Abstract
Verticillium wilt is a severe disease of cotton crops in Xinjiang and affecting yields and quality, due to the continuous cotton cropping in the past decades. The relationship between continuous cropping and the changes induced on soil microbiome remains unclear to date. In this study, the culture types of 15 isolates from Bole (5F), Kuitun (7F), and Shihezi (8F) of north Xinjiang were sclerotium type. Only isolates from field 5F belonged to nondefoliating pathotype, the others belonged to defoliating pathotype. The isolates showed pathogenicity differentiation in cotton. Fungal and bacterial communities in soils had some difference in alpha-diversity, relative abundance, structure and taxonomic composition, but microbial groups showed similarity in the same habitat, despite different sampling sites. The fungal phyla Ascomycota, and the bacterial phyla Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria and Gemmatimonadetes were strongly enriched. Verticillium abundance was significantly and positively correlated with AN, but negatively correlated with soil OM, AK and pH. Moreover, Verticillium was correlated in abundances with 5 fungal and 6 bacterial genera. Overall, we demonstrate that soil microbiome communities have similar responses to long-term continuous cotton cropping, providing new insights into the effects of continuous cotton cropping on soil microbial communities.
Subject terms: Microbiology, Plant sciences
Introduction
Continuous cropping regimes can cause crop yield reduction, soil-borne plant pathogen accumulation1–3, and soil microbial community disruption4–6. In spite of these negative feedbacks, this regime is still common in many agricultural production systems. Cotton is an important economic crop of China, mainly cultivated in Xinjiang, where long-term continuous cropping results in lower yields, also caused by a severe incidence of Verticillium wilt disease, which hinders the development of the national cotton industry.
Verticillium wilt is a catastrophic disease, caused by Verticillium dahliae7. Soil-borne disease are considered as influenced also by changes affecting the below ground microbial communities, a situation summarized by the term “microbiome disease”8. If bacteria, fungi, and nematode pathogens act with a synergistic effect, it may become very difficult to control root colonization and insurgence of such a disastrous disease9,10. Under the cotton continuous cropping system, the increase in prevalence of soil-borne diseases is directly linked to the increase of the pathogen abundance4, and the occurrence of different V. dahliae pathotypes11. Due to the variety of physiological types of V. dahliae, understanding how they affect the soil microbial community among different regions may be useful for an effective disease management.
The soil microbiome provides several services, including organic matter dynamics, nutrient cycling and suppression or regulation of soil-borne diseases12, underpinning soil quality and function in the agroecosystem13. In recent years, the important role of soil microbiome in regulating agricultural production has been elucidated. Upon infection, plants can recruit microbes that promote disease resistance and plant growth with an effect on the composition of the root microbiome14. For example, a rhizosphere Flavobacterium sp. was found to enhance wilt resistance in tomato15. A number of links are active among diseased plants and the community composition and function of the soil microbiome16–18. However, the majority studies have focused on either bacterial or fungal communities alone.
Compared to cultivated soil, the non-rhizosphere one is capable to keep soil aggregates stable sustaining resistance to unfavorable environmental conditions such as drought19. In fact, non-rhizosphere soils have an important role for the next generation of crops. Few studies focused thus far on the effect of continuous cropping regimes on the non-rhizosphere or bulk soil microbiomes20.
In this study, we tested pathotype, growth rate, spore production and pathogenicity of 15 representative V. dahliae isolates, and analyzed fungal and bacterial community compositions of 15 bulk soils, from three cotton producing regions, using Ilumina MiSeq sequencing. The study aim was to (i) determine the diversity and pathogenic differentiation of V. dahliae from cotton in north Xinjiang; (ii) compare the structure and composition of fungal and bacterial communities in soils of three cotton fields; and (iii) to explore potential links between Verticillium wilt prevalence and soil properties or microbial communities.
Results
Pathotype, and pathogenic differentiation of Verticillium dahliae isolates
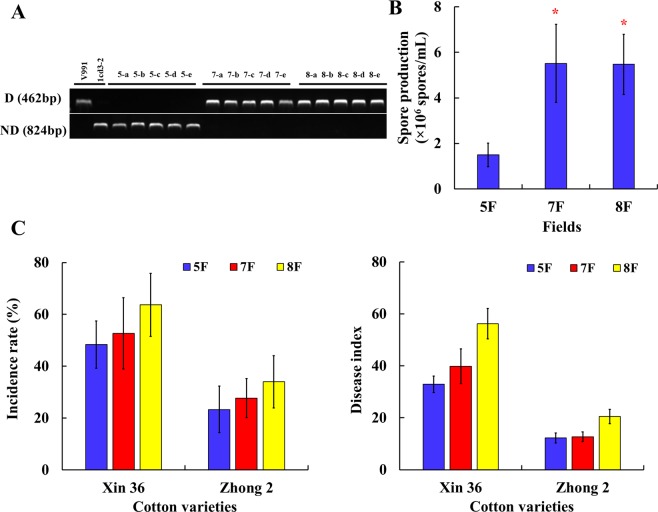
The pathotypes of 15 V. dahliae isolates proceeding from the three cotton fields was determined via a specific PCR assay. Isolates from fields 7F and 8F belonged to the defoliating (D) pathotype (462 bp specific fragment), whereas those from field 5F belonged to the nondefoliating (ND) one (824 bp specific fragment) (Fig. 1A and S1). Morphological observations showed that all isolates were sclerotium type (Fig. S2). Their growth exhibited no significant difference among the three cotton fields (P = 0.783), and also between the D and ND pathotypes, whereas the spore production of D pathotype as significantly higher than that of ND pathotype (5F vs 7F, P = 0.047; 5F vs 8F, P = 0.049) (Fig. 1B). The results of the pathogenicity assay indicated different levels of disease resistance response in cotton varieties (Xinluzao 36, DI = 42.96, P = 0.029; Zhongzhimian 2, DI = 15.14, P = 0.038) vs the 15 isolates (Fig. 1C). The isolates had evident pathogenicity differentiations on the two varieties (Fig. 2), included the two reference strains (Fig. S3). The pathogenicity of the isolates from the three cotton fields were 8F > 7F > 5F (Table. S1). Moreover, the growth rate of all isolates showed no correlation with disease index, however, spore production (R = 0.534*, P = 0.027) showed a significant and positive correlation with the average disease index (Fig. S4).
Figure 1.
The results of the pathotypes, cultural characters and pathogencity of Verticillium dahliae. (A) Identification of the defoliating and non-defoliating pathotypes of all isolates with the corresponding molecular marker (see Fig. S1 for the full-length gels). (B) The spore production of 15 isolates. Red asterisk represents the spore production of D pathotype as significantly higher than that of ND pathotype. (5F vs 7F, P = 0.047; 5F vs 8F, P = 0.049; *P < 0.05). (C) The results of pathogenic assay, including the statistics of the incidence rate and disease index.
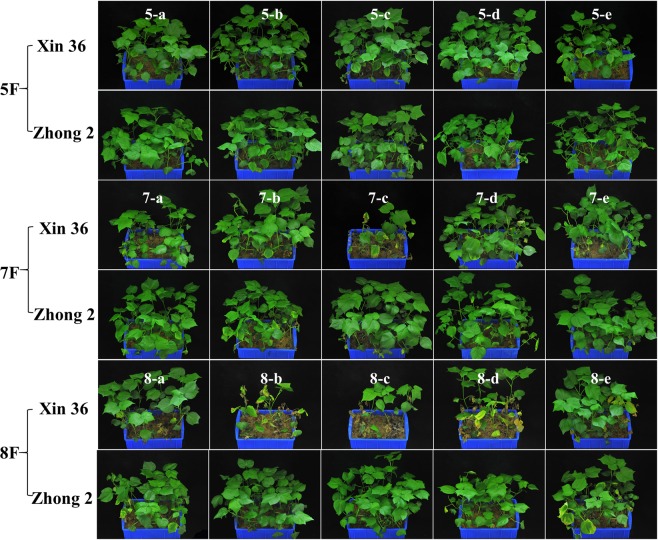
Figure 2.
Phenotypes of two cotton varieties after inoculation with the 15 isolates tested. Each isolate was inoculated with 30 seedlings of two cotton varieties, respectively. Sterile distilled water was used as control. Replicated twice. When the disease index reached 50.0 after inoculation with V991 on Xinluzao 36, the disease indexes were counted.
Alpha-diversity of fungal and bacterial species in soils under long-term continuous cotton cropping
To profile soil microbiome of the three cotton fields under continuous cropping regime in north Xinjiang, 15 soil samples were sequenced by Illumina MiSeq. A total of 1407,987 ITS1 and 484,724 V3-V4 16S rRNA high quality sequencing reads were analyzed. Coverages (> 99% for fungi; > 92% for bacteria) suggested that the identified sequences represented most fungi and bacteria present in the soil samples (Table 1). The high-quality sequences were gathered into 1344 fungal OTUs and 4717 bacterial OTUs at 97% sequence identity, respectively.
Table 1.
Alpha-diversity indices of soil microbiomes among three cotton fields. (Values are the means ± standard deviation, n = 5).
| Sample fields | Coverage | OTUs | Shannon | Chao1 | Evenness | Phylogenetic diversity | |
|---|---|---|---|---|---|---|---|
| Fungal Community | 5F | 0.999 ± 0.0001 | 871 | 4.84 ± 0.09 | 524.45 ± 1.40 | 0.55 ± 0.01 | 98.98 ± 4.55 |
| 7F | 0.998 ± 0.0001 | 842 | 4.22 ± 0.09 | 568.37 ± 1.29 | 0.48 ± 0.01 | 86.78 ± 3.03 | |
| 8F | 0.998 ± 0.0002 | 863 | 3.58 ± 0.26 | 531.44 ± 2.75 | 0.41 ± 0.03 | 88.68 ± 4.70 | |
| Bacterial Community | 5F | 0.923 ± 0.0009 | 3812 | 9.41 ± 0.06 | 3379.91 ± 3.13 | 0.84 ± 0.005 | 135.49 ± 1.01 |
| 7F | 0.928 ± 0.0003 | 3657 | 9.52 ± 0.01 | 3194.69 ± 9.38 | 0.86 ± 0.001 | 132.98 ± 1.04 | |
| 8F | 0.925 ± 0.0011 | 4717 | 9.43 ± 0.02 | 3309.12 ± 3.38 | 0.85 ± 0.002 | 129.61 ± 0.75 |
The diversity (Shannon), richness (Chao1), evenness and phylogenetic diversity indices of fungal and bacterial communities are shown in Table 1. There were significant differences in Shannon (P = 0.004) and evenness index (P = 0.005) of fungi, chao1 (P = 0.007), evenness index (P = 0.008) and phylogenetic diversity (P = 0.006) of bacteria among the three sample groups. Shannon (P = 0.032) index of bacteria showed significant differences among the soil samples of the three sample groups. Significantly higher diversity and evenness in 5F samples were found for fungi, with a lower score for bacteria.
Soil microbial community structures of long-term continuous cotton cropping fields
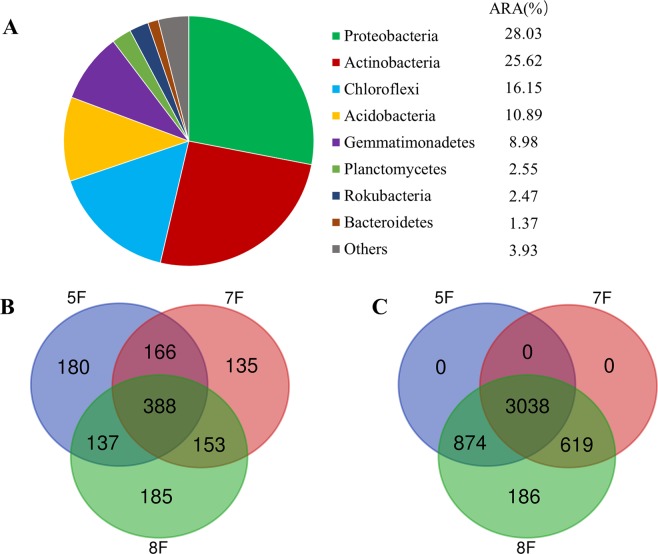
According to 97% species similarity, 11 eukaryotic and 29 prokaryotic phyla were identified from fungal ITS and 16S rRNA gene sequences, respectively. Fungal OTUs were predominantly composed of phyla Ascomycota (93.04%), Basidiomycota (2.15%) and Mortierellomycota (1.71%). Bacterial OTUs mainly consisted of Proteobacteria (28.01%), Actinobacteria (25.60%), Chloroflexi (16.14%), Acidobacteria (10.89%), Gemmatimonadetes (8.98%), Planctomycetes (2.54%), Rokubacteria (2.47%) and Bacteroidetes (1.37%) (Fig. 3A).
Figure 3.
The microbiome compositions of bacterial (A) taxa at phylum level, and Venn diagrams of fungal (B) and bacterial (C) OTUs in soil samples from the three fields. (Five samples in each group, phyla with average relative abundance (ARA) < 1% were merged and indicated as “Others”).
Further investigation was conducted through Venn diagram to identify the dominant OTUs present in the three cotton fields. The fungal and bacterial OTUs shared among the three fields represented 28.87% and 64.41% of total reads, respectively (Fig. 3B,C). For fungi, 13.39%, 10.04% and 13.76% unique OTUs were found in 5F, 7F and 8F, respectively, whereas 3.94% unique OTUs were found only in field 8F for bacteria.
Difference in soil microbial community compositions
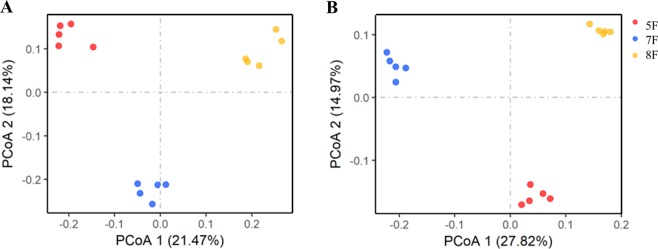
The soil microbiome compositions of different cotton fields at phylum level and UniFrac-unweighted principal coordinate analysis (PCoA) based on the OTU level were analyzed. In the 11 fungal and 29 bacterial phyla identified, 3 fungal and 8 bacterial phyla had a relative abundance above 1% in the three groups. At the phylum level, the dominant phyla (relative abundance above 5% at least in one sample) in three fields were Acomycota, Proteobacteria, Acitnobacteria, Chloroflexi, Acidobacteria and Gemmatimonadetes (Fig. 3A). PCoA analysis showed variations among the 15 soil samples for fungi and bacteria. Both fungal and bacterial communities from three cotton fields could be distinctly separated from each other (Fig. 4A,B).
Figure 4.
Unweighted unifrac- PCoA plots of fungal (A) and bacterial communities (B) at OTU level in three groups.
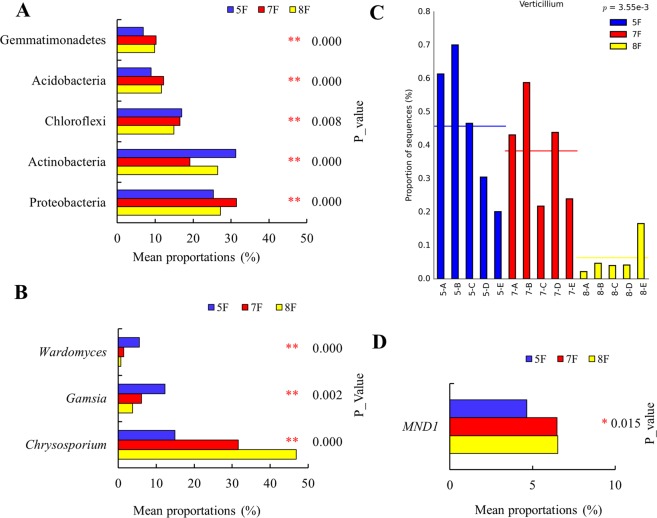
At the phylum and genus level, statistically significant differences were found in both fungal and bacterial taxa among three cotton fields. The dominant phylum in fungi was Ascomycota (5F = 91.13 ± 4.38%, 7F = 95.79 ± 0.51% and 8F = 92.19 ± 6.97%, P = 0.379), with no significant difference among the three fields. Dominant bacterial phyla were Proteobacteria (P = 0.000), Actinobacteria (P = 0.000), Chloroflexi (P = 0.008), Acidobacteria (P = 0.000) and Gemmatimonadetes (P = 0.000), which exhibited significant differences among the three sample groups (Fig. 5A).
Figure 5.
Comparison for abundance of fungal and bacterial sequences in the three sample groups. Bacterial (A) abundance at the phylum level. Fungal (B) and bacterial (D) abundances at the genus level. (C) Verticillium abundance in the three sample groups. (*P < 0.05; **P < 0.01).
253 fungal genera and 603 bacterial genera were identified, of which 47 (fungal) and 269 (bacterial) were differentially represented (P < 0.05). The dominant fungal genera were Chrysosporium, Gamsia and Wardomyces, with significant differential abundance among the three sample groups (Fig. 5B). Additionally, the relative abundance of Verticillium showed extremely statistically significant differences among the three groups (Fig. 5C). The dominant bacterial genera were an unidentified Gemmatimonadetes (5F = 6.61 ± 0.9%, 7F = 7.01 ± 0.51% and 8F = 10.11 ± 0.89%, P = 0.000), MND1 and an unidentified member of Proteobacteria (5F = 2.55 ± 0.29%, 7F = 9.34 ± 2.27% and 8F = 1.00 ± 0.26%, P = 0.000) (Fig. 5D).
Soil physicochemical properties and microbiome
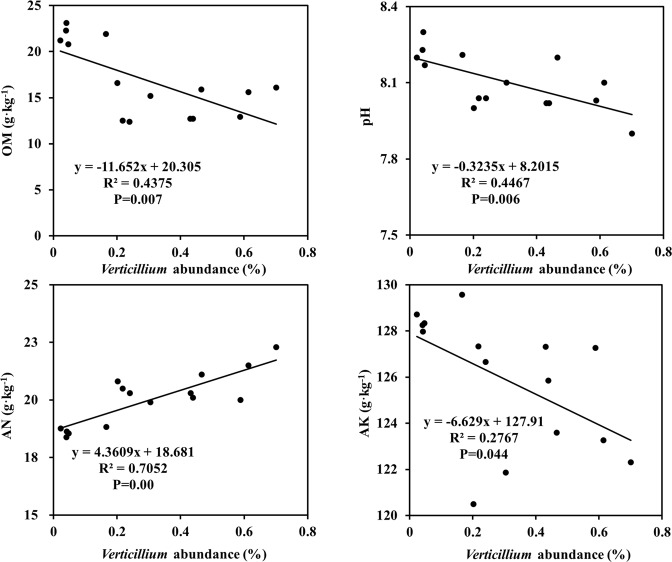
In this study, all soil variables but total phosphorus (TP) exhibited significant differences among the three cotton fields (Table 2). The results of Mantel tests revealed that fungal and bacterial community structures were connected to soil physicochemical properties (Table 3). Pearson correlation coefficient also showed alpha diversity indices had some significant positive or negative correlations with some of the measured soil variables (Table 4). Fungal Shannon and evenness indices were only positively correlated with soil alkali-hydrolyzable nitrogen (AN), but negatively correlated with organic matter (OM), available potassium (AK) and pH. In contrast, bacterial Chao1 was positive correlated with EC and negatively correlated with available phosphorus (AP), whereas evenness showed a positive correlation with AP. Besides, bacterial phylogenetic diversity showed positive correlation with AN, and a negative correlation with OM, AK and pH. Meanwhile, Verticillium abundance was significantly and positively correlated with soil AN, but negatively correlated with OM, AK and pH (Fig. 6).
Table 2.
Soil physicochemical properties of three cotton fields. (Values are means ± standard deviation. The same letter means no significant difference).
| Cotton fields |
Organic Matter (OM) |
Alkali-hydrolyzable nitrogen (AN) | Available phosphorus (AP) |
Available potassium (AK) | pH | Electrical conductivity (EC) | Total nitrogen (TN) | Total phosphorus (TP) |
|---|---|---|---|---|---|---|---|---|
| 5F | 15.88 ± 0.23b | 21.12 ± 0.40a | 7.41 ± 0.05c | 122.30 ± 0.55c | 8.06 ± 0.05b | 0.15 ± 0.00a | 0.04 ± 0.00b | 0.03 ± 0.00a |
| 7F | 12.64 ± 0.09c | 20.24 ± 0.09b | 15.15 ± 0.01a | 126.88 ± 0.29b | 8.03 ± 0.00b | 0.12 ± 0.00c | 0.05 ± 0.00ab | 0.02 ± 0.00b |
| 8F | 21.86 ± 0.41a | 18.63 ± 0.08c | 8.18 ± 0.02b | 128.57 ± 0.28a | 8.22 ± 0.02a | 0.14 ± 0.00b | 0.05 ± 0.00a | 0.02 ± 0.00ab |
Table 3.
Correlations between soil physicochemical properties and fungal or bacterial community by Mantel test. (*P < 0.05; **P < 0.01 n = 15, see Table 2 for variable acronyms).
| Soil variables | OM | AN | AP | AK | PH | EC | TN | TP |
|---|---|---|---|---|---|---|---|---|
| Fungal community | 0.3492** | 0.5591** | — | 0.7904** | 0.2746* | 0.4593** | 0.2303* | — |
| Bacterial community | 0.7461** | 0.3596** | 0.8382** | 0.3626** | 0.4005** | 0.5006** | — | — |
Table 4.
Correlations between soil physicochemical properties and fungal or bacterial community indices. (* P < 0.05; ** P < 0.01 n = 15, see Table 2 for variable acronyms).
| Soil variables | Fungi | Bacteria | ||||||
|---|---|---|---|---|---|---|---|---|
| Shannon | Richness | Evenness | Phylogenetic diversity | Shannon | Richness | Evenness | Phylogenetic diversity | |
| OM | -0.582* | -0.343 | -0.582* | -0.085 | −0.315 | 0.325 | -0.306 | -0.560* |
| AN | 0.693** | -0.122 | 0.717** | 0.172 | 0.24 | 0.038 | 0.126 | 0.753** |
| AP | -0.071 | 0.447 | −0.083 | -0.368 | 0.509 | -0.772** | 0.630* | -0.007 |
| AK | -0.780** | 0.149 | −0.799** | -0.505 | 0.193 | -0.481 | 0.349 | -0.713** |
| PH | -0.518* | -0.244 | -0.529* | -0.044 | -0.433 | 0.225 | -0.367 | -0.626* |
| EC | 0.419 | -0.386 | 0.446 | 0.345 | -0.338 | 0.650** | -0.488 | 0.410 |
| TN | -0.513 | 0.070 | −0.505 | -0.587* | 0.127 | -0.397 | 0.291 | -0.563* |
| TP | 0.395 | 0.094 | 0.388 | 0.535* | 0.104 | 0.477 | -0.098 | 0.346 |
Figure 6.
Correlations between Verticillium abundance and soil physicochemical properties (see Table 2 for variable acronyms).
Correlations between Verticillium and other genera abundance
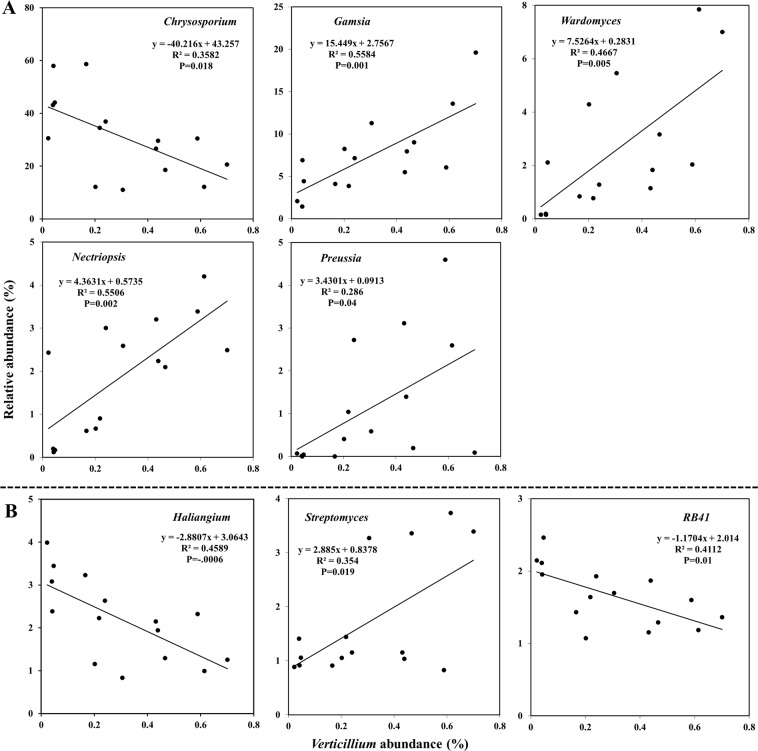
5 fungal genera and 17 bacterial genera were found at an average relative abundance > 1%. To determine which genera were closely associated to Verticillium, correlation analysis with other genera abundance was conducted. In total, 5 fungal and 6 bacterial genera showed significant correlations to Verticillium (Fig. 7). Considering fungi, Verticillium was significantly and positively correlated with the abundance of Chrysosporium, and negatively correlated with Gamsia, Wardomyces, Nectriopsis and Preussia (Fig. 7A). It was positively correlated with Streptomyces, but significantly and negatively correlated to Haliangium and RB41, among bacteria (Fig. 7B).
Figure 7.
Correlations between abundances of Verticillium and other fungal (A) or bacterial (B) genera. The genera selected with the standard of average relative abundance > 1% over 15 samples and P < 0.05.
Discussion
Verticillium wilt is widespread in many cotton-growing areas, and is the most important cotton disease in China21. The current cultivation system (such as long-term continuous cropping, straw returning to the field, drip irrigation, etc.) is very conducive for the occurrence of Verticillium wilt in Xinjiang. In this study, all the tested isolates belonged to the sclerotium type (Fig. S2), with defoliating and nondefoliating pathotypes accounting for 66.7% and 33.3%, respectively. A higher pathogenicity was found for the defoliating isolates, with an average disease index of 32.3, whereas the nondefoliating ones showed an average disease index of 22.6. The strong, moderate and weak pathogenicity isolates accounted for 33.33%, 53.33% and 13.33%, respectively. The results confirmed the importance of Verticillium wilt in Xinjiang, likely related to the high incidence of more and more defoliating and strong pathogenicity isolates. The cultural characteristics and pathogenicity of 15 isolates were significantly different, even when the isolates proceeded from the same cotton field (Table. S1). Some previous studies revealed that there was no correlation between isolate properties and pathogenicity22–24. However, we found that spore production was significantly and positively correlated with pathogenicity (Fig. S4).
Data on the relationship between Verticillium species and belowground microbiome under long-term continuous cotton cropping system appear important to understand the disease ecology. Due to the limited arable land area in Xinjiang, continuous cotton cropping is a common cultivation pattern. Besides, cotton is usually grown on a large scale in the region. Therefore, it was difficult to find an appropriate field to be used as comparison, nearby the selected cotton fields. Although cotton fields have similar fertilization and tillage patterns across the three regions, both fungal and bacterial soil communities showed differences in alpha- diversity, relative abundance, structure and taxonomic composition, as shown by Venn (Fig. 3B,C) and PCoA (Fig. 4A,B) analysis. The main reason for these differences may likely be related to the different cotton varieties used and to soil properties.
Microbial groups from the same habitat appeared similar, despite proceeding from different sampling sites. For fungi, Ascomycota was the most abundant fungal phylum under continuous cotton cropping, suggesting an enrichment and ubiquity in the cotton monoculture soil ecosystem, consistent with the results of previous studies carried out on soybean25, peanut26, and vanilla27 under continuous cropping regimes. Proteobacteria, Acitnobacteria, Chloroflexi, Acidobacteria and Gemmatimonadetes were the most common bacterial phyla in continuous cotton cropping soils (Fig. 3A). Proteobacteria abundance was highest agreeing with data from several previous studies16,27–30, with an important role in the global cycles of carbon, iron, nitrogen and sulphur31–34. Actinobacteria take part in the global carbon cycle35 and break down soil organic matter36. Thus, members of both phyla in continuous cotton cropping soils may have a role in the homeostasis of the soil microbiome.
Many soil properties may have affected microbial communities, according to results (Table 3). Verticillium abundance was negatively correlated with OM, AK and pH, and positively correlated only with AN (Fig. 6). Previous studies also revealed the correlations between OM and Fusarium abundance2,37, pH and tobacco disease rate16. Farmers in China applied large amounts of commercial nitrogen and phosphatic fertilizers to achieve higher yields, neglecting the role of organic matter and potash fertilizer for ages20. Excessive N and P fertilizers may be a disadvantage for plant growth38,39. A variety of soil factors exhibit potential synergistic effects and eventually influence the soil microbial community40, which probably underpin the Verticillium enrichment in soils.
Moreover, five fungal and six bacterial genera were found significantly correlated with the abundance of Verticillium (Fig. 7). The average relative abundance of these related fungal genera in total was 44.11%, much higher than those of the bacterial genera (16.18%), suggesting an important role of fungal communities in keeping V. dahliae infection. Furthermore, results also suggest that long-term continuous cotton cropping regime may affect the whole structure of soil microbiome primarily through abundance and depletion of certain fungal and bacterial taxa.
Besides the fungal pathogenic Verticillium, another soilborne pathogen Fusarium was detected. The introduced cotton varieties and the intensive cropping history were associated with the occurrence of Verticillium and Fusarium wilt in the cotton fields41. Although the average abundance of Fusarium (5.99%) was higher than that of Verticillium (0.30%), it showed no significant difference among the three fields. No evident Fusarium wilt could be found when we collected samples, which may due to the popular application of Fusarium wilt-resistant cotton cultivars.
Conclusion
In this study, we ascertained the cultural characteristics and pathogenicity differentiation of Verticillium dahliae in three cotton fields, from three regions in north Xinjiang. The results showed that sclerotium type, defoliating pathotype and moderate pathogenicity isolates were predominated in long-term continuous cotton cropping fields. The isolates showed pathogenicity differentiation, with spore production linked to pathogenicity. Meanwhile, the composition of fungal and bacterial taxa showed significant differences among the three cotton fields. Verticillium abundance was negatively correlated with organic matter, available potassium and pH, and positively correlated only with alkali-hydrolyzable nitrogen. Verticillium was also positively correlated with abundances of Chrysosporium and Streptomyces, and negatively correlated with fungi such as Gamsia, Wardomyces, Nectriopsis and Preussia, and bacteria such as Haliangium, RB41 and other 3 unidentified genera. These correlations suggested that fungal and bacterial communities around the cotton rhizosphere exhibited similar responses to long-term continuous cropping in north Xinjiang.
Materials and Methods
Sample collection and processing
All samples used in this study were collected from cotton fields in north Xinjiang on July 2017 (at the boll stage). One field only was chosen for each of the three main cotton producing cities, Bole (5F, N 44°25′57.035″; E 84°55′26.676″), Kuitun (7F, N 44°20′3.20″; E 86°03′31.85″), and Shihezi (8F, N 44°52′33.326″; E 82°08′43.828″), which have a long-term continuous cotton cropping history and show serious Verticillium wilt disease incidence.
The five V. dahliae isolates were randomly collected from five diseased cotton plants showing severe Verticillium wilt symptoms in each field. Single spore suspensions were collected and stored in 20% glycerol solution at −80 °C. Meanwhile, five bulk soil samples (15–20 cm in depth) around the roots of diseased cotton plants at the same sites were collected from each field, then put in separate plastic bags. A part of each soil sample was stored at −80 °C for subsequent DNA extraction, the other part being used for physicochemical analysis after air-drying according to previous methods27,42.
Extraction of fungal DNA and pathotype identification
Single spore isolates were grown on potato dextrose agar (PDA) plates for 3 days at 25 °C and then transferred into 20 mL Czapek Dox liquid medium. The mycelia were collected after 3 days to extract DNA using CTAB43,44. The DNA quality and concentration were assessed with 1% agarose gel electrophoresis and NANODROP 2000 spectrophotometer (Thermo SCIENTIFIC, USA). The pathotype was checked by PCR assays using two primer pairs, INTD2f (5′-ACTGGGTATGGATGGCTTTCAGGACT-3′)/INTD2r (5′-TCTCGACTATTGGAAAATCCAGCGAC-3′) and INTND2f (5′-CTCTTCGTACATGGCCATAGATGTGC-3′)/INTND2r (5′-CAATGACAATGTCCTGGGTGTGCCA-3′)45. PCR procedure: 95 °C for 4 min, followed by 30 cycles of 95 °C for 1 min, 60 °C for 1 min, 72 °C for 30 s, and a final extension of 72 °C for 5 min.
Cultural characteristics and pathogenicity of V. dahliae
Isolates V991 and 1cd3-2, previously characterized as defoliating (D) and nondefoliating (ND)46,47, were used as reference isolates, respectively. The growth of fungi was monitored by inoculated a plug (0.8 cm diameter) in the centre of PDA plate at 25 °C, measuring the colony diameters at 10 dpi, then photographed at 15 dpi. Each strain consisted of 5 replicates. To estimate conidia production, each isolate was incubated in 20 mL Bilay’s medium with 100 μL of a 2 × 105 spores/mL spore suspension, and incubated on a shaker (180 rpm/min) at 25 °C for 3 days48,49, in three replicates. The conidia number were counted under light microscope (BX52, Olympus, Tokyo, Japan).
Two differential hosts were used for pathogenicity assays in a greenhouse at 25 °C, with a 16 h light/8 h dark period. One resistant cultivar, Zhongzhimian 2 and a susceptible one, Xinluzao 36, were used. For each isolate, 30 cotton seedlings were inoculated, by immersing in 10 mL 1 × 107 spores/mL conidial suspension for each seedling. Sterile distilled water was used as control. Disease index (DI) was used to evaluate the severity of Verticillium wilt scaling from 0 to 450, using the following formula: DI = [Σ (disease grades × number of infected plants)/(total checked plants × 4)] × 10051. This experiment repeated twice, the data were presented as means ± standard deviation, significance difference was calculated by SPSS (v.17.0).
Soil DNA extraction, PCR amplification and illumina MiSeq sequencing
Total soil DNA was extracted from 15 samples using the DNeasy PowerSoil Kit (QIAGEN, Germany) according to the instructions. The fungi-specific primers pairs: ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′)52 and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′)53 were used to amplify the ITS1 region. The bacteria-specific primer pairs: 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805 R (5′-GACTACHVGGGTATCTAATCC-3′) were used to amplify the V3-V4 hypervariable region of 16S rRNA gene18. PCR reaction system and program were performed following a previous study27. PCR products were purified with Gel Extraction Kit (OMEGA, USA) and pooled in equimolar concentrations. Then, paired-end sequencing of fungal and bacterial amplicons was carried out on an Illumina MiSeq platform at Personal Biotechnology Co., Ltd (Shanghai, China)54.
Statistical analyses
After discarding the no-target amplicon sequence variants (ASVs) and low- abundance ASVs (<5 total counts)55, the dataset was normalized to the minimum number of read counts, and the analyses of alpha and beta diversity were performed with QIIME 256. Alpha-diversity analyses included Shannon, Chao1, richness, evenness, phylogenetic diversity and coverage, calculated using Mothur (v.1.34.4)57. Beta diversity was calculated by weighted UniFrac distance and analyzed by Principal coordinate analyses (PCoA)29. The Kruskal-Wallis test and permutational multivariate analysis of variance (PERMANOVA) with 999 random permutations were respectively used to analyze statistical differences in alpha-diversity and beta diversity58. Venn diagrams were implemented online to show unique and shared OTUs (Operational Taxonomic Units) (http://bioinformatics.psb.ugent.be/webtools/Venn/). Correlations between soil chemical properties and soil microbiome community were calculated by Mantel tests to construct dissimilarity matrices using R package (v. 3.5.3) via Bray-Curtis and Euclidean distance for bacterial and fungal community, respectively59. One-way analysis of variance with Turkey-Kramer test were used for the multiple comparison analyses using STAMP (v.2.0.0)60–62. The differences of growth rate, spore production, disease index and soil chemical properties were determined by one-way analysis of variance (ANOVA) or t tests in SPSS (v.17.0). Pearson correlations coefficients was used to test the correlation significance between spore production and disease index, or soil chemical properties and alpha-diversity indices or Verticillium abundance, or fungal/bacterial community and Verticillium abundance.
Supplementary information
Acknowledgements
This work was supported by National Natural Science Foundation of China (U1703231).
Author contributions
Longfu Zhu and Hui Xi initiated and designed the research. Hui Xi, Jili Shen, Dingyi Yang performed the experiments and Shiming Liu collected the data. Zheng Qu analyzed the raw data of ITS1 and 16S rRNA amplicon sequencing. Hui Xi analyzed the data and wrote the manuscript, Longfu Zhu and Xinhui Nie revised the manuscript. All authors reviewed and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54771-1.
References
- 1.Yang JI, Ruegger PM, McKenry MV, Becker JO, Borneman J. Correlations between root-associated microorganisms and peach replant disease symptoms in a California soil. PLoS One. 2012;7:e46420. doi: 10.1371/journal.pone.0046420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, et al. Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. PLoS One. 2014;9:e86610. doi: 10.1371/journal.pone.0086610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, et al. Response of soil fungal community structure to long-term continuous soybean cropping. Front Microbiol. 2018;9:3316. doi: 10.3389/fmicb.2018.03316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Elsas JD, Garbeva P, Salles J. Effects of agronomical measures on the microbial diversity of soils as related to the suppression of soil-borne plant pathogens. Biodegradation. 2002;13:29–40. doi: 10.1023/A:1016393915414. [DOI] [PubMed] [Google Scholar]
- 5.Garbeva P, van Veen JA, van Elsas JD. Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- 6.Klein E, Katan J, Minz D, Ofek M, Gamliel A. Soil suppressiveness against Fusarium crown and root rot of cucumber in organic-amended soil: Occurrence and possible mechanisms. Phytopathology. 2011;101(6):S92–S92. [Google Scholar]
- 7.Inderbitzin P, et al. Phylogenetics and taxonomy of the fungal vascular wilt pathogen Verticillium, with the descriptions of five new species. PLoS One. 2011;6:e28341. doi: 10.1371/journal.pone.0028341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Elsas JD, et al. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci USA. 2012;109:1159–1164. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oerke EC. Crop losses to pests. J Agr Sci. 2005;144:31–43. doi: 10.1017/s0021859605005708. [DOI] [Google Scholar]
- 10.Bennett AJ, Bending GD, Chandler D, Hilton S, Mills P. Meeting the demand for crop production: the challenge of yield decline in crops grown in short rotations. Biol Rev Camb Philos Soc. 2012;87:52–71. doi: 10.1111/j.1469-185x.2011.00184.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhu HQ, Jian GL. S X. X. Community structure of pathogenic type of Verticillium dahliae Kleb. in cotton field. Cotton. Science. 2004;16(3):147–151. [Google Scholar]
- 12.van der Heijden MG, Bardgett RD, van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 13.Sharma Sushil K., Ramesh Aketi, Sharma Mahaveer P., Joshi Om Prakash, Govaerts Bram, Steenwerth Kerri L., Karlen Douglas L. Sustainable Agriculture Reviews. Dordrecht: Springer Netherlands; 2010. Microbial Community Structure and Diversity as Indicators for Evaluating Soil Quality; pp. 317–358. [Google Scholar]
- 14.Berendsen RL, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12:1496–1507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwak MJ, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018;36:1110–1117. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 16.She S, et al. Significant relationship between soil bacterial community structure and incidence of bacterial wilt disease under continuous cropping system. Arch Microbiol. 2017;199:267–275. doi: 10.1007/s00203-016-1301-x. [DOI] [PubMed] [Google Scholar]
- 17.Xiong W, et al. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol and Biochem. 2017;107:198–207. doi: 10.1016/j.soilbio.2017.01.010. [DOI] [Google Scholar]
- 18.Shi W, et al. The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome. 2019;7:14. doi: 10.1186/s40168-019-0629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbl, J. A. K. U. B., & Záhora, J. A. R. O. S. L. A. V. The comparison of microbial activity in rhizosphere and non-rhizosphere soil stressed by drought. Proceedings of the Mendel Net, 234–240. 10.13140/RG.2.1.1243.5921 (2014).
- 20.Li Z, et al. Different responses of rhizosphere and non-rhizosphere soil microbial communities to consecutive piper nigrum L. monoculture. Sci Rep. 2016;6:35825. doi: 10.1038/srep35825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Y, et al. Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review. Afr. J. Biotechnol. 2009;8(25):7363–7372. [Google Scholar]
- 22.Jin LR, et al. The study on the pathogenicity differentiation of Verticillium dahliae in Hubei province. Cotton. Science. 2011;23(6):566–572. [Google Scholar]
- 23.Lin L, et al. Cultural characteristics and pathogenicity differentiation among strains of Verticillium dahliae from cotton in Jiangsu province. Cotton. Science. 2012;24(3):199–206. [Google Scholar]
- 24.Wang GN, et al. Pathogenicity and ISSR genetic differentiation of Verticillium dahliae isolates from cotton growing areas of Hebei province. Cotton Science. 2012;24(4):348–357. [Google Scholar]
- 25.Li C, Li X, Kong W, Wu Y, Wang J. Effect of monoculture soybean on soil microbial community in the Northeast China. Plant and Soil. 2009;330:423–433. doi: 10.1007/s11104-009-0216-6. [DOI] [Google Scholar]
- 26.Li XG, Ding CF, Zhang TL, Wang XX. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol Biochem. 2014;72:11–18. doi: 10.1016/j.soilbio.2014.01.019. [DOI] [Google Scholar]
- 27.Xiong W, et al. Different continuous cropping spans significantly affect microbial community membership and structure in a Vanilla-grown soil as revealed by deep pyrosequencing. Microb ecol. 2015;70(1):209–218. doi: 10.1007/s00248-014-0516-0. [DOI] [PubMed] [Google Scholar]
- 28.Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 29.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y, Gao L. Bacterial diversity in the rhizosphere of cucumbers grown in soils covering a wide range of cucumber cropping histories and environmental conditions. Microb Ecol. 2014;68:794–806. doi: 10.1007/s00248-014-0461-y. [DOI] [PubMed] [Google Scholar]
- 31.Hedrich S, Schlomann M, Johnson DB. The iron-oxidizing proteobacteria. Microbiology. 2011;157:1551–1564. doi: 10.1099/mic.0.045344-0. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, et al. Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl Microbiol Biotechnol. 2015;99:3291–3302. doi: 10.1007/s00253-014-6262-x. [DOI] [PubMed] [Google Scholar]
- 33.Klase G, et al. The microbiome and antibiotic resistance in integrated fishfarm water: Implications of environmental public health. Sci Total Environ. 2019;649:1491–1501. doi: 10.1016/j.scitotenv.2018.08.288. [DOI] [PubMed] [Google Scholar]
- 34.Rosenzweig N, Tiedje JM, Quensen JF, Meng Q, Hao JJ. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 2012;96(5):718–725. doi: 10.1094/PDIS-07-11-0571. [DOI] [PubMed] [Google Scholar]
- 35.Fradin EF, Thomma BP. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol. 2006;7:71–86. doi: 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- 36.Upchurch R, et al. Differences in the composition and diversity of bacterial communities from agricultural and forest soils. Soil Biol Biochem. 2008;40:1294–1305. doi: 10.1016/j.soilbio.2007.06.027. [DOI] [Google Scholar]
- 37.Shen Z, et al. Banana Fusarium wilt disease incidence is influenced by shifts of soil microbial communities under different monoculture spans. Microb Ecol. 2018;75:739–750. doi: 10.1007/s00248-017-1052-5. [DOI] [PubMed] [Google Scholar]
- 38.Malhia SS, Nyborgb M, Harapiak JT. Effects of long-term N fertilizer-induced acidification and liming on micronutrients in soil and in bromegrass hay. Soil Till Res. 1998;48:91–101. doi: 10.1016/S0167-1987(98)00097-X. [DOI] [Google Scholar]
- 39.Schroder JL, et al. Soil acidification from long-term use of nitrogen fertilizers on winter wheat. Soil Sci. Soc. Am. J. 2011;75:957–964. doi: 10.2136/sssaj2010.0187. [DOI] [Google Scholar]
- 40.Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Bio Fert Soils. 2012;48:489–499. doi: 10.1007/s00374-012-0691-4. [DOI] [Google Scholar]
- 41.Li X, Zhang Y, Ding C, Xu W, Wang X. Temporal patterns of cotton Fusarium and Verticillium wilt in Jiangsu coastal areas of China. Sci Rep. 2017;7:12581. doi: 10.1038/s41598-017-12985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Z, et al. Induced soil microbial suppression of banana Fusarium wilt disease using compost and biofertilizers to improve yield and quality. Eur J Soil Biol. 2013;57:1–8. doi: 10.1016/j.ejsobi.2013.03.006. [DOI] [Google Scholar]
- 43.Lee SB, Milgroom MG, Taylor JW. A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genet Rep. 1988;35(1):23. doi: 10.4148/1941-4765.1531. [DOI] [Google Scholar]
- 44.Wu Z, Wang T, Huang W, Qu Y. A simplified method for chromosome DNA preparation from filamentous fungi. Mycosystema. 2001;20:575. [Google Scholar]
- 45.Pérez-Artés E, Mercado-Blanco J, Ruz-Carrillo AR, Rodríguez-Jurado D, Jiménez-Díaz RM. Detection of the defoliating and nondefoliating pathotypes of Verticillium dahliae in artificial and natural soils by nested PCR. Plant Soil. 2005;268:349–356. doi: 10.1007/s11104-004-0378-1. [DOI] [Google Scholar]
- 46.Zhang WW, et al. Cotton gene expression profiles in resistant Gossypium hirsutum cv. Zhongzhimian KV1 responding to Verticillium dahliae strain V991 infection. Mol Biol Rep. 2012;39:9765–9774. doi: 10.1007/s11033-012-1842-2. [DOI] [PubMed] [Google Scholar]
- 47.Liu LL, et al. Resistance of cotton and tomato to Verticillium dahliae from cotton is independent on Ve1. Scientia sinica Vitae. 2014;44:803–814. doi: 10.1360/052014-90. [DOI] [Google Scholar]
- 48.Booth, C. The genus fusarium commonwealth mycological institute. Kew, Surrey, 237 (1971).
- 49.Ren L, et al. The components of rice and watermelon root exudates and their effects on pathogenic fungus and watermelon defense. Plant Signal Behav. 2016;11:e1187357. doi: 10.1080/15592324.2016.1187357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W-W, Jiang T-F, Cui X, Qi F-J, Jian G-L. Colonization in cotton plants by a green fluorescent protein labelled strain of Verticillium dahliae. Eur J of Plant Pathol. 2012;135:867–876. doi: 10.1007/s10658-012-0131-1. [DOI] [Google Scholar]
- 51.Zhang W, et al. Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis. PLoS One. 2017;12:e0181609. doi: 10.1371/journal.pone.0181609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delventhal R, et al. A comparative analysis of nonhost resistance across the two Triticeae crop species wheat and barley. BMC Plant Biol. 2017;17:232. doi: 10.1186/s12870-017-1178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White TJ. Amplification and direct seqencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;18(1):315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 54.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112:E911–920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson MJ. A new method for non‐parametric multivariate analysis of variance. Austral ecology. 2001;26(1):32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 59.Bonnet E, Van de Peer Y. zt: A sofware tool for simple and partial mantel tests. J Stat softw. 2002;7(10):1. doi: 10.18637/jss.v007.i10. [DOI] [Google Scholar]
- 60.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- 62.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.