Abstract
The food additive titanium dioxide (TiO2), or E171, is a white food colorant. Recent studies showed after E171 ingestion a significantly increased number of colorectal tumours in a colorectal cancer mouse model as well as inflammatory responses and dysregulation of the immune system in the intestine of rats. In the mouse colon, E171 induced gene expression changes related to oxidative stress, impairment of the immune system, activation of signalling and cancer-related processes. E171 comprises nanoparticles (NPs) and microparticles (MPs). Previous in vitro studies showed that E171, NPs and MPs induced oxidative stress responses, DNA damage and micronuclei formation. This study aimed to investigate the relative contribution of the NPs and MPs to effects of E171 at the transcriptome level in undifferentiated Caco-2 cells by genome wide microarray analysis. The results showed that E171, NPs, and MPs induce gene expression changes related to signalling, inflammation, immune system, transport and cancer. At the pathway level, metabolism of proteins with the insulin processing pathway and haemostasis were specific to E171 exposure. The gene expression changes associated with the immune system and inflammation induced by E171, MPs, and NPs suggest the creation of a favourable environment for colon cancer development.
Subject terms: Cancer genomics, Cancer genomics
Introduction
Titanium dioxide (TiO2) is widely used as a food colorant in, among others, sweets, cookies, cake icing, and salad dressings1–3. TiO2 has been approved as a food additive in 1969 by the European Union (EU) under the name of E1714. This approval was based on a report written by the Joint WHO/FAO expert committee of Food Additives (JECFA) referring to the results of 5 studies published in 1927, 1928, 1950, 1955 and 1963 and one unpublished study in 1962 assessing TiO2 absorption and toxicity in animals after ingestion5. Based on the results of these studies, JECFA concluded that TiO2 was free from toxic effects on account of its insolubility and inertness6. Therefore the EU classified E171 as a non-active compound and there is no limit of authorised concentration in food products. In the USA, the Food and Drug Administration (FDA) approved the use of TiO2 as a food additive in 1966 and TiO2 should not exceed 1% by total weight of the product7.
E171 is a mixture of different particles with sizes in both the nano- and the micro-range. Characterisation of E171 by our group showed that E171 comprises 39% of nanoparticles (NPs) and 61% of microparticles (MPs)8. These results are in line with the size distribution observed in commercially purchased E171 from different manufacturers and in E171 extracted from food products by Scanning Electron Microscopy (SEM) or Transmission Electron Microscopy (TEM)1,9–11. These results showed that, in E171, the NPs fraction comprised 25 to 40% of all material, the remainder being MPs.
More recently, a limited number of animal studies have been published that provide evidence on the potential adverse effects after ingestion of E171 (2 in mice and 2 in rats). The first described effects of E171 ingestion in a murine colorectal cancer model12. After 10 weeks of intragastric ingestion of 5 mg/kg bw/day of E171 in combination with the genotoxicant azoxymethane (AOM) and the irritant dextran sodium sulphate (DSS), the number of tumours in the colon was significantly increased as compared to the control (AOM/DSS). The second in vivo study aimed to investigate the effects of E171 on the immune system in rats13. Bettini et al. showed that, after intragastric exposure of 10 mg/kg bw/day of E171 for 7 days, a decrease of regulatory T and Th cells and an increase production of IFN-γ in Peyer’s patches were observed which would eventually lead to an impairment of the immune system. A third study applying a long term (100 days) exposure, in a chemically induced carcinogenesis rat model, showed that E171 (10 mg/kg bw/day) promoted colon carcinogenesis by inducing preneoplastic lesions as well as the growth of aberrant crypt foci and mucosal low-grade inflammation13. The fourth in vivo study was performed on a normal BALB/c mouse model in which intragastric ingestion by oral gavage of 5 mg/kg bw/day of E171 was executed for 2, 7, 14, and 21 days14. Gene expression changes were observed in the colon which indicated oxidative stress responses, an impairment of the immune system, activation of signalling, and cancer-related genes.
More studies have been performed after ingestion of the smallest fraction of TiO2, the NPs fraction, indicating the translocation of the TiO2 NPs to the blood stream and other organs. Several groups showed that after intragastric administration of TiO2 NPs, these particles were found in the brain, blood, liver, and kidneys15–18. In the gut, inflammatory response via cytokine production or NLRP3 inflammasome, and dysregulation of the innate and adaptive immune system were identified after ingestion of TiO2 NPs13,19,20.
Two in vivo studies have been performed on MPs to assess translocation and toxicity. After intragastric ingestion, the particles were found in spleen, brain, while an inflammatory response in the small intestine was observed detected by an increased inflammatory cytokine production and T CD4+ cell proliferation16,20.
Few in vitro studies show the potential of E171 as well as TiO2 NPs to induce oxidative stress in Caco-2, Caco-2/HT29-MTX, human epidermal, and human amnion epithelial (WISH) cells9,21–23. In the exact same cell lines, DNA damage was also observed after exposure to E171 and NPs of TiO2. These results are in line with the results observed by our group where oxygen radical formation was detected in Caco-2 cells by the MPs of TiO2 and by E171 and NPs of TiO2 in a cell-free environment. Additionally, induction of DNA damage and micronuclei was measured after exposure to E171, NPs, and MPs of TiO2 in Caco-2 cells and HCT116 respectively8. In addition, induction of micronuclei was observed in HCT116 cells8. Another in vitro study showed that E171 extracted from gum disrupted the normal arrangement of constituent microvilli of the Caco-2BBe1 brush border24.
The aim of our study was to investigate the relative contribution of the NPs and MPs fractions to the effects of E171 at the transcriptome level. This investigation was performed using in vitro exposure of undifferentiated Caco-2 cells to E171 as well as the NPs and MPs fractions of TiO2 and assessing genome wide gene expression analysis. We specifically performed this study on undifferentiated Caco-2 cells because it was shown that these cells better reflect a gene expression pattern of human colon cancer cells in comparison to differentiated Caco-2 cells25. The set-up of the study design and transcriptomics analysis was linked to data on undifferentiated Caco-2 cells regarding dose-dependent cytotoxicity, oxidative stress DNA damage8. Undifferentiated enterocytes also form a subpopulation of cells in the colon and a comparison showed that undifferentiated Caco-2 cells are more sensitive towards particle toxicity and are therefore a preferred target in this type of studies21. We assumed that because of its small size and higher surface area, the NPs fraction would show the most pronounced effect on gene expression. Therefore, we first performed a time course study (2, 4, and 24 h of exposure) with this fraction to identify the most relevant time point at which the highest number of genes and related biological processes would be influenced. Next, we exposed undifferentiated Caco-2 cells to E171, and the MPs fraction at the optimal duration (24 h) to establish the relative contribution of each fraction to the various gene expression changes and related biological processes.
Results
Time course responses to TiO2 NPs exposure
We first exposed undifferentiated Caco-2 cells to TiO2 NPs for 2, 4, and 24 h to identify what time point would show the most abundant and pronounced transcriptomic response.
Differentially expressed genes
After LIMMA, DEG were identified per time point and shown in Table 1 including different fold changes (FC) cut-offs. Only with a FC ≥ 1.5 and a p-value < 0.05, a gradual increase of the number of DEG was observed. Furthermore, when using a less stringent FC like 1.2, noise might be introduced to the gene expression data while with a more stringent FC (1.8), part of the biological effect might be lost. Therefore, the value of 1.5 was used as a cut-off value for FC. Due to the absence of DEG when including an adjusted p-value, the analysis was further performed with a regular p-value of 0.05 and a FC ≥ 1.5. Under these conditions, 234 DEG were found after 2 h exposure, which slightly increased to 242 DEG after 4 h and to 281 DEG after 24 h.
Table 1.
DEG per time point after Caco-2 cells exposure to TiO2 NPs.
| FC ≥ 1.2 | FC ≥ 1.5 | FC ≥ 1.8 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 h | 4 h | 24 h | 2 h | 4 h | 24 h | 2 h | 4 h | 24 h | |
| |FC| | 3827 | 5220 | 3916 | 806 | 797 | 623 | 253 | 155 | 248 |
| Upregulated | 2004 | 2051 | 1978 | 595 | 503 | 234 | 214 | 63 | 169 |
| Downregulated | 1823 | 3169 | 1938 | 211 | 294 | 389 | 39 | 92 | 79 |
| p.val < 0.05 | 532 | 1991 | 470 | 532 | 470 | 1991 | 532 | 1991 | 470 |
| adj.p.val < 0.05 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 3 | 0 |
| |FC| and p.val | 434 | 1532 | 432 | 234 | 242 | 281 | 121 | 88 | 129 |
| |FC| and adj.p.val | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
A LIMMA test was used in R with a fold change (FC) of > = 1.2, 1.5 or 1.8 and a p-value < 0.05. DEG used for the analysis are shown in bold. |FC| = Fold Change, p.val = p-value, adj.p.val = adjusted p-value.
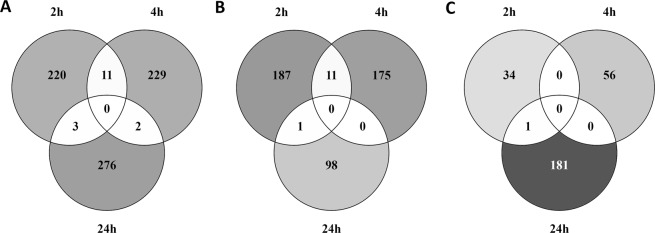
The overlap of DEG between the three time points was visualized using Venn diagrams, showing the overlap of all DEG (Fig. 1A), the overlap of upregulated DEG (Fig. 1B), and downregulated DEG (Fig. 1C) after exposure to TiO2 NPs for 2, 4, and 24 hours. Not a single gene was differentially expressed at all time points (Fig. 1A). There were 11 DEG overlapping between 2 and 4 h of exposure, all of which were upregulated (Fig. 1B). Two DEG were overlapping between 4 and 24 h of which 1 was upregulated and 1 downregulated (Fig. 1C) at both time points. Three DEG were overlapping between 2 and 24 h of exposure, none of these genes were expressed in the same direction at both time points. All the other DEG were time specific.
Figure 1.
Overlap of DEG over time after exposure to TiO2 NPs for 2, 4 or 24 hours. Venn diagrams representing the overlap of (A) all DEG, (B) upregulated DEG and (C) downregulated DEG.
Pathway and gene analyses
Using CPDB, the DEG for each time point were integrated in known pathways from different databases. The exposure to TiO2 NPs led to changes in gene expression that indicate different types of biological processes (Table 2). The result of this analysis is further described below.
Table 2.
Pathway over-representation analysis (ORA) of the DEG.
| Time of exposure | Biological function | Name pathway | p-value |
|---|---|---|---|
| 2 h | Signal Transduction | Olfactory receptor activity | 3.03E-09 |
| Olfactory transduction | 4.73E-09 | ||
| Olfactory Signalling Pathway | 8.72E-09 | ||
| GPCR downstream Signalling | 6.47E-07 | ||
| Signalling by GPCR | 1.60E-06 | ||
| GPCRs, Class A Rhodopsin-like | 0.00518 | ||
| Signal Transduction | 0.0125 | ||
| Immune system | Staphylococcus aureus infection | 0.0113 | |
| Dectin-2 family | 0.0125 | ||
| Metabolism | Vitamin A and Carotenoid Metabolism | 0.0297 | |
| Retinol metabolism | 0.0468 | ||
| Transport | Cation-coupled Chloride cotransporters | 0.0125 | |
| 4 h | Signal Transduction | Olfactory Signalling Pathway | 1.57E-05 |
| Olfactory receptor activity | 9.53E-05 | ||
| Olfactory transduction | 9.53E-05 | ||
| GPCR downstream Signalling | 0.000714 | ||
| Signalling by GPCR | 0.000807 | ||
| GPCR Signalling-G alpha i | 0.0223 | ||
| GPCR Signalling-pertussis toxin | 0.0223 | ||
| VEGF binds to VEGFR leading to receptor dimerization | 0.0223 | ||
| VEGF ligand-receptor interactions | 0.0223 | ||
| GPCR Signalling-cholera toxin | 0.0234 | ||
| GPCR Signalling-G alpha s Epac and ERK | 0.0234 | ||
| GPCR Signalling-G alpha q | 0.0234 | ||
| VEGF and VEGFR Signalling network | 0.0234 | ||
| Signal Transduction | 0.0234 | ||
| GPCR Signalling-G alpha s PKA and ERK | 0.0234 | ||
| Metabolism | Serotonin and melatonin biosynthesis | 0.0127 | |
| Bile acid and bile salt metabolism | 0.029 | ||
| Peptide hormone metabolism | 0.029 | ||
| Synthesis of bile acids and bile salts via 24-hydroxycholesterol | 0.029 | ||
| Biogenic Amine Synthesis | 0.037 | ||
| Metabolism of Angiotensinogen to Angiotensins | 0.0389 | ||
| Primary bile acid biosynthesis | 0.0407 | ||
| Amine-derived hormones | 0.044 | ||
| Transport | Ligand-gated ion channel transport | 0.00701 | |
| Transport of organic anions | 0.0234 | ||
| Transmembrane transport of small molecules | 0.029 | ||
| Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds | 0.0389 | ||
| Neuronal system | Serotonin and anxiety | 0.029 | |
| Serotonergic synapse | 0.0407 | ||
| Neuroactive ligand-receptor interaction | 0.0446 | ||
| 24 h | Signal Transduction | Gastrin-CREB Signal Transduction pathway via PKC and MAPK | 0.108 |
| Peptide ligand-binding receptors | 0.108 | ||
| Class A/1 (Rhodopsin-like receptors) | 0.108 | ||
| G alpha (i) Signal Transduction events | 0.128 | ||
| Immune system | Negative regulation of MAPK pathway | 0.131 | |
| Metabolism | Glucocorticoid Receptor Pathway | 0.00221 | |
| Glycine, serine, alanine and threonine metabolism | 0.0207 | ||
| Creatine metabolism | 0.108 | ||
| Prostaglandin Synthesis and Regulation | 0.108 | ||
| Metabolism of polyamines | 0.128 | ||
| Transport | Bile salt and organic anion SLC transporters | 0.128 | |
| Neuronal system | TarBasePathway | 0.128 | |
| HCN channels | 0.03 | ||
| Cancer | Photodynamic therapy-induced HIF-1 survival Signalling | 0.128 | |
| Gene expression (Transcription) | TP53 Regulates Transcription of Cell Cycle Genes | 0.128 | |
| Developmental biology | Endoderm Differentiation | 0.128 | |
| Developmental Biology | 0.108 | ||
| Haemostasis | Hemostasis | 0.0382 |
Pathways related to the DEG after ORA with ConsensusPathDB. The pathways were grouped per time points and per biological function.
After 2 h of exposure to TiO2 NPs, ORA showed that the modulated DEG were involved in pathways related to signal transduction with 7 pathways involved in olfactory, G protein-coupled receptor (GPCR) and other signal transduction. In addition, 2 pathways were associated with the innate immune system containing genes coding for C-type lectin domain, Fc fragment, HLA, and complement system. Two other pathways, vitamin A and retinol metabolism were affected after 2 h exposure. These 2 pathways were also implicated in increasing oxidative stress26. DEG after 2 h exposure were also involved in transport of molecules with HTR3B, GABRG2, and 2 solute carrier genes.
After 4 h of exposure to TiO2 NPs, 30 pathways associated to the DEG were observed after ORA. Different biological processes were identified such as signal transduction with 15 pathways involved in olfactory, GPCR, and VEGF signalling and metabolism with 8 pathways related to serotonin, bile acid, peptide hormones, and angiotensin metabolism. In addition, neuronal system was observed with 3 pathways associated to serotonin activity and neuroactive ligand-receptor interaction. Transport of small molecules was a biological function observed after 4 h exposure and contained 4 pathways involved in ligand-gated ion channel transport, transport of organic anion, and transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds.
After 24 h exposure, ORA identified 18 pathways affected after exposure to TiO2 NPs. These pathways were involved in a various number of biological processes such as signal transduction with 4 pathways related to gastrin-CREB, peptide ligand binding, and GPCR signalling. Metabolism was also observed with 5 pathways including 2 pathways (creatinine and prostaglandin metabolism) involved in a response to oxidative stress27,28. Neuronal system biological function was also detected with 2 pathways containing genes involved in HCN channels. In addition, 2 pathways were observed in developmental biology including endoderm differentiation, 1 in immune system: negative regulation of MAPK pathway, 1 in cancer: photodynamic therapy-induced HIF-1 survival signalling, 1 in haemostasis, 1 in gene expression (transcription): TP53 regulates transcription of cell cycle genes, and 1 in transport of small molecules: bile salt and organic anion SLC transporters.
Comparison of the different time points
No pathways after ORA were common to all time points but 7 were overlapping between 2 and 4 h of exposure to TiO2 NPs and were all related to signal transduction and transport of small molecules (Table 2). No affected pathways were overlapping between 4 and 24 h and between 2 and 24 h of exposure. Pathways classified according to their biological function showed that signal transduction, metabolism and transport of small molecules were common to all time points. In addition, between 2 and 24 h molecular changes were observed in the immune system.
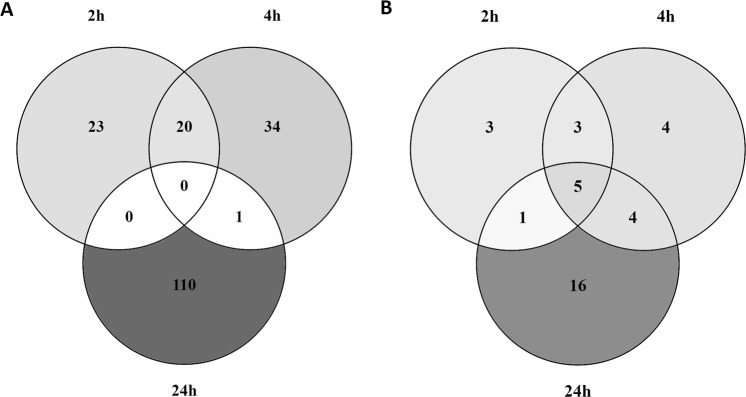
The same affected biological processes were observed when the GO terms associated to the DEG were analysed. Over time, the number of GO terms observed increased from 43 GO terms after 2 h of exposure, to 54 GO terms after 4 h and 111 GO terms after 24 h (Fig. 2A). Overlap was observed between 2 and 4 h with 20 GO terms involved in cellular response to stimulus, membrane, nervous system, receptor activity, signal transduction, and system process. Only one GO term was overlapping between 4 and 24 h, regulation of secretion. However, when the GO terms were classified according to their biological function, more overlap was observed. Figure 2B shows the different biological processes per time point with 12 after 2 h exposure, 12 after 4 h and 26 after 24 h. Of these 26 biological processes after 24 h exposure, 5 were in common with 2 and 4 h: cellular response to stimulus, nervous system, membrane, metabolism, immune system, 4 GO terms between 4 and 24 h: transport of small molecules, response to stress, secretion, and response to stimulus, and 1 GO term between 2 and 24 h: reproduction.
Figure 2.
Overlap of GO terms and their biological processes over time. Venn diagram representing (A): the number of GO terms in common between 2, 4, and 24 h exposure to TiO2 NPs, and (B): the associated biological processes to the GO terms in common between 2, 4, and 24 h exposure to TiO2 NPs.
The results show the most abundant and diverse gene expression and pathway changes after 24 h exposure, for which reason we choose this time point to establish gene expression responses following MPs and E171 exposure.
Comparison of transcriptome responses to different sizes of TiO2
Differentially expressed genes
In order to compare the gene expression responses induced by different fractions of E171, undifferentiated Caco-2 cells were also exposed to E171 and MPs for 24 h. Table 3 shows the number of DEG identified after LIMMA with the same FC and p-value cut-offs used for the time course with TiO2 NPs. After exposure to E171, the number of DEG was 3 times larger than after MPs and NPs exposure. As explained in the Materials and Methods, a new pre-processing was done on NPs with its time-matched control.
Table 3.
DEG after exposure of Caco-2 cells to different TiO2 particles for 24 h.
| NPs | MPs | E171 | |
|---|---|---|---|
| |FC| > = 1.5 | 415 | 365 | 1040 |
| Upregulated | 148 | 136 | 63 |
| Downregulated | 267 | 229 | 977 |
| p.val < 0.05 | 664 | 459 | 1198 |
| adj.p.val < 0.05 | 0 | 0 | 0 |
| |FC| and p.val | 121 | 121 | 365 |
| |FC| and adj.p.val | 0 | 0 | 0 |
A LIMMA test was used in R with a fold change (FC) of >=1.5 and a p-value < 0.05 after in vitro exposure to E171, MPs, and NPs of TiO2. The number of DEG used for further analysis is shown in bold. |FC| = Fold Change, p.val = p-value, adj.p.val = adjusted p-value.
Figure 3 shows that the DEG after exposure to the different sizes of TiO2 were mostly specific to each fraction, only a few genes were in common. One DEG was in common after MPs and NPs exposure: LGALS14, a galectin gene. Two DEG were in common between the NPs and E171 exposure: TCP11X2, a T-complex family gene and KCCAT333, a renal clear cell carcinoma-associated transcript but the exact function of these 2 genes remains unknown. When comparing the DEG after MPs and E171 exposure, 3 DEG were observed: WDR78, LOC401478, and ANKRD26P3. WDR78 gene is shown to be a target of the hedgehog activity. LOC401478 and ANKRD26P3 have unknown functions.
Figure 3.
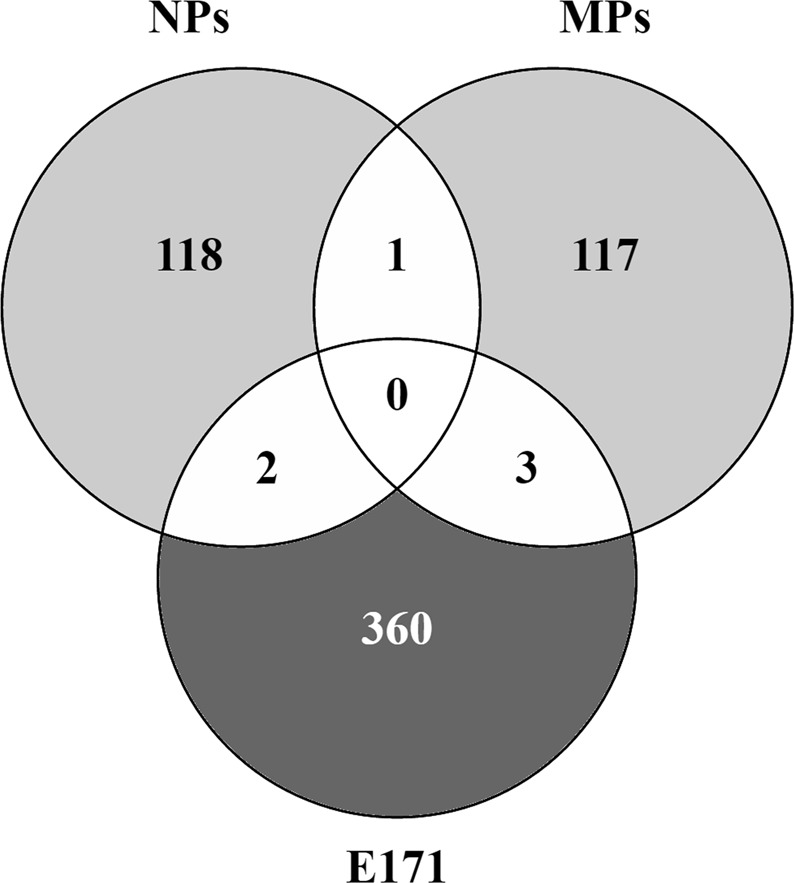
Unique and common DEG between E171 and the different size fractions of TiO2. Venn diagram showing the overlap of DEG after exposure of undifferentiated Caco-2 cells to E171, MPs, and NPs for 24 h.
Pathway and gene analyses
E171 exposure
ORA showed that after E171 exposure, 9 pathways involved in signal transduction were observed; 4 of them were related to GPCR, 3 to olfactory signalling, 1 to signal transduction and 1 to bone morphogenic protein signalling (Table 4). In addition, gene expression was affected in one pathway involved in haemostasis (endothelins), one in neuronal system (dopaminergic neurogenesis), one in metabolism of proteins (insulin processing), and another one in transport of small molecules (zinc efflux by solute carriers family).
Table 4.
Pathways associated with the DEG after NPs, MPs and E171 exposure.
| Time of exposure | Biological function | Name pathway | p-value |
|---|---|---|---|
| E171 | Signal Transduction | GPCR downstream signalling | 1.09E-08 |
| Signalling by GPCR | 3.75E-07 | ||
| Olfactory Signalling Pathway | 2.05E-06 | ||
| Olfactory transduction - Homo sapiens (human) | 3.36E-05 | ||
| Olfactory receptor activity | 6.69E-05 | ||
| GPCR ligand binding | 0.000236 | ||
| Signal Transduction | 0.000461 | ||
| G alpha (q) signalling events | 0.00222 | ||
| Bone Morphogenic Protein (BMP) Signalling and Regulation | 0.00845 | ||
| Transport of small molecules | Zinc efflux and compartmentalization by the SLC30 family | 0.00406 | |
| Neuronal system | Dopaminergic Neurogenesis | 0.0004 | |
| Developmental Biology | Interaction between L1 and Ankyrins | 0.0041 | |
| Haemostasis | Endothelins | 0.00989 | |
| Metabolism of proteins | Insulin processing | 0.00406 | |
| MPs | Signal Transduction | Olfactory Signalling Pathway | 0.0579 |
| Olfactory transduction - Homo sapiens (human) | 0.0579 | ||
| Developmental Biology | CDO in myogenesis | 0.0654 | |
| Myogenesis | 0.0654 | ||
| Netrin-1 signalling | 0.0775 | ||
| NPs | Metabolism | Glycine, serine, alanine and threonine metabolism | 0.000543 |
| Creatine metabolism | 0.000789 | ||
| Urea cycle and metabolism of amino groups | 0.00514 | ||
| Glucocorticoid Receptor Pathway | 0.00541 | ||
| Prostaglandin Synthesis and Regulation | 0.00861 | ||
| Neuronal system | HCN channels | 8.60E-05 | |
| TarBasePathway | 0.00126 | ||
| Potassium Channels | 0.00689 | ||
| Transport of small molecules | Amino acid transport across the plasma membrane | 0.00926 |
ORA analysis performed with CPDB on all the DEG after NPs, MPs and E171 exposure in vitro for 24 h.
In addition, gene analysis showed biological effects on the immune system with 34 DEG including BCL10, CD274, COLEC10, defensin, and IFI27 genes. Gene expression in metabolism was also influenced by E171 exposure with a total of 22 DEG including genes involved in ATP binding, cytochrome P450, bile acid, and prostaglandin. With 13 DEG involved in cholinergic receptor, calcium channels, potassium channels, and glutamate receptor, neuronal system was also affected by E171 exposure. The gene expression (transcription) biological function was altered by 12 DEG such as MTERF1 (mitochondrial transcription), SMURF1 (ubiquitin related gene), HIST1H3I (histone cluster), zinc finger genes, and TP53I11 (tumour protein p53 inducible protein 11). With 10 DEG each, 2 biological processes were modulated, cellular responses to external stimuli with genes coding for prostaglandin protein, histone cluster, and heat shock protein and disease with NAG20, NRG1, SLA2, CSNK1A1L (signalling by WNT in cancer). E171 exposure also affected, with less than 10 DEG each, gene expression in other biological processes such as metabolism of RNA, DNA repair, cell cycle, muscle contraction, extracellular matrix organization, organelle biogenesis and maintenance, cell-cell communication, chromatin organization, circadian clock, DNA replication, programmed cell death, and reproduction.
MPs exposure
ORA showed that after MPs exposure, signal transduction was affected by the modulation of gene expression in 2 olfactory pathways. In addition, pathways involved in developmental biology were observed, mostly involved in the myogenesis (Table 4).
Gene analysis showed that after exposure to MPs for 24 h, gene expression was also affected in the immune system with defensins, interleukins, THRIL, BTC, TIMP4, and MPEG1 genes. In addition, 8 genes were related to transport of small molecules and vesicle-mediated transport (solute carriers, ABCG4, CLIC2, KIF25, MYH13 and LCN9). One DEG, up-regulated after MPs exposure, was found to be up-regulated in presence of oxidative stress (UCP1)29. Cancer development was observed by the modulation of genes involved in cell death (HIST1H1T, CASP14, and DNAJC3), cancer related genes (WNT8A and CASC8), gene expression (transcription) (ZNF570 and ZFP42), and metabolism of proteins (CALB2 and KLHDC3). Three DEG were found to be related to the extracellular matrix organisation (COL17A1, ARC, and TIMP4). Six others were modulated in the developmental biology biological function (CDH4, KRT14, POU3F3, NTN3, RGMA, and HFE2), 3 DEG in metabolism (ARG1, TPH2, and PNPLA2), and one DEG in neuronal system (CHRNA3).
NPs exposure
As shown in Table 3, the new pre-processing was statistically more stringent. More than 90% of the DEG of the pre-processing of 24 h only was also present in the time course analysis. A pathway and gene analysis were performed with the 121 DEG of the new pre-processing and compared to the one performed with the 281 DEG of the time course analysis. The biological response after NPs exposure after 24 h exposure was similar to the 24 h time point described in the time course with pathways involved in HCN channels, transport of small molecules, and 5 metabolism pathways including creatinine and prostaglandin also involved in response to oxidative stress response (Table 4).
Common and specific biological reactions
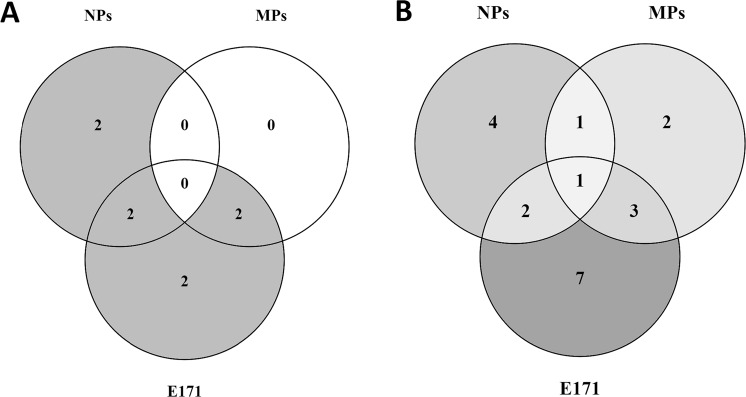
Pathways extracted after ORA showed no common response between all the particles, all pathways were specific to each fraction. However, Fig. 4A shows that, after E171 and NPs exposure, 2 biological processes were in common: transport of molecules and neuronal system. After E171 and MPs exposure, signal transduction and developmental biology were common processes affected by gene expression changes. Both signal transduction pathways are related to the olfactory transduction.
Figure 4.
Overlap of the biological processes between the different sizes of TiO2. Venn diagram showing after exposure of undifferentiated Caco-2 cells to E171, MPs, and NPs for 24 h (A): the overlap of biological processes after ORA, (B): the overlap of the biological processes related to the GO terms.
Specific biological processes related to the DEG were also observed after NPs exposure: metabolism and response to stress. All biological processes after MPs exposure were common to E171 exposure. After E171 exposure, 2 specific biological reactions were observed: haemostasis and metabolism of proteins.
Figure 4B shows that one biological process extracted from the GO terms was common between the different types of TiO2, which is cellular response to stimuli. Furthermore, after E171 and NPs exposure gene expression was affected in 2 common biological processes: Transport and Membrane. Other common effects on gene expression were identified after E171 and MPs exposure in signal transduction, system processes, and neuronal system. After MPs and NPs exposure DEG were observed in the Binding biological process.
After NPs exposure, specific biological processes extracted from the GO terms of the DEG were related to metabolism, haemostasis, oxidative stress, and regulation of biological process. Two specific biological processes were observed after MPs exposure: reproduction and extracellular matrix organisation. The number of biological processes observed after E171 exposure was increased compared to NPs and MPs with receptor activity, immune system, DNA repair, muscle adaptation, catalytic activity, organ development, and transcription factor.
Discussion
The aim of this study was to assess the gene expression changes in colonic cells after in vitro exposure to E171 and the contribution of the NPs and MPs fractions to these changes based on transcriptome analysis in undifferentiated Caco-2 cells. A transcriptomics study confirmed that in comparison to differentiated Caco-2 cells, undifferentiated Caco-2 cells better reflect a gene expression pattern of human colon cancer cells25. So this cell model shows a higher compatibility with our previous study in a mouse model of colon cancer that showed that E171 increased the number of tumours in the colon specifically12.
Although the majority of the DEG were not common between time points, this does not imply that the biological responses observed at earlier time points are transient. Interpretation of the biological functions of the genes using e.g. ORA and GO terms identified immune response, signal transduction and cancer signalling to be modulated after 2, 4, and 24 h. These dynamic biological responses are involved in consecutive mechanisms towards an increased risk of colon cancer development. First the optimal time point for the evaluation of gene expression responses was established to be 24 hours because this time point showed the highest number of DEG and affected pathways. Subsequently, cells were also exposed for 24 hours to MPs and E171.
A previous study performed by our group showed that TiO2 NPs induced ROS formation only in a cell-free environment8. In the current study, at the gene level, oxidative stress and related responses were observed: After 4 h the up-regulation of a protein coding gene (GML) involved in cellular response to stress and heat shock response and after 24 h with 2 DEG (TUBB2B and SCML4) involved in cellular response to stress, oxidative stress induced senescence and in a heat shock protein (HSP90) chaperone cycle. These results confirm previous studies which showed the capacity of TiO2 NPs to induce significant levels of oxidative stress in a cell-free environment as well as in vitro in Caco-2 and in human amnion epithelial (WISH) cells9,21–23. Oxidative stress is one of the factors that can induce DNA damage. Previously, we reported that TiO2 NPs induced significant levels of DNA damage after exposure to TiO2 NPs in undifferentiated Caco-2 cells as well as induction of micronuclei in HCT116 cells8. DNA damage was also shown by other studies performed in Caco-2, Caco-2/HT29-MTX co-culture system, and WISH cells after exposure to TiO2 NPs9,21–23. Here we show that, after 4 h exposure, 4 DEG (HAP1, GML, SPIDR, and REM1) were significantly expressed that are involved in DNA damage and repair. Two DEG were involved in the mechanisms of DNA repair activity either via a p53-target gene (GML)30 or via homologous recombination (SPIDR). Furthermore, HAP1 is known to be activated after DNA damage31 and REM1 expression is also shown to be modulated in CRC samples32.
Exposure to TiO2 NPs also induced gene expression changes related to the immune system. These changes start to manifest after 2 h via modulation of the expression of C4B and C3AR1 genes from the complement system as well as HLA-DRB5 gene involved in TCR signalling. After 4 h, no pathways involved in immune system were found, however, at the GO term level, cytokine production and tumour necrosis factor were observed. After 24 h exposure to TiO2 NPs, similar effects to 2 and 4 h exposure on the immune system were observed like on the innate immune system. In addition, specific gene expression changes related to IL-1 production, leukocyte migration, and chemokine signalling were observed, suggesting an impact on the innate and adaptive immune system. Overall 24 h exposure was the most pronounced time point regarding effects on the immune system because the majority of the 2 and 4 h exposure effects were found, including specific 24 h effects. TiO2 NPs exposure in vivo demonstrated impact on the immune response via a potent Th1/Th17 immune response after exposure to 10 mg/kg bw/day in rats of P25 TiO213. Our results show that the induction of the immune response by TiO2 NPs might not only be induced via immune related organs, as seen in vivo, but also as a consequence of direct exposure of intestinal epithelium cells.
Furthermore, transport of small molecules and vesicle-mediated transport were observed by gene expression changes of solute carrier genes from 2 h to 4 h and 24 h exposure with 3, 7 and 7 DEG respectively. In addition, 2 cadherin genes, which play a role with cargo proteins in clathrin-independent endocytosis33, differentially expressed (4 h: CDH18, 24 h: PCDH9). Induction of genes in transport pathways may be explained by the small size and high surface area of TiO2 NPs. Previous experiments showed that TiO2 NPs can enter the cells as well as the nucleus22,23,34 and suggest that solute carriers and endocytosis might be two mechanisms by which the particles enter the cells34–36.
Overall, Fig. 2 shows that after 24 h exposure to TiO2 NPs, the gene expression changes were partly similar to other time points but also more diverse. Therefore, 24 h exposure was chosen to be the optimal time of exposure for the comparison of the different particles of TiO2. Effects of MPs and E171 at the molecular levels as well as the contribution of each fraction to these effects will be described below.
Like for the NPs, we have shown that MPs also induce oxidative stress after 1 h exposure in undifferentiated Caco-2 cells8. We found a significant up-regulation of UCP1, a gene with a very strong link to mitochondrial oxidative stress and DNAJC3, related to endoplasmic reticulum stress. UCP encodes for uncoupling proteins which limits ROS production in the mitochondrial respiratory chain after stimulation by O2.−37,38. DNAJC3 encodes for a protein involved in the unfolded protein response during endoplasmic reticulum stress39. Oxidative stress and cellular stress observed after MPs exposure might explain the induction of DNA damage and micronuclei previously observed after exposure of undifferentiated Caco-2 cells8. Here, gene expression change of one histone gene (HIST1H1T) was observed, which is involved in DNA repair40,41. In addition, after exposure to TiO2 MPs, DEG related to cancer, cell death, and gene transcription were observed like WNT8A and CASC8 which are highly correlated with CRC development and progression42,43. Furthermore, CASP14, a potent pro-apoptotic gene but also potentially involved in cell differentiation44 and RD3 a down-regulated gene normally expressed in the colon, while loss is associated with the development of neuroblastoma and other cancers45, were observed. So TiO2 MPs induced gene expression changes seem to indicate molecular responses that could reflect facilitation of cancer development.
Like exposure to TiO2 NPs, MPs also affected gene expression changes in relation to the immune system with the down-regulation of pro-inflammatory cytokine receptor IL-6R involved in the MAPK1 and MAPK3 activation and also in response to oxidative stress46. Other genes involved in the innate immune system such as defensins, interleukins, THRIL, BTC, TIMP4, and MPEG1 were additionally affected. TiO2 MPs ingested in the diet via E171 induced a pro-inflammatory and immune response which could increase the symptoms of inflammatory bowel disease via the NLRP3 inflammasome47 or when MPs are bound to Pathogen Associated Molecular Patterns (PAMPs) in the gastrointestinal lumen48.
Seven DEG were observed involved in transport of small molecules and vesicle-mediated transport. This included ABCG4, coding for a protein in the ATP-binding cassette (ABC) transporter superfamily which transport various molecules across extra- and intra-cellular membranes, and LCN9,involved in the transport of sugars, bile salts and organic acids, metal ions and amine compounds and were down- and up-regulated respectively. This suggests a misbalance of the efflux pumps which, correlated to induction of oxidative stress, may lead to accumulation of O2.− 49.
After exposure to E171, the modulation of 2 genes (PTGES3 and HIST1H3I) related to cellular response to stress, oxidative stress induced senescence and cellular response to heat stress was observed. E171 additionally induced gene expression changes towards the development of inflammation which can also lead to DNA damage. As described for NPs and MPs, E171 also induced DNA damage in undifferentiated Caco-2 cells as well as induction of micronuclei in HCT116 cells8. Here, 4 DEG (POLB, NEIL3, POLD2 and LIG4) involved in DNA repair were modulated, like POLB involved in base excision and repair and POLD2 in DNA replication and repair. NEIL3 encodes for DNA glycosylases which cleaves bases damaged by ROS50. The protein coded by LIG4 is essential in DNA double-strand break repair through non-homologous end joining. Cancer related genes, which can be modulated as a consequence of DNA damage, were also observed, like down-regulation of the tumour suppressor CELF251. In addition, NRG1 is involved in constitutive signalling by aberrant PI3K in cancer, CSNK1A1L in signalling by WNT in cancer, and MUCL1 in defective GALNT12 causes colorectal cancer.
E171 resulted in 31 DEG associated with the innate and adaptive immune system. These effects of E171 are in line with previous in vivo studies in which impairment of the immune system was observed such as that after ingestion of 5 mg/kg bw/day of E171 for 2, 7, 14 and 21 days, based on gene expression, BALB/c mice showed an impairment of the immune system14. One immune-related gene was commonly down-regulated in both in vitro and in the in vivo experiment, BCL10. It is involved in different processes in the immune system but also in protein ubiquitination and haemostasis. Another study described an impairment of the immune system via a Th1/Th7 response after exposure to 10 mg/kg bw/day in rats for 7 and 100 days13. The dysregulation of the immune system is a known factor that can lead to the development of cancer.
As described, several biological processes observed after E171 exposure were commonly induced by the MPs and NPs fraction such as gene expression changes in oxidative stress, DNA repair, immune system, and transport. Furthermore, other common effects are associated to signal transduction. In regards to the oxidative stress and DNA damage, the current results confirm the results previously observed via Electron Spin Resonance and the Comet assay in undifferentiated Caco-2 cells8.
In addition to common effects in gene expression in the immune system with the 3 types of TiO2, specific effects of NPs and E171 were observed in TLR cascade, advanced glycolysation, the MHC class I antigen processing and presentation and MHC class II presentation, and cytokine signalling with IL-1, IL-17, and IL-20.
In addition, 3 DEG involved in potassium channels were modulated. Changes in potassium channel expression are associated with colon carcinogenesis, however, it is not clearly defined whether those changes are inducing the development of cancer or a consequence reflecting dedifferentiation and ongoing proliferation52.
GO term analysis showed that Membrane was also observed as a common biological process after the NPs and E171 exposure. Within this biological process, 1 DEG after NPs and 1 DEG after E171 exposure were both coding for a late cornified envelope protein and both down-regulated. The cornified envelope is a structure which is responsible for the major barrier function of the skin53. Whether it plays a role in other epithelia barrier functions such as in the intestines is unknown. A dysregulation of the genes coding for these proteins might have a consequence on the integrity of the barrier in the gut and allow particles to pass through.
Exposure to NPs and E171 also induced gene expression changes in the cell cycle. The up-regulation of TUBB1 after NPs exposure involved in G2-G2/M and M phases and the down-regulation of HIST1H3I after E171 exposure also involved in M phase show a potential incidence of the TiO2 exposure on the regulation of cell cycle. Effects of titanium dioxide on the G2/M phase were already observed by Wu et al. who showed that anatase TiO2 NPs (25, 50, 100, and 200 μg/ml) in neuronal cells induced p53 and JNK activation in G2/M cell cycle arrest and apoptosis54.
After E171 and MPs exposure, specific biological processes and 3 DEG were commonly observed. Two out of three DEG have unknown function but WDR78 gene is shown to be a target of the Hedgehog activity and a promotor of FOXJ1 activity in zebrafish55. FOXJ1 is involved in the production of the motile cilia. In relation to the immune system, MPs and E171 affected gene expression in relation with the α-defensins including DEFA6 after MPs exposure and DEFA4 after E171 exposure were up- and down-regulated respectively. Normally present in epithelial cells, the defensins are antimicrobial peptides contributing to the mucosal host defence by the action of permeabilization of the microbe membrane56. Gene expression changes after MPs and E171 exposure were associated with signalling, notably olfactory signalling.
After MPs exposure, 2 DEG involved in cell junction organisation were modulated and 1 DEG after E171 exposure. More precisely 2 DEG (E171: CDH18 and MPs: CDH4) were both coding for cadherin surface proteins. These proteins not only play an important role in endocytosis but also cellular adhesion and intercellular liaison between the cells57. These results might identify one of the mechanisms by which TiO2 can enter the cell. In the neuronal system, 2 CHRNA genes, 1 after MPs and 1 after E171 exposure, were modulated and coding for cholinergic receptors. Cholinergic receptors are normally present in Caco-2 cells and after production of acetylcholine by the cells, a common effect in colorectal cells, the cholinergic receptor are affected and its dysregulation might be associated with inflammation and tumour development58,59.
Overall, the majority of the effects after exposure to E171 could be associated to the effects of the NPs and MPs fractions. However, some effects could only be attributed to the E171 itself. At the pathway level, metabolism of proteins with the insulin processing pathway and haemostasis were specific to E171 exposure. Effects in the expression of insulin genes were also observed in vivo by several groups14,60. At the GO term level, receptor activity, muscle adaptation, catalytic activity, organ development, and transcription factor were specific to E171 exposure. In addition, at the gene expression level, changes were observed in metabolism of RNA, chromatin organization, circadian clock, and programmed cell death. For these specific effects of E171, it should be taken into consideration that the NPs and MPs used do not fully cover the range of E171. The gene expression changes associated with oxidative stress support the detection of ROS formation in undifferentiated Caco-2 cells8 and in vivo transcriptomics analysis on colons of mice after oral administration of E17114.
Our study on ingestion of E171 in the mouse14 also found an effect on the mRNA levels in the colon related to oxidative stress, DNA repair, immune system, transport as well as signal transduction, neuronal system, and extracellular matrix organisation. In addition, the expression of seven orthologue genes affected after in vivo exposure were also found in vitro and were representative of the major effects of E171: immune system (BCL10, TNFAIP8L2, LRRC15), inflammation (LRRC15), signal transduction (TAS2R3), metabolism (BAAT, TNFAIP8L2, and BCL10), and cancer (WDR78). The WDR78 gene has been shown to be in the top 20 of hyper-methylated genes in CRC61, so the downregulation of WDR78 observed in our study could be caused by this. Furthermore, WDR78 is also significantly downregulated after exposure to TiO2 MPs, indicating that these MPs could also induce gene expression changes in relation to cancer.
To determine the relevance of our in vitro study for human exposure conditions, an estimation of human intake of TiO2 has been made8. According to several studies, humans are on average exposed to 1 mg/kgbw/day, implying that a 70 kg adult would ingest 70 mg of TiO2 per day. Assuming the production of 250 g faeces per day, the concentration of TiO2 would be 0.28 mg of TiO2/g of faeces. As faeces contain 75% of water, the density of faeces is approximately the same as water, thus the concentration of TiO2 would be 0.28 mg/mL of faeces. Translocation to other organs in the human body indicates a direct contact between the cellular wall of the intestines and E17162, Therefore, if we assume that only 1% is biologically available for exposure, 0.0028 mg/mL of TiO2 is potentially reaching the colon cells. The concentration used during the experiment is 0.01 mg/ml which is in the same order as the estimated concentration of TiO2 in the colon.
The present study provides an overview of the major effects on the transcriptome after exposure to E171 and the NPs and MPs fractions of TiO2. E171 causes the highest transcriptional impact both at the gene expression and pathway level, all pointing towards the facilitation of colon cancer development. Although both the NPs and MPs show lower but comparable levels of deregulated genes, biological interpretations showed that these fractions cause distinct cellular effects, So E171 shows a synergetic effect and is more than the added effect of the different size fractions. Taken together, these results indicate that the classification of E171 as an inert particle may need to be reconsidered and that NPs induce a more pronounced effect at the transcriptome level than MPs.Because a major part of our observations, like the immune responses, are still based on gene expression levels only, more mechanistically studies to support these observations are needed. Although changes at the level of gene expression or pathways cannot always be translated straightforward into changes at the level of proteins or specific markers e.g. the immune response. Based on our observations, we suggest performing a follow-up study focussing on phenotypical anchoring of the immune responses in which analyses a broad panel of cytokines and interleukins is included.
But since an increasing number of animal studies point out that E171 facilitate harmful effects in the colon including cancer development, the gene expression changes observed in this human colon cancer cell line delivers stronger evidence that exposure might also form an increased risk for humans. Although more evidence is needed from e.g. human intervention studies, suggestion by food industry and regulatory organisations to reduce the harmful effect by removing the NP fraction from E171, could be worthwhile.
Materials and Methods
Cell culture
The human colon carcinoma cell line Caco-2 (American Type Culture Collection ATCC, HTB-37) a frequently used colon cell line for in vitro toxicity studies, was cultured in an incubator under standard cell culture conditions (37 °C, 5% CO2) in Dulbecco’s Modified Eagles Medium (DMEM) (Sigma-Aldrich, the Netherlands). DMEM was supplemented with 10% FBS, 1% non-essential amino acids, 1% sodium pyruvate and 1% penicillin/streptomycin. Cells were passaged at 80 to 90% confluency.
Characterisation of particles
E171 was kindly donated by Sensient Technologies, Mexico. TiO2 NPs were purchased at Io-Li-Tec, Germany, with 99.5% anatase particles. TiO2 MPs were customarily manufactured by PlasmaChem (Germany). Characterisation of the primary size of the particles was derived by SEM as well as a dynamic light scattering and zeta potential8. The similarity of the three-dimensional structures of E171, TiO2 NPs and MPs was confirmed by SEM. E171 contains slightly to fully rounded anatase particles with a proportion of 39% NPs and 61% MPs8. For NPs, the particle size ranged from 10 to 30 nm8, while the manufacturer indicated 15–25 nm, and a surface area of 50 to 150 m2/g. MPs had a crystal structure of anatase, a size higher than 100 nm and an average size of 622 nm8.
Dynamic light scattering results showed that the NPs and MPs had a zeta potential and a polydispersity index in the same range as E171. At a concentration of 1.10−3 mg/mL, E171 had the lowest hydrodynamic size in DMEM (316.8 ± 282.4 d.nm) compared to MPs (1385.83 ± 38.85 d.nm) and NPs the highest size (1942.17 ± 61.12 d.nm). The presence of serum reduced the agglomeration and aggregation of the particles8. Zeta potential results also showed that the addition of serum leads to more colloidal stability which indicates a decrease in the aggregation and agglomeration8. However, all results show that the suspension remained unstable.
Particle exposure of Caco-2 cells
Caco-2 cells were used with a passage number ranging from 27 to 37. Each exposure was performed in triplicate and within each experiment, all the samples were in duplicate. Three days before exposure, ± 1.106 Caco-2 cells were seeded in 21 cm2 culture dishes with 3 ml supplemented DMEM (Sigma Aldrich) and grown to reach 80 to 90% confluency.
E171 or TiO2 NPs or TiO2 MPs, were sterilised at 121 °C for 20 minutes prior to use, and subsequently dispersed in glass tubes at a concentration of 1 mg/ml in supplemented DMEM where FBS was replaced by 0.05% BSA in order to reduce the antioxidant effect of FBS. The solutions were vortexed and subsequently sonicated for 30 minutes at 40 KHz in a bath sonicator (Branson 2200). After sonication, the particles were further dissolved to 0.01 mg/ml in 0.05% BSA supplemented DMEM. Prior to exposure, cells were washed twice with Hank’s Balanced Salt Solution (HBSS) without Ca2+and Mg2+ (Sigma-Aldrich, The Netherlands). Cells were exposed to 3 ml of 0.01 mg/ml TiO2 NPs, MPs or E171 for 2, 4 or 24 hours which gives a final concentration of 1.43 µg/cm2. This concentration was previously shown by our group to be the lowest non-cytotoxic8. Timepoints were based on a previous study63 that showed internalisation of TiO2 NPs after 2 h and reached saturation after 8 h, so that in our study we investigated early as well as prolonged effects of internalisation. After exposure, cells were washed with HBSS (without Ca2+and Mg2+) once and lysed using 1 ml Qiazol Lysis Reagent (Qiagen, The Netherlands). The Qiazol cell solution was stored at −80 °C until RNA isolation.
RNA isolation
Total RNA was isolated from undifferentiated Caco-2 cells exposed to TiO2 NPs, TiO2 MPs, and E171 in Qiazol (Qiagen, The Netherlands) using the miRNeasy Mini Kit (Qiagen, The Netherlands) according to the manufacturer’s protocols for Animal Cells and Animal Tissues including a DNase treatment64. Total RNA yield as well as the 260/230 and the 260/280 were measured on a Nanodrop® ND-1000 spectrophotometer (Thermo Fischer, The Netherlands). Samples with a 260/230 ratio between 1.8 and 2.0 and a 260/280 ratio between 1.9 and 2.1 were checked for their integrity. The integrity of total RNA was checked using RNA Nanochips on a 2100 Bioanalyzer (Agilent Technologies, The Netherlands). All samples had on average a RNA integrity number (RIN) of 9.5 ± 0.4, which is above 6, the number at which each sample is approved for microarray analysis.
cRNA synthesis, labelling and hybridization
Total RNA was converted into cRNA and labelled according to the One-Color Microarray-Based Gene Expression Analysis protocol version 6.6 (Agilent Technologies, The Netherlands)65 as described previously66. In brief, 200 ng of total RNA was diluted in Spike Mix. The primer and the template were denaturated by using a thermocycler (Tprofessional, Biometra, Germany) at 65 °C for 15 minutes. cDNA was synthetized by incubating each sample with cDNA Master Mix in a thermocycler at 40 °C for two hours and 70 °C for 15 minutes. Afterwards, cRNA was synthetized and labelled by incubating Transcription Master Mix, containing the cyanine 3-CTP dye, with the cDNA at 40 °C on a thermocycler for two hours. The RNeasy Mini Kit was used to purify the amplified cRNA samples according to the manufacturer’s protocol of Agilent. Subsequently, the cRNA was quantified using a spectrophotometer with the Microarray Measurement function. The yield and specific activity of Cy3 bound to the cRNA was determined using the formulas in the manufacturer’s protocol of Agilent.
After incubating cRNA samples with Gene Expression Blocking Agent and Fragmentation Buffer (Agilent Technologies) at 60 °C for 30 minutes, hybridization was performed on SurePrint G3 Human GE 8 × 60k V2 slides and a hybridization oven (Shel Lab, United States) was used at 65 °C for 17 hours. After hybridization, the microarray slides were washed with Gene Expression wash buffers (Agilent Technologies, The Netherlands) and scanned using an Agilent DNA Microarray Scanner with Surescan High-resolution Technology (Agilent Technologies, The Netherlands) with scanner settings to Dye Channel: G, Profile: AgilentG3_GX_1Color, Scan region: Agilent HD (61 × 21.3 mm), Scan resolution 3 µm, Tiff file dynamic range: 20 bit, Red PMT gain: 100%, and Green PMT gain: 100%.
Pre-processing and data analysis of microarrays
Pre-processing methods used were described previously14. In short, all samples met the quality criteria of the Feature extraction software (FES) (version 10.7.3.1) from Agilent. An in-house QC pipeline was developed and published (github.com/BiGCAT-UM/arrayQC_Module) to thoroughly check the quality and normalize the data as follows: local background correction, flagging of bad spots, controls and spots without adequate intensity, log2 transformation and quantile normalization. Five samples out of 71 did not pass the quality control the first time. These samples were re-run and were validated by the QC the second time. Raw data that has both expression values and genes were selected for data analysis based on flags and missing values (GEO accession: GSE110410).
First pre-processing was performed with the samples of the time course experiment with NPs. For this, six groups were defined: NPs 2 h, 4 h, and 24 h for the exposed samples and control 2 h, 4 h, and 24 h for the controls. The second pre-processing was split in 2 parts in which the first part was a pre-processing of the NPs 24 h with its time-matched control and the second part was a pre-processing of E171 24 h and MPs 24 h with their time-matched control. The second pre-processing was split in 2 because the experiments were performed independently.
After defining the different groups for the pre-processing, the next steps were exactly the same for all groups. Within each group, probes were flagged for bad spots and at least 30% of all samples had to have good spots. Spot identifiers were deleted when more than 40% of samples in each group had a missing value. Background correction was performed by removing the spot identifiers that had an average expression less than four in all groups. Missing values were imputed by k-nearest neighbours using the standard settings of the GenePattern ImputeMissingValues.KNN module v1367. Spot identifiers were annotated to Agilent probe identifiers and merging of exact same probe identifiers with the median method for pre-processing data was performed in Babelomics 568. Next, Agilent probe identifiers were re-annotated to EntrezGeneIDs. Merging of the expression data for all genes with an identical EntrezGeneIDs was done with Babelomics 5 using the median method for pre-processing data.
Differentially expressed genes (DEG) were identified by using a Linear Mixed Model Analysis for Microarrays (LIMMA) (version 1.0) R-script with cut-off values of a fold-change (FC) of 1.2, 1.5, and 1.8 and a p-value of 0.0569. The data was paired for replicates. Furthermore, according to the Benjamini-Hochberg method with a threshold at 0.05, the false discovery rate was calculated. With the LIMMA analysis, the number of DEG were identified from each sample; data of the exposed samples were corrected to the data from its time matched control sample.
Pathway analyses
Over-representation gene set analysis (ORA) was performed with Consensus Pathway Database (CPDB) with the DEG of each time point70. CPDB is an aggregate of 16 different databases developed by the Max Planck institute and its functionalities have been explained in Nature Protocols71. CPDB has a focus on molecular interactions, and identifies pathways associated with DEG. For computing the significance of the over-representation of the annotation sets with respect to user-input molecules, CPDB applies Fisher’s exact test. For each annotation set, a p-value is calculated. As many annotation sets are tested, CPBD corrects for multiple hypothesis testing using the false discovery rate procedure within each type of annotation set70,71. All the available databases from CPDB were used (release 32, 1 Nov. 2017) with settings in the “pathways as defined by pathway databases” with a minimum overlap of input list of 2 and a p-value cut-off of p < 0.01. As shown previously, the pathway analysis is limited to the number of genes that can be mapped to a pathway which is often less than 50% of the input list14. In order to retrieve the maximal amount of relevant biological information from our gene lists, the corresponding GO terms were also determined.
Gene Ontology (GO) term classification
GO categories related to the DEG were identified by CPBD. In order to retrieve more biological information, GO terms of level 2, 3, 4, and 5 were selected with a p-value cut off of 0.01. Biological processes of each GO term were retrieved based on GO terminology.
Author contributions
H.v.L., J.B. and T.d.K. designed the study. H.P. and M.J. performed the experiments. H.P., M.J. and M.J.J. performed the initial analysis of the data, that was further interpreted by H.P., M.J., M.J.J., J.B., T.d.K., and H.v.L. All authors commented on the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weir A, Westerhoff P, Fabricius L, Hristovski K, von Goetz N. Titanium dioxide nanoparticles in food and personal care products. Environ Sci Technol. 2012;46:2242–2250. doi: 10.1021/es204168d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters RJ, et al. Characterization of titanium dioxide nanoparticles in food products: analytical methods to define nanoparticles. J Agric Food Chem. 2014;62:6285–6293. doi: 10.1021/jf5011885. [DOI] [PubMed] [Google Scholar]
- 3.Lomer MCE, Thompson RPH, Commisso J, Keen CL, Powell JJ. Determination of titanium dioxide in foods using inductively coupled plasma optical emission spectrometry. Analyst. 2000;125:2339–2343. doi: 10.1039/b006285p. [DOI] [PubMed] [Google Scholar]
- 4.EU. Regulation No 231-2012 on food additives. Official Journal of the European Union (2012).
- 5.FAO & WHO. Toxicological Evaluation of Some Food Colours, Emulsifiers, Stabilizers, Anti-Caking Agents and Certain Other Substances. FAO Nutrition Meetings Report Series No. 46 A WHO/FOOD ADD/70.36 (1969). [PubMed]
- 6.FAO & WHO. Specification for the identity and purity of food additives and their toxicological evaluation. World Health Organization technical report series No. 445, FAO nutrition meetings report series No. 46 (1970). [PubMed]
- 7.FDA. Listing of color additives exempt from certification. TITLE 21–FOOD AND DRUGS; CHAPTER I–FOOD AND DRUG ADMINISTRATION. Code of Federal Regulations 21CFR73.1575 (2014).
- 8.Proquin H, et al. Titanium dioxide food additive (E171) induces ROS formation and genotoxicity: contribution of micro and nano-sized fractions. Mutagenesis. 2017;32:139–149. doi: 10.1093/mutage/gew051. [DOI] [PubMed] [Google Scholar]
- 9.Dorier, M. et al. Continuous in vitro exposure of intestinal epithelial cells to E171 food additive causes oxidative stress, inducing oxidation of DNA bases but no endoplasmic reticulum stress. Nanotoxicology, 1–54, 10.1080/17435390.2017.1349203 (2017). [DOI] [PubMed]
- 10.Dudefoi W, Moniz K, Allen-Vercoe E, Ropers MH, Walker VK. Impact of food grade and nano-TiO2 particles on a human intestinal community. Food Chem Toxicol. 2017;106:242–249. doi: 10.1016/j.fct.2017.05.050. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, et al. Characterization of Food-Grade Titanium Dioxide: The Presence of Nanosized Particles. Environmental Science & Technology. 2014;48:6391–6400. doi: 10.1021/es500436x. [DOI] [PubMed] [Google Scholar]
- 12.Urrutia-Ortega IM, et al. Food-grade titanium dioxide exposure exacerbates tumor formation in colitis associated cancer model. Food Chem Toxicol. 2016;93:20–31. doi: 10.1016/j.fct.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Bettini S, et al. Food-grade TiO2 impairs intestinal and systemic immune homeostasis, initiates preneoplastic lesions and promotes aberrant crypt development in the rat colon. Sci Rep. 2017;7:40373. doi: 10.1038/srep40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proquin H, et al. Gene expression profiling in colon of mice exposed to food additive titanium dioxide (E171) Food Chem Toxicol. 2018;111:153–165. doi: 10.1016/j.fct.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, et al. Oral administration of nano-titanium dioxide particle disrupts hepatic metabolic functions in a mouse model. Environ Toxicol Pharmacol. 2017;49:112–118. doi: 10.1016/j.etap.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Acute toxicity and biodistribution of different sized titanium dioxide particles in mice after oral administration. Toxicol Lett. 2007;168:176–185. doi: 10.1016/j.toxlet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Brun E, et al. Titanium dioxide nanoparticle impact and translocation through ex vivo, in vivo and in vitro gut epithelia. Part Fibre Toxicol. 2014;11:13. doi: 10.1186/1743-8977-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, et al. Effect of titanium dioxide nanoparticles on glucose homeostasis after oral administration. J Appl Toxicol. 2018;38:810–823. doi: 10.1002/jat.3589. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz PA, et al. Titanium dioxide nanoparticles exacerbate DSS-induced colitis: role of the NLRP3 inflammasome. Gut. 2017;66:1216–1224. doi: 10.1136/gutjnl-2015-310297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nogueira CM, et al. Titanium dioxide induced inflammation in the small intestine. World J Gastroenterol. 2012;18:4729–4735. doi: 10.3748/wjg.v18.i34.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerloff K, Albrecht C, Boots AW, Forster I, Schins RPF. Cytotoxicity and oxidative DNA damage by nanoparticles in human intestinal Caco-2 cells. Nanotoxicology. 2009;3:355–364. doi: 10.3109/17435390903276933. [DOI] [Google Scholar]
- 22.Saquib Q, et al. Titanium dioxide nanoparticles induced cytotoxicity, oxidative stress and DNA damage in human amnion epithelial (WISH) cells. Toxicol In Vitro. 2012;26:351–361. doi: 10.1016/j.tiv.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Shukla RK, et al. ROS-mediated genotoxicity induced by titanium dioxide nanoparticles in human epidermal cells. Toxicol In Vitro. 2011;25:231–241. doi: 10.1016/j.tiv.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Faust JJ, Doudrick K, Yang Y, Westerhoff P, Capco DG. Food grade titanium dioxide disrupts intestinal brush border microvilli in vitro independent of sedimentation. Cell Biol Toxicol. 2014;30:169–188. doi: 10.1007/s10565-014-9278-1. [DOI] [PubMed] [Google Scholar]
- 25.Saaf AM, et al. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Molecular biology of the cell. 2007;18:4245–4260. doi: 10.1091/mbc.e07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dal-Pizzol F, Klamt F, Benfato MS, Bernard EA, Moreira JCF. Retinol supplementation induces oxidative stress and modulates antioxidant enzyme activities in rat sertoli cells. Free Radical Research. 2001;34:395–404. doi: 10.1080/10715760100300331. [DOI] [PubMed] [Google Scholar]
- 27.Basu S, Michaelsson K, Olofsson H, Johansson S, Melhus H. Association between oxidative stress and bone mineral density. Biochemical and Biophysical Research Communications. 2001;288:275–279. doi: 10.1006/bbrc.2001.5747. [DOI] [PubMed] [Google Scholar]
- 28.Araujo, M. B. et al. Creatine supplementation and oxidative stress in rat liver. Journal of the International Society of Sports Nutrition10, doi:Artn 5410.1186/1550-2783-10-54 (2013). [DOI] [PMC free article] [PubMed]
- 29.Bouillaud F, Alves-Guerra MC, Ricquier D. UCPs, at the interface between bioenergetics and metabolism. Biochimica Et Biophysica Acta-Molecular Cell Research. 2016;1863:2443–2456. doi: 10.1016/j.bbamcr.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 31.Walker LJ, Craig RB, Harris AL, Hickson ID. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barresi V, et al. Chromosomal instability analysis and regional tumor heterogeneity in colon cancer. Cancer Genetics. 2017;210:9–21. doi: 10.1016/j.cancergen.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai K, et al. Correlation of the cytotoxicity of TiO2 nanoparticles with different particle sizes on a sub-200-nm scale. Small. 2011;7:3026–3031. doi: 10.1002/smll.201101170. [DOI] [PubMed] [Google Scholar]
- 35.Steffansen B, et al. Intestinal solute carriers: an overview of trends and strategies for improving oral drug absorption. Eur J Pharm Sci. 2004;21:3–16. doi: 10.1016/j.ejps.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 36.des Rieux A, Fievez V, Garinot M, Schneider YJ, Preat V. Nanoparticles as potential oral delivery systems of proteins and vaccines: A mechanistic approach. Journal of Controlled Release. 2006;116:1–27. doi: 10.1016/j.jconrel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Rial E, Zardoya R. Oxidative stress, thermogenesis and evolution of uncoupling proteins. J Biol. 2009;8:58. doi: 10.1186/jbiol155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins D, Zhao Y. New aspects of mitochondrial Uncoupling Proteins (UCPs) and their roles in tumorigenesis. Int J Mol Sci. 2011;12:5285–5293. doi: 10.3390/ijms12085285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunt CR, et al. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res. 2013;179:383–392. doi: 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossetto D, Avvakumov N, Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–1108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao, K. H., Hua, L., Wei, L. S., Meng, J. M. & Hu, J. H. Correlation Between CASC8, SMAD7 Polymorphisms and the Susceptibility to Colorectal Cancer An Updated Meta-Analysis Based on GWAS Results. Medicine94, doi:ARTN e1884 10.1097/MD.0000000000001884 (2015). [DOI] [PMC free article] [PubMed]
- 43.Polakis P. The many ways of Wnt in cancer. Current Opinion in Genetics &. Development. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Pistritto G, et al. Expression and transcriptional regulation of caspase-14 in simple and complex epithelia. Cell Death Differ. 2002;9:995–1006. doi: 10.1038/sj.cdd.4401061. [DOI] [PubMed] [Google Scholar]
- 45.Aravindan S, et al. Retinal Degeneration Protein 3 (RD3) in normal human tissues: Novel insights. Sci Rep. 2017;7:13154. doi: 10.1038/s41598-017-13337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atreya R, et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat Med. 2000;6:583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- 47.Becker HM, Bertschinger MM, Rogler G. Microparticles and their impact on intestinal immunity. Dig Dis. 2012;30(Suppl 3):47–54. doi: 10.1159/000342602. [DOI] [PubMed] [Google Scholar]
- 48.Powell JJ, Thoree V, Pele LC. Dietary microparticles and their impact on tolerance and immune responsiveness of the gastrointestinal tract. British Journal of Nutrition. 2007;98:S59–S63. doi: 10.1017/S0007114507832922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorier M, et al. Impact of anatase and rutile titanium dioxide nanoparticles on uptake carriers and efflux pumps in Caco-2 gut epithelial cells. Nanoscale. 2015;7:7352–7360. doi: 10.1039/c5nr00505a. [DOI] [PubMed] [Google Scholar]
- 50.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/S1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 51.Ramalingam S, Ramamoorthy P, Subramaniam D, Anant S. Reduced Expression of RNA Binding Protein CELF2, a Putative Tumor Suppressor Gene in Colon Cancer. Immunogastroenterology. 2012;1:27–33. doi: 10.7178/ig.1.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heitzmann D, Warth R. Physiology and pathophysiology of potassium channels in gastrointestinal epithelia. Physiological Reviews. 2008;88:1119–1182. doi: 10.1152/physrev.00020.2007. [DOI] [PubMed] [Google Scholar]
- 53.Kalinin AE, Kajava AV, Steinert PM. Epithelial barrier function: assembly and structural features of the cornified cell envelope. Bioessays. 2002;24:789–800. doi: 10.1002/bies.10144. [DOI] [PubMed] [Google Scholar]
- 54.Wu J, Sun JA, Xue Y. Involvement of JNK and P53 activation in G2/M cell cycle arrest and apoptosis induced by titanium dioxide nanoparticles in neuron cells. Toxicology letters. 2010;199:269–276. doi: 10.1016/j.toxlet2010.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- 56.Bevins CL, Martin-Porter E, Ganz T. Defensins and innate host defence of the gastrointestinal tract. Gut. 1999;45:911–915. doi: 10.1136/gut.45.6.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng K, et al. Acetylcholine release by human colon cancer cells mediates autocrine stimulation of cell proliferation. Am J Physiol Gastrointest Liver Physiol. 2008;295:G591–597. doi: 10.1152/ajpgi.00055.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelissier-Rota M, Lainé M, Ducarouge B, Jacquier-Sarlin BBM. Role of Cholinergic Receptors in Colorectal Cancer: Potential Therapeutic Implications of Vagus Nerve Stimulation? Journal of Cancer Therapy. 2013;4:1116–1131. doi: 10.4236/jct.2013.46128. [DOI] [Google Scholar]
- 60.Hu H, et al. Titanium dioxide nanoparticles increase plasma glucose via reactive oxygen species-induced insulin resistance in mice. J Appl Toxicol. 2015;35:1122–1132. doi: 10.1002/jat.3150. [DOI] [PubMed] [Google Scholar]
- 61.Kim YH, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol. 2011;18:2338–2347. doi: 10.1245/s10434-011-1573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heringa MB, et al. Detection of titanium particles in human liver and spleen and possible health implications. Part Fibre Toxicol. 2018;15:15. doi: 10.1186/s12989-018-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gitrowski C, Al-Jubory AR, Handy RD. Uptake of different crystal structures of TiO(2) nanoparticles by Caco-2 intestinal cells. Toxicology letters. 2014;226:264–276. doi: 10.1016/j.toxlet.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 64.Qiagen. RNeasy Mini Handbook. https://www.qiagen.com/us/resources/resourcedetail?id=14e7cf6e-521a-4cf7-8cbc-bf9f6fa33e24&lang=en (2012).
- 65.Agilent. One-Color Microarray-Based Gene Expression Analysis. http://www.agilent.com/cs/library/usermanuals/Public/G4140-90040_GeneExpression_OneColor_6.9.pdf (2015).
- 66.Jiang J, et al. Omics-based identification of the combined effects of idiosyncratic drugs and inflammatory cytokines on the development of drug-induced liver injury. Toxicol Appl Pharmacol. 2017;332:100–108. doi: 10.1016/j.taap.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 67.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 68.Alonso R, et al. Babelomics 5.0: functional interpretation for new generations of genomic data. Nucleic Acids Res. 2015;43:W117–121. doi: 10.1093/nar/gkv384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smyth, G. K. in Bioinformatics and computational biology solutions using R and Bioconductor 397-420 (Springer, 2005).
- 70.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11:1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]