Clin. Trials. 2018;71:33–39. doi: 10.1016/j.cct.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toker L, Belmaker RH, Agam G. Gene-expression studies in understanding the mechanism of action of lithium. Expert review of neurotherapeutics. 2012;12:93–97. doi: 10.1586/ern.11.184. [DOI] [PubMed] [Google Scholar]
- 16.Mauer S, Vergne D, Ghaemi SN. Standard and trace-dose lithium: a systematic review of dementia prevention and other behavioral benefits. Aust. N. Z. J. Psychiatry. 2014;48:809–818. doi: 10.1177/0004867414536932. [DOI] [PubMed] [Google Scholar]
- 17.Ebstein R, Belmaker R, Grunhaus L, Rimon R. Lithium inhibition of adrenaline-stimulated adenylate cyclase in humans. Nature. 1976;259:411–413. doi: 10.1038/259411a0. [DOI] [PubMed] [Google Scholar]
- 18.Belmaker RH. Receptors, adenylate cyclase, depression, and lithium. Biol. Psychiatry. 1981;16:333–350. [PubMed] [Google Scholar]
- 19.Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 20.Hedgepeth CM, et al. Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev. Biol. 1997;185:82–91. doi: 10.1006/dbio.1997.8552. [DOI] [PubMed] [Google Scholar]
- 21.Manji HK, Lenox RH. Ziskind-Somerfeld Research Award. Protein kinase C signaling in the brain: molecular transduction of mood stabilization in the treatment of manic-depressive illness. Biol. Psychiatry. 1999;46:1328–1351. doi: 10.1016/S0006-3223(99)00235-8. [DOI] [PubMed] [Google Scholar]
- 22.Manji HK, Moore GJ, Chen G. Lithium up-regulates the cytoprotective protein Bcl-2 in the CNS in vivo: a role for neurotrophic and neuroprotective effects in manic depressive illness. J. Clin. Psychiatry. 2000;61(Suppl 9):82–96. [PubMed] [Google Scholar]
- 23.Arendt T, Lehmann K, Seeger G, Gartner U. Synergistic effects of tetrahydroaminoacridine and lithium on cholinergic function after excitotoxic basal forebrain lesions in rat. Pharmacopsychiatry. 1999;32:242–247. doi: 10.1055/s-1999-7961. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez G, et al. Lithium protects cultured neurons against beta-amyloid-induced neurodegeneration. FEBS Lett. 1999;453:260–264. doi: 10.1016/S0014-5793(99)00685-7. [DOI] [PubMed] [Google Scholar]
- 25.Moore GJ, et al. Lithium increases N-acetyl-aspartate in the human brain: in vivo evidence in support of bcl-2′s neurotrophic effects? Biol. Psychiatry. 2000;48:1–8. doi: 10.1016/S0006-3223(00)00252-3. [DOI] [PubMed] [Google Scholar]
- 26.Gould TD, Manji HK. The Wnt signaling pathway in bipolar disorder. Neuroscientist. 2002;8:497–511. doi: 10.1177/107385802237176. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Rajkowska G, Du F, Seraji-Bozorgzad N, Manji HK. Enhancement of hippocampal neurogenesis by lithium. J. Neurochem. 2000;75:1729–1734. doi: 10.1046/j.1471-4159.2000.0751729.x. [DOI] [PubMed] [Google Scholar]
- 28.Alda M. Who are excellent lithium responders and why do they matter? World psychiatry: official journal of the World Psychiatric Association (WPA) 2017;16:319–320. doi: 10.1002/wps.20462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern S, Santos R, Marchetto M C, Mendes A P D, Rouleau G A, Biesmans S, Wang Q-W, Yao J, Charnay P, Bang A G, Alda M, Gage F H. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Molecular Psychiatry. 2017;23(6):1453–1465. doi: 10.1038/mp.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertens J, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobe BTD, et al. Probing the lithium-response pathway in hiPSCs implicates the phosphoregulatory set-point for a cytoskeletal modulator in bipolar pathogenesis. Proc. Natl. Acad. Sci. USA. 2017;114:E4462–e4471. doi: 10.1073/pnas.1700111114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosetti F, et al. Analysis of gene expression with cDNA microarrays in rat brain after 7 and 42 days of oral lithium administration. Brain research bulletin. 2002;57:205–209. doi: 10.1016/S0361-9230(01)00744-4. [DOI] [PubMed] [Google Scholar]
- 33.McQuillin A, Rizig M, Gurling HM. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenetics and genomics. 2007;17:605–617. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- 34.Seelan RS, Khalyfa A, Lakshmanan J, Casanova MF, Parthasarathy RN. Deciphering the lithium transcriptome: microarray profiling of lithium-modulated gene expression in human neuronal cells. Neuroscience. 2008;151:1184–1197. doi: 10.1016/j.neuroscience.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 35.Chetcuti A, Adams LJ, Mitchell PB, Schofield PR. Microarray gene expression profiling of mouse brain mRNA in a model of lithium treatment. Psychiatric genetics. 2008;18:64–72. doi: 10.1097/YPG.0b013e3282fb0051. [DOI] [PubMed] [Google Scholar]
- 36.Peter-Ross EM. Molecular hypotheses to explain the shared pathways and underlying pathobiological causes in catatonia and in catatonic presentations in neuropsychiatric disorders. Med. Hypotheses. 2018;113:54–64. doi: 10.1016/j.mehy.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Ragan C, et al. Small non-coding RNA expression from anterior cingulate cortex in schizophrenia shows sex specific regulation. Schizophr. Res. 2017;183:82–87. doi: 10.1016/j.schres.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Gibbons Andrew, Udawela Madhara, Dean Brian. Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia. Non-Coding RNA. 2018;4(2):11. doi: 10.3390/ncrna4020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung KN, Vallero RO, DuBose AJ, Resnick JL, LaSalle JM. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum. Mol. Genet. 2009;18:4227–4238. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamova B, Ander BP, Barger N, Sharp FR, Schumann CM. Specific Regional and Age-Related Small Noncoding RNA Expression Patterns Within Superior Temporal Gyrus of Typical Human Brains Are Less Distinct in Autism Brains. J. Child Neurol. 2015;30:1930–1946. doi: 10.1177/0883073815602067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munoz-Culla M, et al. Progressive changes in non-coding RNA profile in leucocytes with age. Aging (Albany NY) 2017;9:1202–1218. doi: 10.18632/aging.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andres-Benito P, et al. Locus coeruleus at asymptomatic early and middle Braak stages of neurofibrillary tangle pathology. Neuropathol. Appl. Neurobiol. 2017;43:373–392. doi: 10.1111/nan.12386. [DOI] [PubMed] [Google Scholar]
- 43.Scrucca L, Fop M, Murphy TB, Raftery A. E. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. The R journal. 2016;8:289–317. doi: 10.32614/RJ-2016-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheather SJ, Jones C. A reliable data-based bandwidth selection method for kernel density estimation. J. R. Stat. Soc. Series B (Method.) 1991;53:683–690. [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B (Method.) 1995;57:289–300. [Google Scholar]
- 46.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annual review of biochemistry. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 47.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/S0092-8674(00)80238-X. [DOI] [PubMed] [Google Scholar]
- 48.Lv J, et al. Identification and characterization of long non-coding RNAs related to mouse embryonic brain development from available transcriptomic data. PloS one. 2013;8:e71152. doi: 10.1371/journal.pone.0071152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene expression. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- 50.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews. Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 51.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 52.Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. International journal of molecular sciences. 2013;14:16010–16039. doi: 10.3390/ijms140816010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bratkovic T, Rogelj B. The many faces of small nucleolar RNAs. Biochimica et biophysica acta. 2014;1839:438–443. doi: 10.1016/j.bbagrm.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 54.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Powell WT, et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Human molecular genetics. 2013;22:4318–4328. doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreira J, Geoffroy PA. Lithium and bipolar disorder: Impacts from molecular to behavioural circadian rhythms. Chronobiol. Int. 2016;33:351–373. doi: 10.3109/07420528.2016.1151026. [DOI] [PubMed] [Google Scholar]
- 57.Geoffroy PA, et al. Lithium response in bipolar disorders and core clock genes expression. World J. Biol. Psychiatry. 2018;19:619–632. doi: 10.1080/15622975.2017.1282174. [DOI] [PubMed] [Google Scholar]
- 58.McCarthy MJ. Missing a beat: assessment of circadian rhythm abnormalities in bipolar disorder in the genomic era. Psychiatr. Genet. 2019;29:29–36. doi: 10.1097/ypg.0000000000000215. [DOI] [PubMed] [Google Scholar]
- 59.McCarthy MJ, et al. Chronotype and cellular circadian rhythms predict the clinical response to lithium maintenance treatment in patients with bipolar disorder. Neuropsychopharmacology. 2019;44:620–628. doi: 10.1038/s41386-018-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 61.Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harbor symposia on quantitative biology. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 62.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gandal Michael J., Zhang Pan, Hadjimichael Evi, Walker Rebecca L., Chen Chao, Liu Shuang, Won Hyejung, van Bakel Harm, Varghese Merina, Wang Yongjun, Shieh Annie W., Haney Jillian, Parhami Sepideh, Belmont Judson, Kim Minsoo, Moran Losada Patricia, Khan Zenab, Mleczko Justyna, Xia Yan, Dai Rujia, Wang Daifeng, Yang Yucheng T., Xu Min, Fish Kenneth, Hof Patrick R., Warrell Jonathan, Fitzgerald Dominic, White Kevin, Jaffe Andrew E., Peters Mette A., Gerstein Mark, Liu Chunyu, Iakoucheva Lilia M., Pinto Dalila, Geschwind Daniel H. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420):eaat8127. doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agam G, Shaltiel G. Possible role of 3′(2′)-phosphoadenosine-5′-phosphate phosphatase in the etiology and therapy of bipolar disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2003;27:723–727. doi: 10.1016/s0278-5846(03)00125-8. [DOI] [PubMed] [Google Scholar]
- 66.Dos Santos MCT, et al. miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget. 2018;9:17455–17465. doi: 10.18632/oncotarget.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers R. H. microRNA Profiles in Parkinson’s Disease Prefrontal. Cortex. Front. Aging Neurosci. 2016;8:36. doi: 10.3389/fnagi.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoss AG, et al. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genomics. 2015;8:10. doi: 10.1186/s12920-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marques TM, et al. MicroRNAs in Cerebrospinal Fluid as Potential Biomarkers for Parkinson’s Disease and Multiple System Atrophy. Mol. Neurobiol. 2017;54:7736–7745. doi: 10.1007/s12035-016-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller M, Kuiperij HB, Claassen JA, Kusters B, Verbeek MM. MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol. Aging. 2014;35:152–158. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 71.Delay C, et al. MicroRNAs targeting Nicastrin regulate Abeta production and are affected by target site polymorphisms. Front. Mol. Neurosci. 2014;7:67. doi: 10.3389/fnmol.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou R, et al. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiu CT, Wang Z, Hunsberger JG, Chuang DM. Therapeutic potential of mood stabilizers lithium and valproic acid: beyond bipolar disorder. Pharmacol Rev. 2013;65:105–142. doi: 10.1124/pr.111.005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lei P, et al. Lithium suppression of tau induces brain iron accumulation and neurodegeneration. Mol. Psychiatry. 2017;22:396–406. doi: 10.1038/mp.2016.96. [DOI] [PubMed] [Google Scholar]
- 75.Long Justin M., Maloney Bryan, Rogers Jack T., Lahiri Debomoy K. Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: Implications in Alzheimer’s disease. Molecular Psychiatry. 2018;24(3):345–363. doi: 10.1038/s41380-018-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rogers JT, et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J. Biol. Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- 77.Rogers JT, et al. A role for amyloid precursor protein translation to restore iron homeostasis and ameliorate lead (Pb) neurotoxicity. J. Neurochem. 2016;138:479–494. doi: 10.1111/jnc.13671. [DOI] [PubMed] [Google Scholar]
- 78.Frisch S, Grunwald F, Friedrichs B. Cognitive sequelae of lithium intoxication: a case report. Int. Psychogeriatr. 2017;29:1747–1751. doi: 10.1017/s1041610217000540. [DOI] [PubMed] [Google Scholar]
- 79.Nunes MA, Viel TA, Buck HS. Microdose lithium treatment stabilized cognitive impairment in patients with Alzheimer’s disease. Curr Alzheimer Res. 2013;10:104–107. doi: 10.2174/1567205011310010014. [DOI] [PubMed] [Google Scholar]
- 80.Devanand DP, et al. Low-dose Lithium Treatment for Agitation and Psychosis in Alzheimer Disease and Frontotemporal Dementia: A Case Series. Alzheimer Dis. Assoc. Disord. 2017;31:73–75. doi: 10.1097/wad.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nunes MA, et al. Chronic Microdose Lithium Treatment Prevented Memory Loss and Neurohistopathological Changes in a Transgenic Mouse Model of Alzheimer’s Disease. PLoS One. 2015;10:e0142267. doi: 10.1371/journal.pone.0142267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Budni J, et al. Lithium and memantine improve spatial memory impairment and neuroinflammation induced by beta-amyloid 1-42 oligomers in rats. Neurobiol. Learn. Mem. 2017;141:84–92. doi: 10.1016/j.nlm.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 83.Li D, et al. Decreased hippocampal cell proliferation correlates with increased expression of BMP4 in the APPswe/PS1DeltaE9 mouse model of Alzheimer’s disease. Hippocampus. 2008;18:692–698. doi: 10.1002/hipo.20428. [DOI] [PubMed] [Google Scholar]
- 84.Xu H, et al. The function of BMP4 during neurogenesis in the adult hippocampus in Alzheimer’s disease. Ageing research reviews. 2013;12:157–164. doi: 10.1016/j.arr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 85.Kwak YD, Hendrix BJ, Sugaya K. Secreted type of amyloid precursor protein induces glial differentiation by stimulating the BMP/Smad signaling pathway. Biochem. Biophys. Res. Commun. 2014;447:394–399. doi: 10.1016/j.bbrc.2014.03.139. [DOI] [PubMed] [Google Scholar]
- 86.Sastre M, et al. Nonsteroidal anti-inflammatory drugs repress beta-secretase gene promoter activity by the activation of PPARgamma. Proc. Natl. Acad. Sci. USA. 2006;103:443–448. doi: 10.1073/pnas.0503839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S, et al. Gene- Gene Interaction between PPARG and APOE Gene on Late-Onset Alzheimer’s Disease: A Case- Control Study in Chinese Han Population. J. Nutr. Health Aging. 2017;21:397–403. doi: 10.1007/s12603-016-0794-y. [DOI] [PubMed] [Google Scholar]
- 88.Martinez-Mir A, et al. Genetic study of neurexin and neuroligin genes in Alzheimer’s disease. J. Alzheimers Dis. 2013;35:403–412. doi: 10.3233/jad-122257. [DOI] [PubMed] [Google Scholar]
- 89.Naito Y, Tanabe Y, Lee AK, Hamel E, Takahashi H. Amyloid-beta Oligomers Interact with Neurexin and Diminish Neurexin-mediated Excitatory Presynaptic Organization. Sci. Rep. 2017;7:42548. doi: 10.1038/srep42548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bot N, Schweizer C, Ben Halima S, Fraering PC. Processing of the synaptic cell adhesion molecule neurexin-3beta by Alzheimer disease alpha- and gamma-secretases. J. Biol. Chem. 2011;286:2762–2773. doi: 10.1074/jbc.M110.142521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reinbold CS, et al. Analysis of the Influence of microRNAs in Lithium Response in Bipolar Disorder. Frontiers in psychiatry. 2018;9:207. doi: 10.3389/fpsyt.2018.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ghosh C, Song W, Lahiri DK. Efficient DNA transfection in neuronal and astrocytic cell lines. Mol. Biol. Rep. 2000;27:113–121. doi: 10.1023/A:1007173906990. [DOI] [PubMed] [Google Scholar]
- 93.Bailey JA, Maloney B, Ge YW, Lahiri DK. Functional activity of the novel Alzheimer’s amyloid beta-peptide interacting domain (AbetaID) in the APP and BACE1 promoter sequences and implications in activating apoptotic genes and in amyloidogenesis. Gene. 2011;488:13–22. doi: 10.1016/j.gene.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nciri R, et al. Neuroprotective effects of chronic exposure of SH-SY5Y to low lithium concentration involve glycolysis stimulation, extracellular pyruvate accumulation and resistance to oxidative stress. Int. J. Neuropsychopharmacol. 2013;16:365–376. doi: 10.1017/s1461145712000132. [DOI] [PubMed] [Google Scholar]
- 95.Nciri R, et al. Chronic neuroprotective effects of low concentration lithium on SH-SY5Y cells: possible involvement of stress proteins and gene expression. Neural regeneration research. 2014;9:735–740. doi: 10.4103/1673-5374.131578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alural B, Ozerdem A, Allmer J, Genc K, Genc S. Lithium protects against paraquat neurotoxicity by NRF2 activation and miR-34a inhibition in SH-SY5Y cells. Front. Cell. Neurosci. 2015;9:209. doi: 10.3389/fncel.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karro JE, et al. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 2007;35:D55–60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yates B, et al. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017;45:D619–d625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stelzer G, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Current protocols in bioinformatics. 2016;54(1):30.31–31.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 100.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heinze, G. & Ploner, M. logistf: Firth’s bias-reduced logistic regrssion: version 1.23, https://CRAN-R-project.org/package=logistf (2018).
- 102.Barton, K. MuMIn: Multi-model inference version 1.42.1, https://CRAN.R-project.org/package=MuMIn (2018).
- 103.Salkind, N. Effect Coding, https://methods.sagepub.com/Reference/encyc-of-research-design/n127.xml (2010).
- 104.Lenth, R. emmeans: Estimated marginal means, aka least-squares means. R package version 1.1.3, https://CRAN.R-project.org/package=emmeans (2018).
- 105.Lee S, et al. TCSBN: a database of tissue and cancer specific biological networks. Nucleic Acids Res. 2018;46:D595–d600. doi: 10.1093/nar/gkx994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Basha O, Shpringer R, Argov CM, Yeger-Lotem E. The DifferentialNet database of differential protein-protein interactions in human tissues. Nucleic Acids Res. 2018;46:D522–d526. doi: 10.1093/nar/gkx981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 108.Gene Ontology Resource The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–d338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang J, et al. Noggin and BMP4 co-modulate adult hippocampal neurogenesis in the APP(swe)/PS1(DeltaE9) transgenic mouse model of Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2009;385:341–345. doi: 10.1016/j.bbrc.2009.05.067. [DOI] [PubMed] [Google Scholar]
- 110.Puthiyedth N, Riveros C, Berretta R, Moscato P. Identification of Differentially Expressed Genes through Integrated Study of Alzheimer’s Disease Affected Brain Regions. PLoS One. 2016;11:e0152342. doi: 10.1371/journal.pone.0152342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li A, et al. Silencing of the Drosophila ortholog of SOX5 leads to abnormal neuronal development and behavioral impairment. Hum. Mol. Genet. 2017;26:1472–1482. doi: 10.1093/hmg/ddx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davidsson P, Bogdanovic N, Lannfelt L, Blennow K. Reduced expression of amyloid precursor protein, presenilin-1 and rab3a in cortical brain regions in Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 2001;12:243–250. doi: 10.1159/000051266. [DOI] [PubMed] [Google Scholar]
- 113.Bereczki E, et al. Synaptic proteins predict cognitive decline in Alzheimer’s disease and Lewy body dementia. Alzheimers Dement. 2016;12:1149–1158. doi: 10.1016/j.jalz.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 114.Cruchaga C, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Calderari S, et al. Molecular genetics of the transcription factor GLIS3 identifies its dual function in beta cells and neurons. Genomics. 2018;110:98–111. doi: 10.1016/j.ygeno.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 116.Stopschinski BE, et al. Specific glycosaminoglycan chain length and sulfation patterns are required for cell uptake of tau versus alpha-synuclein and beta-amyloid aggregates. J. Biol. Chem. 2018;293:10826–10840. doi: 10.1074/jbc.RA117.000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huttenrauch M, et al. Glycoprotein NMB: a novel Alzheimer’s disease associated marker expressed in a subset of activated microglia. Acta Neuropathol Commun. 2018;6:108. doi: 10.1186/s40478-018-0612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carballo-Pacheco M, Ismail AE, Strodel B. Oligomer Formation of Toxic and Functional Amyloid Peptides Studied with Atomistic Simulations. J. Phys. Chem. B. 2015;119:9696–9705. doi: 10.1021/acs.jpcb.5b04822. [DOI] [PubMed] [Google Scholar]
- 119.Haspel N, et al. Conformational exploration of two peptides and their hybrid polymer conjugates: potentialities as self-aggregating materials. J. Phys. Chem. B. 2012;116:13941–13952. doi: 10.1021/jp3043363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roesler R, et al. Molecular mechanisms mediating gastrin-releasing peptide receptor modulation of memory consolidation in the hippocampus. Neuropharmacology. 2006;51:350–357. doi: 10.1016/j.neuropharm.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 121.Zhu Y, et al. Serum Hepatocyte Growth Factor Is Associated with Small Vessel Disease in Alzheimer’s Dementia. Front. Aging Neurosci. 2018;10:8. doi: 10.3389/fnagi.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wright JW, Harding JW. The Brain Hepatocyte Growth Factor/c-Met Receptor System: A New Target for the Treatment of Alzheimer’s Disease. J. Alzheimers Dis. 2015;45:985–1000. doi: 10.3233/jad-142814. [DOI] [PubMed] [Google Scholar]
- 123.Sharma S. Hepatocyte growth factor in synaptic plasticity and Alzheimer’s disease. TheScientificWorldJournal. 2010;10:457–461. doi: 10.1100/tsw.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fenton H, et al. Hepatocyte growth factor (HGF/SF) in Alzheimer’s disease. Brain Res. 1998;779:262–270. doi: 10.1016/S0006-8993(97)00958-X. [DOI] [PubMed] [Google Scholar]
- 125.Hinney A, et al. Genetic variation at the CELF1 (CUGBP, elav-like family member 1 gene) locus is genome-wide associated with Alzheimer’s disease and obesity. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014;165b:283–293. doi: 10.1002/ajmg.b.32234. [DOI] [PubMed] [Google Scholar]
- 126.Xu M, et al. A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer’s disease. Alzheimers Dement. 2018;14:215–229. doi: 10.1016/j.jalz.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 127.Wang H, et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-beta type II receptor. Brain Res. 2010;1357:166–174. doi: 10.1016/j.brainres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 128.Velez JI, et al. Pooling/bootstrap-based GWAS (pbGWAS) identifies new loci modifying the age of onset in PSEN1 p.Glu280Ala Alzheimer’s disease. Mol. Psychiatry. 2013;18:568–575. doi: 10.1038/mp.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ronn, T. et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum. Mol. Genet., 10.1093/hmg/ddv124 (2015). [DOI] [PubMed]
- 130.Pedros I, et al. Early alterations in energy metabolism in the hippocampus of APPswe/PS1dE9 mouse model of Alzheimer’s disease. Biochim. Biophys. Acta. 2014;1842:1556–1566. doi: 10.1016/j.bbadis.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 131.Aberg D, et al. Increased Cerebrospinal Fluid Level of Insulin-like Growth Factor-II in Male Patients with Alzheimer’s Disease. J. Alzheimers Dis. 2015;48:637–646. doi: 10.3233/jad-150351. [DOI] [PubMed] [Google Scholar]
- 132.Lane EM, Hohman TJ, Jefferson AL. Insulin-like growth factor binding protein-2 interactions with Alzheimer’s disease biomarkers. Brain imaging and behavior. 2017;11:1779–1786. doi: 10.1007/s11682-016-9636-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McLimans KE, Webb JL, Anantharam V, Kanthasamy A, Willette AA. Peripheral versus Central Index of Metabolic Dysfunction and Associations with Clinical and Pathological Outcomes in Alzheimer’s Disease. J. Alzheimers Dis. 2017;60:1313–1324. doi: 10.3233/jad-170263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fugistier P, et al. Comparison of frailty of primary neurons, embryonic, and aging mouse cortical layers. Neurobiol. Aging. 2014;35:322–330. doi: 10.1016/j.neurobiolaging.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 135.Morgan AR, et al. Association studies of 23 positional/functional candidate genes on chromosome 10 in late-onset Alzheimer’s disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144b:762–770. doi: 10.1002/ajmg.b.30509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Authors agree to make materials, data and associated protocols promptly available to readers without undue qualifications in material transfer agreements. Raw RNA sequence data is in process of deposition in GEO, accession # pending.