Abstract
Single-cell RNA sequencing can reveal the transcriptional state of cells, yet provides little insight into the upstream regulatory landscape associated with open or accessible chromatin regions. Joint profiling of accessible chromatin and RNA within the same cells would permit direct matching of transcriptional regulation to its outputs. Here, we describe droplet-based single-nucleus chromatin accessibility and mRNA expression sequencing (SNARE-seq), a method that can link a cell’s transcriptome with its accessible chromatin for sequencing at scale. Specifically, accessible sites are captured by Tn5 transposase in permeabilized nuclei to permit, within many droplets in parallel, DNA barcode tagging together with the mRNA molecules from the same cells. To demonstrate the utility of SNARE-seq, we generated joint profiles of 5,081 and 10,309 cells from neonatal and adult mouse cerebral cortices. We reconstructed the transcriptome and epigenetic landscapes of major and rate cell types, uncovered lineage-specific accessible sites especially for low-abundance cells, and connected the dynamics of promoter accessibility with transcription level during neurogenesis.
RNA sequencing of single cells or nuclei reveals their transcription state, whereas chromatin accessibility sequencing uncovers the associated regulatory landscape. Current strategies1,2, which involve profiling these modalities separately followed by computational integration, may not fully recapitulate the true biological state. Joint profiling of two layers of -omics information within the same cells would enable a direct matching of transcriptional regulation to its output, allowing for more accurate reconstruction of the molecular processes underlying a cell’s physiology.
To enable highly parallel profiling of chromatin accessibility and mRNA from individual nuclei, we developed SNARE-seq, implemented on a micro-droplet platform3. In this method, the accessible chromatin in permeabilized nuclei is captured by the Tn5 transposase, prior to droplet generation. We reason that, without heating or detergent treatment, binding of transposases to its DNA substrate after transposition could maintain the contiguity of the original DNA strands4, allowing for the co-packaging of accessible genomic sites and mRNA from individual nuclei in the same droplets. As such, we designed a splint oligonucleotide with sequence complementary to the adapter sequence inserted by transposition (5’ end) and the poly A bases (3’ end) allowing capture by oligo-dT-bearing barcoded beads. After encapsulation of nuclei, their mRNAs and fragmented chromatin can be released by heating the droplets, allowing access to splint oligos and adaptor coated beads having a shared cellular barcode (Fig. 1a). A pair of RNA-seq and chromatin accessibility libraries can then be generated for sequencing (see Methods). The resulting data can then be connected by their shared cellular barcodes, without the need for probabilistic mapping of single-cell clusters from separate analyses. While SNARE-seq shows similarities to sci-CAR5 conceptually, our method was implemented on a widely accessible Drop-seq platform and provides denser chromatin information due to a design that captures chromatin information first, then linking it to the transcriptome.
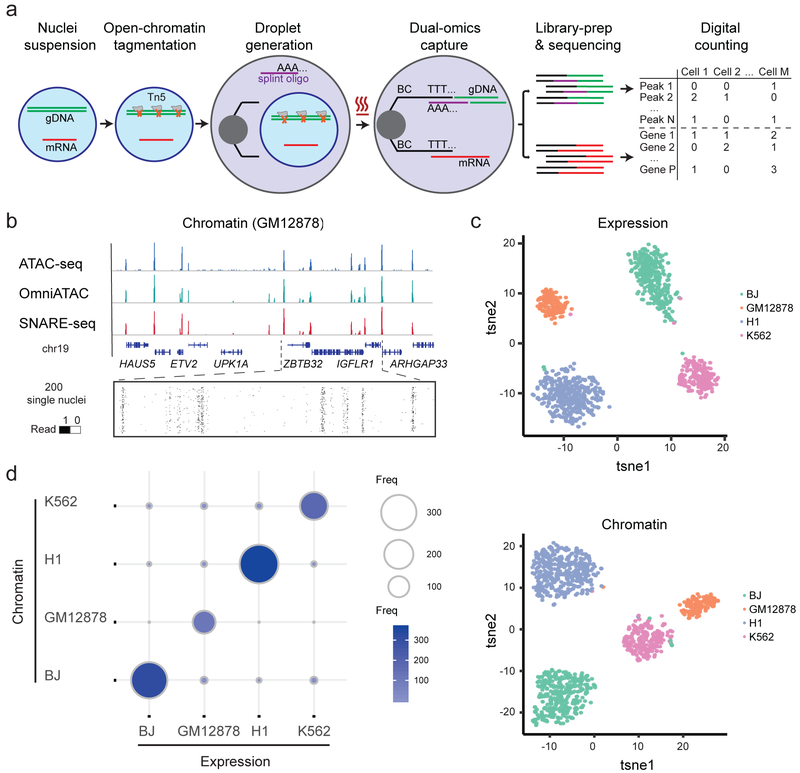
Figure 1.
Linked single-nucleus transcriptome and chromatin accessibility sequencing of human cell mixtures. a, Workflow of SNARE-seq. Key steps are outlined in the main text. b, Aggregate single-nucleus chromatin accessibility profiles recaptured published profiles of ATAC-seq and Omni-ATAC in GM12878 cells. c, t-SNE visualization of SNARE-seq paired gene expression (upper panel) and chromatin accessibility (lower panel, n=1,047) data from BJ, GM12878, H1 and K562 cell mixture. Cellular identities are colored based on independent clustering results with either expression or chromatin data. d, Inter-assay identity agreement reveals consistent linked transcriptome and chromatin accessibility profiles of SNARE-seq data. The size and color depth of each circle represents the number of cellular barcodes that were identified by the different assays.
To evaluate SNARE-seq’s ability to capture accessible chromatin, we first performed a proof-of-concept experiment on lymphoblastoid cell line GM12878, which have extensively characterized chromatin landscapes. Ensemble profiles of SNARE-seq accessibility data showed a signal-to-noise ratio similar to ATAC-seq6 and Omni-ATAC7 (Fig. 1b). The aggregate SNARE-seq data also showed the expected periodical nucleosome pattern and a strong enrichment of fragments within canonical promoter regions (Fig. S1a, c), which are typical characteristics of bulk ATAC-seq data. We validated the peaks called from the SNARE-seq data by overlapping them with those of published bulk ATAC-seq and Omni-ATAC data (Fig. S1b) and found that 85.9% of ATAC-seq peaks were shared among all the three assays, and that 87.6% of Omni-ATAC peaks were shared between Omni-ATAC and SNARE-seq. After filtering out low quality data, we obtained a median of 2720 accessible sites per nuclei, which is comparable with several published single cell/nuclei ATAC-seq methods and roughly 4–5 folds less dense than the 10X Genomics sc-ATAC-seq method (Fig. S1d and S2a).
To assess the accuracy of SNARE-seq in identifying cell types, we performed SNARE-seq on mixtures of cultured human BJ, H1, K562 and GM12878 cells, and collected 1,047 paired profiles (median 500 UMIs; median 805 accessible sites, Fig. S2a,d). Separate clustering of expression and chromatin accessibility data showed clear separation into four distinct clusters (Fig. 1c). Differential expression of maker genes (Fig. S3a) validated these cluster identities. The classification results from both profiles were in good agreement (kappa coefficient of 0.92, Fig. 1d). Notably, we found that transcription factors JUN, IRF8, POU5F1 and GATA1, which showed enriched expression in BJ, GM12878, H1 and K562 cells, respectively (Fig. S3c), also exhibited a similar pattern of preferential binding to chromatin sequences captured by SNARE-seq accessibility assay (Fig. S3b,d), consistent with previous observations8. We improved the detection sensitivity on chromatin further by using NP40 based Nuclei EZ buffer to boost tagmentation efficiency and adding RNase inhibitor combination9 to protect RNA from degradation. From the mixed cell lines we acquired 1,043 paired profiles with median number of 1159 UMIs and 2254 accessible sites captured (Fig. S2a, d). We then compared SNARE-seq expression and chromatin data with those generated from snDrop-seq or SNARE-seq chromatin only experiments, and observed consistent clustering (Fig. S4a,b), high level correlation of raw reads (Fig. S4d), as well as efficient recovery rate (Fig. S4e, 66% recovery of RNA and 100% recovery of chromatin). Furthermore, the species-mixing experiment indicated a high purity and low doublet rate (6%, Fig. S4f) of SNARE-seq. Therefore, on simple cell mixture, SNARE-seq can effectively separate cell types based on both their chromatin signatures and transcriptomes, with a high level of concordance.
We next applied SNARE-seq to mouse neonatal cerebral cortex (postnatal day 0, n=5) and recovered 5,081 nuclei that had linked transcriptome (median 357 UMIs) and chromatin accessibility (median 2583 accessibility sites) data after QC filtering (Fig. S2a, d). Correlation analysis of expression or chromatin profiles demonstrated great reproducibility between independent SNARE-seq experiments (Fig. S5a,b). Among all RNA reads, 94% aligned to the genome, with 37% of these mapped to exons and 42% mapped to introns (Fig. S5c), reflecting the enrichment of nascent transcripts in the nucleus1. In comparison, despite a similar mapping rate (>91%), the chromatin accessibility data showed a larger fraction of reads (34%) mapped to intergenic regions. There was also enrichment of accessibility reads in close proximity to the transcription start site (10%) and low coverage in exons, suggestive of enhancer and promoter sequences present in those noncoding regions. Therefore, both RNA and chromatin reads showed expected genome distributions comparable to the snDrop-seq1 and snATAC data10.
Unsupervised clustering of cerebral cortex transcriptomes identified 19 cell clusters, including: astrocytes/radial glia (Ast/RG); intermediate progenitor cells (IP); excitatory neurons (Ex); migrating inhibitory neurons (In); and Cajal-Retzius cells (CR). We further detected several non-neuronal cell types, including: oligodendrocyte progenitor cells (OPC); endothelial cells (End); pericytes (Peri); and microglia (Mic). These cell clusters ranged in size from 37 (0.7%) to 542 (10.7%) cells (Fig. S6a), and were independent of batch or sequencing depth (Fig. S6b–e). Uniform Manifold Approximation and Projection (UMAP) revealed a trajectory extending from the progenitor states reflective of the sequential development of cortical cell fates. Consistently, nuclei occurring adjacent to intermediate progenitors represented those of the late born neurons of the superficial layers (proceeding deep layer neurons) and glial cell types associated with the onset of gliogenesis that is expected at this time point (Fig. 2a). We compared SNARE-seq transcriptome data with a recently published single-cell RNA-seq dataset of the mouse cortex at a similar developmental time point that was generated by SPLiT-seq9. Despite a lower number of detected UMIs, the cell types and their signatures were reasonably well correlated (Fig. S7a–c). Notably, we captured finer distinctions between closely related cellular states and identified three sub-clusters of intermediate progenitor cells: cluster IP-Hmgn2, expressing Mki67, Top2a and Kif23 (Fig. 2b, Fig. S7d and Table S1), representing cycling progenitors; cluster IP-Gadd45g, which was enriched for Gadd45g, representing apical progenitors that exited from cell-cycle11; and cluster IP-Eomes, representing basal progenitors that show early commitment to the neuronal lineage. Cell-type and layer identities of our clusters were further validated by expected expression of known marker genes and in situ staining of makers discovered here (Fig. 2b, S8 and Table S1).
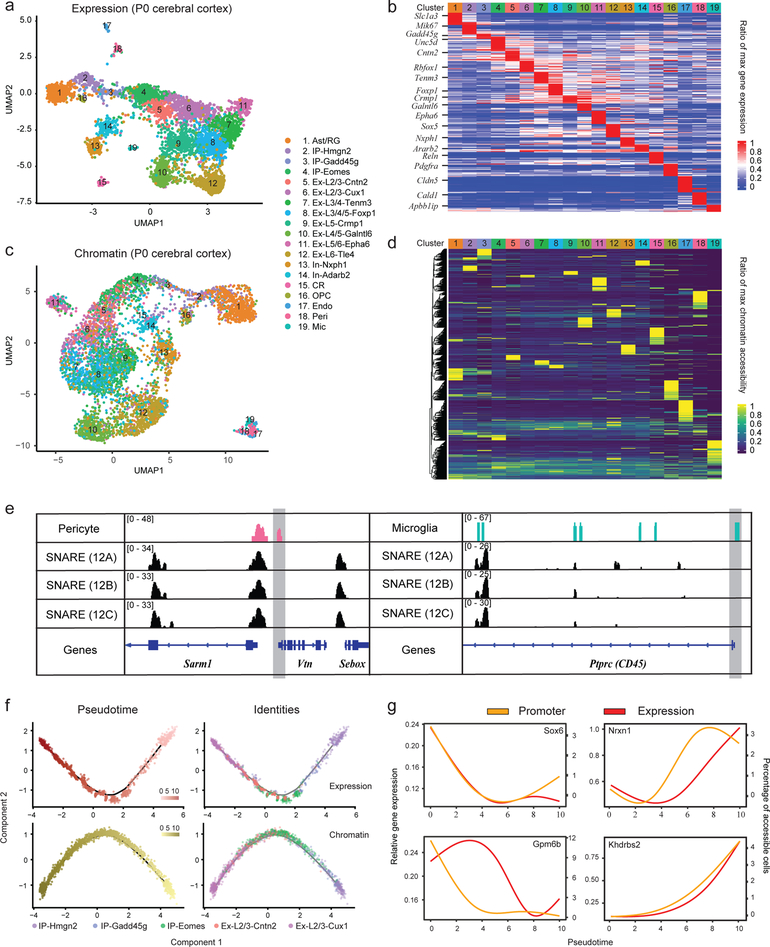
Figure 2.
Dual-omics profiling of neonatal mouse cerebral cortex with SNARE-seq (n=5 replicates). a, UMAP projection of 5,081 SNARE-seq expression data of mouse cerebral cortex nuclei. Cell types were assigned based on known markers. b, Heatmap showing the normalized expression of cell type-specific genes relative to the maximum expression level across all cell types. c, UMAP projection of SNARE-seq chromatin accessibility data of mouse cerebral cortex nuclei. Cells are labeled with the same color codes for cell types identified by the linked expression data. d, Heatmap showing the normalized chromatin accessibility of type-specific accessible sites relative to the maximum accessibility across all cell types. e, Chromatin accessibility tracks generated from cell-type specific or batch aggregated (batch code 12A, 12B and 12C) chromatin accessibility data at pericyte (left) and microglia (right) marker gene loci (Vtn and CD45 respectively). For better visualization, the promoter regions were shaded in gray. f, Pseudotime trajectories constructed with SNARE-seq expression (upper panels) and chromatin accessibility (lower panels) profiles for 1,469 nuclei (214 IP-Hmgn2, 99 IP-Gadd45g, 437 IP-Eomes, 177 Ex-L2/3-Cntn2 and 542 Ex-L2/3-Cux1) from the mouse cerebral cortex. Cells are colored according to pseudotime score (left panels) or cellular identities (right panels). g, Promoter accessibility (yellow) and gene expression (red) changes of Sox6, Gpm6b, Nrxn1 and Khdrbs2 across pseudotime during early neurogenesis. Misc, cells of miscellaneous clusters.
We compared aggregated SNARE-seq chromatin accessibility profiles with published bulk ATAC-seq ENCODE data on neonatal mouse brain cortex and found a strong concordance between these two methods (Fig. S7e–f). To cluster co-assayed cells based on their chromatin accessibility profiles, we used their corresponding transcriptional profiles to aggregate chromatin accessibility signals for each cluster separately, followed by peak calling and the probabilistic topic modeling method implemented in cisTopic12. After projecting onto lower dimensions using UMAP, most single-nuclei chromatin accessibility clusters (Fig. 2c), corresponded to the same cell types resolved from the corresponding expression data (Fig. 2a). Notably, the chromatin accessibility of deep layer excitatory neurons and migrating inhibitory neurons, which differentiated earlier in the cerebral cortex and ganglionic eminences, respectively, showed well-separated clusters, whereas those of late-generated superficial layer excitatory neurons were less distinct. Those diffuse boundaries identified by expression profile were also clustered as subtypes based on chromatin information (Fig. S9a–b). Those subtypes may represent datasets with insufficient clustering power due to the sparsity of chromatin data and/or dynamic epigenetic states that are still undergoing maturation. Cell-type identities of the major clusters were further supported by the specific accessibility in the promoter region for marker gene loci Hes5 (Ast/RG), Gadd45g (IP), Meg3 (Neurons), Pdgfra (OPC), Vtn (Peri) and Apbb1ip (Mic) (Fig. S9d). We found that the promoter accessibility of lineage markers Vtn and CD45 (for pericyte and microglia representing 1% and 0.7% of total cells) were present only in cell-type aggregated chromatin profiles that were identified de novo with transcriptome data. In contrast, chromatin data analyzed based on the accessible peaks called from the batch-aggregated profiles, the current default strategy for analyzing sc-ATAC-seq data, failed to recover these accessible peaks unique to rare cells in the presence of background noise from other more abundant cells (Fig. 2e). Consistent with this notion, clustering of chromatin profiles generated without using any expression information yielded less clear cell type boundary and many of those low-abundant cell types were largely undetected (Fig. S9c). Therefore, a priori knowledge of cell type identity in chromatin accessibility data using the linked gene expression profiles permits more sensitive detection of accessible chromatin region. This underscores the strength of our SNARE-seq dual-omic assay over independent single-cell RNA and chromatin accessibility sequencing methods for detecting cell-type and subtype specific gene expression and accessible chromatin.
Differential accessibility (DA) test of SNARE-seq chromatin profiles identified 35,166 sites (p<0.05) across the 19 murine cerebral cortex cell types (Fig. 2d and Table S2). Of all 35,166 differential accessible sites, 2,835 (8%) located within promoter regions, and 128 also showed differential gene expression between clusters (Fig. S10a). For theses 128 genes, the expression levels and their promoter accessibilities across all cell types were mostly positively correlated (median r 0.34, Fig. S10b), indicating a direct linkage of chromatin accessibilities to the corresponding transcriptomes. To further characterize the DA sites, we performed gene ontology enrichment and motif discovery analysis using GREAT and HOMER, respectively (Fig. S11). Notably, genomic elements that were mostly associated with Ast/RG and OPC cells fell into the biological processes regulating stem cell maintenance and differentiation. These sites were further enriched for binding motifs of LHX2 and SOX2, both of which are known regulators of neurogenesis and gliogenesis13,14. We also found that differential accessible sites of the IP-Gadd45g cells (representing 1.9% of the total cells) were enriched for the Wnt signaling pathway components, consistent with the role or this pathway in regulating cell cycle exit and promoting neuronal differentiation of intermediate progenitors15. Therefore, linking chromatin accessibility profiles to transcriptomic data directly allowed us to effectively identify cell-type specific transcription regulatory mechanisms.
Next, we focused on the transition of intermediate progenitors to upper layer excitatory neurons. Using Monocle, we ordered gene-expression profiles of 1,469 nuclei along a pseudotime trajectory based on the top differential expressed genes (qval<0.05, Fig. 2f, upper panel). From transcription kinetics, we found a clear pattern originating from a cell-cycle exited state (Mki67 and Gadd45g), that progressed from neuroblast stages (Eomes and Unc5d) to Foxp1 and Cux1-expressing upper layer neurons16,17 (Fig. S12a). We further oriented accessibility profiles of the same nuclei along a separate trajectory (Fig. 2f, lower panel) based on a set of 1,332 sites that showed differential accessibility (qval<0.1). These separately constructed developmental trajectories showed high correlation (r=0.87) along pseudotime. From these differential accessible sites, 103 were found within promoter regions and 21 associated genes were also differentially expressed by pseudotime. Most of these genes presented similar directional changes in promoter accessibility and expression level (Fig. S12b–c). For example, Sox6, a transcription factor required for maintenance of neural precursor cells18, and membrane protein-encoding Mlc1 showed a decline along neuronal differentiation, while Khdrbs2 (SLM1), an RNA-binding protein participating in alternative splicing, and its regulating target Nrxn119 showed similar directional raise along neurogenesis (Fig. 2g, S12c,d). Thus, SNARE-seq provided linked expression and chromatin accessibility profiles that enables construction of regulatory dynamics during developmental programs, as well as detailed characterization of epigenetic state for the cell clusters (Fig. S13).
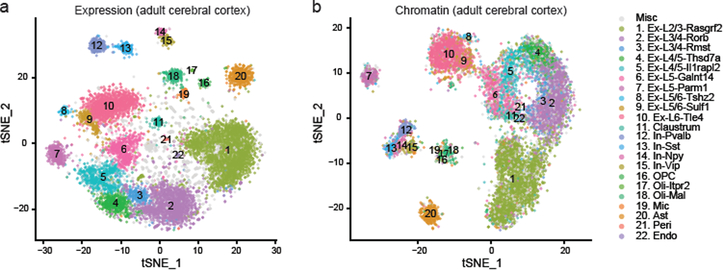
We finally applied SNARE-seq to adult mouse cerebral cortex and obtained 10,309 paired profiles (median 1,332 RNA UMIs and median 2,000 chromatin accessibility sites per nucleus) after QC filtering (Fig. S2a,d). Unsupervised clustering of the 10,309 transcriptomes revealed 22 cell clusters, including 10 excitatory neuron types, 4 inhibitory neuron types (Pvalb, Sst, Npy and Vip-expressing) and oligodendrocyte progenitor cells (OPC), newly-formed Itpr2-expressing oligodendrocyte (Oli-Itpr2) and mature oligodendrocyte (Oli-Mal), as well as other non-neuronal cells (Fig. 3a). Most of the clusters can be identified with existing lineage or cortical layer markers. Those marker genes expressed in a similar but more specific pattern (Fig. S8, 14a–d and Table S1) in the cell clusters. To investigate the epigenetic patterns of each cell cluster, we aggregated SNARE-seq chromatin data of adult cerebral cortex, which showed high similarity to bulk ATAC-seq data (Fig. S14e), based on cell-type identifies defined by de novo clustering of linked transcriptome data. We then performed peak-calling and clustering using the topic modeling method12. The cell clusters were more cleanly and distinctly separated, compared to chromatin profile of neonatal cortex, probably due to the more discrete cell states of adult brain. We next performed gene ontology and motif enrichment analysis on those differential accessible peaks identified across all cell clusters (Table S3). Although some clusters, such as astrocyte and microglia showed similar enrichment of biological process and transcription factors (Fig. S15), most other clusters revealed regulation features different from the corresponding cells in the developing mouse cortex, which might reflect the postnatal maturation within brain.
Figure 3.
SNARE-seq profiling of adult mouse cerebral cortex. a, tSNE projection of SNARE-seq expression data of mouse cerebral cortex 10,309 nuclei (n=12 replicates). Cell types were assigned based on known markers. b, tSNE projection of SNARE-seq chromatin accessibility data of adult mouse cerebral cortex nuclei. Cells were labeled with the same color codes for cell types identified by the linked expression data.
Overall, SNARE-seq is a robust method allowing the joint measurement of transcriptome and chromatin accessibility in single cells or nuclei. Due to a simple design that does not rely on proprietary reagents, SNARE-seq can be widely implemented. Compared to the recently reported sci-CAR5, SNARE-seq detects RNA molecules at a sensitivity comparable to other single nuclei RNA-seq methods (Fig. S2d,e), and captures 4–5x more accessible sites (Fig. S2a,b), which improved the discovery of differentially accessible sites by ~2-fold and provided a better separation of cell clusters (Fig. S13). Finally, the throughput of this assay can be further improved through an integration with a cellular combinatorial indexing strategy10. SNARE-seq represents a valuable tool for characterizing tissue complexity on both the inputs and outcomes of transcriptional regulation units, and would be especially useful for creating cell atlases of human tissues and clinical samples.
Online Methods
Ethics.
The human embryonic stem cell line H1 was purchased from WiCell and the related study was approved by UCSD Embryonic Stem Cell Research Oversight (ESCRO) Committee.
Cell culture.
BJ and K562 cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. GM12878 cells were maintained in 1640 medium supplemented with 15% fetal bovine serum. H1 human embryonic stem cell line was maintained in feeder-free mTeSR medium and passaged with ReLeSR according to manufacturer’s instruction.
Nuclei preparation.
GM12878 nuclei were extracted with ATAC-Resuspension Buffer containing 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin as described previously7. Nuclei from human cell line mixture were extracted with either nuclear extraction buffer (NEB) (0.32 M sucrose, 5 mM CaCl2, 3 mM Mg(Ac)2, 0.1 mM EDTA, 20 mM Tris-HCl (pH=8), and 0.1% Triton X-100) or ice-cold Nuclei EZ Prep buffer (Cat # NUC101). To extract nuclei from mouse cerebral cortex (C57BL/6 mouse cortex at postnatal day 0 and 2 months, purchased from BrainBits (Cat # C57PCX)), the pair of tissue samples were chopped into small pieces with a razor blade and were homogenized using a glass Dunce tissue grinder (10 times with pastel A and 20 times with pastel B) in 2 ml ice-cold Nuclei EZ Prep buffer. Nuclei were then passed through a 30-μm filter (Sysmex Partec), spun down for 10 min at 900g, and then washed and resuspended in PBS supplemented with 1% fatty-acid-free BSA.
Tn5 tagmentation of nuclei.
Nuclei were counted with an automated cell counter and approximately 200,000 nuclei were used for tagmentation. Nuclei pellets were resuspended in a total of 50 μL reaction mix containing 25 μL 2X Nextera Tagment DNA Buffer, 8 μL TDE1 Tagment DNA Enzyme and 1 μL NxGen RNase inhibitor (or 1 μL Enzymatics RNase-In and 1 μL Superase-In for human cell line mix and adult mouse cerebral cortex experiments) and incubated at 37 °C for 30 min with shaking at 500 rpm. After tagmentation, nuclei were resuspended and washed with PBS containing 1% BSA and kept on 4 °C until droplet generation.
Nuclei barcoding.
Droplet generation was performed as described previously1, with a few modifications. Briefly, tubing and syringes were coated with 1% BSA to prevent nonspecific binding and then rinsed with PBS prior to experiment. Ficoll PM-400 was added in nuclei suspension buffer instead of lysis buffer to mitigate nuclei settling. To capture released chromatin fragments with barcoded beads, 1 μL splint oligo (Nextera-R1-rc-polyA, 10 μM, Table S3) was added into Drop-seq lysis buffer. Nuclei suspension at a concentration of 100 nuclei/μl were co-encapsulated with barcoded beads (from ChemGenes, Cat # Macosko201110) in droplets. When encapsulation was complete, microfluidic emulsion collected in Falcon tubes were overlaid with a layer of mineral oil and then transferred to a 72 °C water bath to lyse nuclei and release binding of Tn5 with genomic DNA. After 5 minutes of incubation, collection tubes were moved from the water bath to ice.
Sequencing library preparation.
Droplets were broken by perfluoro-octanol, after which beads were harvested and washed with 6X SSC containing 10 μL blocking oligos (Nextera-R1-bk and Nextera-Ad2-bk, 100 μM). After washing beads with 6X SSC again and RT buffer once, beads were resuspended in 200 μL reverse transcription/ligation mix (2X T7 ligation buffer, 50 mM KCl, 2% FicoII, 1 nM dNTP, 2.5 μM Template Switch Oligo, 10 mM DTT, 5 μl RNase inhibitor, 12.5 μL Hemo Klentaq, 2.5 μL T7 ligase and 2.5 μL reverse transcriptase), and incubated at room temperature for 30 minutes and at 42 °C for 90 minutes, followed by treatment with Exonuclease I at 37 °C for 45 minutes. Then an aliquot of 10,000 beads were spun down and library was then PCR amplified using primer pair Nextera-R2/Tso-PCR for a total of 16 cycles. After column purification, PCR products were split into two halves for either cDNA or chromatin library amplification. To prepare the cDNA sequencing library, 0.6X bead size selected PCR products were amplified with primer Tso-PCR alone to enrich cDNA library, following by another round of 0.6X bead size selection. Sequencing libraries were constructed with Nextera XT kit as described previously3. To prepare the chromatin sequencing library, primer pair P5XX-Tso/Ad2.X (Supplementary Table 4) were used to add indexes and P5/P7 sequences, and the DNA library with fragment sizes between 225 to 1000 bp was carefully excised from PAGE gel and purified using column purification.
NGS Sequencing.
SNARE-seq cDNA libraries were sequenced on an Illumina HiSeq2500 instrument with Read1CustSeqB and HP11 for priming of read 1 (30 bp) and read 2 (80 bp) respectively. SNARE-seq chromatin libraries were sequenced on the same instrument with Read1CustSeqB for priming of read 1 (30 bp), HP10 for priming of index 1 (75 bp), and HP11 for priming of read 2 (75 bp), with 8 bp index 2 read for de-multiplexing.
Sequencing data preprocessing.
Paired-end sequencing reads of cDNA libraries were processed exactly as described previously3. First, reads with less than six T bases in the last nine bases of read 1 or a poor quality score (<10) were filtered out to remove any contaminated or low quality reads. Cell barcode and UMI information were then inferred from the first 20 bases. After trimming away any portion of the SMART adaptor sequence or large stretches of poly(A) tails, read 2 were then aligned to the human (hg38) or mouse genome (mm10) with STAR v2.5 using default parameter settings. Reads that mapped to intronic or exonic regions of genes were recorded and digital expression matrix was then generated with genes as rows and cells as columns. UMI counts for each gene of each cell were assigned by collapsing UMI reads that had only 1 edit distance. To process chromatin sequencing results, cell barcode and UMI were assigned in a similar way as aforementioned. Paired chromatin reads (read2 and read3) were processed using ENCODE ATAC-seq pipeline (https://github.com/kundajelab/atac_dnase_pipelines), and peaks were called with hg38 or mm10 as reference and using default settings. Peak files were then converted to Picard style interval list file and overlapped with each mapped reads to assign reads with peak names. Digital chromatin accessibility count matrix was then generated with peak names as rows and cells as columns.
Sample correlation analyses.
For expression data, Pearson correlation was calculated with log normalized transcriptional reads aggregated by samples. For chromatin data, pairwise genomic read coverage was calculated using multiBamSummary with consecutive bins of equal size (10 kb) across genome, and the resulting correlation matrices were used to compute the overall similarity between samples.
Expression data clustering.
For human cell line mixture, barcodes with fewer than 200 UMIs or more than 2,000 UMIs (Triton-X lyzed)/ 5,000 UMIs (Nuclei EZ lyzed) were omitted, and barcodes with both transcriptome and chromatin accessibility profiles were selected. The expression count matrix was then normalized in PAGODA2 package (https://github.com/hms-dbmi/pagoda2). Winsorization procedure was employed to cap the magnitude of the ten most extreme values for each gene. Variance of each gene were modeled as dependency on the expression magnitude (log scale) as a smoothed generalized additive model with smoothing term k = 10 (mgcv package in R). The observed-to-expected variance ratio for each gene was modeled by F distribution using the degrees of freedom corresponding to the number of successful gene observations. To normalize the contribution of each gene in the subsequent principal component analysis, we rescaled the variance of each gene to match the tail probability obtained from the F distribution under a standard normal sampling process. Cell clusters were determined from an approximate k-nearest-neighbors graph based on a cosine distance of the top 10 principal components derived from the top 1,000 variable genes from the variance-adjusted expression matrix, using the Infomap community detection algorithm (as implemented in the igraph R package). Cell clusters were visualized by t-distributed stochastic neighbor embedding (t-SNE). For postnatal day 0 mouse cerebral cortex experiments, 6663 barcodes with more than 200 UMIs and less than 1200 UMIs were retained, and 5488 (82.4%) barcodes were left after a second round filtration to remove those with fewer than 250 accessible sites and fraction of reads in peak lower than 0.4. The expression count matrices were combined across independent experiments and were batch corrected, and normalized in PAGODA2 package. Expression variance was adjusted as aforementioned. Then top 2,000 variable genes were used to derive top 50 principal components, and cell clusters were determined from KNN graph. Cell clusters with fewer than 25 cells were omitted from further analysis and resulting 5081(76.3%) cells were re-clustered and visualized by UMAP projection on the top 20 principal components. Genes that were differentially expressed between cell types were identified using Wilcoxon rank sum test in Seurat (v2.3.4, https://satijalab.org/seurat/). Cell clusters were annotated manually on the basis of known markers for the cerebral cortex and gene expression pattern from DropViz (http://dropviz.org/). 10,309 adult mouse cerebral cortex expression datasets were recovered and clustered in a similar way but using different cutoffs (minimum 200 genes, maximum 2500 genes and fraction of reads in peak higher than 0.5).
Comparison of SNARE-seq expression data with SPLiT-seq and DroNc-seq data.
Top 20 genes from the statistically significant principal components differentiating cell types, as well as the top 50 differentially expressed genes associated with each cell type, were identified by Seurat and cluster-averaged expression values were used for correlation analysis between SNARE-seq P0 and SPLiT-seq P2 mouse cerebral cortex expression dataset, and between SNARE-seq and DroNc-seq adult mouse cerebral cortex dataset.
Cell Cycle Phase Assignments.
Each cell was scored using CellCycleScoring function in Seurat based on its expression of G2/M and S phase marker genes. Cells with high G2/M or S scores were assigned as G2/M phase or S phase respectively while cells expressing neither are assigned as G0/G1 phase.
Clustering of chromatin accessibility data.
To cluster chromatin accessibility data from the human cell mixture, the count matrix was first binarized and peaks with fewer than overall 5 counts or expressing in more than 10% of cells were removed. Probability of a region-topic distribution and topic-cell distribution were calculated using latent Dirichlet allocation model with a collapsed Gibbs sampler in cisTopic (v0.1, https://github.com/aertslab/cisTopic). The number of topics with the highest likelihood were picked and principal component analysis were performed for all topics and clustering was visualized by UMAP projection of PCA scores. For mouse cerebral cortex accessibility datasets, cell clusters identified by expression data were used and raw chromatin reads associated with barcodes from the same cell types were aggregated together and cluster-specific peaks were called with bulk ATAC-seq pipeline for each identified cluster. Peaks lists were then merged and the accessibility count matrices were generated by overlapping reads with the merged list. The accessibility count matrices were combined across experiments and clustering was done in a same way in cisTopic as aforementioned. Cell clusters were visualized by UMAP projection of the principal components scores of top 25 topics.
Identification of differential accessible sites.
To identify cluster-specific accessible sites, differential accessible probabilities (p-value) for each peak in each cluster were calculated using Fisher’s exact test. P-values were then converted to q-values by the Benjamini-Hochberg procedure, and peaks with p-values lower than 0.05 in each cluster were kept. The cluster-specific peak counts per cell were then aggregated and normalized by cell-specific library size factors computed separately by estimateSizeFactorsForMatrix in Monocle (v2.10, http://cole-trapnell-lab.github.io/monocle-release/) and visualized using heatmap.
Developmental ordering of early neurogenesis subset.
To order cells according to their developmental trajectory of early neurogenesis based on expression data, we selected 1,498 expression datasets for cells from the mouse cerebral cortex identified as IP-Hmgn2, IP-Gadd45g, IP-Eomes, Ex-L2/3-Cntn2 and Ex-L2/3-Cux1 by the previous PAGODA2 clustering-based approaches. Differentially expressed genes across cell types were identified with the differentialGeneTest function of Monocle and 503 most significant genes (qval < 0.001) were retained to construct the pseudotime trajectory. Cells were ordered according to their value along the trajectory tree. The gene expression along pseudotime was calculated in the same way and genes passing significant test (qval < 0.05) and gene expression kinetics were visualized using the plot_genes_in_pseudotime function in Monocle. Chromatin accessibility dynamics along pseudotime were calculated similarly with gene expression. Briefly, peaks within 10 kb distance were merged in Cicero and differential accessible sites across cell types were tested. After ranking accessible sites by significance (as reported by differentialGeneTest), the top 1,300 most significant sites (qval<0.1) were used to construct the pseudotime trajectory. To select the differentially accessible promoters along pseudotime, we first selected the differential accessible sites within 2 kb of a gene’s transcriptional start site and intersected with the list of differential expressed genes obtained from the step above. Promoter accessibilities were then visualized with the plot_accessibility_in_pseudotime function in Monocle and a natural spline was used to fit the promoter accessibilities along pseudotime with percentage of accessible cells as a covariate.
Annotation of genomic elements.
The GREAT algorithm (http://great.stanford.edu/public/html/) was used to annotate differential accessible sites using the following settings:1 kb upstream and 1 kb downstream, up to 500-kb max extension. The HOMER package (v4.10, http://homer.ucsd.edu/homer/) was used to determine motif enrichment using default setting.
External data.
Published Omni-ATAC (SRP103230), scATAC-seq (GSE65360), snATAC (GSE100033), SPLiT-seq (GSE110823), sci-ATAC(GSE68103), sci-CAR (GSE117089), DroNc-seq (https://portals.broadinstitute.org/single_cell) and ATAC-seq (ENCODE, https://www.encodeproject.org/experiments/ENCSR310MLB/ and https://www.encodeproject.org/experiments/ENCSR889WQX/) data were reprocessed. RNA in situ hybridization images for marker genes was taken from the Allen Institute Brain Atlas.
Data availability.
Raw and processed data is available at Gene Expression Omnibus database under the accession number GSE126074.
Code availability.
Custom script for processing single nucleus chromatin accessibility reads is available at https://github.com/chensong611/SNARE_prep.
Supplementary Material
Supplementary Figure 1. Quality metrics of SNARE-seq chromatin profiles of GM12878. a, DNA fragment size distribution showing nucleosomal periodicity. (Inset) log-transformed histogram. b, Representative Venn diagram showing the numbers of overlapping accessible sites generated using ATAC-seq, Omni-ATAC and SNARE-seq chromatin assays. c, Enrichment of chromatin accessibility signals around transcription start sites (TSS). Top: enrichment around individual TSS. Bottom: aggregated enrichment across all TSSs. X axis indicates the relative distance to TSSs. d, Total number of accessible sites versus fraction of reads in open chromatin peaks (FRiP) within GM12878 cells (n = 3,787 after filtration). Dotted lines (2,000 and 49%) represent cutoffs used for downstream analysis.
Supplementary Figure 2. Comparison of the number of accessible sites, genes and transcripts detected per nucleus by SNARE-seq with different single-cell/nucleus chromatin accessibility or RNA-seq methods. a, Histogram showing the numbers of accessible sites captured by SNARE-seq chromatin profiles. b, Histogram showing the numbers of accessible sites detected per nucleus with different single-cell/nucleus ATAC-seq methods. The processed peak count matrices of published reports were downloaded from GEO (scATAC, GSE65360; sci-ATAC, GSE68103; snATAC, GSE100033; sci-CAR, GSE117089) and binarized. c, Histogram showing the fraction of reads in peaks (FRiP) within GM12878 or postnatal day 0 mouse cerebral cortex SNARE-seq chromatin accessibility data. GM12878, GM; Human cell lines mixture (BJ, GM12878, H1 and K562), lyzed by Triton-X, HuMix; Human cell lines mixture, lyzed by Nuclei EZ Prep, HuMix2; Postnatal day 0 mouse cerebral cortex, P0-brain; Adult mouse cerebral cortex, Ad-brain. d, Histogram showing the numbers of UMIs and genes captured by SNARE-seq expression profiles. e, Histogram showing the number of UMIs and genes detected per nucleus with different single-cell/nucleus RNA-seq methods. The UMI count matrices of published reports were downloaded from GEO (snDrop-seq, GSE97942; SPLiT-seq, GSE110823; sciCAR, GSE117089). Adult human brain cortex, Brain (H); Postnatal day 2 mouse cerebral cortex, Brain (M).
Supplementary Figure 3. SNARE-seq identified cell types within a human cell line mixture (n=1,047). a, Feature plot showing the marker gene expression of individual cell lines within each cluster. b, Biplot showing the contribution of accessible peak topics (n=11) identified by cisTopic in classifying cell types with chromatin data. c, Dot plot showing the expression of transcription factors (TF) in individual clusters. The size of the dot represents the percentage of nuclei within a cell type expressing the transcription factor and the color indicates the average expression level. d, Motif analysis identified the level of significance (in p-value) of transcription factor binding within differential accessible peak topics (n=404,665 fragments) as mentioned above. One-tailed Fisher's exact test was used to calculate significance, and Bonferroni correction was made for multiple testing. p-value of marker TF for each cell type is colored in red.
Supplementary Figure 4. Comparison of SNARE-seq dual-omics assay (n=1,043) with single-omic expression (snDrop-seq, n=591) and chromatin (chromatin only, n=494) methods. a, Clustering of snDrop-seq and SNARE-seq combined expression profiles of human cell line mixture. Cells were labeled by cell type (left) or method (right). b, Clustering of SNARE-seq chromatin profiles (dual or chromatin-only assay) of human cell line mixture. Cells were labeled by cell type (left) or method (right). c, Distribution of transcripts and accessible chromatin peaks detected by SNARE-seq method in individual cell types d, Pearson correlation of gene expression (n=34,828 genes) and chromatin profiles (n=309,891 genomic regions) between dual- and single-omic assays. Aggregated transcript reads and chromatin reads were log10 normalized. e, Distribution of transcripts and chromatin peaks detected by dual- and single-omic assays. The median numbers of transcripts detected by snDrop-seq and SNARE-seq are 1747 and 1159 respectively and the median number of chromatin peaks detected by SNARE-seq single- and dual-omic assay are 2254 and 1960 respectively. In box plots, center lines indicate the median, box limits correspond to the first and third quartiles and whiskers indicate 1.5x interquartile range. f, Species-mixing experiment showing the transcript and chromatin reads detected by SNARE-seq and proportion of human reads in each barcodes.
Supplementary Figure 5. Reproducibility of SNARE-seq (n=5 replicates). a, Pair-wise correlation of gene expression profiles between individual replicates of postnatal day 0 sample. Aggregated transcript reads were log10 normalized. b, Pair-wise correlation of chromatin accessibility profiles between individual replicates. Aggregated genome coverage was log10 normalized. c, Proportion of sequencing reads mapped to different genomic features. Top, mapping of reference expression reads, chromatin reads and accessible peaks. Bottom, mapping of SNARE-seq expression reads, chromatin reads and accessible peaks of mouse cerebral cortex data. For this analysis, total expression reads of snDrop-seq and SNARE-seq are 32,059,445 and 8,238,261, respectively. Total chromatin reads and peaks called are 180,548,727 and 140,102, 428,942,515 and 175,298 for snATAC and SNARE-seq, respectively.
Supplementary Figure 6. Robustness of SNARE-seq. a, Barplot showing the numbers of nuclei recovered for each cell type. UMAP projection of mouse cerebral cortex expression data (n=5,081) as in Fig. 2a showing batch identity (b), and UMI read depth (c). UMAP projection of chromatin accessibility data (n=5,081) as in Fig. 2c showing batch identity (d), and peak read depth (e).
Supplementary Figure 7. Neonatal mouse cerebral cortex SNARE-seq profiles are correlated with published expression and chromatin data. a, Pearson correlation heatmap of mouse cerebral cortex cell types identified with SNARE-seq expression data (n=4,768) compared with previously identified cell types using SPLiT-seq (n=28,384). b, Intra-assay pair-wise Pearson correlation heatmap of cell types identified with SNARE-seq expression data (n=5,081). c, Intra-assay pair-wise Pearson correlation heatmap of cell types identified with SPLiT-seq expression data (n=28,384). d, Proportion of nuclei in different cell cycle phases showing cell cycle exit of late intermediate progenitor cells. e, Representative Venn diagram showing the number of overlap of common peaks between bulk ATAC-seq (ENCODE) data and SNARE-seq chromatin profiles of mouse brain cortex. f, Aggregated SNARE-seq chromatin profiles (bottom two tracks) agree with bulk ATAC-seq (ENCODE, top two tracks) and are consistent between independent experiments.
Supplementary Figure 8. SNARE-seq expression data identified cell type specific markers in mouse neonatal cerebral cortex. a, Dot plot showing the expression of known and novel marker genes (Table S1) in each cell type. b, RNA in situ hybridization (ISH) stains (Allen Human Brain) of postnatal (day 4) mouse cerebral cortex showing layer-specific marker gene expression.
Supplementary Figure 9. Clustering of SNARE-seq single-cell chromatin accessibility on mouse neonatal cerebral cortex (n=5,081). a, UMAP projection of SNARE-seq chromatin data generated with aggregating chromatin reads by transcriptome labeling first, same as Figure 2c. Cells are labeled with the same color codes for cell types identified by the linked expression data. b, Same UMAP projection of SNARE-seq chromatin as a, but cells are labeled with results of independent clustering with each cell's Principal Component scores of topic information calculated by cisTopic. c, Different from a and b, chromatin peak count matrix were generated without using any expression information, and data was clustered independently by cisTop c. Cells are labeled with corresponding cell types identified by expression profile. d, Aggregate chromatin accessibility profiles at loci of cell type-specific marker genes. For better visualization, tracks of differential expressing genes in respective cell types are shaded in gray.
Supplementary Figure 10. SNARE-seq links the promoter accessibility with expression level of cell type-specific genes. a, Top, dot plot showing the differential expression of genes in each cell type. The size of the dot represents the percentage of nuclei within a cell type expressing the gene and the color depth indicates the average expression level. Bottom, dot plot showing the promoter accessibility of markers in each cell type. The size of the dot represents the percentage of nuclei within a cell type that is accessible in promoter regions of corresponding genes and the color depth indicates the average accessibility level. b, Pearson correlation of promoter accessibility and expression levels (n=128 genes) across all cell types for the lineage-specific genes.
Supplementary Figure 11. SNARE-seq chromatin data (n=5,081) from neonatal mouse cerebral cortex uncovers lineage-specific regulatory information. Gene ontology of top biological process was analyzed using a binomial test over 1 kb upstream and 1 kb downstream, up to 500-kb max extension of differential accessible genomic regions in GREAT and transcription factor motifs were discovered using one-tailed Fisher's exact test and Bonferroni correction in HOMER for each cell type. Interesting examples are shaded in gray.
Supplementary Figure 12. Pseudotime analysis reveals dynamics of gene expression in early neurogenesis. a, Heatmap of selected genes involved in neurogenesis showing the expression changes along developmental trajectory. b, Promoter accessibility and gene expression dynamics showing mostly similar directional changes along pseudotime. c, Relative expression of gene markers identified by the pseudotime trajectory analysis. The relative expression is calculated by first model read count with the negative binomial and then normalize with cell size factor estimated by estimateSizeFactors function in Monocle. Smoothed pseudotime-dependent gene expression curve as shown in Figure 2g is colored in red. d, Histogram showing the percentage of cells that had accessible promoters calculated by aggregating chromatin signals into bins (bins=30) along pseudotime, and smoothed curve as shown in Figure 2g is colored in orange.
Supplementary Figure 13. Sensitivity of SNARE-seq chromatin data. a, Total called peaks, recovered promoter regions, median numbers of accessible sites detected per nuclei and total differential accessible sites across all cell types after downsampling raw reads 20-, 15-, 10- and 5-folds. Each dot represents a random downsampling test. Sci-CAR data have a median of ~700 accessible sites/nucleus (indicated by the dash line), which is close to the 15x down-sampled SNARE-seq data. b, Representative UMAP projection of cisTopic clustering result of SNARE-seq chromatin data (n=5,081) at varying down-sampled depths. Cells are labeled with the same color codes for cell types identified by the linked expression data and those discernible clusters are labeled with cluster identities showing on the right.
Supplementary Figure 14. The SNARE-seq profiles of adult mouse cerebral cortex are correlated with published gene expression and chromatin accessibility data. a, The expression level of marker genes (Table S1) for each cell type in adult mouse cerebral cortex. b, Pearson correlation heatmap of mouse cerebral cortex cell types identified with SNARE-seq expression data (n=9,119) compared with previously identified cell types using DroNc-seq (n=4,596). c, Intra-assay pair-wise pearson correlation heatmap of cell types identified with SNARE-seq expression data. d, Intra-assay pair-wise pearson correlation heatmap of cell types identified with DroNc-seq expression data. e, Pair-wise correlation of chromatin accessibility profiles between adult mouse frontal cortex replicates (ENCODE, n=2) and SNARE-seq replicates (n=2). Aggregated genome coverage was log10 normalized. f, In silico downsampling showing the change of genes and UMIs detected by SNARE-seq expression on different sequencing depth.
Supplementary Figure 15. SNARE-seq chromatin data (n=10,309) reveals lineage-specific regulatory information in adult mouse cerebral cortex. Gene ontology of top biological process (binomial test over 1 kb upstream and 1 kb downstream, up to 500-kb max extension of differential accessible genomic regions in GREAT) and transcription factor motifs analysis (using one-tailed Fisher's exact test and Bonferroni correction in HOMER) of each cell type in adult mouse cerebral cortex.
Acknowledgments.
This project was supported by NIH grants U01MH098977, R01HL123755, U54HL145608 to K.Z.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Lake BB. et al. Nat Biotechnol 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duren Z. et al. Proc Natl Acad Sci U S A 115, 7723–7728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macosko EZ. et al. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amini S. et al. Nat Genet 46, 1343–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao J. et al. Science 361, 1380–1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buenrostro JD et al. Nat Methods 10, 213–8 (2013). [Google Scholar]
- 7.Corces MR. et al. Nat Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamanighomi M. et al. Nat Commun 9, 2410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg AB. et al. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preissl S. et al. Nat Neurosci 21, 432–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuzwa SA. et al. Cell Rep 21, 3970–3986 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Bravo González-Blas C. et al. Nat Methods 16, 397–400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian L. et al. Proc Natl Acad Sci U S A 108, E265–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S. et al. Mol Neurobiol 55, 9001–9015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison-Uy SJ, Pleasure SJ. Cold Spring Harb Perspect Biol 4, a008094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artegiani B. et al. Cell Rep 21, 3271–3284 (2017). [DOI] [PubMed] [Google Scholar]
- 17.La Manno G. et al. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KE. et al. Proc Natl Acad Sci U S A 111, 2794–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iijima T. et al. Cell 147, 1601–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Quality metrics of SNARE-seq chromatin profiles of GM12878. a, DNA fragment size distribution showing nucleosomal periodicity. (Inset) log-transformed histogram. b, Representative Venn diagram showing the numbers of overlapping accessible sites generated using ATAC-seq, Omni-ATAC and SNARE-seq chromatin assays. c, Enrichment of chromatin accessibility signals around transcription start sites (TSS). Top: enrichment around individual TSS. Bottom: aggregated enrichment across all TSSs. X axis indicates the relative distance to TSSs. d, Total number of accessible sites versus fraction of reads in open chromatin peaks (FRiP) within GM12878 cells (n = 3,787 after filtration). Dotted lines (2,000 and 49%) represent cutoffs used for downstream analysis.
Supplementary Figure 2. Comparison of the number of accessible sites, genes and transcripts detected per nucleus by SNARE-seq with different single-cell/nucleus chromatin accessibility or RNA-seq methods. a, Histogram showing the numbers of accessible sites captured by SNARE-seq chromatin profiles. b, Histogram showing the numbers of accessible sites detected per nucleus with different single-cell/nucleus ATAC-seq methods. The processed peak count matrices of published reports were downloaded from GEO (scATAC, GSE65360; sci-ATAC, GSE68103; snATAC, GSE100033; sci-CAR, GSE117089) and binarized. c, Histogram showing the fraction of reads in peaks (FRiP) within GM12878 or postnatal day 0 mouse cerebral cortex SNARE-seq chromatin accessibility data. GM12878, GM; Human cell lines mixture (BJ, GM12878, H1 and K562), lyzed by Triton-X, HuMix; Human cell lines mixture, lyzed by Nuclei EZ Prep, HuMix2; Postnatal day 0 mouse cerebral cortex, P0-brain; Adult mouse cerebral cortex, Ad-brain. d, Histogram showing the numbers of UMIs and genes captured by SNARE-seq expression profiles. e, Histogram showing the number of UMIs and genes detected per nucleus with different single-cell/nucleus RNA-seq methods. The UMI count matrices of published reports were downloaded from GEO (snDrop-seq, GSE97942; SPLiT-seq, GSE110823; sciCAR, GSE117089). Adult human brain cortex, Brain (H); Postnatal day 2 mouse cerebral cortex, Brain (M).
Supplementary Figure 3. SNARE-seq identified cell types within a human cell line mixture (n=1,047). a, Feature plot showing the marker gene expression of individual cell lines within each cluster. b, Biplot showing the contribution of accessible peak topics (n=11) identified by cisTopic in classifying cell types with chromatin data. c, Dot plot showing the expression of transcription factors (TF) in individual clusters. The size of the dot represents the percentage of nuclei within a cell type expressing the transcription factor and the color indicates the average expression level. d, Motif analysis identified the level of significance (in p-value) of transcription factor binding within differential accessible peak topics (n=404,665 fragments) as mentioned above. One-tailed Fisher's exact test was used to calculate significance, and Bonferroni correction was made for multiple testing. p-value of marker TF for each cell type is colored in red.
Supplementary Figure 4. Comparison of SNARE-seq dual-omics assay (n=1,043) with single-omic expression (snDrop-seq, n=591) and chromatin (chromatin only, n=494) methods. a, Clustering of snDrop-seq and SNARE-seq combined expression profiles of human cell line mixture. Cells were labeled by cell type (left) or method (right). b, Clustering of SNARE-seq chromatin profiles (dual or chromatin-only assay) of human cell line mixture. Cells were labeled by cell type (left) or method (right). c, Distribution of transcripts and accessible chromatin peaks detected by SNARE-seq method in individual cell types d, Pearson correlation of gene expression (n=34,828 genes) and chromatin profiles (n=309,891 genomic regions) between dual- and single-omic assays. Aggregated transcript reads and chromatin reads were log10 normalized. e, Distribution of transcripts and chromatin peaks detected by dual- and single-omic assays. The median numbers of transcripts detected by snDrop-seq and SNARE-seq are 1747 and 1159 respectively and the median number of chromatin peaks detected by SNARE-seq single- and dual-omic assay are 2254 and 1960 respectively. In box plots, center lines indicate the median, box limits correspond to the first and third quartiles and whiskers indicate 1.5x interquartile range. f, Species-mixing experiment showing the transcript and chromatin reads detected by SNARE-seq and proportion of human reads in each barcodes.
Supplementary Figure 5. Reproducibility of SNARE-seq (n=5 replicates). a, Pair-wise correlation of gene expression profiles between individual replicates of postnatal day 0 sample. Aggregated transcript reads were log10 normalized. b, Pair-wise correlation of chromatin accessibility profiles between individual replicates. Aggregated genome coverage was log10 normalized. c, Proportion of sequencing reads mapped to different genomic features. Top, mapping of reference expression reads, chromatin reads and accessible peaks. Bottom, mapping of SNARE-seq expression reads, chromatin reads and accessible peaks of mouse cerebral cortex data. For this analysis, total expression reads of snDrop-seq and SNARE-seq are 32,059,445 and 8,238,261, respectively. Total chromatin reads and peaks called are 180,548,727 and 140,102, 428,942,515 and 175,298 for snATAC and SNARE-seq, respectively.
Supplementary Figure 6. Robustness of SNARE-seq. a, Barplot showing the numbers of nuclei recovered for each cell type. UMAP projection of mouse cerebral cortex expression data (n=5,081) as in Fig. 2a showing batch identity (b), and UMI read depth (c). UMAP projection of chromatin accessibility data (n=5,081) as in Fig. 2c showing batch identity (d), and peak read depth (e).
Supplementary Figure 7. Neonatal mouse cerebral cortex SNARE-seq profiles are correlated with published expression and chromatin data. a, Pearson correlation heatmap of mouse cerebral cortex cell types identified with SNARE-seq expression data (n=4,768) compared with previously identified cell types using SPLiT-seq (n=28,384). b, Intra-assay pair-wise Pearson correlation heatmap of cell types identified with SNARE-seq expression data (n=5,081). c, Intra-assay pair-wise Pearson correlation heatmap of cell types identified with SPLiT-seq expression data (n=28,384). d, Proportion of nuclei in different cell cycle phases showing cell cycle exit of late intermediate progenitor cells. e, Representative Venn diagram showing the number of overlap of common peaks between bulk ATAC-seq (ENCODE) data and SNARE-seq chromatin profiles of mouse brain cortex. f, Aggregated SNARE-seq chromatin profiles (bottom two tracks) agree with bulk ATAC-seq (ENCODE, top two tracks) and are consistent between independent experiments.
Supplementary Figure 8. SNARE-seq expression data identified cell type specific markers in mouse neonatal cerebral cortex. a, Dot plot showing the expression of known and novel marker genes (Table S1) in each cell type. b, RNA in situ hybridization (ISH) stains (Allen Human Brain) of postnatal (day 4) mouse cerebral cortex showing layer-specific marker gene expression.
Supplementary Figure 9. Clustering of SNARE-seq single-cell chromatin accessibility on mouse neonatal cerebral cortex (n=5,081). a, UMAP projection of SNARE-seq chromatin data generated with aggregating chromatin reads by transcriptome labeling first, same as Figure 2c. Cells are labeled with the same color codes for cell types identified by the linked expression data. b, Same UMAP projection of SNARE-seq chromatin as a, but cells are labeled with results of independent clustering with each cell's Principal Component scores of topic information calculated by cisTopic. c, Different from a and b, chromatin peak count matrix were generated without using any expression information, and data was clustered independently by cisTop c. Cells are labeled with corresponding cell types identified by expression profile. d, Aggregate chromatin accessibility profiles at loci of cell type-specific marker genes. For better visualization, tracks of differential expressing genes in respective cell types are shaded in gray.
Supplementary Figure 10. SNARE-seq links the promoter accessibility with expression level of cell type-specific genes. a, Top, dot plot showing the differential expression of genes in each cell type. The size of the dot represents the percentage of nuclei within a cell type expressing the gene and the color depth indicates the average expression level. Bottom, dot plot showing the promoter accessibility of markers in each cell type. The size of the dot represents the percentage of nuclei within a cell type that is accessible in promoter regions of corresponding genes and the color depth indicates the average accessibility level. b, Pearson correlation of promoter accessibility and expression levels (n=128 genes) across all cell types for the lineage-specific genes.
Supplementary Figure 11. SNARE-seq chromatin data (n=5,081) from neonatal mouse cerebral cortex uncovers lineage-specific regulatory information. Gene ontology of top biological process was analyzed using a binomial test over 1 kb upstream and 1 kb downstream, up to 500-kb max extension of differential accessible genomic regions in GREAT and transcription factor motifs were discovered using one-tailed Fisher's exact test and Bonferroni correction in HOMER for each cell type. Interesting examples are shaded in gray.
Supplementary Figure 12. Pseudotime analysis reveals dynamics of gene expression in early neurogenesis. a, Heatmap of selected genes involved in neurogenesis showing the expression changes along developmental trajectory. b, Promoter accessibility and gene expression dynamics showing mostly similar directional changes along pseudotime. c, Relative expression of gene markers identified by the pseudotime trajectory analysis. The relative expression is calculated by first model read count with the negative binomial and then normalize with cell size factor estimated by estimateSizeFactors function in Monocle. Smoothed pseudotime-dependent gene expression curve as shown in Figure 2g is colored in red. d, Histogram showing the percentage of cells that had accessible promoters calculated by aggregating chromatin signals into bins (bins=30) along pseudotime, and smoothed curve as shown in Figure 2g is colored in orange.
Supplementary Figure 13. Sensitivity of SNARE-seq chromatin data. a, Total called peaks, recovered promoter regions, median numbers of accessible sites detected per nuclei and total differential accessible sites across all cell types after downsampling raw reads 20-, 15-, 10- and 5-folds. Each dot represents a random downsampling test. Sci-CAR data have a median of ~700 accessible sites/nucleus (indicated by the dash line), which is close to the 15x down-sampled SNARE-seq data. b, Representative UMAP projection of cisTopic clustering result of SNARE-seq chromatin data (n=5,081) at varying down-sampled depths. Cells are labeled with the same color codes for cell types identified by the linked expression data and those discernible clusters are labeled with cluster identities showing on the right.
Supplementary Figure 14. The SNARE-seq profiles of adult mouse cerebral cortex are correlated with published gene expression and chromatin accessibility data. a, The expression level of marker genes (Table S1) for each cell type in adult mouse cerebral cortex. b, Pearson correlation heatmap of mouse cerebral cortex cell types identified with SNARE-seq expression data (n=9,119) compared with previously identified cell types using DroNc-seq (n=4,596). c, Intra-assay pair-wise pearson correlation heatmap of cell types identified with SNARE-seq expression data. d, Intra-assay pair-wise pearson correlation heatmap of cell types identified with DroNc-seq expression data. e, Pair-wise correlation of chromatin accessibility profiles between adult mouse frontal cortex replicates (ENCODE, n=2) and SNARE-seq replicates (n=2). Aggregated genome coverage was log10 normalized. f, In silico downsampling showing the change of genes and UMIs detected by SNARE-seq expression on different sequencing depth.
Supplementary Figure 15. SNARE-seq chromatin data (n=10,309) reveals lineage-specific regulatory information in adult mouse cerebral cortex. Gene ontology of top biological process (binomial test over 1 kb upstream and 1 kb downstream, up to 500-kb max extension of differential accessible genomic regions in GREAT) and transcription factor motifs analysis (using one-tailed Fisher's exact test and Bonferroni correction in HOMER) of each cell type in adult mouse cerebral cortex.
Data Availability Statement
Raw and processed data is available at Gene Expression Omnibus database under the accession number GSE126074.