Abstract
Cells grown in three dimensions (3D) within natural extracellular matrices or synthetic scaffolds more closely recapitulate the phenotype of those cells within tissues in regard to normal developmental and pathobiological processes. This includes degradation of the surrounding stroma as the cells migrate and invade through the matrices. As 3D cultures of tumor cells predict efficacy of, and resistance to, a wide variety of cancer therapies, we employed tissue-engineering approaches to establish 3D pathomimetic avatars of human breast cancer cells alone and in the context of both their cellular and pathochemical microenvironments. We have shown that we can localize and quantify key parameters of malignant progression by live-cell imaging of the 3D avatars over time (4D). One surrogate for changes in malignant progression is matrix degradation, which can be localized and quantified by our live-cell proteolysis assay. This assay is predictive of changes in spatio-temporal and dynamic interactions among the co-cultured cells and changes in viability, proliferation, and malignant phenotype. Furthermore, our live-cell proteolysis assay measures the effect of small-molecule inhibitors of proteases and kinases, neutralizing or blocking antibodies to cytokines and photodynamic therapy on malignant progression. We suggest that 3D/4D pathomimetic avatars in combination with our live-cell proteolysis assays will be a useful preclinical screening platform for cancer therapies. Our ultimate goal is to develop 3D/4D avatars from an individual patient’s cancer in which we can screen “personalized medicine” therapies using changes in proteolytic activity to quantify therapeutic efficacy.
Keywords: Spatio-temporal modeling, Proteolysis, Live-cell imaging, 3D cultures
1. Introduction
Elegant studies by Bissell, Brugge and colleagues [1–3], in which they cultured breast cells in three dimensions (3D) in natural extracellular matrices, have established that the 3D context is essential to the formation of breast structures that can undergo development and malignant progression and be predictive of clinical outcome. The American Cancer Society predicts that there will be 268,600 new cases of breast cancer diagnosed in women in the USA in 2019 [4]. This number does not include the ~ 63,000 predicted new cases of ductal carcinoma in situ (DCIS). A diagnosis of DCIS is problematic as DCIS may remain indolent or may progress, yet DCIS that will progress to life-threatening invasive ductal carcinoma (IDC) cannot at present be distinguished from DCIS that would remain indolent. As a result, almost all women who are diagnosed with DCIS undergo aggressive treatment.
3D cultures of cells derived from premalignant breast lesions such as DCIS can model malignant progression and predict resistance and sensitivity to cytotoxic and targeted therapies [2, 5–8]. We used a series of isogenic cell lines derived from MCF-10A non-transformed human breast epithelial cells to build 3D pathomimetic avatars that replicate the transition of DCIS to IDC (for review, see [9]). The hallmark of this transition is progression from a preinvasive phenotype to an invasive phenotype. The first sign that DCIS is undergoing transition is the presence of DCIS with microinvasion, a state in which a few DCIS cells have invaded through the underlying basement membrane (for more detailed information on DCIS, please see a series of recent reviews [10]). Proteases of aspartic, cysteine, metallo, serine, and threonine catalytic classes have been implicated in the local microinvasion of DCIS and the invasive phenotype of IDC ([11, 12] and also see reviews in this issue). Invasion at local and distant sites involves DCIS and breast cancer cells as well as other cell types found in the tumor microenvironment. This includes lymphocytes that are predictive of recurrence in DCIS [13]; macrophages that promote invasion and yet are anti-tumor [14] or that reduce response to chemotherapy [15] and fibroblasts that promote invasion [16]. Proteolysis is a common underlying mechanism and in particular proteolysis of extracellular matrix [17], which is consistent with the high levels of expression of proteases in these tumor-associated cells [15, 16]. We predict that establishing 3D pathomimetic avatars of human breast cancer cells that also incorporate tumor-associated cells will provide a robust and tractable model system in which the transition of DCIS to IDC can be analyzed, and the role(s) of proteolysis in this transition can be evaluated. In this review, we discuss the strategies we used to develop (1) pathomimetic avatars for breast cancer progression (Sect. 2), (2) live-cell imaging protocols for quantitative analysis of the pathomimetic avatars (Sect. 3), and (3) a live-cell proteolysis assay that can localize and quantify degradation in real-time over extended periods of time (Sect. 4). Importantly, this assay uses a protein substrate and thus, rather than measuring activity of a single protease, captures the overall action of proteolytic networks.
2. Pathomimetic avatars for modeling breast cancer progression in vitro
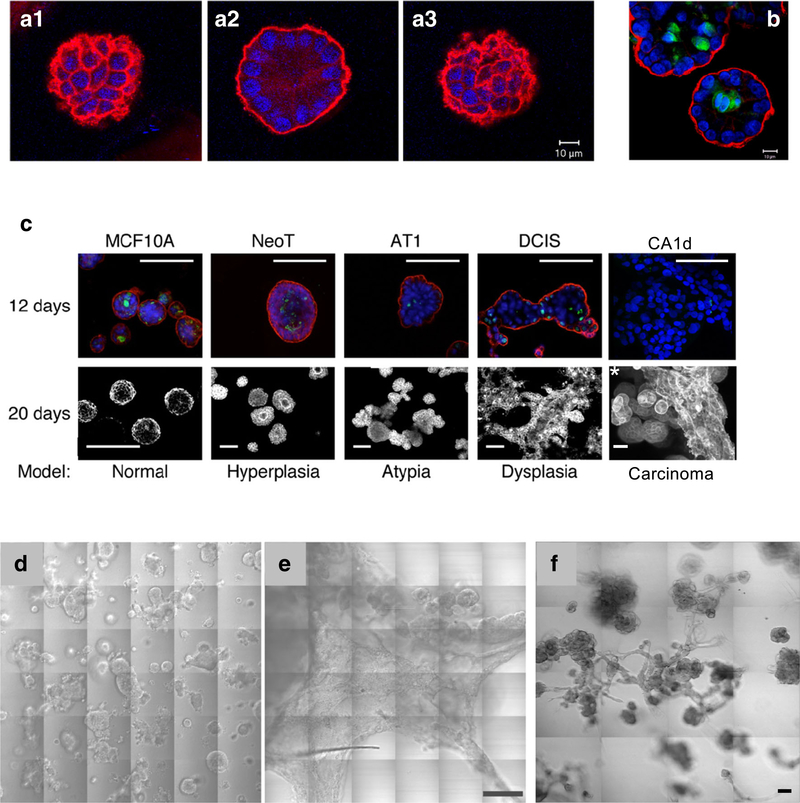
3D mono-cultures MCF-10A human breast epithelial cells, when seeded sparsely in reconstituted basement membrane (rBM) overlay cultures, develop over time into grape-like clusters of acini linked by tube-like structures resembling those present in the human breast (Fig. 1). Individual acini polarize in the rBM overlay cultures and undergo lumen formation at least in part due to apoptosis of cells within the lumen (Fig. 2a, b). Brugge and colleagues have conducted a series of studies showing that 3D rBM overlay cultures of 10A cells can be used to study tumor initiation and oncogenesis in vitro [1]. We expanded on those observations, using a series of isogenic 10A variants developed by Fred Miller [18,19], to establish 3D mono-culture models for various stages of breast cancer progression (Fig. 2c). These models recapitulate the morphology of various stages in progression, including dramatic changes in growth and invasiveness (Fig. 2d, e). The 10.DCIS variant has proved of particular interest as it is poised to become invasive both when grown as a xenograft in nude mice [20] and when grown in 3D (Fig. 2d). In vitro there is heterogeneity with both delimited and invasive DCIS structures present (Fig. 2d). This is comparable to the heterogeneity observed in vivo. Growing 10A variants in 3D cultures results in histopathologies and morphologies comparable to those observed in vivo in xenograft models and in genomic and transcriptomic profiles observed in primary breast cancers. Maguire et al. [8] have found driver alterations in the 10A series only when they are grown in 3D culture, further confirming that 3D mono-culture models will be useful for identifying and testing druggable pathways targeting breast cancer progression.
Fig. 1.
MCF-10A human breast epithelial cells develop over time into clusters of acini linked by tubules when grown in 3D rBM overlay monocultures. Image is 3D reconstruction of optical sections taken through entire volume on a Zeiss LSM-510 META confocal microscope. Structures were fixed and processed at 21 days of culture and stained for human laminin 332 to indicate production of human basement membrane proteins in these cultures (green) and Hoechst 33342 as a marker of nuclei (blue). One grid is 45 μm
Fig. 2.
Pathomimetic avatars of MCF10 variants. Individual acini of MCF-10A human breast epithelial cells are polarized when grown for 12 days in 3D rBM overlay mono-cultures. This is indicated by staining for the basal polarity marker, α6 integrin (red), in confocal sections at the top (A1), middle (A2), and bottom (A3) of this acinus. Apoptosis results in lumen formation, as shown in this equatorial plane of an acinus stained for cleaved caspase 3 (B, green). Nuclei are stained with DAPI (blue). Size bars, 10 μm. C 3D rBM overlay cultures of MCF10 variants model hyperplasia, atypical hyperplasia, dysplasia, and carcinoma. Images in upper row represent a single confocal section at the equatorial plane of 12-day cultures that were fixed and stained for the basal polarity marker, α6 integrin (red); the apoptotic marker, cleaved caspase 3 (green); and nuclei with DAPI (blue); size bars, 50 μm. Lower row shows differential interference contrast images of 20-day live cultures with the exception of the *CA1d sample, which was fixed and processed at 8 days; size bars, 50 μm. In 3D rBM overlay cultures, DCIS (D) and CA1d (E) variants of the MCF10 series develop into large preinvasive dysplastic structures and extensive invasive tumoroids, respectively. This is illustrated in 36 contiguous tiled areas representing a 1200 × 1200 μm field; size bar, 200 μm. Regions of tumoroid invasion into the matrix are revealed by change in focus in panel E. F Co-culture of human breast carcinoma-associated fibroblasts (CAFs) with DCIS cells confers an invasive phenotype. In 3D rBM overlay co-cultures of DCIS and CAFs at a ratio of 5:1, extensive multicellular invasive outgrowths are present (compare with DCIS cells in 3D mono-cultures in panel D). Differential interference contrast images of live cultures were obtained at 8 days of culture and presented here as 16 contiguous tiled fields. Scale bar, 90 μm. Imaging was conducted on a Zeiss LSM-510 META confocal microscope
3D co-cultures The tumor microenvironment is an active, rather than a passive, participant in development and progression of breast cancer. 3D models such as the mono-cultures described above comprised of only breast cancer cells do not recapitulate interactions with other cell types in the tumor microenvironment such as stromal, lymphovascular, and immune cells. Therefore, 3D mono-cultures do not model paracrine pathways that impact malignant progression and cannot be used to screen responses to therapies that target those pathways. To this end, we include in our 3D pathomimetic avatars additional cell types that are present in vivo in the microenvironment of premalignant breast lesions and breast cancers [21]. We have optimized growth conditions for live-cell imaging and molecular and biochemical analyses of phenotypic changes induced as the breast cancer cells interact with the other cell types in real-time, i.e., 4D (3D + time) [21].
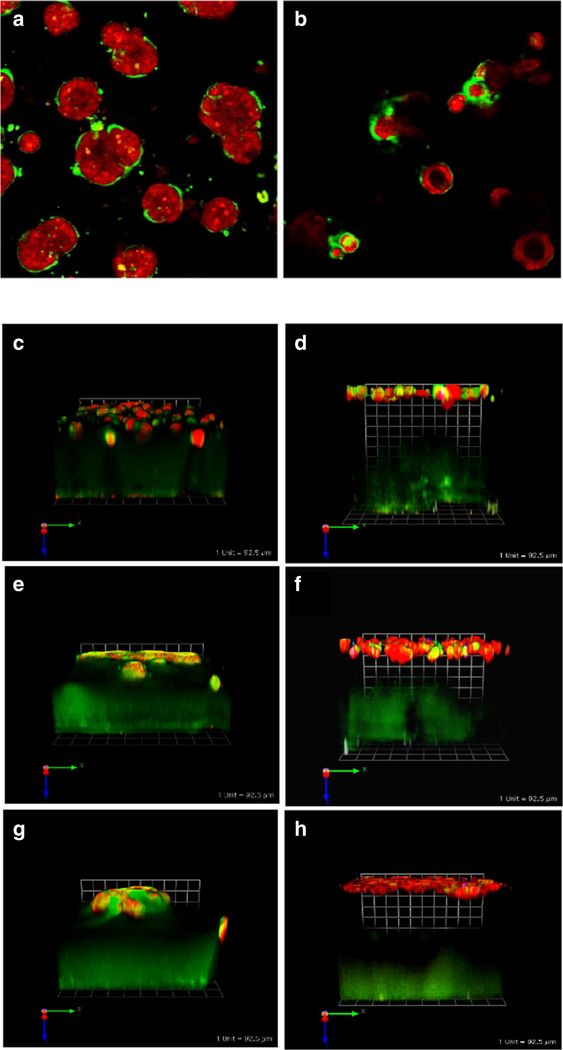
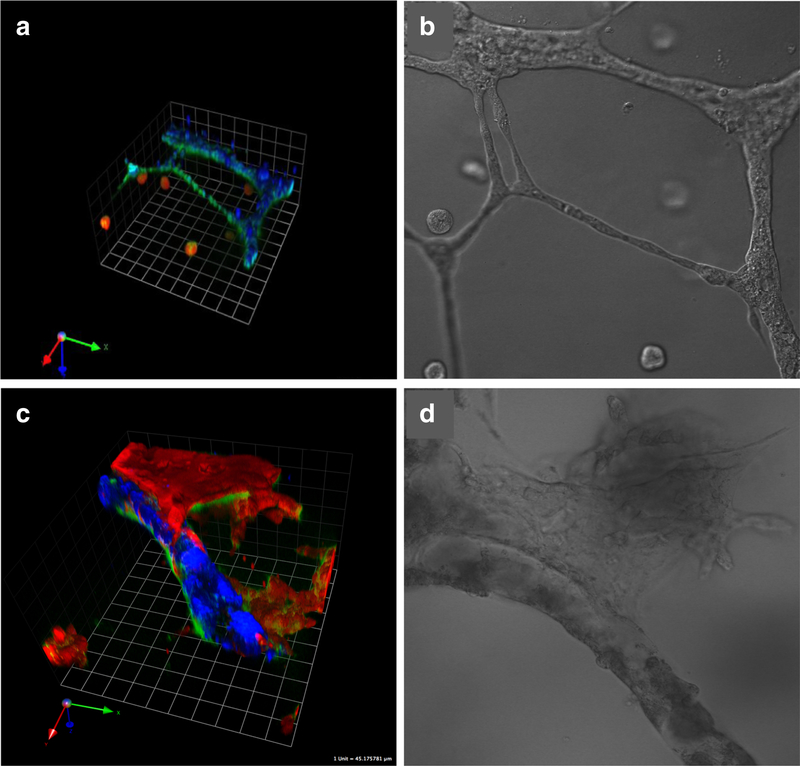
To 3D rBM overlay cultures of human MCF-10 breast variants, we have added human breast myoepithelial cells and human breast carcinoma-associated fibroblasts (CAFs). The myoepithelial cells, which surround the epithelial cells in normal breast acini in vivo, have been hypothesized to play a tumor-suppressor role [22]. Myoepithelial cells are lost during the transition from DCIS to IDC. In 3D rBM overlay co-cultures, we have observed that myoepithelial cells confer a less dysplastic phenotype on the structures formed by DCIS cells [[21] and (Fig. 3a, b)]. The presence of myoepithelial cells is sufficient to induce reversion of dysplastic DCIS structures, in some cases to acini with central lumens. CAFs in contrast increase transition of DCIS to an invasive phenotype (Figs. 2f and 3e) and the invasiveness of structures formed by MCF-10A and MCF10.CA1d cells (Fig. 3c, g). Direct physical interactions between CAFs and cancer cells exert a pulling force on the cancer cells that is mediated by cadherins on the membranes of the two cell types and that results in collective invasion by the cancer cells [23]. This has been established in 3D cultures of epidermoid cancers and in lung and vulvar tumors in vivo. An involvement of cadherins in CAF-induced invasion of breast cancer has not yet been demonstrated; however, our live-cell images are consistent with direct physical interactions between CAFs and DCIS cells leading collective migration of the DCIS cells in the 3D co-cultures (Fig. 4). Myoepithelial cells are able to suppress development of an invasive phenotype by DCIS structures even when the transition to invasiveness is induced by co-culture with CAFs. This is strikingly apparent from the greater height of the co-cultures when myoepithelial cells are present (Fig. 3d, f, h). Similar effects on lesions/tumors are observed in vivo when the various cell types are implanted into nude mice [21], consistent with 3D co-cultures being able to phenocopy breast cancer progression and being suitable for identifying and validating paracrine pathways that impact progression [24].
Fig. 3.
Cell/cell interactions in pathomimetic avatars. SUM 225 DCIS cells [42] grow into disorganized dysplastic structures without a central lumen (a). However, in co-cultures with N1ME myoepithelial cells (2 DCIS:1 myoepithelial cell), the structures formed are more organized with some central lumens (b) (cf. with 10A acini in Fig. 2). Cultures were imaged live at 12 days. Two hours before imaging cells were stained with CellTracker Orange (red) to visualize morphology of the structures. Green fluorescence represents degradation products of DQ-collagen IV. Confocal image stacks, obtained on a Zeiss LSM-510 confocal microscope, were rendered in 3D with Volocity software with the equatorial plane shown here; magnification, × 40. Myoepithelial cells also reduce CAF-induced increases in invasiveness and associated degradation of extracellular matrices by MCF-10A, MCF10.DCIS, and MCF10.CA1d grown in tripartite 3D co-cultures. Tripartite cultures consist of CAFs (WS12Ti) plated in collagen I spiked with DQ-collagen I, covered with rBM spiked with DQ-collagen IV that contains cells of MCF10 variants ± myoepithelial (N1ME) cells, and followed by an rBM overlay: c 10A + CAFs, d 10A + CAFs + myoepithelial cells, e 10.DCIS + CAFs, f 10.DCIS + CAFs + myoepithelial cells, g CA1d+ CAFs, and h CA1d + CAFs + myoepithelial cells. Co-cultures were imaged live at day 8. Two hours before imaging cells were stained with CellTracker Orange (red) to visualize morphology of the structures. Green fluorescence represents degradation products of DQ-collagens. Confocal image stacks, obtained on a Zeiss LSM-510 confocal microscope, were rendered in 3D with Volocity software; one grid is 92 μm
Fig. 4.
A single CAF exerts a pulling effect on a 3D DCIS structure as has been observed in other types of cancers. Prior to seeding, CAFs were labeled with CellTrace CFSE (green). Cultures were imaged live. Confocal image stacks, obtained on a Zeiss LSM-510 confocal microscope, were rendered in 3D with Volocity software. Prior to imaging, nuclei were labeled with Hoechst 33342. One grid is 45 μm
Other cells found in the microenvironment of breast cancers also alter DCIS progression in 3D co-cultures. Perhaps most dramatic are the effects observed in co-cultures of DCIS cells and endothelial cells. In mono-cultures, human umbilical vein endothelial cells grow into 3D networks of tubules [25]. This is also seen initially in co-cultures with DCIS cells (Fig. 5). Within 5 days, however, there have been extensive morphological changes. The endothelial tubules have developed into tubes with lumens. There is dramatic expansion of the DCIS structures with some attaching to and intravasating into the endothelial tubes. Interestingly, microvascular endothelial cells of lymphatic origin induce more rapid migration of DCIS cells than do those of blood vessel origin (data not shown). This would be consistent with the first site of breast cancer metastasis being the lymphatics and suggests that such 3D co-cultures may serve as screens for anti-metastatic therapies.
Fig. 5.
Human DCIS cells migrate toward the tubular networks formed by human umbilical vein endothelial cells over time in 3D rBM overlay co-cultures, attach and intravasate into the tubules. In movies of 3D reconstructions of optical sections, individual DCIS cells can be seen within the tubules. There is in parallel a striking increase in the growth and invasive phenotype of DCIS structures from 2 days in culture (a, b) to 5 days (c, d). DCIS cells were transduced with LentiRFP (red) and endothelial cells labeled with CellTrace Far Red (pseudocolored blue). Green fluorescence represents degradation products of DQ-collagen IV. Images of live co-cultures were obtained on a Zeiss LSM-510 META confocal microscope and confocal image stacks reconstructed in 3D with Volocity software. One grid is 45 μm
3. Live-cell quantitative imaging of pathomimetic avatars
We and others have established 3D culture and co-culture models that can be used to identify and test mechanisms that mediate breast cancer progression (see Sect. 2). These models clearly show changes in morphology and cell/cell interactions associated with progression. The changes with progression are dynamic, take place in three dimensions, and can involve reciprocal changes as cells interact, whether they be breast cancer cells or breast cancer cells interacting with CAFs, myoepithelial cells, endothelial cells, etc. Therefore, to obtain a more complete understanding, one needs to be able to monitor and record the cultures in 3D in real time and over the time periods necessary for progression to occur, i.e., in 4D. This type of model system and the live-cell imaging provide an opportunity for testing interventions at specific periods during progression, e.g., cytotoxic agents at the point when the cancer cells are rapidly proliferating or anti-angiogenic drugs as endothelial cells are migrating. Changes in morphology and invasiveness can be assessed over time in optical sections acquired throughout the entire volume of 3D cultures and illustrated in 3D reconstructions of the cultures generated from the optical sections. This includes (1) cell number/proliferation, (2) cell migration, (3) cell/cell interactions, (4) structure volumes, (5) invasive outgrowths (volumes or lengths), and (5) complexity of 3D structures, including shape, ratio of surface area to volume, and deviation from sphericity. However, to obtain accurate measurements of structure volumes, the cells need to be labeled. This can be done, prior to seeding, by lentiviral transduction with fluorescent proteins (Fig. 5) and, for experiments over periods of time that will not be affected by cells proliferating, labels such as CellTracker or CellTrace dyes can be used (Figs. 3, 4, and 5). Labeling also allows one to follow interactions of one cell type with another, such as the intravasation of DCIS cells into an endothelial tube (Fig. 5). Labels of course need to be selected for distinct fluorescence properties that allow simultaneous confocal detection of multiple cell types.
Imaging can produce a beautiful picture, but how can one make sure their results are reproducible, prevent bias in selection of images, and quantify their findings? We address reproducibility by acquiring optical Z-sections throughout the entire depth of our cultures/co-cultures and routinely doing so in 16 contiguous fields. Thus, we are acquiring data in 3D from 16 fields. This reduces the possibility of selecting single fields for analysis that do not reflect the heterogeneous nature of a sample. We use Volocity and/or Huygens software for 3D reconstruction of optical Z-sections, volume rendering of the image stacks, image analysis and quantification, intensity measurements, morphometric analyses, and labeling and counting nuclei so that quantification can be conducted on a per cell basis.
4. Live-cell functional imaging of pathomimetic avatars
The pathomimetic avatars described are proving useful for identifying and validating druggable pathways that mediate breast cancer progression (for example, see [21, 24, 26]). There is a need for non-destructive assays that measure a function such as activity in 3D over time (4D). To this end, we have developed a live-cell proteolysis assay [27, 28]. Our rationale was that proteases have been widely implicated in cancer progression (for review, see [11, 12, 29]). The proteases implicated in the invasive phenotype of cancers do not act alone, but act as part of an interactive proteolytic network or cancer degradome consisting of multiple families of proteases and their endogenous inhibitors. This suggested to us that using a natural protein substrate from the matrices into which cancer cells invade might identify proteolytic networks that could be targeted as opposed to using substrates highly selective for individual proteases or protease classes. The substrates that we use in our live-cell proteolysis assays are dye-quenched (DQ)-collagens IV and I originally developed by Molecular Probes and now available from ThermoFisher. We spike the DQ-collagen IV into rBM as illustrated in Figs. 3 and 5. DQ-collagen I is spiked into collagen I as in the bottom layer of the tripartite cultures illustrated in Fig. 3. This is a gain-of-function assay in which proteolysis results in formation of fluorescent degradation products of the collagens that can be quantified. Using the DQ-collagens, we have shown that when CAFs are co-cultured with breast cancer cells, there is an increase in proteolysis and, when myoepithelial cells are present, there is a decrease in proteolysis [21]. In tripartite cultures, proteolysis by MCF10 variant structures in the upper layers of the cultures is reduced in the presence of myoepithelial cells whereas proteolysis by CAFs in the lower collagen I layer is not affected (Fig. 3). The live-cell proteolysis assay is thus able to detect spatial differences in DQ-collagen degradation. In general, the fluorescence representing degradation products of DQ-collagen IV is observed on the surface of structures, whether they be DCIS structures (Figs. 3 and 5), MCF10 variant structures (Fig. 3), or endothelial tubules/tubes (Fig. 5). Indeed, live-cell imaging over a 16-h time period demonstrated that degradation products of DQ-collagen IV localize along the surface of endothelial cells as they migrate toward one another and coalesce into tubes, including on the surface of the sprouting tips of the endothelial cells [25]. The live-cell proteolysis assay also localizes degradation products within cells [25, 30] due to an alternative intracellular degradation pathway dependent on receptor-mediated collagen uptake into lysosomes via uPARAP/Endo180 [31]. In addition, the live-cell proteolysis assay can detect effects on proteolytic pathways of pathochemical changes in the tumor microenvironment such as acidosis. Acidosis, which is part of the emerging cancer hallmark of reprogramming of energy metabolism [32], results in an increase in degradation of DQ-collagen by breast cancer cells [30] and colon cancer cells in parallel with an increase in invasiveness [33].
Other functional imaging assays that can be used to detect active proteases in live cells use small-molecule optical probes that are either activity-or substrate-based (for discussion of advantages and disadvantages of the two types of probes, see [34]). We have used activity-based probes to identify and localize active forms of cysteine cathepsins both in vitro and in vivo [30, 35–37]. Since cysteine cathepsins sit upstream in many proteolytic networks [11], probes that detect active cysteine cathepsins can serve as surrogate markers for an invasive phenotype. Specificity of the probes is an issue however if one wants to identify an individual cysteine cathepsin or an individual protease of any class. This is being addressed by Drag and colleagues by use of unnatural amino acids in the probes such as those they developed for detection of active serine proteases in neutrophils [38]. We have used activity-based probes for cysteine cathepsins, as we did the live-cell proteolysis assay, to detect effects of acidosis in the tumor microenvironment [30]. This technology revealed an increase in active cysteine cathepsins extracellularly in breast cancer cells exposed to an acidic microenvironment, a finding that is consistent with earlier studies by us and others in which trafficking of lysosomes to the cell surface and lysosomal enzyme secretion had been found [39, 40].
5. Conclusions
In Nature Reviews Drug Discovery, Moffat et al. [41] contended that there is an unmet need for mechanism-informed phenotypic screening in cancer drug discovery that integrates “three-dimensional cell-culture systems, high-throughput confocal microscopy and three-dimensional image analysis tools” as well as “sophisticated cell models” that include tumor cells and cells of the in vivo tumor microenvironment. The pathomimetic avatars we have described here are such sophisticated 3D cell models. In addition, we have shown that these pathomimetic avatars can be used for livecell quantitative imaging in real time of spatial, temporal, and dynamic changes in breast cancer progression. We have established that our models and technologies can identify, validate, and track proteolytic pathways that mediate breast cancer progression. We suggest they can be used for comparable studies in a wide variety of cancers in which proteases play causal roles in progression and metastasis and thus will meet the need for mechanism-informed phenotypic screening in cancer drug discovery.
Acknowledgments
Funding information This work was supported in part by National Institute of Health grants R01 CA131990 (RRM and BFS) and R21 CA1759331 (BFS), a Department of Defense Breast Cancer Research Program Postdoctoral Fellowship Award (W81XWH-12-1-0024; KO), and an award from the President’s Research Enhancement Program of Wayne State University (BFS). Imaging was performed in the Microscopy, Imaging and Cytometry Resources Core (KM), which is supported, in part, by National Institutes of Health Center grant P30 CA022453 to the Karmanos Cancer Institute at Wayne State University, and the Perinatology Research Branch of the National Institute of Child Health and Development at Wayne State University.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no competing interests.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Debnath J, & Brugge JS (2005). Modelling glandular epithelial cancers in three-dimensional cultures. Nature Reviews. Cancer, 5(9), 675–688. 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 2.Martin KJ, Patrick DR, Bissell MJ, & Fournier MV (2008). Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One, 3(8), e2994. 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigelt B, Ghajar CM, & Bissell MJ (2014). The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Advanced Drug Delivery Reviews, 69–70, 42–51. 10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, & Jemal A (2019). Cancer statistics, 2019. CA: a Cancer Journal for Clinicians, 69(1), 7–34. 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 5.Li Q, Mullins SR, Sloane BF, & Mattingly RR (2008). p21-activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia, 10(4), 314–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Chow AB, & Mattingly RR (2010). Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. The Journal of Pharmacology and Experimental Therapeutics, 332(3), 821–828. 10.1124/jpet.109.160390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam JM, Onodera Y, Bissell MJ, & Park CC (2010). Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer Research, 70(13), 5238–5248. 10.1158/0008-5472.CAN-09-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire SL, Peck B, Wai PT, Campbell J, Barker H, Gulati A, Daley F, Vyse S, Huang P, Lord CJ, Farnie G, Brennan K, & Natrajan R (2016). Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. The Journal of Pathology, 240(3), 315–328. 10.1002/path.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brock EJ, Ji K, Shah S, Mattingly RR, & Sloane BF (2019). In vitro models for studying invasive transitions of ductal carcinoma in situ. Journal of Mammary Gland Biology and Neoplasia, 24(1), 1–15. 10.1007/s10911-018-9405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herschkowitz JI, & Behbod F (2018). Advances in DCIS research and treatment. [journal issue]. Journal of Mammary Gland Biology and Neoplasia, 23 & 24, 1–301. 10.1007/s10911-018-9419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards D, Hoyer-Hansen G, Blasi F, & Sloane BF (2008). The cancer degradome: protease and cancer biology. New York: Springer. [Google Scholar]
- 12.Sloane BF, List K, Fingleton B, & Matrisian L (2013). Proteases: structure and function. New York: Springer. [Google Scholar]
- 13.Darvishian F, Ozerdem U, Adams S, Chun J, Pirraglia E, Kaplowitz E, Guth A, Axelrod D, Shapiro R, Price A, Troxel A, Schnabel F, & Roses D (2019). Tumor-infiltrating lymphocytes in a contemporary cohort of women with ductal carcinoma in situ (DCIS). Annals of Surgical Oncology, 26(10), 3337–3343. 10.1245/s10434-019-07562-x. [DOI] [PubMed] [Google Scholar]
- 14.Grugan KD, McCabe FL, Kinder M, Greenplate AR, Harman BC, Ekert JE, van Rooijen N, Anderson GM, Nemeth JA, Strohl WR, Jordan RE, & Brezski RJ (2012). Tumor-associated macrophages promote invasion while retaining Fc-dependent anti-tumor function. Journal of Immunology, 189(11), 5457–5466. 10.4049/jimmunol.1201889. [DOI] [PubMed] [Google Scholar]
- 15.Shree T, Olson OC, Elie BT, Kester JC, Garfall AL, Simpson K, Bell-McGuinn KM, Zabor EC, Brogi E, & Joyce JA (2011). Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes & Development, 25(23), 2465–2479. 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdogan B, & Webb DJ (2017). Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochemical Society Transactions, 45(1), 229–236. 10.1042/BST20160387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu SQ, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, & Schroder CP (2018). Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treatment Reviews, 70, 178–189. 10.1016/j.ctrv.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Dawson PJ, Wolman SR, Tait L, Heppner GH, & Miller FR (1996). MCF10AT: a model for the evolution of cancer from proliferative breast disease. The American Journal of Pathology, 148(1), 313–319. [PMC free article] [PubMed] [Google Scholar]
- 19.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, & Miller FR (2001). Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Research and Treatment, 65(2), 101–110. [DOI] [PubMed] [Google Scholar]
- 20.Miller FR, Santner SJ, Tait L, & Dawson PJ (2000). MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. Journal of the National Cancer Institute, 92(14), 1185–1186. 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 21.Sameni M, Cavallo-Medved D, Franco OE, Chalasani A, Ji K, Aggarwal N, Anbalagan A, Chen X, Mattingly RR, Hayward SW, & Sloane BF (2017). Pathomimetic avatars reveal divergent roles of microenvironment in invasive transition of ductal carcinoma in situ. Breast Cancer Research, 19(1), 56 10.1186/s13058-017-0847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polyak K, & Hu M (2005). Do myoepithelial cells hold the key for breast tumor progression? Journal of Mammary Gland Biology and Neoplasia, 10(3), 231–247. 10.1007/s10911-005-9584-6. [DOI] [PubMed] [Google Scholar]
- 23.Labernadie A, Kato T, Brugues A, Serra-Picamal X, Derzsi S, Arwert E, et al. (2017). A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nature Cell Biology, 19(3), 224–237. 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sameni M, Tovar EA, Essenburg CJ, Chalasani A, Linklater ES, Borgman A, Cherba DM, Anbalagan A, Winn ME, Graveel CR, & Sloane BF (2016). Cabozantinib (XL184) inhibits growth and invasion of preclinical TNBC models. Clinical Cancer Research, 22(4), 923–934. 10.1158/1078-0432.CCR-15-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavallo-Medved D, Rudy D, Blum G, Bogyo M, Caglic D, & Sloane BF (2009). Live-cell imaging demonstrates extracellular matrix degradation in association with active cathepsin B in caveolae of endothelial cells during tube formation. Experimental Cell Research, 315(7), 1234–1246. 10.1016/j.yexcr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osuala KO, Sameni M, Shah S, Aggarwal N, Simonait ML, Franco OE, Hong Y, Hayward SW, Behbod F, Mattingly RR, & Sloane BF (2015). Il-6 signaling between ductal carcinoma in situ cells and carcinoma-associated fibroblasts mediates tumor cell growth and migration. BMC Cancer, 15, 584 10.1186/s12885-015-1576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jedeszko C, Sameni M, Olive MB, Moin K, & Sloane BF (2008). Visualizing protease activity in living cells: from two dimensions to four dimensions. Current Protocols in Cell Biology, 39(1), 4.20.1–4.20. 10.1002/0471143030.cb0420s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalasani A, Ji K, Sameni M, Mazumder SH, Xu Y, Moin K, et al. (2017). Live-cell imaging of protease activity: assays to screen therapeutic approaches. Methods in Molecular Biology, 1574, 215–225. 10.1007/978-1-4939-6850-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji K, Mayernik L, Moin K, & Sloane BF (2019). Acidosis and proteolysis in the tumor microenvironment. Cancer Metastasis Reviews, 38(1–2), 103–112. 10.1007/s10555-019-09796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothberg JM, Bailey KM, Wojtkowiak JW, Ben-Nun Y, Bogyo M, Weber E, Moin K, Blum G, Mattingly RR, Gillies RJ, & Sloane BF (2013). Acid-mediated tumor proteolysis: contribution of cysteine cathepsins. Neoplasia, 15(10), 1125–1137. 10.1593/neo.13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curino AC, Engelholm LH, Yamada SS, Holmbeck K, Lund LR, Molinolo AA, Behrendt N, Nielsen BS, & Bugge TH (2005). Intracellular collagen degradation mediated by uPARAP/Endo180 is a major pathway of extracellular matrix turnover during malignancy. The Journal of Cell Biology, 169(6), 977–985. 10.1083/jcb.200411153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, Johnson J, Gatenby RA, & Gillies RJ (2013). Acidity generated by the tumor microenvironment drives local invasion. Cancer Research, 73(5), 1524–1535. 10.1158/0008-5472.CAN-12-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgington LE, Verdoes M, & Bogyo M (2011). Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Current Opinion in Chemical Biology, 15(6), 798–805. 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, et al. (2005). Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nature Chemical Biology, 1(4), 203–209. 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- 36.Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K, Sloane BF, Anderson RL, Bogyo MS, & Parker BS (2012). Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Research, 72(5), 1199–1209. 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duivenvoorden HM, Rautela J, Edgington-Mitchell LE, Spurling A, Greening DW, Nowell CJ, Molloy TJ, Robbins E, Brockwell NK, Lee CS, Chen M, Holliday A, Selinger CI, Hu M, Britt KL, Stroud DA, Bogyo M, Moller A, Polyak K, Sloane BF, O’Toole SA, & Parker BS (2017). Myoepithelial cell-specific expression of stefin A as a suppressor of early breast cancer invasion. The Journal of Pathology, 243(4), 496–509. 10.1002/path.4990. [DOI] [PubMed] [Google Scholar]
- 38.Kasperkiewicz P, Altman Y, DAngelo M, Salvesen GS, & Drag M (2017). Toolbox of fluorescent probes for parallel imaging reveals uneven location of serine proteases in neutrophils. Journal of the American Chemical Society, 139(29), 10115–10125. 10.1021/jacs.7b04394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heuser J (1989). Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. The Journal of Cell Biology, 108(3), 855–864. 10.1083/jcb.1083.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rozhin J, Sameni M, Ziegler G, & Sloane BF (1994). Pericellular pH affects distribution and secretion of cathepsin B in malignant cells. Cancer Research, 54(24), 6517–6525. [PubMed] [Google Scholar]
- 41.Moffat JG, Rudolph J, & Bailey D (2014). Phenotypic screening in cancer drug discovery - past, present and future. Nature Reviews. Drug Discovery, 13(8), 588–602. 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZQ, Albertson D, & Ethier SP (2004). Genomic organization of the 8p 11-p12 amplicon in three breast cancer cell lines. Cancer Genetics and Cytogenetics, 155(1), 57–62. 10.1016/j.cancergencyto.2004.03.013. [DOI] [PubMed] [Google Scholar]