Summary
The regulation of the proliferation and polarity of neural progenitors is crucial for the development of the brain cortex. Animal studies have implicated glycogen synthase kinase 3 (GSK3) as a pivotal regulator of both proliferation and polarity, yet the functional relevance of its signaling for the unique features of human corticogenesis remains to be elucidated. We harnessed human cortical brain organoids to probe the longitudinal impact of GSK3 inhibition through multiple developmental stages. Chronic GSK3 inhibition increased the proliferation of neural progenitors and caused massive derangement of cortical tissue architecture. Single-cell transcriptome profiling revealed a direct impact on early neurogenesis and uncovered a selective role of GSK3 in the regulation of glutamatergic lineages and outer radial glia output. Our dissection of the GSK3-dependent transcriptional network in human corticogenesis underscores the robustness of the programs determining neuronal identity independent of tissue architecture.
Key words: human brain organoids, single cell transcriptomics, GSK3, corticogenesis, outer radial glia
Graphical Abstract
Highlights
-
•
Cortical organoids recapitulate stereotypical neurogenic trajectories
-
•
GSK3 inhibition disrupts neuroepithelium polarity and cortical tissue organization
-
•
GSK3 activity controls oRG production and neurogenesis
In this article, Testa and colleagues show that the chronic inhibition of GSK3 in human cortical organoids causes early disruption of neural progenitor proliferation and polarity, which turns at later stages into a pronounced defect in neurogenesis and outer radial glia production.
Introduction
Neurogenesis is initiated by the formation of a neuroepithelium, composed of neural stem cells (NSCs) organized in apico-basal orientation that divide symmetrically leading to a rapid expansion of the NSC pool (Taverna et al., 2014). The polarization of the neuroepithelium precedes the differentiation of NSCs into radial glia cells (RGCs), and triggers the elongation of cytoplasmic processes that form a migratory scaffold for newborn neurons (Betizeau et al., 2013). Such polarization is in turn necessary for the acquisition of the key properties that define tissue organization (Johansson et al., 2010).
Glycogen synthase kinase 3 alpha and beta (GSK3α and β) are serine/threonine kinases encoded by two different genes, which function as integrating hubs for multiple proliferation and differentiation signals due to their central role in the receptor tyrosine kinase, Wnt and sonic hedgehog signaling pathways (Kim et al., 2009, McCubrey et al., 2016). GSK3 is involved in neurodevelopment through the phosphorylation of a broad set of substrates, including transcription factors essential for brain development, such as CREB (Grimes and Jope, 2001a), neurogenin2 (Ma et al., 2008), β-catenin (Aberle et al., 1997), and multiple microtubule-associated proteins (Fumoto et al., 2008, Hur and Zhou, 2010). Studies in animal models have provided a wealth of evidence linking GSK3 activity to the regulation of early and late neurogenesis in a stage-wise fashion. Its activity is required to maintain the overall polarity of the radial glia scaffold (Yokota et al., 2010). Genetic ablation of both GSK3α and GSK3β in RGCs results in a massive increase in neural progenitor proliferation, with marked suppression of intermediate progenitor cells (IPCs) and postmitotic neurons (Kim et al., 2009). However, paralog-specific knockdown at later stages results in distinct outcomes, with the loss of GSK3β markedly decreasing the production of IPCs and upper-layer Cux1-positive neurons (Ma et al., 2017).
Despite the abundant evidence connecting GSK3 to neurogenesis in various animal models, much less is known of its role in the far more complex human context, mostly due to the lack of models that efficiently recapitulate the human-specific range of progenitor subpopulations. Seminal work conducted in human embryonic stem cells (hESCs) demonstrated their proficiency to form 3D aggregates containing self-organized apico-basally polarized cortical tissues with neurogenic properties (Watanabe et al., 2005). These 3D aggregates are able to generate features usually absent in monolayer cultures, such as specific progenitor subpopulations and organizing centers (Eiraku et al., 2008, Kadoshima et al., 2013), constituting the precursors of current brain organoids protocols. Brain organoids have emerged as the most promising alternative to model neurodevelopment under a strictly human genetic background (Di Lullo and Kriegstein, 2017). Their ontogeny recapitulates most of the salient features of early to mid-fetal brain development, including progenitor populations and distribution of cell domains (Lancaster et al., 2013, Mariani et al., 2015, Mariani et al., 2012, Paşca et al., 2015). Single-cell transcriptional profiling of organoids revealed a cellular diversity that closely matches in composition and transcriptional landscape the human fetal brain and confirmed the presence of human progenitor populations responsible for neocortical expansion (Amiri et al., 2018, Camp et al., 2015, Quadrato et al., 2017).
Here we explore the role of GSK3 on early to middle corticogenesis through the chronic and specific inhibition of its activity in human cortical organoids. By combining morphological characterization with massive parallel RNA sequencing (RNA-seq) on bulk and single cells, we uncover the molecular pathways modulated by GSK3 and break down its effect on distinct cell subpopulations of the developing human cortex, revealing a differential impact on neuronal progenitor subtypes and outer radial glia (oRG) output.
Results
GSK3 Is Essential for Cortical Organoid Morphogenesis
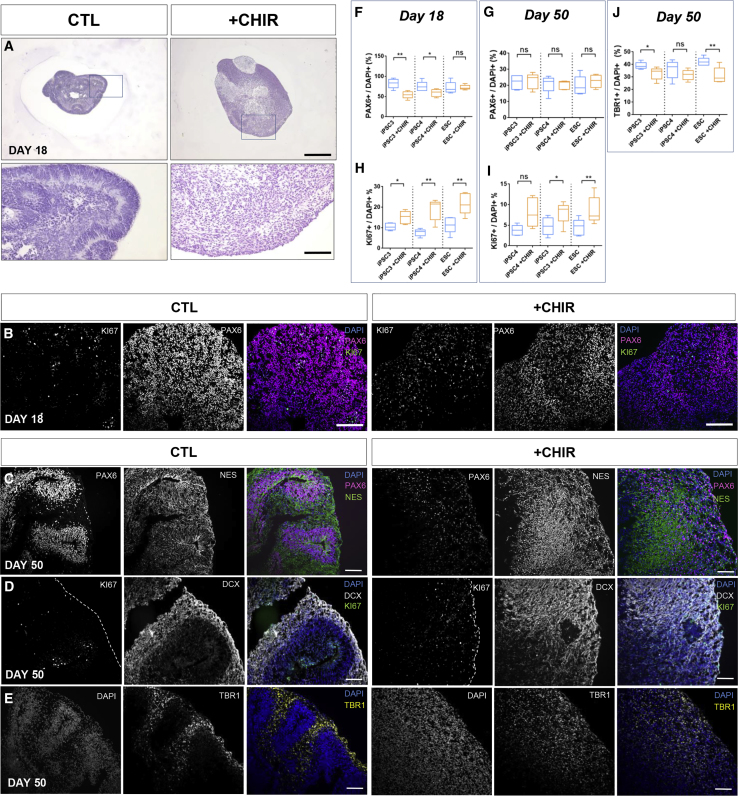
Patterned cortical organoids follow a stereotypical morphogenesis beginning with RGCs aligned in three dimensions around ventricle-like structures (VLS), mimicking the hierarchical organization of the ventricular zone of the dorsal telencephalon. Polarized RGCs are evident by day 18 (Figure 1A), with NESTIN+/PAX6+ cells comprising about 80% of the population (Figure 1B). This proportion decreases over time and becomes restricted to well-confined proliferative domains that gradually generate the neurons of the cortical plate (Paşca et al., 2015). To investigate the role of GSK3 activity throughout corticogenesis, we chronically exposed cortical organoids to the most specific GSK3 inhibitor available CHIR99021 (termed CHIR hereafter) at a 1 μM concentration, selected below the threshold for endodermal or mesodermal lineage induction in hPSCs (Patsch et al., 2015). Chronic GSK3 inhibition resulted in an increase in organoid size (Figures 1A and 2F) concomitant with a virtually complete loss of VLS (Figures 1A and 1B), with a stronger effect by day 50 (Figures 1C–1E). Unexpectedly, this difference in organoid size and radial organization was accompanied by a marginal decrease of PAX6+ cells at day 18 (Figure 1F) (CTL 76 ± 3.3, CHIR 65 ± 2.7), as well as of its expression levels (Figure S1A), while no significant difference was observed at day 50 in the proportion of PAX6+ cells (Figure 1G) (CTL 20 ± 1.2, CHIR 22 ± 1.0). Moreover, staining of early neuron markers TBR1 (layer V–VI neuronal progenitors) (Figure 1E) and DCX (Figure 1D) revealed a profound disarray in tissue architecture with only a slight reduction in TBR1+ cells (CTL 39 ± 1.3, CHIR 32 ± 1.3) (Figure 1J), indicating that GSK3 activity is critical for the correct morphogenesis of the developing cortex.
Figure 1.
Morphogenetic Alterations Caused by Chronic GSK3 Inhibition
(A) Representative captions of cortical organoids at day 18, stained with H&E (magnifications: upper, 10×; lower, 63×). Scale bars, 200 and 10 μm, respectively.
(B) Representative images from day 18 organoids immunostained with anti-PAX6 (red), anti-KI67 (green), and DAPI (blue), wide-field fluorescence images. Scale bar, 50 μm.
(C–E) Representative images from day 50 organoids immunostained with: (C) anti-PAX6 (red), anti-Nestin (green), and DAPI (blue), (D) anti-DCX (white), anti-KI67 (green), and DAPI (blue), and (E) anti-TBR1 (yellow) and DAPI (blue). Scale bars, 50 μm.
(F and G) Quantification of the proportion of PAX6+ nuclei relative to total nuclei (DAPI) at (F) day 18 and (G) day 50 of differentiation.
(H and I) Quantification of the proportion of KI67+ nuclei relative to total nuclei (DAPI) at (H) day 18 and (I) day 50 of differentiation.
(J) Quantification of the proportion of TBR1+ nuclei relative to total nuclei (DAPI) in day 50 organoids. All quantifications were performed in five organoids per line, three independent hPSC lines (N = 15, n = 3), unpaired t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
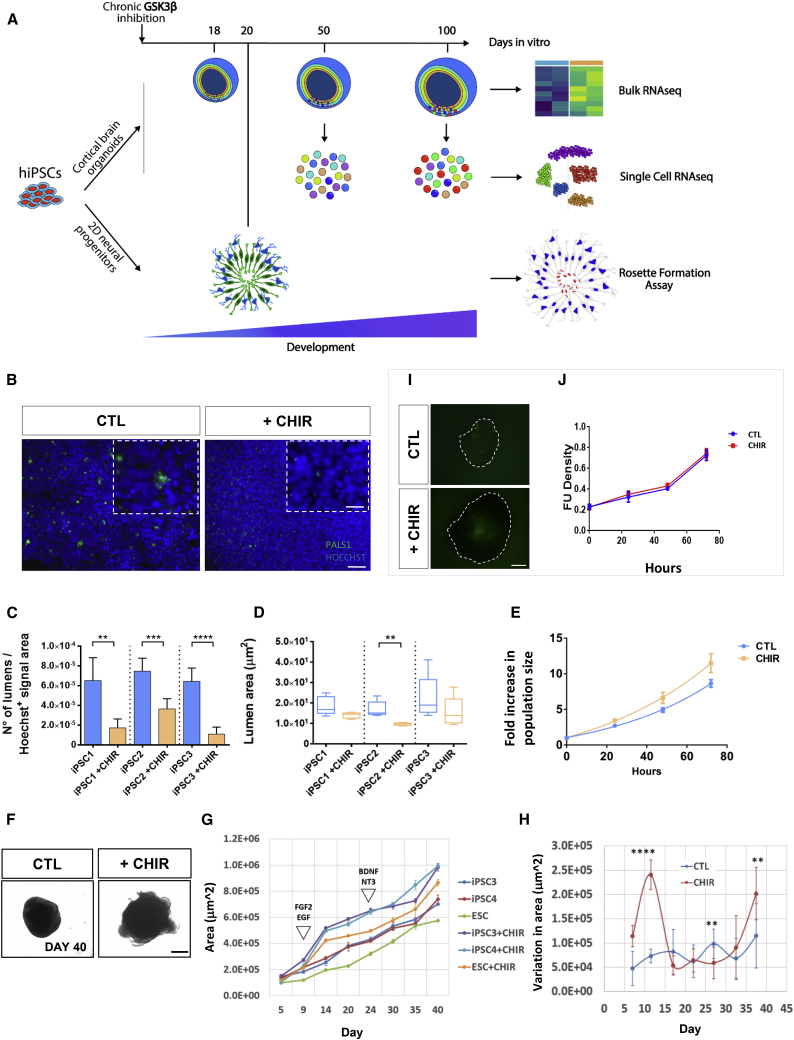
Figure 2.
GSK3 Inhibition Disrupts Lumen Organization and Progenitor Proliferation Rate
(A) Experimental design: hPSCs differentiated following two parallel protocols, in 3D (up) cortical organoids or 2D (down) dual-smad inhibition. In both cases, parallel rounds were either exposed or not to GSK3 inhibitor CHI99021 (1 μM) starting from day 0 until the indicated sample collection time point.
(B) Representative captions from immunostaining performed for anti-Pals1(green)/Hoechst(blue), wide-field fluorescence images acquisition, 20×. Scale bar, 50 μm (zoom panel, 10 μm).
(C) Bar plots represent the average lumen number ± SD of five independent images for lines/conditions.
(D) Lumen area quantification was performed by CellProfiler software; unpaired t test; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(E) Cell proliferation rate was estimated by CellTiter-Glo luminescence assay, with measurements every 24 h for 96 h in triplicate, three independent hPSC lines (n = 3).
(F) Representative bright-field captions of day 40 organoids. Scale bar, 500 μm.
(G) Growth curve performed in organoids differentiated for 40 days. Bright-field captions were taken at days 5, 9, 14, 20, 24, 30, 35, and 40. Points represent the average of 4 organoids per line. Size quantifications were performed with a custom-made FIJI function.
(H) Derivative of the growth-rate delta between time points. The delta was computed on the average of untreated or treated samples. SD was calculated as a cumulative SD across all replicates and samples from each condition of three independent hPSC lines (n), four organoids measured per line/time point (N) (N = 72, n = 3), unpaired t test; ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(I) Representative images from day 50 organoids stained with CellTox green as a marker of cytotoxicity after 72 h of growth factor starvation (depletion B27 and growth factors). Scale bar, 200 μm.
(J) Quantification of CellTox fluorescence at 0, 24, 48, and 72 h after growth factor starvation.
To dissect the mechanisms underlying this morphogenic defect, we adopted a two-tiered strategy: (1) a validation in a classic 2D model attuned to quantify essential properties of NSCs such as polarity and proliferation (Conforti et al., 2018); and (2) transcriptional profiling of patterned cortical organoids at 18, 50 and 100 days for a dynamic characterization of early corticogenesis (Figure 2A). We followed the emergence of 2D neural rosettes until 20 days in vitro, when rosettes are typically PASL1+ at the apical end. GSK3 inhibition drastically reduced rosette number (Figures 2B and 2C) as well as the average size of formed rosette lumens (Figure 2D). Likewise, a growth curve for 72 h revealed an increase in proliferation rate up to 1.5-fold compared with control (Figure 2E), indicating that both the morphogenic disarray and the increased size observed in early cortical organoids arise from aberrations already present during the transition from pluripotency to NSCs. In addition, we quantified the impact of GSK3 inhibition on organoid size through a growth measurement over a 40-day time course. We found a sustained increase in organoid size upon chronic GSK3 inhibition (Figures 2F–2H), with a robust surge in pace between days 10 and 15, subsequent to the time point when cortical organoids begin to be exposed to proliferation-boosting factors, fibroblast growth factor 2 and epidermal growth factor (Figure 2H), in agreement with the increased proliferation rate observed at the first 72 h of neural induction (Figure 2E) and pointing to a growth factor-mediated proliferation process.
A previous report suggested that the sustained application of CHIR99021 in forebrain organoids after day 12 of differentiation decreased the number of Caspase 3+ core cells (Qian et al., 2016), which could explain the increase in organoid size. To test whether chronic CHIR application had an overall effect on cell viability, we performed luminescent quantitation of membrane permeability in day 50 organoids every 24 h, during 72 h of growth factor starvation. Quantification showed no changes in cell death as a result of GSK3 inhibition, either in baseline (0 h) or after 72 h of starvation (Figures 2I and 2J), indicating that chronic GSK3 inhibition has no impact on basal or starvation-mediated cell death, hence excluding reduced apoptosis as a plausible cause of the increased organoid size. Instead, quantification of the proliferation marker Ki67 showed a marked increase of actively dividing cells in organoids at day 18 (CTL 10 ± 0.7, CHIR 19 ± 1.3) (Figure 1H) and day 50 (CTL 4.3 ± 0.4, CHIR 8.0 ± 0.8) (Figure 1I) pointing to the regulation of progenitor proliferation as a core effect of GSK3 activity.
Transcriptional Regulation Associated to GSK3 Activity throughout Cortical Development
Abundant evidence supports an active role of GSK3 in neurogenesis (Hur and Zhou, 2010); however, since complete GSK3 ablation causes systemic failure and early embryonic lethality (Hoeflich et al., 2000), its levels/activity are usually manipulated both in vitro and in vivo after the early stages of neurodevelopment have already been completed. The surprisingly mild effect of chronic GSK3 inhibition on neural maturation prompted us to dissect its role on gene expression by bulk RNA-seq at three critical time points recapitulating relevant stages of human cortical development (i.e., day 18: abundant apical radial glia; day 50: presence of intermediate progenitors and early neurons; and day 100: presence of lower layer neurons and beginning of astrogenesis) (Paşca et al., 2015).
A dissection of the most influential genes on component 1 of principal-component analysis identified genetic drivers of cell proliferation as main source of variability (Figures 3A and S3A) and highlighted the stronger impact of CHIR treatment at days 50 and 100. A survey of gene signatures defining specific stages of differentiation (active proliferation, early and late RGs, early and late neurons) (Figure S1) confirmed a sustained increase of the proliferation marker MKI67 upon CHIR treatment at days 18 and 50 (Figure S1A–S1C), along with an upregulation in genes controlling distinct aspects of cell-cycle progression at days 50 and 100. Notably, we observed a robust upregulation of the anaphase promoting complex/cyclosome (APC/C), CDC20 (Figures S1B and S1C), which has been implicated in the modulation of the synaptic connectivity regulator NEUROD2 levels through the regulation of its ubiquitination (Yang et al., 2009). Intriguingly, despite the strong proliferation effect, the expression levels of canonical neuronal markers (DCX, MEF2C, STMN2) (Figures S1B and S1C), cortical layer makers (POU3F2, CUX1, BCL11B), and ventral fate determinants (DLX1-2, ARX, GAD1-2, ASCL1) (Figures S2A–S2C) showed either mild or no differences, whereas the neuron-specific microtubule-related genes showed differential modulation upon CHIR inhibition (i.e., upregulation of TUBB3 and downregulation MAP2) (Figures S1B and S1C). Likewise, we observed a strong downregulation of NEUROD2 expression at days 50 and 100 (Figures S1B and S1C), which could in turn explain the downregulation of several glutamate channel subunits observed at day 100 (Figures S1C and S1D).
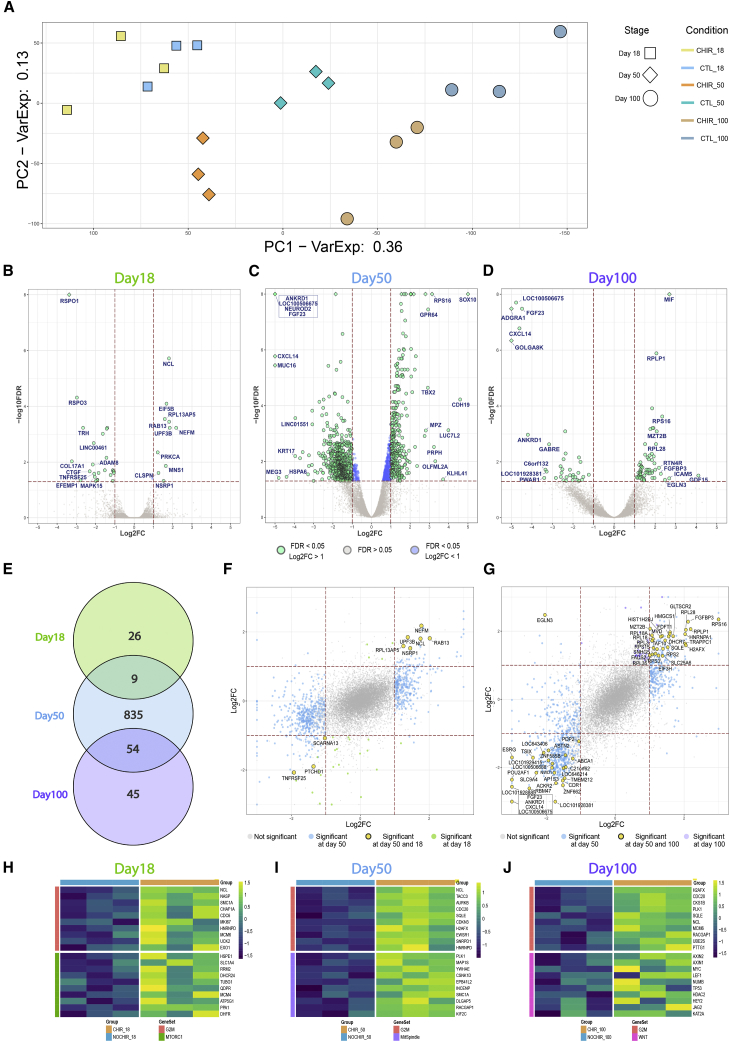
Figure 3.
Impact of GSK3 Inhibition on the Transcriptional Landscape
(A) Principal component analysis performed on the whole transcriptome of untreated and CHIR-treated brain organoids at three stages of development: days 18, 50, and 100.
(B–D) Volcano plots illustrating the differential expression analysis for CHIR treatment at day 18 (B), day 50 (C), and day 100 (D). Results are reported for the pool of tested genes as –log10 false discovery rate (FDR) and log2 fold change (FC). Genes identified as significantly modulated (FDR < 5% and absolute log2FC > 1) are shown in green, while those respecting only the FDR threshold are depicted in blue and not significant genes (FDR > 5%) in gray. Gene symbols highlight the top 10 upregulated and downregulated genes (ranked by fold change) in each stage.
(E) Venn diagram depicting the overlap of modulated genes across developmental stages.
(F and G) Scatterplots representing the relationship of the fold change induced by GSK3 inhibition at day 50 on the x axis and day 18 on the y axis (F) or day 50 on the x axis and day 100 on the y axis (G), for the subset of genes tested in both conditions. DEGs shared between the two examined conditions are reported in yellow, while DEGs specific for days 18, 50, or 100 are in green, blue, or violet, respectively. Correlation coefficient calculated according the Spearman metrics––0.45 in (F) and 0.62 in (G).
(H–J) Gene expression profiles for selected gene sets significantly associated with CHIR treatment by GSEA: (H) day 18; (I) day 50; and (J) day 100. Expression levels (as Z score) for the top 10 ranking genes in the leading edge are visualized for each gene set. All analyses were done in three independent hPSC lines/time point (N = 18, n = 3).
To address the functional implications of chronic CHIR exposure, we performed stage-wise differential expression analysis (DEA) (Figures 3B–3D). DEA confirmed the strongest CHIR effect at day 50, with 898 differentially expressed genes (DEGs) (Figure 3C) (full list of DEGs, Table S2). Ontology analysis revealed a persistent upregulation of categories linked to cell proliferation and DNA replication (nucleosome assembly) and a downregulation of cell surface components, as well as ion channels, including AMPA subunits (Figures S3C and S3D). While there were no DEGs preserved at all stages upon CHIR treatment, we identified 9 DEGs consistently dysregulated in the day 18 → day 50 transition and 54 DEGs in the day 50 → day 100 transition (Figure 3E). Interestingly, day 18 → day 50 shared DEGs included the upregulation of neurite outgrowth factors (NEFM, NCL, and RAB13), and downregulation of the brain morphogenesis modulator (PTCHD1) (Figure 3F), whereas the day 50 → day 100 dysregulation featured an abundant upregulation of protein translation and replication regulators (Figure 3G). The overall comparison of gene modulation across stages showed a stronger preservation in the fold changes trends between day 50 → day 100 (R = 0.62) compared with day 18 → day 50 (R = 0.45), despite the longer time span and increased cell-type diversity at later stages (Figures 3F and 3G), suggesting a more robust dependency of GSK3 activity occurring during this transition.
To gain deeper insight into the coordinated modulation of functionally related genes induced by GSK3 inhibition, we performed gene set enrichment analysis (GSEA). This approach confirmed a robust enrichment for pathways targeted by GSK3 activity (MTORC1, Myc signaling) and narrowed down the upregulation of G2-M transition modulators as a persistent feature of GSK3 transcriptional impact at all stages (Figures 3H–3J). Likewise, several of the identified GSEA categories (complete list in Table S3) are directly linked to the observed phenotypes, including the increased progenitor production (mammalian target of rapamycin complex 1 pathway), changes of cell polarity (mitotic spindle orientation) and neuronal fate (Wnt-β-catenin pathway). Interestingly, both gene ontology analysis at day 50 and GSEA (at all stages) point to the downregulation of inflammatory pathways by GSK3 inhibition, in line with a recognized role of GSK3 as mediator of neuroinflammation (Grimes and Jope, 2001b).
Cortical Organoids Recapitulate the Main Features of Mid-fetal Human Corticogenesis at the Single-Cell Level
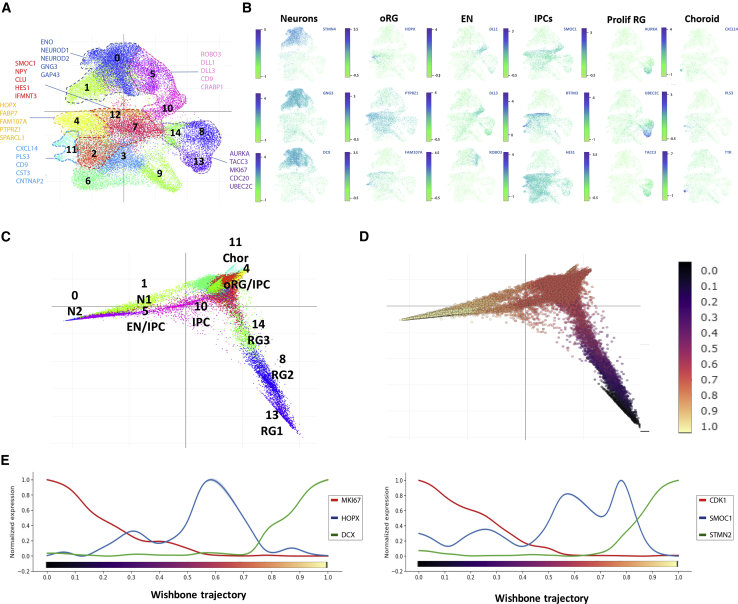
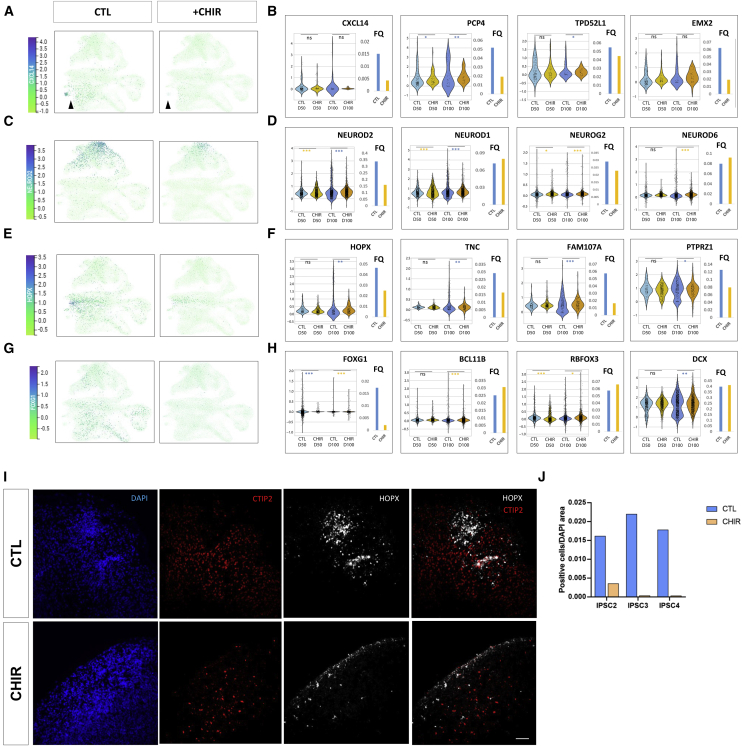
Given the greater transcriptional impact of GSK3 observed at the day 50 → day 100 transition, we harnessed single-cell transcriptomics combined with distance-based analytical tools to break down the effects of CHIR exposure in terms of population frequencies and developmental trajectories found during this time frame. We carried out droplet-based single-cell mRNA-seq to profile over 30,000 cells (N = 33293) in 11 biological samples from unexposed and exposed cortical organoids at day 50 and day 100 of differentiation, attaining 15 unsupervised cell clusters defined using the Louvain modularity algorithm (Šubelj and Bajec, 2011). By projecting the expression levels of canonical population markers over uniform manifold approximation and projection (UMAP) (Figures 4A and 4B), combined with the overlap of cluster-specific genetic signatures from single-cell RNA-seq datasets of fetal human brain samples (Nowakowski et al., 2017) (Figure S4) and considering their position over diffusion map (Figure 4C), we grouped the Louvain clusters into five main population identities, including proliferating RG, IPCs (including a discernible subset of oRG), early neurons, neurons, and choroid. We then investigated the underlying developmental trajectories using a diffusion map algorithm (Coifman et al., 2005) (Figures 4C and 4D). This confirmed the presence of a hierarchical progression from RGs to postmitotic neurons linked by intermediate populations (Figure 4C). The application of pseudotime for lineage-branching reconstruction (Haghverdi et al., 2016, Setty et al., 2016), with an origin anchored in the early progenitors, reproduced the organization of populations from RG to neurons (Figure 4D). Likewise, the distribution of expression of specific population markers confirmed the presence of a lineage path from RGs through the IPC-oRG transition and into early and late neural maturation stages (Figure 4E). Together, these results confirm that patterned cortical organoids recapitulate cardinal features of human corticogenesis, including the presence of early and intermediate progenitors, the emergence of oRGs and their hierarchical positioning in neurogenic trajectories.
Figure 4.
Cortical Organoids Recapitulate the Main Features of Cortical Development
(A) Louvain clusters in UMAP plot colored by cluster identity; lines depict population areas defined by contrast with markers obtained from human fetal brain dataset (radial glia, 8, 13, and 14; intermediate progenitor cells, 2, 7, and 12; outer radial glia, 4; early neurons, 5 and 10; neurons, 0 and 1; choroid 11).
(B) UMAP plots. For each sub-panel, cells (represented as dots) are colored according to the expression levels of representative cell-type markers: STMN4, GNG3, DCX, neurons; HOPX, PTPRZ1, FAM107A, outer radial glia; DLL1, DLL3, ROBO3, early neurons; SMOC1, IFITM3, HES1, intermediate progenitors; AURKA, UBEC2C, TACC3, proliferating progenitors; CXCL14, PLS3, TTR, choroid.
(C and D) Diffusion map representing the developmental trajectory of the system. Cells (dots) are colored according to cluster identity (C) and to pseudotime trajectory (D), from origin in black to terminal state in light yellow according to wishbone algorithm.
(E) Visualization of the expression levels of representative genes along pseudotime: CDK1 and MKI67, proliferating progenitors; DCX and STMN2, neurons; HOPX, outer radial glia; SMOC1, intermediate progenitors. Analyses done in 33,293 cells from 6 hPSC lines from day 50 and 5 hPSC lines from day 100.
GSK3 Inhibition Differentially Affects Specific Domains of Corticogenesis
A comparative analysis of subpopulations revealed a selective impact of GSK3 inhibition on the relative proportion of specific cell subtypes (Figures 5A–5H). A salient effect was the almost complete loss of a subpopulation characterized by the expression of genes expressed in choroid cells (CXCL14, TPD52L1, PCP4, EMX2) (Figures 5A and 5B), as well as a noticeable decrease in frequency (FQ) of NEUROD2-expressing cells at day 100 in neuronal clusters (0, 1, 5) without affecting NEUROD6- or NEUROG2-expressing cells (Figures 5C and 5D), underscoring the selectivity of GSK3 activity on these neurogenic pathways. It is worth noting that analysis of the frequency and distribution of cells expressing a canonical oRG gene signature (HOPX, TNC, FAM107A, PTPRZ1) (Pollen et al., 2015) (Figures 5E and 5F), showed a marked reduction in both frequency and expression levels upon CHIR treatment at day 100, which was further confirmed by a reduction of HOPX+ cells in the tissue (Figure 5I). Given the cardinal role of oRG on neuronal production and cortical expansion, we checked the distribution of expression of telencephalic markers, including forebrain determinant (FOXG1), early neurons (DCX), lower-layer (BCL11B, RBFOX3), and upper-layer (SATB2, POU3F2) markers. Whereas upper-layer markers were either not detected or present at very low frequencies, consistent with their surge at later stages of organoid corticogenesis, the expression of lower layer and general neuronal identity markers was unchanged by CHIR (Figure 5H), whereas FOXG1 was significantly reduced at day 50 (Figures 5G and 5H). Interestingly, despite the mild changes frequency and expression levels of BCL11B at day 50 and day 100 (Figures 5H and S2), staining for its protein product (CTIP2)+ cells showed a drastic reduction in day 100 CHIR-treated organoids (Figure 5I), pointing to regulatory mechanisms at the post-transcriptional level reported to affect CTIP2 activity and stability (Selman et al., 2018, Zhang et al., 2012).
Figure 5.
Effects of GSK3 Inhibition at a Single-Cell Level in Day 50 and 100 Cortical Organoids
(A–H) Visualization of normalized expression levels for genes identifying specific populations either in UMAP divided by CHIR-treated and control samples (A, C, E, and G). Sub-sampled cluster-specific expression levels (violin plots), Fisher test; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. FQ, normalized cell frequencies of cells expressing detectable levels/total population (bar plots) (B, D, F, and H). The following genes are examined: (A and B) CXCL14 in UMAP and CXCL14, PCP4, TDP52L1, EMX2 in violin plots on cluster 11 for choroid population; (C and D) NEUROD2 in UMAP and NEUROD2, NEUROD1, NEUROG2, and NEUROD6 on clusters 0, 1, and 5 for neuronal markers; (E and F) HOPX in UMAP and HOPX, TNC, FAM107A, and PTPRZ1 on cluster 4 for oRG markers; (G and H) FOXG1 in UMAP and FOXG1 (total) and BCL11B, RBFOX3, and DCX on clusters 0, 1, and 5 for dorsal telencephalon markers. Sub-sampling has been applied in order to randomly compare an equal number of cells for each category.
(I) Representative wide-field fluorescence images from day 100 organoids immunostained with anti-CTIP2 (red), anti-HOPX (gray), and DAPI (blue). Scale bar, 20 μm.
(J) Semiquantitative estimation of HOPX positive cells/DAPI area. Quantification corresponds to three organoids per line, three independent hPSC lines. Analyses done in 33,293 cells from 6 hPSC lines from day 50 and 5 hPSC lines from day 100.
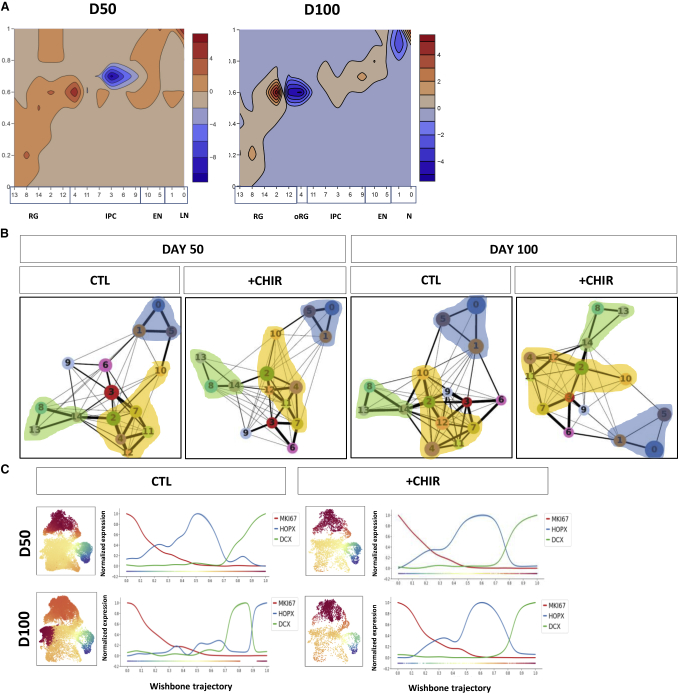
Next, to study the temporal associations between populations, we performed pseudotime analysis. The analysis of cell-type distribution across pseudotime uncovered a higher proportion of cells in both progenitor and mature areas upon chronic GSK3 inhibition at day 50 (Figure 6A). This result was compatible with two possibilities: (1) the cells identified as mature by predefined gene signatures could actually turn out to display mixed identities, co-expressing both progenitor and maturation markers as a result of a fundamental derangement of developmental hierarchies; or (2) the highly proliferative progenitors from CHIR-treated organoids undergo a faster transition through the intermediate stages or even skip them altogether. To test these hypotheses, we plotted proliferating/maturity signatures into UMAP and found no evidence of overlap in CHIR-treated organoids (Figure S5A); likewise, we did not observe an overlapping distribution between Ki67+ and DCX+ cells (Figure 1D), thus ruling out the possibility of a prevalent mixed identity. Instead, the contour plot (Figure 6A) pointed to a reduction in the intermediate populations in CHIR-treated organoids at days 50 and 100, consistent with the higher proportion of mature cells found at the same developmental stage (Figure 6A). Next, to further investigate the relations between populations, we used partition-based graph abstraction as a way to estimate the strength of connectivity across the cells belonging to each cluster, thus inferring transitional links among subpopulations. This approach confirmed a decrease of connections between intermediate progenitor clusters (yellow), including oRG, upon GSK3 inhibition, particularly evident at day 100 (Figure 6B), corroborating a CHIR-dependent overall reduction of the intermediate to maturity transition.
Figure 6.
Effects of GSK3 Inhibition at a Single-Cell Level in Day 50 and Day 100 Cortical Organoids
(A) Contour plot representing the difference in frequency distribution in clusters and pseudotime intervals of treated versus untreated cells at days 50 and 100. Red depicts values higher in CHIR-treated cells, while blue depicts values higher in CTL-treated cells.
(B) Partition-based graph abstraction analysis applied stage- and treatment-wise on the 15 clusters identified on the complete dataset. Circle diameter represents the fraction of cells assigned to each cluster; edge thickness visualizes the strength of connections across cells of the related clusters. Shadows highlight areas: progenitors (green), neurons (blue), and intermediate progenitors (yellow).
(C) Over-imposition of pseudotime analyses performed separately for each experimental condition as a color scale on a UMAP calculated on the complete system. Blue (origin) and dark red (terminal state) according to wishbone trajectories. Visualization of the expression levels of representative genes along the condition-specific pseudotime. Analyses done in 33,293 cells from 6 hPSC lines from day 50 and 5 hPSC lines from day 100.
Finally, the application of pseudotime, independently to all conditions and stages, revealed two developmental trajectories, one that goes from RG to neurons through a subset of IPCs and one that becomes apparent by day 100 and ends in an oRG identity (Figure 6C). Strikingly, GSK3 inhibition resulted in a complete loss of the oRG-generating trajectory at day 100 (Figure 6C), an effect that could be reproduced bidirectionally by decomposing each trajectory into first and second components (Figure S5B). In agreement, HOPX distribution of expression on pseudotime peaked at day 100 in control organoids, while it remained stalled in day 100 CHIR-treated organoids (Figure 6C). Together, these results suggest that GSK3 inhibition results in a severe reduction of HOPX-expressing cells with the attending impact on oRG-dependent lineages.
Discussion
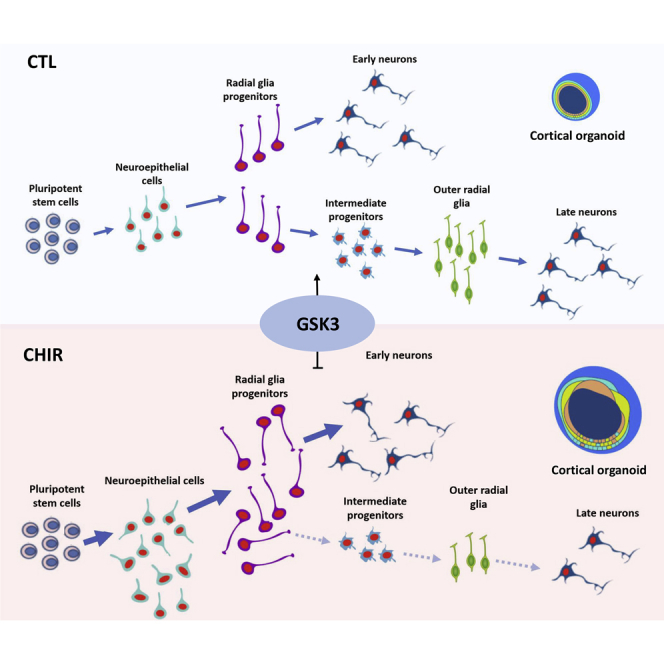
This work provides the first dissection of the role of GSK3 activity throughout early to mid-fetal corticogenesis in a human background. Specifically, our integrated analysis of tissue and transcriptional regulation at single-cell resolution uncovers a key role for GSK3 activity in cortical tissue architecture, with a differential impact on neuronal output vis-a-vis a strong decrease of oRG production.
GSK3 activity has been implicated in the regulation of neocortical neuron production, with pronounced impact at late stages (Li et al., 2012), consistent with the increased transcriptional dysregulation that we observe in days 50 to 100 cortical organoids. Our in-depth transcriptional analysis indicates that not all aspects of cortical neurogenesis are equally affected, even in the face of the early increase in progenitor proliferation and massive morphologic disarray, which emphasizes the resilience and degree of cell autonomy of the transcriptional programs involved in neuronal identity, even against the disruption of stereotypical tissue architecture. In particular, early neuronal driver NEUROD2 was strongly downregulated in bulk transcriptomes and remained reduced both, in terms of expression levels and frequency in single cells, concomitantly with a reduced number of cells positive for low-layer markers TBR1 and CTIP2, while the frequency of cells expressing neurogenic drivers (NEUROG2), fetally expressed pan-neuronal markers (DCX, STMN2), and low-layer determinants (RBFOX3, CUX1) was either unchanged or only slightly differed in expression levels among very low cell frequencies. Interestingly, we found a decrease in FOXG1 levels at day 18 bulk transcriptomes and in the frequency of FOXG1-expressing cells at day 50. These changes in FOXG1 expression resulted in no evident posteriorizing effects, likely reflecting the minute fraction of FOXG1-positive cells at days 50 to 100 (consistently with its expression peaking at day 25; Birey et al., 2017) and the cortical organoid patterning that drives toward a telencephalic fate (Paşca et al., 2015). In light of the emerging disconnect between transcript and protein levels of several neurogenic transcription factors due to post-transcriptional regulation (Yoon et al., 2017), it will be interesting to determine the short- to long-term impact of such transcriptional alterations on the respective protein effectors.
Depletion of GSK3 activity using pharmacological inhibition or knockdown causes drastic changes in radial glial organization (Yokota et al., 2010). As polarization of radial glia is a prerequisite for cortical scaffolding, neuronal migration, and layering (Shah et al., 2017), its alterations result in impaired cortical plate formation (Beattie et al., 2017, Shah et al., 2017). The effects of GSK3 in radial glia organization constitute fast cellular responses triggered by phosphorylation of targets such as CRMP2, MAP1B, and CLASP, which in turn affects modifications and structural changes in microtubule cytoskeleton (Hur and Zhou, 2010, Yokota et al., 2010). We did not observe any differences in expression of canonical polarity markers or determinants of radial glia organization (data not shown); thus, in the human setting, the sustained morphological defects caused by GSK3 activity inhibition are not caused by transcriptional regulation of polarity determinants. Rather, the most salient transcriptional global effect was the upregulation of modulators of cell replication and, in particular, the G2M transition emerged as a recurrent target at all stages, indicating that the modulation of the G2M is a persistent feature of GSK3 activity throughout human corticogenesis.
During human corticogenesis, the bulk of neuronal production comes via IPCs that originate from RGs and are located in the subventricular zone (Bystron et al., 2008, Florio and Huttner, 2014). Spatiotemporal regulation of GSK3 activity is required for an appropriate transition from the proliferative to the neurogenic phase occurring during brain development (Kim et al., 2009). In our chronic setting, starting from a pluripotent state (day 0), GSK3 inhibition caused increased proliferation and polarity defects in NSCs, reflected also in larger organoid size. The dramatic increase in CHIR-dependent transcriptional dysregulation at day 50, compared with day 18, indicates that the early defects in NSCs are amplified in intermediate and committed progenitors, and suggests a higher reliance on GSK3 activity at later stages and stronger in IPCs. In agreement, the population breakdown by single-cell analysis revealed a decrease of IPCs and in particular oRGs.
Pseudotime trajectories showed that cortical organoids recapitulate the co-existence of a direct neurogenic trajectory, visible by day 50, that is juxtaposed to an indirect trajectory reaching an oRG identity by day 100, in agreement with a protracted wave of neurogenesis dependent on oRG production, known to be a salient feature of primate corticogenesis and responsible for lateral expansion of the human cortex (Bershteyn et al., 2017, Florio and Huttner, 2014). In sum, the disappearance of the indirect neurogenic trajectory at day 100 upon CHIR treatment points to a key role of GSK3 in the establishment of IPC/oRG populations. Our results thus indicate that, in humans, while GSK3 activity selectively contributes to different early RG-derived neuronal identities, it is pivotal for the later generation of oRG with the ensuing outcome in terms of oRG-dependent lineages.
Experimental Procedures
hPSC Culture
Five different hPSC lines from different backgrounds (one ESC and four iPSCs) were used across all conducted experiments (relevant ethics approvals are referred to in the original publications reporting their first use and/or derivation). The culture conditions of hPSCs are described in Supplemental Experimental Procedures. Distribution of lines across experiments is delineated in Table S1.
Neural Induction in 2D and Lumen Quantification
Three biological replicates were analyzed for untreated and treated cells. From day 0 of neural induction, CHIR99021 was added to the medium at 1 μM concentration in parallel with neural induction with DMSO until day 20. At day 12, cells were moved to neural medium containing DMEM/F12 (Life Technologies) with N2 and B27 plus retinoic acid and 30 ng/mL brain-derived neurotrophic factor. Differentiation process and immunostaining procedure are described in Supplemental Experimental Procedures.
Proliferation Assay
Number of viable proliferating cells was estimated by luminescence assay CellTiter-Glo (Promega). In brief, 2 × 103 cells/well were plated in 96-well flat bottom plates (Corning) with 4 replicates per condition and left to proliferate for 96 h. Measurements were performed every 24 h. Each data point was normalized to a blank from unseeded wells.
Cortical Organoid Differentiation and Inhibition
Cortical organoids were differentiated as described previously (Paşca et al., 2015). A detailed protocol is given in Supplemental Experimental Procedures. Chronic GSK3 inhibition was performed by adding CHIR99021 (Merck SML1046) to the medium at day 0 (1 μM) and kept throughout the differentiation process until reaching the respective collection time points.
Growth Curve
Organoids were moved at day 0 to 96-well U-bottom ultra-low attachment plates (Corning) and kept individually to avoid fusions for image acquisition until day 12 of differentiation. From day 12 onward, organoids were moved individually to 24-well ultra-low attachment plates (Corning). Images were acquired with an EVOS Cell imaging System XL (Thermo) at the indicated differentiation days. Organoid size was calculated using an in-house developed custom-script (by C.E.V.) for FIJI software (v.1.49; NIH, USA).
Tissue Preservation and Staining
Organoids were fixed in 4% (vol/vol) paraformaldehyde for a minimum of 2 h for day 18 organoids to overnight for day 50 and day 100 organoids. Fixed organoids were washed twice with PBS and mounted on OCT cryopreservation medium on dry ice. Cryoblocks were preserved at −80°C until the moment of sectioning. Cryosections were prepared using Leica CM 1900 instrument with 5 μm thickness. Sections were incubated with 10 mM sodium citrate buffer (Normapur) for 45 min at 95°C + Tween 20 0.5% for simultaneous antigen retrieval and permeabilization. Antibody incubation details and image acquisition and quantification details are given in Supplemental Experimental Procedures.
Total RNA Extraction and Sequencing
Total RNA was isolated with the RNeasy Micro Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. RNA was quantified with Nanodrop and then the integrity was evaluated with Agilent 2100 Bioanalyzer. A TruSeq Stranded Total RNA LT Sample Prep Kit (Illumina) was used for library preparation starting from 500 ng of total RNA for each sample. Sequencing was performed with the Illumina NOVAseq 6000 platform, with an average depth of 35 million 50 bp paired-end reads per sample.
Bulk Transcriptome Analysis
Three biological replicates were analyzed for untreated and treated organoids at each time point, for a total of 18 samples subjected to bulk RNA-seq. The detailed analytical pipeline is described in Supplemental Experimental Procedures.
Single-Cell Suspension, cDNA Synthesis, Library Preparation, and Sequencing
Organoids were collected at days 50 or 100. Three to 5 organoids per condition were dissociated by incubation with a solution of 0.5 mg/mL trypsin + 0.22 mg/mL EDTA (Euroclone) with 10 μL of DNaseI 1,000 U/mL (Zymo Research) for 30–45 min according to organoid size. Digested suspensions were passed once through 0.4-μm Flowmi cell strainers, resuspended in PBS and counted using a TC20 automatic cell counter (Bio-Rad). Droplet-based single-cell partitioning and single-cell RNA-seq libraries were generated using the Chromium Single-Cell 3′ Reagent v2 Kit (10× Genomics, Pleasanton, CA) following the manufacturer's instructions. Detailed library preparation is given in Supplemental Experimental Procedures.
Single-Cell Transcriptome Analysis
Eleven biological samples (day 50, 3 untreated and 2 treated; day 100, 4 untreated and 2 treated) were examined by single-cell analysis, for a total of 33,293 cells and a median of 1,733 features for cells. Libraries from single-cell sequencing were aligned relying on the CellRanger v2.1 pipeline and using hg38 as reference. Array express accession number: E-MTAB-8337. Detailed single-cell transcriptome analysis is given in Supplemental Experimental Procedures.
Statistical Analysis
Statistical analyses were done using PRISM (GraphPad, version 6.0). Statistical significance was tested with the unpaired t test, considering each hPSC line as biological replicates (n) and total number of organoids per line (N). Treatments tested as variables. p values and significance, n and N are reported in each figure and legend. All results were expressed as means ± SD. No data points were excluded from the reported analyses.
Author Contributions
A.L.-T., S.T., N.C., M.L., and B.M.C. organoid differentiation and sample collection for the different experiments. C.E.V. and C.C. bioinformatic analysis. A.L.-T., S.T., and N.C. single-cell experimental procedures. A.L.-T. and M.L. organoid growth curve. A.L.-T., N.C., S.T., and P.L.R. stainings, quantifications, and statistical analysis. P.C., R.I., and E.C. neural induction experiments in 2D. M.T.R. proliferation assay in 2D. E.T. and F.T. cortical organoid generation and library preparation. M.D.S., I.L.-N., G.M., and M.P. contributed with funding and expertise. A.L.-T. wrote the manuscript with contributions from all authors. G.T. conceived, designed, and supervised the study.
Acknowledgments
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) (IG 2014-2018 to G.T.); EPIGEN Flagship Project of the Italian National Research Council (CNR) (to G.T., G.M., and M.P); the European Research Council (ERC DISEASEAVATARS no. 616441 to G.T.); Fondazione Cariplo (2017-0886 to A.L.-T.); Fondazione Italiana per la Ricerca sul Cancro (FIRC) and Fondazione Istituto Europeo di Oncologia - Centro Cardiologico Monzino (IEO-CCM) (to P.L.R.); the AIRC grant no. IG2016-ID18575 (to M.P.) and the ERC Consolidator grant no. 617978 (to M.P.) and the IEO Single Cell Program. S.T. and N.C. are PhD students within the European School of Molecular Medicine (SEMM). M.P. is a founder, shareholder and a member of the scientific advisory board of CheckmAb s.r.l. We are grateful to Pierre-Luc Germain for providing scripts that facilitated the analysis, to Andreas Püschel (University of Münster, Germany) for critical comments on the introduction, to Stefano Piccolo (University of Padua, Italy) for sharing of expertise, and to Federica Pisati from the tissue processing facility and the IEO genomic unit team.
Published: October 10, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.09.005.
Accession Numbers
The accession number for the bulk RNA seq reported in this paper is ArrayExpress: E-MTAB-8325. The accession number for the single-cell RNA seq reported in this paper is ArrayExpress: E-MTAB-8337.
Supplemental Information
References
- Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A., Coppola G., Scuderi S., Wu F., Roychowdhury T., Liu F., Pochareddy S., Shin Y., Safi A., Song L. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362:eaat6720. doi: 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie R., Postiglione M.P., Burnett L.E., Laukoter S., Streicher C., Pauler F.M., Xiao G., Klezovitch O., Vasioukhin V., Ghashghaei T.H. Mosaic analysis with double markers reveals distinct sequential functions of Lgl1 in neural stem cells. Neuron. 2017;94:517–533.e3. doi: 10.1016/j.neuron.2017.04.012. [DOI] [PubMed] [Google Scholar]
- Bershteyn M., Nowakowski T.J., Pollen A.A., Di Lullo E., Nene A., Wynshaw-Boris A., Kriegstein A.R. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20:435–449.e4. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betizeau M., Cortay V., Patti D., Pfister S., Gautier E., Bellemin-Ménard A., Afanassieff M., Huissoud C., Douglas R.J., Kennedy H. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I., Blakemore C., Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman R.R., Lafon S., Lee A.B., Maggioni M., Nadler B., Warner F., Zucker S.W. Geometric diffusions as a tool for harmonic analysis and structure definition of data: multiscale methods. Proc. Natl. Acad. Sci. U S A. 2005;102:7432–7437. doi: 10.1073/pnas.0500896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti P., Besusso D., Bocchi V.D., Faedo A., Cesana E., Rossetti G., Ranzani V., Svendsen C.N., Thompson L.M., Toselli M. Faulty neuronal determination and cell polarization are reverted by modulating HD early phenotypes. Proc. Natl. Acad. Sci. U S A. 2018;115:E762–E771. doi: 10.1073/pnas.1715865115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Florio M., Huttner W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- Fumoto K., Lee P.C., Saya H., Kikuchi A. AIP regulates stability of Aurora-A at early mitotic phase coordinately with GSK-3β. Oncogene. 2008;27:4478–4487. doi: 10.1038/onc.2008.92. [DOI] [PubMed] [Google Scholar]
- Grimes C.A., Jope R.S. Creb DNA binding activity is inhibited by glycogen synthase kinase-3?? and facilitated by lithium. J. Neurochem. 2001;78:1219–1232. doi: 10.1046/j.1471-4159.2001.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes C.A., Jope R.S. The multifaceted roles of glycogen synthase kinase 3β in cellular signaling. Prog. Neurobiol. 2001;65:391–426. doi: 10.1016/s0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Haghverdi L., Büttner M., Wolf F.A., Buettner F., Theis F.J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- Hoeflich K.P., Luo J., Rubie E.A., Tsao M.S., Jin O., Woodgett J.R. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- Hur E.M., Zhou F.Q. GSK3 signalling in neural development. Nat. Rev. Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P.A., Cappello S., Götz M. Stem cells niches during development––lessons from the cerebral cortex. Curr. Opin. Neurobiol. 2010;20:400–407. doi: 10.1016/j.conb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Wang X., Wu Y., Doble B.W., Patel S., Woodgett J.R., Snider W.D. GSK-3 is a master regulator of neural progenitor homeostasis. Nat. Neurosci. 2009;12:1390–1397. doi: 10.1038/nn.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mattar P., Zinyk D., Singh K., Chaturvedi C.-P., Kovach C., Dixit R., Kurrasch D.M., Ma Y.-C., Chan J.A. GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 2012;32:7791–7805. doi: 10.1523/JNEUROSCI.1309-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E., Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wang X., Chen J., Li B., Hur E.-M., Saijilafu Differential roles of glycogen synthase kinase 3 subtypes alpha and beta in cortical development. Front. Mol. Neurosci. 2017;10:391. doi: 10.3389/fnmol.2017.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.C., Song M.R., Park J.P., Henry Ho H.Y., Hu L., Kurtev M.V., Zieg J., Ma Q., Pfaff S.L., Greenberg M.E. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58:65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Vittoria M., Palejev D., Tomasini L., Coppola G., Szekely A.M., Horvath T.L., Vaccarino M.V. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Coppola G., Zhang P., Abyzov A., Provini L., Tomasini L., Amenduni M., Szekely A., Palejev D., Wilson M. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey J.A., Rakus D., Gizak A., Steelman L.S., Abrams S.L., Lertpiriyapong K., Fitzgerald T.L., Yang L.V., Montalto G., Cervello M. Effects of mutations in Wnt/β-catenin, hedgehog, Notch and PI3K pathways on GSK-3 activity—diverse effects on cell growth, metabolism and cancer. Biochim. Biophys. Acta. 2016;1863:2942–2976. doi: 10.1016/j.bbamcr.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.-Y., O’Rourke N.A., Nguyen K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsch C., Challet-Meylan L., Thoma E.C., Urich E., Heckel T., O’Sullivan J.F., Grainger S.J., Kapp F.G., Sun L., Christensen K. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A.A., Nowakowski T.J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C.R., Shuga J., Liu S.J., Oldham M.C., Diaz A. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Yang S.M., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman W.H., Esfandiari E., Filtz T.M. Alteration of Bcl11b upon stimulation of both the MAP kinase- and Gsk3-dependent signaling pathways in double-negative thymocytes. Biochem. Cell Biol. 2018;97:201–213. doi: 10.1139/bcb-2018-0132. [DOI] [PubMed] [Google Scholar]
- Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P., Choi K., Bendall S., Friedman N., Pe’Er D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah B., Lutter D., Tsytsyura Y., Glyvuk N., Sakakibara A., Klingauf J., Püschel A.W. Rap1 GTPases are master regulators of neural cell polarity in the developing neocortex. Cereb. Cortex. 2017;27:1253–1269. doi: 10.1093/cercor/bhv341. [DOI] [PubMed] [Google Scholar]
- Šubelj L., Bajec M. Unfolding communities in large complex networks: combining defensive and offensive label propagation for core extraction. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2011;83 doi: 10.1103/PhysRevE.83.036103. [DOI] [PubMed] [Google Scholar]
- Taverna E., Götz M., Huttner W.B. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Kamiya D., Nishiyama A., Katayama T., Nozaki S., Kawasaki H., Watanabe Y., Mizuseki K., Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Yang Y., Kim A.H., Yamada T., Wu B., Bilimoria P.M., Ikeuchi Y., De La Iglesia N., Shen J., Bonni A. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Eom T.-Y., Stanco A., Kim W.-Y., Rao S., Snider W.D., Anton E.S. Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development. 2010;137:4101–4110. doi: 10.1242/dev.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.J., Vogel W.K., Liu X., Topark-Ngarm A., Arbogast B.L., Maier C.S., Filtz T.M., Leid M. Coordinated regulation of transcription factor bcl11b activity in thymocytes by the mitogen-activated protein kinase (MAPK) pathways and protein sumoylation. J. Biol. Chem. 2012;287:26971–26988. doi: 10.1074/jbc.M112.344176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.