Abstract
Bone morphogenetic proteins (BMPs) are secreted ligands of the transforming growth factor-β (TGF-β) family that control embryonic patterning, as well as tissue development and homeostasis. Loss of function mutations in the type II BMP receptor BMPR2 are the leading cause of pulmonary arterial hypertension (PAH), a rare disease of vascular occlusion that leads to high blood pressure in the pulmonary arteries. To understand the structural consequences of these mutations, we determined the crystal structure of the human wild-type BMPR2 kinase domain at 2.35 Å resolution. The structure revealed an active conformation of the catalytic domain that formed canonical interactions with the bound ligand Mg-ADP. Disease-associated missense mutations were mapped throughout the protein structure, but clustered predominantly in the larger kinase C-lobe. Modelling revealed that the mutations will destabilize the protein structure by varying extents consistent with their previously reported functional heterogeneity. The most severe mutations introduced steric clashes in the hydrophobic protein core, whereas those found on the protein surface were less destabilizing and potentially most favorable for therapeutic rescue strategies currently under clinical investigation.
Subject terms: Biochemistry, Genetics, Molecular biology, Structural biology, Molecular medicine
Introduction
Pulmonary arterial hypertension (PAH) is a devastating cardiovascular disease that affects up to 15 patients per million population1. The pathology typically presents in individuals aged 30–50 years who suffer vascular occlusion and high pressure in the pulmonary arteries due to the abnormal proliferation of pulmonary artery endothelial cells, pulmonary artery smooth muscle cells and fibroblasts2. Most patients succumb to cardiac failure within 3–5 years of diagnosis.
To date, sequencing studies have identified 16 causative genes for PAH pathology3. Mutations in the gene BMPR2 have emerged as the predominant risk factor accounting for some 53–86% of heritable cases and 14–35% of idiopathic cases4. The majority of these involve autosomal dominant non-sense and frameshift mutations that implicate BMPR2 haploinsufficiency as the pathogenic mechanism3. BMPR2 encodes for the bone morphogenetic protein (BMP) type II receptor kinase (BMPR2), which assembles with type I BMP receptor kinases to transduce BMP ligand signaling through the phosphorylation of transcription factors SMAD1/5/8 (Fig. 1)5. Significantly, loss of function mutations associated with vascular pathology have also been identified in the type I BMP receptor ALK1 (ACVRL1), their co-receptor endoglin (ENG) and their physiological ligands BMP9 (GDF2) and BMP10 (BMP10)3. These factors assemble specifically in endothelial cells to form a functional heteromeric signaling complex suggesting the loss of this signaling pathway as the basis for vascular dysfunction6.
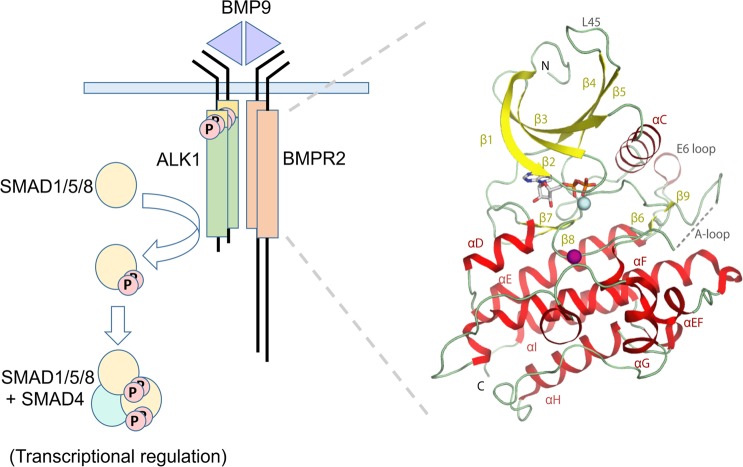
Figure 1.
Structural overview of the BMPR2 complex. Schematic of the canonical BMPR2 signalling complex implicated in PAH and related vascular disorders (left) and crystal structure of the recombinant BMPR2 kinase domain (right). Secreted dimeric ligands such as BMP9 assemble a heteromeric transmembrane receptor complex consisting of two type I receptors (ALK1 shown) and two type II receptors (BMPR2 shown). Ligand binding stimulates BMPR2-mediated phosphorylation of the GS domain in ALK1, which activates ALK1 for the subsequent recruitment and phosphorylation of SMAD1/5/8 proteins. Two of these phosphorylated SMADs then assemble with SMAD4 as a heterotrimer for nuclear translocation where they bind to promoter regions containing SMAD-binding elements for transcriptional regulation. In the BMPR2 crystal structure the secondary structure elements are labelled and shown as ribbons. ADP is shown in stick representation, and Mg2+ ion (cyan) and sulphate molecule (purple) are displayed as spheres. Disordered residues in the activation loop (A-loop) are indicated by a dotted line.
Over 400 different PAH-associated mutations have now been identified in the BMPR2 gene7. Importantly, some 25% of these are missense mutations where a protein product is likely to be expressed without non-sense mediated decay. These mutations are distributed throughout the gene affecting the extracellular ligand-binding domain, the intracellular kinase domain and the extended cytoplasmic tail. Functional studies have revealed dominant negative behavior, as well as heterogeneity in the effects of mutation8–10. Cysteine substitutions disrupting the extracellular cystine knot domain were found to cause retention of mutant BMPR2 within the endoplasmic reticulum, whereas kinase domain mutants were properly trafficked to the plasma membrane but lacked functional signaling through the SMAD1/5/8 pathway. An interesting exception was the kinase domain mutant E503D, which activated a SMAD-dependent transcriptional reporter construct as efficiently as wild-type BMPR210.
To date, structural reports on BMPR2 have been limited to the extracellular domain11. To better understand the structural consequences of PAH-associated missense mutations in the intracellular domain, we determined the crystal structure of the BMPR2 kinase domain at 2.35 Å resolution. The structure revealed three classes of mutations, including those causing buried steric clashes, those removing buried packing interactions and those found on the protein surface. Modelling of the mutants indicated that the surface mutations were the least destabilizing and therefore the most favorable for potential therapeutic rescue strategies under current clinical investigation12.
Results
Structure determination
To enable structural studies, the kinase domain of human wild-type BMPR2 was cloned into a bacterial expression vector providing a non-cleavable C-terminal hexahistidine tag and purified by Ni-affinity, size-exclusion and anion exchange chromatography. Diffracting crystals were obtained in the presence of the ligand ADP and the crystal structure was solved and refined at a resolution of 2.35 Å. Two protein molecules were present in the asymmetric unit. Both chains formed an identical interaction with Mg-ADP, but showed minor differences in the partially disordered activation loop as well as the C-terminus through which crystal contacts were formed. A summary of statistics for data collection and refinement is reported in Table 1.
Table 1.
Data collection and refinement statistics.
| Complex | BMPR2-ADP |
|---|---|
| PDB accession code | 3G2F |
| Synchrotron beamline | SLS, PX10 |
| Wavelength (Å) | 1.0000 |
| Spacegroup | P21212 |
| Unit cell dimensions |
a = 94.5, b = 218.8, c = 44.2 Å α = β = γ = 90.0° |
| Resolution rangea (Å) | 47.35–2.35 (2.47–2.35) |
| No. unique reflectionsa | 39,590 (5,694) |
| Completenessa (%) | 100.0 (100.0) |
| I/σIa | 8.5 (2.1) |
| Rmergea (%) | 12.0 (67.7) |
| Redundancya | 5.5 (5.5) |
| Refinement | |
| No. Protein atoms in refinement | 4934 |
| No. Heteroatoms, organic solvent and water | 204 |
| Rfact (%) | 20.8 |
| Rfree (%) | 24.7 |
| Bf protein (Å2) | 25 |
| Bf ligand (Å2) | 35 |
| rms deviation bond lengthb (Å) | 0.010 |
| rms deviation bond angleb (°) | 1.30 |
| Molprobity | |
| Ramachandran favour | 94.8% |
| Ramachandran allowed | 100.0% |
aValues in brackets show the statistics for the highest resolution shell.
brms indicates root-mean-square.
Conserved features of the BMPR2 kinase domain
BMPR2 shows the typical bilobal architecture of a protein kinase. The structure reveals a conserved fold with other type II receptors with a pattern of specific kinase domain insertions that define the BMP/TGF-β receptor family (Fig. 1). These include the L45 loop and the E6 loop as well as insertions flanking the αF helix and an insertion in the substrate pocket preceding the αG helix.
The BMPR2 kinase domain adopts an active conformation
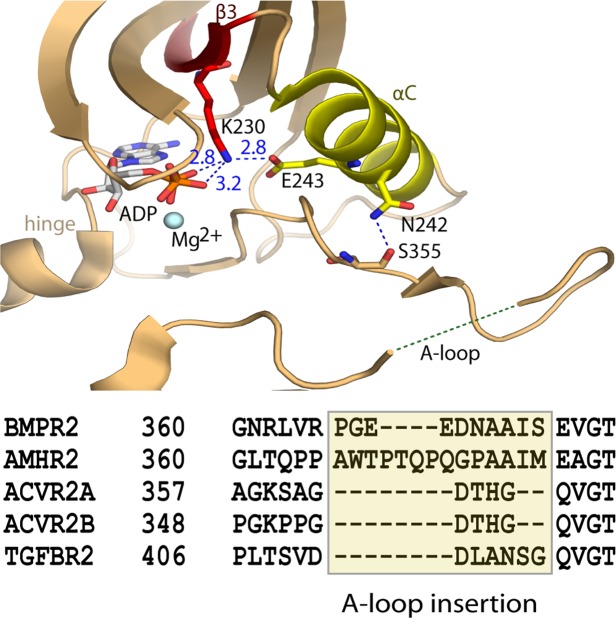
The type II receptor BMPR2 lacks the regulatory GS domain found in BMP type I receptors and is accepted to be constitutively active5. In agreement, the BMPR2 structure displays features characteristic for an active kinase (Fig. 2). The N and C-terminal kinase lobes adopt a closed conformation. The correct positioning of the αC helix is evidenced by the 2.8 Å salt bridge formed between the catalytic residues Lys230 (β3 strand) and Glu243 (αC helix) (Fig. 2). Canonical interactions are also observed with Mg-ADP, including two hydrogen bonds with the kinase hinge region and further hydrogen bonds with the catalytic loop and phosphate-binding loop (β1-β2), also known as the glycine-rich loop (Fig. 3). The activation loop (A-loop) makes surprisingly few contacts with the catalytic domain. Here, interaction with the αC helix is mediated by a hydrogen bond formed between Asn242 and the carbonyl of Ser355 (Fig. 2). The BMP/TGF-β receptor kinases differ from many other kinase families in that they are not regulated by activation loop phosphorylation and therefore lack the common interaction between this phosphate moiety and the αC. For this reason, we predict that further interaction with substrate is necessary to order this A-loop segment. Of note for such interaction, the A-loop in BMPR2 contains a six residue insertion relative to activin type II receptors, such as ACVR2B (also known as ActRIIB)(Fig. 2). By contrast, the loop conformation is well defined in the equivalent ACVR2B structure13 and appears stiffened by three proline residues that are not conserved in BMPR2 (Fig. 2).
Figure 2.
Active conformation of the BMPR2 catalytic domain. (A) BMPR2 adopts an active conformation as shown by a salt bridge between the catalytic residues K230 and E243. Electrostatic hydrogen bond and salt bridge interactions are denoted by a blue dashed line together with their distances (Å). A large insertion in the activation loop is disordered. A sequence alignment reveals insertions here in the activation loops of BMPR2 and AMHR2 compared to other type II receptors.
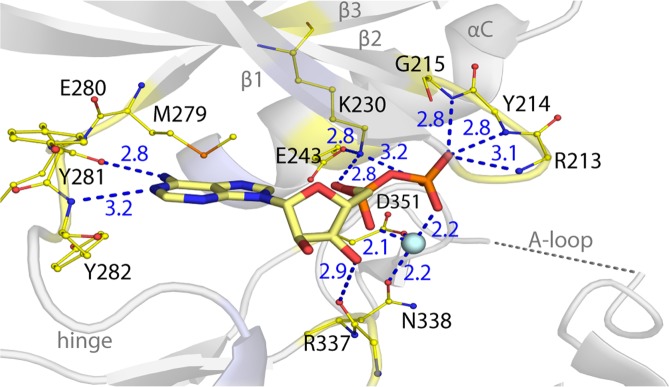
Figure 3.
ATP-pocket interactions of the bound Mg-ADP. BMPR2 forms canonical interactions with ADP and a single Mg2+ ion (cyan sphere). These include two hydrogen bonds with the kinase hinge region as well as further hydrogen bonds with the catalytic loop and phosphate-binding loop (β1-β2).
PAH-associated mutations destabilize the active BMPR2 structure
PAH-associated missense and nonsense mutation sites are located throughout the length of the BMPR2 protein. Due to their large number, we focus our analysis here on the subset of missense mutations reported by Machado et al. that fall within the kinase domain of BMPR2 (Fig. 4)14. The structure of BMPR2 reveals how these PAH-associated mutations effect a loss of function.
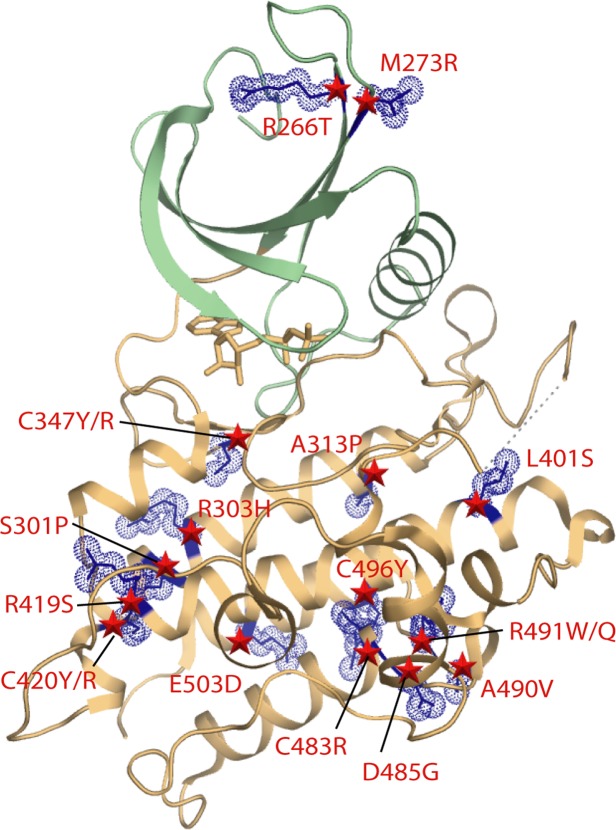
Figure 4.
Locations of PAH-associated missense mutations in the BMPR2 structure. Ribbon representation is shown for the kinase N and C-lobes and coloured green and brown, respectively. The positions of mutations are indicated by blue stick and space-fill representations of the wild-type side chains. The distribution of PAH-associated mutations in BMPR2 is widespread in the kinase C-lobe, but more limited in the N-lobe.
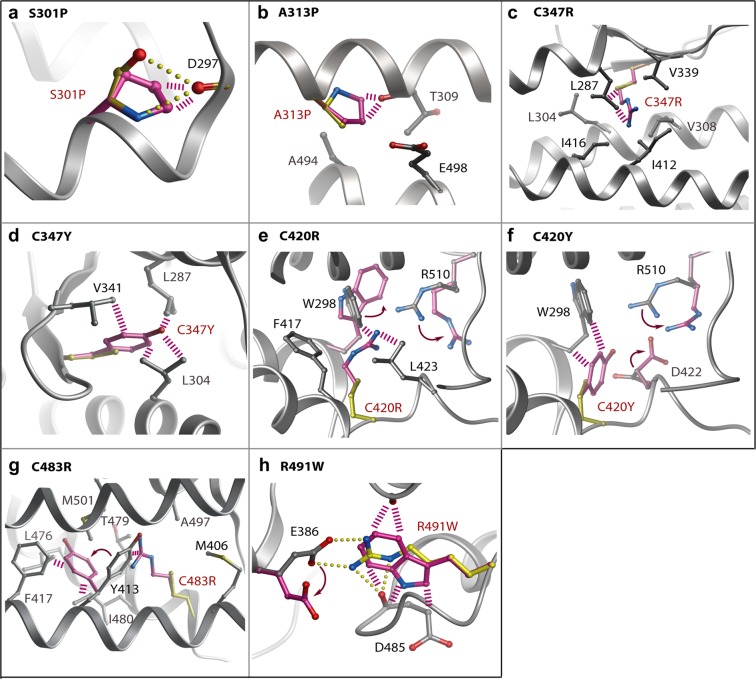
The most frequent missense mutations are buried in the kinase C-lobe. These mutations are predicted to severely disrupt the stability and folding of the catalytic domain (Table 2). Here, the most destabilizing mutations S301P, A313P, C347R/Y, C420R/Y, C483R and R491W are all predicted to introduce severe steric clashes (Fig. 5). Ser301 is located near the start of the long αE helix where it forms backbone and side chain hydrogen bonds with the backbone carbonyl of Asp297 to stabilize both the helix and the preceding loop conformation. Its mutation to proline would break both of these interactions, introduce local steric clashes and truncate the αE helix. Ala313 lies in the same αE where it packs against the C-terminal αI helix. A mutation to proline at this position would similarly truncate the αE helix resulting in the perturbation of the adjacent ATP-binding pocket. Notably, four of the most severe mutations affect cysteine residues in the hydrophobic core of the kinase C-lobe. The C347R and C347Y mutations located within the β8 strand introduce large bulky side chains that would form steric clashes with residues in the surrounding β7 strand, as well as with helices αD, αE and αF. Similar bulky substitutions arising from the C420R and C420Y mutations would affect the end of the αF helix and are predicted to disrupt proper folding through severe steric clashes with Trp298 (αE). The fourth cysteine position Cys483 (αH) packs in the hydrophobic core against residues in the αE and αI helices. Its mutation to arginine would introduce unsatisfied charge into the core and cause a severe clash with Tyr413 (αF). The final severe clash is introduced by the R491W mutation, which occurs within the loop connecting helices αH and αI. This mutation would disrupt a salt bridge with Glu386 in the short αEF helix, as well as hydrogen bonds to the backbone carbonyl of Asp485, another site of disease mutation.
Table 2.
Predicted destabilization of PAH-associated mutants of BMPR2.
| Class | Mutation site | Predicted ΔΔGa |
|---|---|---|
| 1. Buried steric clashes | S301P | N.D.b |
| A313P | >10 | |
| C347R | >10 | |
| C347Y | >10 | |
| C420R | >10 | |
| C420Y | >10 | |
| C483R | >10 | |
| R491W | >10 | |
| 2. Buried broken interactions | L401S | 4.97 |
| R419S | 7.41 | |
| R491Q | 5.94 | |
| 3. Exposed | R266T | 0.78 |
| M273R | 2.08 | |
| R303H | 5.57 | |
| D485G | 7.97 | |
| A490V | 5.56 | |
| C496Y | 4.93 | |
| E503D | 1.66 |
aStructural models and destabilization free energies were calculated using the Eris server (http://dokhlab.unc.edu/tools/eris/). The ΔΔG value defines the difference in the free energy between the wild-type and mutant structures. Values > 0 are destabilizing.
bNot determined. The S301P mutation was too severe for the Eris server to model. The structural model was instead calculated using ICM27.
Figure 5.
Modelling of PAH-associated mutations causing buried steric clashes. Structural models for the indicated mutants (A–H) were calculated using the Eris server (http://dokhlab.unc.edu/tools/eris/)26. Changes in the mutant structure are coloured purple and overlaid onto the wild-type residue coloured yellow. Hydrogen bonds in the wild-type (yellow) and clashes in the mutant structure (purple) are shown by spheres and dashed lines, respectively.
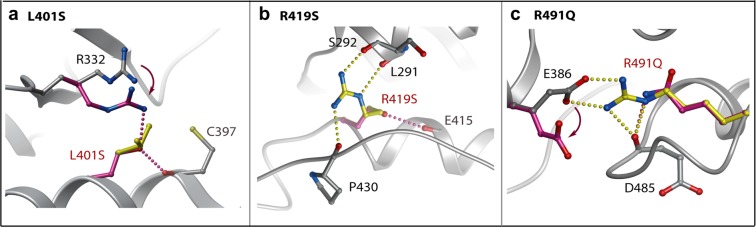
A further three PAH-associated mutations L401S, R419S and R491Q also affect buried sites in the protein structure, but result in smaller side chain substitutions that are predicted to disrupt packing interactions without introducing steric clashes (Fig. 6 and Table 2). Leu401 is located near the start of the large αF helix where it contributes to hydrophobic packing with the β9 strand. Its mutation to serine would break these van der Waals contacts and potentially perturb the proximal catalytic loop by forming a new hydrogen bond to Arg332. This arginine position falls within the highly conserved HRD motif of the eukaryotic kinases, which functions in catalysis by engaging the bound substrate as well as the kinase activation loop. Arg419 packs at the end of the αF helix where it forms 3 hydrogen bonds to tether the large αG-αH loop in close proximity to the shorter αD helix. Introduction of a serine at this position would break these bonds and disrupt the folding around this part of the kinase C-lobe. The R491Q mutation is predicted to be slightly less damaging than the R491W mutation described above, but will nonetheless break the salt bridge with Glu386 (αEF helix) to disrupt folding.
Figure 6.
Modelling of PAH-associated mutations disrupting buried interactions. Structural models for the indicated mutants (A–C) were calculated using the Eris server. Changes in the mutant structure are coloured purple and overlaid onto the wild-type residue coloured yellow. Hydrogen bonds in the wild-type (yellow) and clashes in the mutant structure (purple) are shown by spheres and dashed lines, respectively.
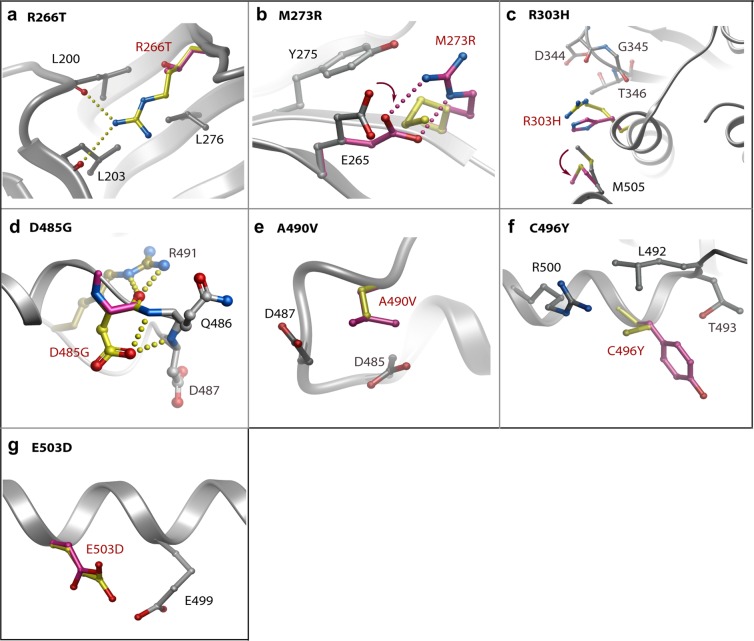
The remaining mutation sites are solvent exposed and generally predicted to be less disruptive (Fig. 7 and Table 2). A notable exception is the mutation D485G which breaks two hydrogen bonds that help to order the αH-αI loop. Perturbation of this loop risks disrupting two further backbone hydrogen bonds to R491, another site of disease mutation described above. R266T and M273R are more unusual in that they are located in the smaller N-lobe of the kinase domain. Arg266 (β4) forms hydrogen bonds with backbone carbonyls in the β1 strand that would be lost upon mutation causing some destabilization of the N-lobe β-sheet conformation. Met273 is an exposed residue in the β5 strand. Mutation to arginine introduces potential hydrogen bonding to Glu265 (β4) which may cause minor perturbation of the surface packing. The R303H (αE), A490V (αH-αI loop), C496Y (αI) and E503D (αI) mutations are not predicted to affect hydrogen bonding. Rather these substitutions may subtly influence the side chain packing at the surface of the kinase domain. The occurrence of apparently benign mutations at the protein surface suggests a potential role for some of these residues in protein-protein interactions. Further functional and structural studies are needed to define these interaction partners and their binding sites.
Figure 7.
Modelling of PAH-associated mutations that are surface exposed in the BMPR2 structure. Structural models for the indicated mutants (A–G) were calculated using the Eris server. Changes in the mutant structure are coloured purple and overlaid onto the wild-type residue coloured yellow. Hydrogen bonds in the wild-type (yellow) and clashes in the mutant structure (purple) are shown by spheres and dashed lines, respectively.
Discussion
Here we present a molecular model for the BMPR2 receptor catalytic domain and its deregulation in disease. BMP/TGF-β signaling is unusual in utilizing two distinct classes of transmembrane receptor kinases, the type II and type I receptors, respectively, which form a heteromeric complex for ligand binding and intracellular signaling. Biochemical assays have suggested that type II receptors such as BMPR2 are constitutively active, whereas the type I receptors require activating phosphorylations within their juxtamembrane GS domains by the type II receptors. Consistent with this view, the kinase domain structure of BMPR2 shows an active conformation as evidenced by the correct positioning of the αC helix, as well as the catalytic residues within the ATP-binding pocket which are captured in complex with Mg-ADP. However, a minor deviation from this state is apparent from some localized disorder in the activation loop. This observation potentially reflects the requirement for this flexible region to bind to the substrate type I receptor as they receptors assemble together upon extracellular ligand binding. Additional structures will be needed to elucidate the precise nature of this most important interaction.
PAH-associated mutations have been shown to abrogate receptor trafficking, assembly and downstream SMAD signaling8–10. The structure reveals how many of the missense mutations break interactions in the core of the BMPR2 structure to disrupt protein folding. Importantly for our understanding of the genotype-phenotype relationship, the structure also explains the contrasting effects of a subset of solvent-exposed mutations. Thus, the functionally defective mutant D485G disrupts a site of four critical hydrogen bonds, while the mutation E503D has no apparent structural consequences and has proven to be functional in cellular SMAD reporter assays10. This class of mutation may potentially interfere with BMPR2 function in alternative ways, for example disrupting other protein-protein interactions, as observed for C-terminal tail mutants which fail to bind to the kinase LIMK115.
Overall, all of the analyzed mutations were predicted to be destabilizing to some degree consistent with the accepted disease model of BMPR2 loss of function. Current rescue strategies under investigation include the use of small molecule chaperones to maximize receptor trafficking, as well the delivery of recombinant BMP9 to bolster the remaining signaling potential6,12. The varying extent of the predicted disruption suggests that different mutations will have different propensities for rescue. In this respect, we identify a subset of surface exposed mutations that would appear to be the most promising for further investigation.
Methods
Protein expression and purification
The kinase domain of human BMPR2 (residues 189–517) was subcloned into the pNIC-CH vector and transformed into E. coli strain BL21(DE3)R3-pRARE2 for expression. Cultures were induced with 1 mM IPTG overnight at 18 °C and the cells harvested and lysed by ultrasonication. Recombinant BMPR2 protein was recovered from the lysate by capture on Ni-IDA resin (Genscript) buffered in 50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 5 mM imidazole, 0.5 mM TCEP. Further purification was achieved by size exclusion chromatography using a S200 HiLoad 16/60 Superdex column (GE Healthcare) buffered in 25 mM HEPES pH 7.5, 300 mM NaCl, 1 mM DTT. A final clean up step was performed using a MonoQ 5/50 GL anion exchange column (GE Healthcare) with an elution gradient from 0 to 1 M NaCl in 50 mM HEPES pH 7.5 buffer. Proteins were stored at 4 °C buffered in 50 mM HEPES pH 7.4, 300 mM NaCl, 10% glycerol, 10 mM DTT, 50 mM L-arginine, 50 mM L-glutamate.
Crystallization
BMPR2 protein was concentrated to 5.8 mg/mL in a 10 kDa cut-off Amicon Ultra-4 filter before addition of 10 mM ADP. Crystallisation trials were performed at 4 °C and 20 °C using the sitting-drop vapour diffusion method. Diffraction quality crystals grew in a 150 nL crystallization drop containing an equal volume of the protein and a reservoir solution containing 25% PEG 8,000, 0.3 M ammonium sulphate, 0.05 M MgCl2, and 0.1 M sodium cacodylate pH 6.0.
Data collection and structure refinement
Crystals were cryoprotected with mother liquor plus 20% ethylene glycol for the BMPR2-ADP complex and vitrified in liquid nitrogen. Diffraction data were collected at Swiss Light Source, station PX10 using monochromatic radiation at wavelength 1.000 Å. Data were processed with MOSFLM16 and subsequently scaled using the program SCALA from the CCP4 suite17. Initial phases were obtained by molecular replacement using the program PHASER18 and the structure of ACVR2B (PDB 2qlu) as a search model. Density modification and NCS averaging were performed using the program DM19, and the improved phases were used in automated model building with the programs ARP/wARP20 and Buccaneer21. The resulting structure solution was refined using REFMAC5 from the CCP4 suite22 and manually rebuilt with COOT23. Appropriate TLS restrained refinement using the tls tensor files calculated from the program TLSMD24 was applied at the final round of refinement. The complete structure was verified for geometric correctness with MolProbity25.
Acknowledgements
The authors would like to thank the Swiss Light Source (SLS) station PX10 for beamtime. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z].
Author contributions
A.N.B. and F.v.D designed the research. C.T. performed protein expression, purification and crystallisation. A.C. solved the structure. A.C. and A.N.B prepared the figures and wrote the initial draft manuscript. All authors approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Humbert M, et al. Pulmonary arterial hypertension in France: results from a national registry. Am J Respir Crit Care Med. 2006;173:1023–1030. doi: 10.1164/rccm.200510-1668OC. [DOI] [PubMed] [Google Scholar]
- 2.Tuder RM, et al. Relevant issues in the pathology and pathobiology of pulmonary hypertension. J Am Coll Cardiol. 2013;62:D4–12. doi: 10.1016/j.jacc.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Southgate, L., Machado, R. D., Graf, S. & Morrell, N. W. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol, 10.1038/s41569-019-0242-x (2019). [DOI] [PubMed]
- 4.Machado RD, et al. Pulmonary Arterial Hypertension: A Current Perspective on Established and Emerging Molecular Genetic Defects. Hum Mutat. 2015;36:1113–1127. doi: 10.1002/humu.22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 6.Long L, et al. Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat Med. 2015;21:777–785. doi: 10.1038/nm.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austin ED, Loyd JE. The genetics of pulmonary arterial hypertension. Circ Res. 2014;115:189–202. doi: 10.1161/CIRCRESAHA.115.303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishihara A, Watabe T, Imamura T, Miyazono K. Functional heterogeneity of bone morphogenetic protein receptor-II mutants found in patients with primary pulmonary hypertension. Mol Biol Cell. 2002;13:3055–3063. doi: 10.1091/mbc.e02-02-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudarakanchana N, et al. Functional analysis of bone morphogenetic protein type II receptor mutations underlying primary pulmonary hypertension. Hum Mol Genet. 2002;11:1517–1525. doi: 10.1093/hmg/11.13.1517. [DOI] [PubMed] [Google Scholar]
- 10.Nasim MT, et al. Stoichiometric imbalance in the receptor complex contributes to dysfunctional BMPR-II mediated signalling in pulmonary arterial hypertension. Hum Mol Genet. 2008;17:1683–1694. doi: 10.1093/hmg/ddn059. [DOI] [PubMed] [Google Scholar]
- 11.Mace PD, Cutfield JF, Cutfield SM. High resolution structures of the bone morphogenetic protein type II receptor in two crystal forms: implications for ligand binding. Biochem Biophys Res Commun. 2006;351:831–838. doi: 10.1016/j.bbrc.2006.10.109. [DOI] [PubMed] [Google Scholar]
- 12.Ormiston ML, Upton PD, Li W, Morrell NW. The promise of recombinant BMP ligands and other approaches targeting BMPR-II in the treatment of pulmonary arterial hypertension. Glob Cardiol Sci Pract. 2015;2015:47. doi: 10.5339/gcsp.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S, Loulakis P, Griffor M, Xie Z. Crystal structure of activin receptor type IIB kinase domain from human at 2.0 Angstrom resolution. Protein Sci. 2007;16:2272–2277. doi: 10.1110/ps.073068407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machado RD, et al. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum Mutat. 2006;27:121–132. doi: 10.1002/humu.20285. [DOI] [PubMed] [Google Scholar]
- 15.Foletta VC, et al. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J Cell Biol. 2003;162:1089–1098. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 17.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr D Biol Crystallogr. 1998;54:487–493. doi: 10.1107/S0907444997011980. [DOI] [PubMed] [Google Scholar]
- 20.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/S0907444901014007. [DOI] [PubMed] [Google Scholar]
- 21.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 25.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 27.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;Suppl 1:215–220. doi: 10.1002/(SICI)1097-0134(1997)1+<215::AID-PROT29>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]