Abstract
Ribosome is a vital molecular machine for protein translation in the cell. Defects in several ribosomal proteins including RPS19, RPL11 and RPS14 have been observed in two types of anemia: Diamond Blackfan Anemia and 5q- syndrome. In zebrafish, deficiency of these ribosomal proteins shows similar anemic phenotype. It remains to be determined if any other ribosome proteins are similarly involved in regulating erythropoiesis. Here we generated mutations in zebrafish rps9, a rarely studied ribosomal protein gene, and investigated its function. Analysis of this mutant demonstrates that rps9 disruption leads to impairment of erythrocyte maturation, resulting in anemia. In addition, the overall phenotype including the anemic state is p53-dependent in rps9 mutants. Furthermore, this anemic state can be partially relieved by the treatment of L-leucine, and dexamethasone, which have been previously used in rescuing the phenotype of other ribosomal protein mutants. Finally, by comparing the phenotype, we show that there are considerable differences in morphology, cytomorphology, and hemoglobin levels for four ribosomal protein mutants in zebrafish. Based on the observed difference, we suggest that the level of anemic severity correlates with the delayed status of erythrocyte maturation in zebrafish models.
Keywords: ribosomal protein, anemia, rps9, p53, zebrafish
As a vital machine for mRNA translation, ribosome comprises a large 60S subunit containing 25S, 5.8S and 5S rRNA with 46 ribosomal proteins and a small 40S subunit containing 18S rRNA with 33 ribosomal proteins (Thomson et al. 2013). Ribosomal proteins play a general but essential role in protein translation and ribosome assembly. However, ribosomal proteins also have specific function such as interaction with p53 to transfer ribosomal stress to the cell, which elicits cell cycle arrest and apoptosis (Zhou et al. 2015).
Due to the pivotal roles of the ribosome, defects in ribosomal proteins can cause many diseases. Several defective ribosomal proteins have been discovered as causal factors of the human disease Diamond-Blackfan anemia, which is a type of anemia characterized by macrocytic anemia and bone defects. Notably, only one deficient ribosomal protein, RPS14, is identified in 5q- syndrome, which is another type of anemia diagnosed with macrocytic anemia and micromegakaryocytes accompanied with a low risk of developing into acute myeloid leukemia (Narla and Ebert 2010). Early studies in zebrafish have demonstrated that p53 activation induced by ribosomal protein deficiency plays a major role in causing anemia, although other p53-independent signaling pathways are also involved in these disorders (Danilova et al. 2008; Torihara et al. 2011; Provost et al. 2012).
Previous genetic studies of ribosomal proteins in model animals such as zebrafish have focused on a few ribosomal protein genes including rps19, rpl11, and rps14 due to their prior identification in human diseases (Danilova et al. 2011; Jia et al. 2013; Zhang et al. 2013; Zhang et al. 2014; Ear et al. 2016). It remains to be determined if the function of other ribosomal protein genes is also involved in regulating erythropoiesis. This question was addressed by transient gene knockdown using morpholino (MO) targeting nearly two dozen of ribosomal protein genes (Uechi et al. 2006; Uechi et al. 2008). Uechi et al. discovered that various degrees of defects in morphology and erythropoiesis were induced by knockdown of different ribosomal protein genes. These results suggest that not all ribosomal proteins are equally involved in regulating erythropoiesis. However, it has been recently appreciated that phenotypic results obtained from studies of morphants and genetic mutants often have discrepancies (Kok et al. 2015). Since p53 activation, which plays a role in phenotype of ribosomal deficiency, can be non-specifically induced by morpholino (Robu et al. 2007), it is better to interrogate ribosomal function using genetic mutants for consistency.
rps9 is a ribosomal protein contained in the 40S subunit of the ribosome. Sporadic studies on rps9 have demonstrated its interaction with nucleophosmin, its role in normal cell proliferation as well as regulation in osteosarcoma cell growth (Lindström and Zhang 2008; Lindström 2012; Cheng et al. 2017). However, no research on the organism level has been reported to elucidate the function of rps9 in vivo. In this study, we generated rps9 mutants by CRISPR/Cas9 gene targeting in zebrafish. Detailed analyses showed that mutation in rps9 could lead to anemia, accompanied by severe morphological abnormalities. This anemic phenotype was due to a block of the terminal maturation of erythroid cells. Further study revealed that the anemia was largely dependent on p53 signaling pathway. Treatment of previously used agents including L-leucine and dexamethasone was able to alleviate the anemia phenotype in rps9 mutant. Moreover, the comparison of different ribosomal protein mutants revealed considerable differences in morphology, erythroid cytomorphology, and blood level for different mutants. Overall, our study suggests that rps9 can be a candidate for human anemia disease.
Materials and Methods
Zebrafish strains and lines
All zebrafish experiments were approved by Institutional Animal Care and Use Committee of Peking University and UCLA. rps9 mutants were generated using CRISPR-Cas9 method in wild-type (AB) strain, while rpl11, rps14, and rps19 mutants were previously described (Danilova et al. 2011; Zhang et al. 2014; Ear et al. 2016).
mRNA and gRNA synthesis, genotyping
pT3TS-nCas9n plasmid (addgene, plasmid #46757) for zebrafish was linearized with XbaI, mRNA was obtained through in vitro transcription using linearized plasmids with T3 mMESSAGE Kit (Ambion). For rps9 mRNA synthesis, coding sequence was amplified using cDNA and cloned into pCSDest2 using gateway technology (Thermo Fisher), then mRNA was transcribed after plasmid linearization using SP6 mMESSAGE Kit (Ambion). Rescue experiment was performed by injecting 400 pg rps9 mRNA to the embryos at one cell stage. For gRNA synthesis, DNA template was amplified using gene specific oligo (rps9 guide F: TAATACGACTCACTATAGCGTATTGGAGTGCTGGATGGTTTTAGAGCTAGAAATAGC), then gRNA was transcribed in vitro using MAXIscript T7 kit (Ambion) and purified using MicroRNA Isolation Kit (BioChain USA). 200 pg of Cas9 mRNA and 100 pg of gRNA were mixed and injected into embryos at the one-cell stage. For genotyping, PCR products were amplified from genomic DNA and subjected to electrophoresis using 8% TBE-PAGE gel with the following primers: rps9 Fwd: 5′-TTCCTCGTCAATGAGGCCAT-3′ and rps9 Rev: 5′-CTTTGCACATGTAGTTAGCA-3′. PCR products showing positive bands were further cloned and sequenced.
Wright-Giemsa staining
Embryos were anesthetized with tricaine, then immersed in PBS/40% FBS (Hyclone) mixed with a final concentration of 5 mM EDTA (Corning). Blood cells were collected by stabbing the heart using a micro-injection needle. Collected cells were transferred on the slides and air dry. After fixing in the methanol for 5 s, cells were soaked in Wright-Giemsa staining solution for 15 s. Finally, slides were rinsed with deionized water. Images were taken with AxioPlan2 after air dry.
In situ hybridization, TUNNEL staining, o-dianisidine staining
cDNA fragment for rps9 was cloned into pUC19 plasmid (Takara) using rps9-probeF: 5′-GGTACCTGTTCGAGCCTGACACGGAC-3′ and rps9-probeR:5′-TAATACGACTCACTATAGCAAGCACTGCTGAGTCCCT-3′. Probes for other genes were used as previously described (Danilova et al. 2011; Ear et al. 2016). Whole mount in situ hybridization was performed as described (Thisse and Thisse 2008).
Embryos for TUNNEL staining were stored as described in whole mount in situ hybridization. Subsequent TUNNEL staining was performed on the embryos following the manufacturer’s protocol (Roche, In Situ Cell Death Detection Kit, TMR red).
For o-dianisidine staining, embryos were collected and fixed at room temperature for 2 h using 4% paraformaldehyde (PFA) in PBS buffer after dechorionation. After the removal of PFA by PBS wash, embryos is incubated in the o-dianisidine (Sigma) staining solution in the dark for 30 min followed by PBS wash. Embryos were then bleached for 20 min. Finally, embryos were immediately imaged after washed by PBS.
Real-time quantitative PCR
Total RNA was isolated using TRIzol Reagent (Life Technologies) from pool of 30 embryos according to the manufacturer’s directions. cDNA was made using ProtoScript II Reverse Transcriptase and random hexamer primers (New England Biolabs). Quantitative PCR was performed on Stratagene Mx3005P (Agilent Technologies) using Luna Universal qPCR Master Mix (New England Biolabs). Each experiment was performed in triplicate. Primers for p53, mdm2, and p21 are previously described (Ear et al. 2015), and primers for rps9 are as follows: rps9-oligoF: 5′-AGAAGGATCCTAAGCGTCTC-3′, rps9-oligoR: 5′-CTCTCCAAGAAATCCTCCAC-3′.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary information files. Figure S1-S2 contains protein alignment and gene targeting illustration. Figure S3-S7 and Movie S1-2 contain blood related information. Figure S8 contains vessel related information. File S1 contains a full description of these 8 supplementary figures. Supplemental material available at figshare: https://doi.org/10.25387/g3.9948260.
Results
Deficiency in rps9 leads to morphological defects
Located in chromosome 16, rps9 gene is highly conserved between human and zebrafish and their proteins share 95% of amino acid similarity (Figure S1). By targeting rps9 with CRISPR/Cas9, three mutant lines were generated. All these three mutant lines had a small deletion in the third exon of rps9 locus. Specifically, two frame-shift mutant lines, rps9la491 and rps9la490, have alleles predicted to encode truncated polypeptides and one mutant line, rps9la489, has an allele producing a protein with one amino acid deletion and a leucine changed to histidine (Figure S2, A and B). Three mutant lines were outcrossed to wildtype fish and maintained for further experiments.
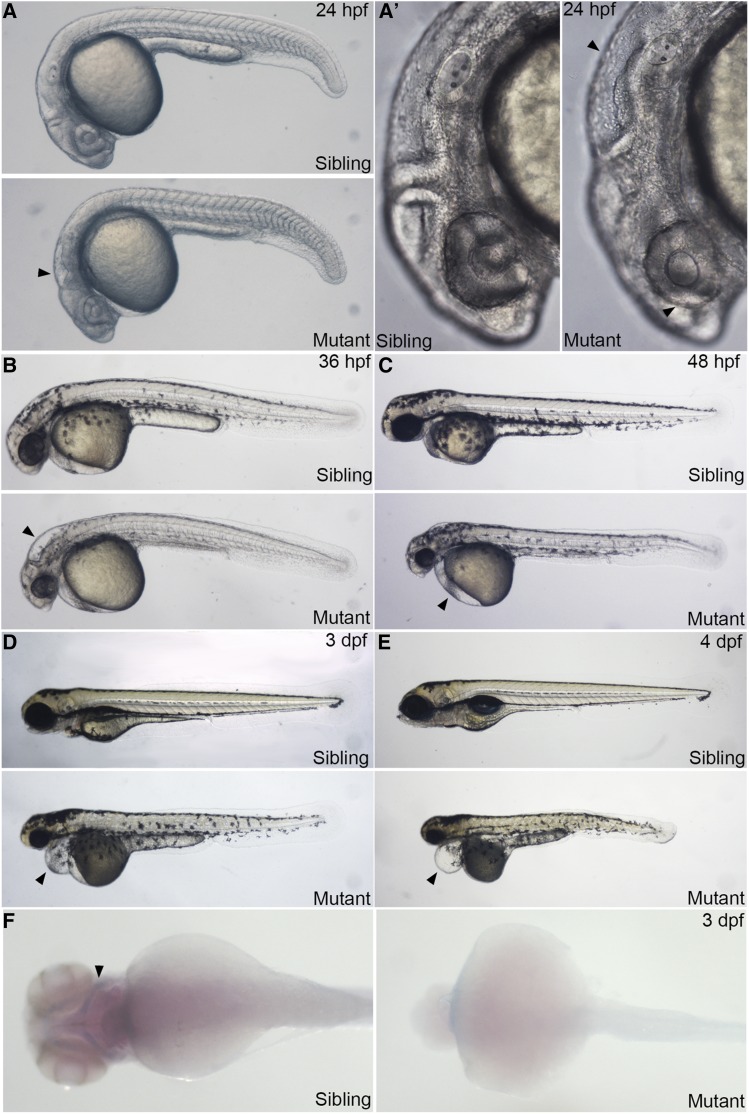
No notable abnormality was observed in offspring from in-crossing the rps9la489/rps9 mutant line. When rps9la491/rps9 and rps9la490/rps9 adult fishes were in-crossed, embryos presumably with homozygous mutation started to show notable abnormality from 21 hr post fertilization (hpf). By 24 hpf, aplasia was seen in the head in rps9-deficient embryos, indicated by small eyes and enlarged hindbrain ventricles (Figure 1A and 1A’). Smaller eyes and enlarged hindbrain were apparent at 36 hpf, accompanied by reduction of pigment (Figure 1B). From 48 hpf to 4 days post fertilization (dpf), the pericardium gradually became enlarged and finally developed edema. The body length also decreased and the trunk displayed apoptosis (Figure 1, C-E). The homozygous embryos eventually died at 5 dpf. Furthermore, the formation of cartilage was blocked when examining with alcian blue staining at 3 dpf, indicated that cartilage development was disrupted in rps9 mutants (Figure 1F). To verify that the morphologically abnormal embryos were linked to rps9 mutation, we genotyped them by direct sequencing. Sequencing results showed that the embryos (N > 5) with morphological defects all had the identified indels in the rps9 locus (Figure S2C). This study confirmed a null deletion in the rps9 gene is associated with the observed phenotype of rps9la491 and rps9la490.
Figure 1.
Embryos with rps9 deficiency display morphological abnormalities. (A-E) Overall morphological phenotype of rps9 deficient embryos. At 24 hpf (A and A’), smaller eyes and abnormal hindbrain (arrowhead) appear in the head region. By 36 hpf (B), enlarged hindbrain (arrowhead) becomes apparent and pigment is reduced. From 48 hpf to 4 dpf (C-E), edema (arrowhead) is gradually developed in the pericardial region, accompanied with shortened trunk. (F) Cartilage fails to grow in the pharyngeal arch in rps9 mutant embryos.
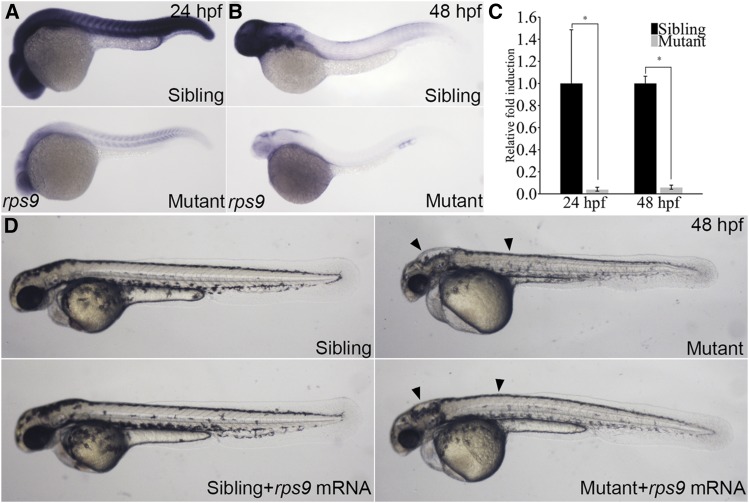
To determine expression level and patterns of rps9, we examined its expression by whole-mount RNA in situ hybridization. At 24hpf, the wild type sibling showed strong expression throughout the body while the mutant displayed a dramatic reduction in expression levels (Figure 2A). Similarly, a remarkable decrease of gene expression was observed in the rps9 deficient embryos by 48 hpf (Figure 2B). This reduction of rps9 transcript in the mutant was also revealed quantitatively by real-time PCR (Figure 2C). To further confirm that reduction of rps9 transcript level was responsible for the morphological abnormalities in mutants, we conducted a rescue experiment using in vitro transcribed rps9 mRNA. Indeed, injection of rps9 mRNA to the mutant embryos improved morphology both in the hindbrain and trunk (Figure 2D). Altogether, these results established that the morphological defects seen in the mutant were due to the deficiency of rps9.
Figure 2.
Phenotype of rps9 deficient embryos is partially rescued by rps9 mRNA injections. (A-B) Whole mount in situ hybridization for rps9 in mutants and siblings at 24 hpf or 48 hpf. (C) qPCR analysis of rps9 transcript levels. *, P < 0.05. (C) In vitro transcribed rps9 mRNA (400 pg) partially rescues phenotype in rps9 mutants. Arrowhead indicates morphological improvement in the hindbrain and trunk.
rps9 is required for normal maturation of erythroid cells
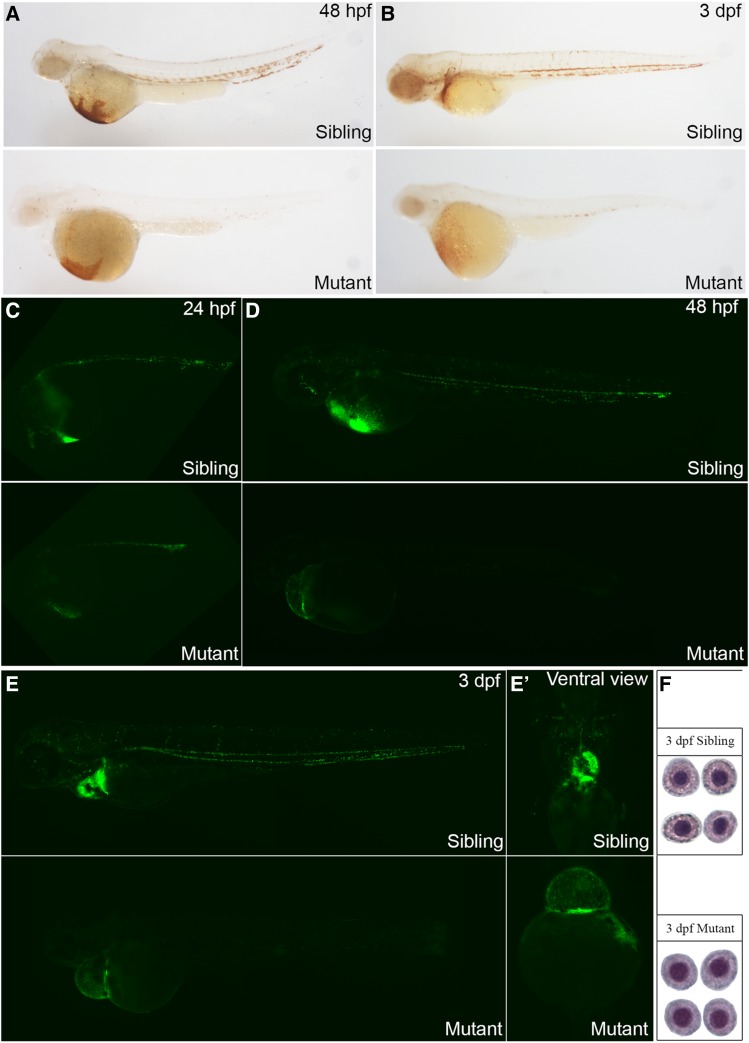
We next evaluated whether the development of red blood cells was affected by rps9 deficiency in the zebrafish mutants. We found that, at both 48 hpf and 3 dpf, o-dianisidine staining level in embryos was severely reduced in the mutants (Figure 3, A and B) and this reduction in hemoglobin levels could be partially restored by the injection of rps9 mRNA (Figure S3), implying that deficiency in rps9 was responsible for the anemic phenotype. To evaluate numbers of red blood cells, we mated rps9 mutant line into the LCR2:EGFP transgenic fish line, whose red blood cells were tagged with EGFP. A moderate decrease of the EGFP positive cells was revealed between the sibling and mutant embryos at 24hpf, and the decline became significant at 48 hpf (Figure 3, C and D). At 3 dpf, EGFP positive cells were only found at the cardiac region (Figure 3, E and E’). Furthermore, blood circulation was diminished at 24 hpf and gradually lost due to the lack of blood cells and malfunctioned heart (Movie S1 and S2).
Figure 3.
Deficiency in rps9 induces anemia in mutant embryos. (A-B) Detection of hemoglobin levels by o-dianisidine staining for rps9 in both siblings and mutants at 48 hpf and 3 dpf. (C-E, E’) Detection of erythroid cells by crossing rps9 mutant into LCR:EGFP transgenic fish line. At 24 hpf, the number of blood cells are reduced slightly; At 48 hpf and 3 dpf, erythrocytes are almost deprived in the trunk, and only a small amount of red blood cells remain in the cardiac region. (F) Wright-Giemsa staining of erythroid cells at 3 dpf in sibling and mutant.
To detect the cytomorphological change of the erythroid cells in rps9 deficiency, we did a Wright-Giemsa staining at 3 dpf. Based on the size and shape of the nucleus as well as staining of the plasma, the developmental stage of the erythroid cells can be identified (Qian et al. 2007). In rps9 mutant, the erythroid cells remained at the polychromatophilic stage, which was usually seen at 2 dpf (Figure 3F). This developmental delay of erythrocyte suggests that the maturation of erythrocyte is impaired in rps9 deficient embryos.
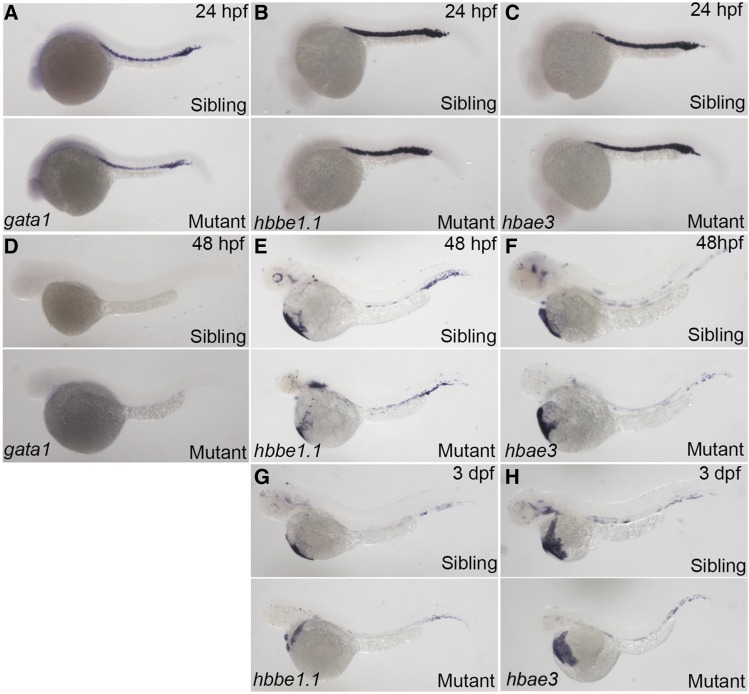
We then accessed the effect of rps9 disruption on erythropoiesis in mutant embryos by detecting the gene expression pattern of three erythroid markers at several developmental stages. As one of the markers for the specification of erythroid lineage, gata1 was used to detect the early stage of erythropoiesis. Two other globin transcripts, i.e., hbbe1.1, and hbae, can be used to mark the mature red blood cells. At 24 hpf, the expression of gata1, hbbe1.1, and hbae3 in mutant embryos was indistinguishable from sibling control embryos, suggesting the specification step of erythroid cells is not affected (Figure 4, A-C). At 48 hpf, both mutant and sibling control embryos displayed a faint expression of gata1, and the expression of hbbe1.1 and hbae3 remained mostly unchanged (Figure 4, D and F). Additionally, the expression of hbee1.1 and hbae3 appeared slightly decreased at 72 hpf due to the shrunken size of mutant embryos (Figure 4, G and H). These results suggest that overall erythroid gene transcription is not significantly downregulated.
Figure 4.
No obvious change was seen for expression of erythroid specific markers in rps9 mutants. (A-F) Expression levels of gata1, hbbe1.1, and hbae3 in mutants remain unchanged at 24 hpf (A-C) and 48 hpf (D-F). (G-H) Because of the shortened size of the mutant embryos, slight decrease in hbbe1.1 and hbae3 is seen at 3 dpf.
To investigate the effect of rps9 deficiency on overall hematopoiesis, we analyzed other non-erythroid markers. A similar expression pattern was observed for three markers of myeloid cells, namely, l-plastin, mpx, and pu.1, indicating that the specification of primitive myeloid cells is not affected (Figure S4, A-C). Likewise, the development of hematopoietic stem cell is not affected as there was no considerable change for the expression level for scl and c-myb, markers for the hematopoietic stem cell emergence (Figure S4, D and E). However, expression of rag1 decreased significantly, suggesting that lymphoid development is disturbed (Figure S4F).
Finally, the expression pattern for two other ribosomal protein genes was also detected to check the expression change for other ribosomal protein genes. The expression of rpl11 and rps14 shares a similar pattern between the mutant and wildtype sibling control, suggesting that the regulation for other ribosomal protein genes is unaffected upon the deficiency of rps9 (Figure S5).
rps9 deficiency leads to p53 upregulation and downregulation of p53 can largely rescue the phenotype
The morphological defect is usually associated with an abnormal increase in the levels of apoptosis. To determine the level change of apoptosis, we conducted a TUNNEL staining for both the mutant and wildtype sibling control. An elevation of the apoptosis level was indeed detected at all time points analyzed, confirming that morphological defect is likely caused by increased activity of apoptosis (Figure S6).
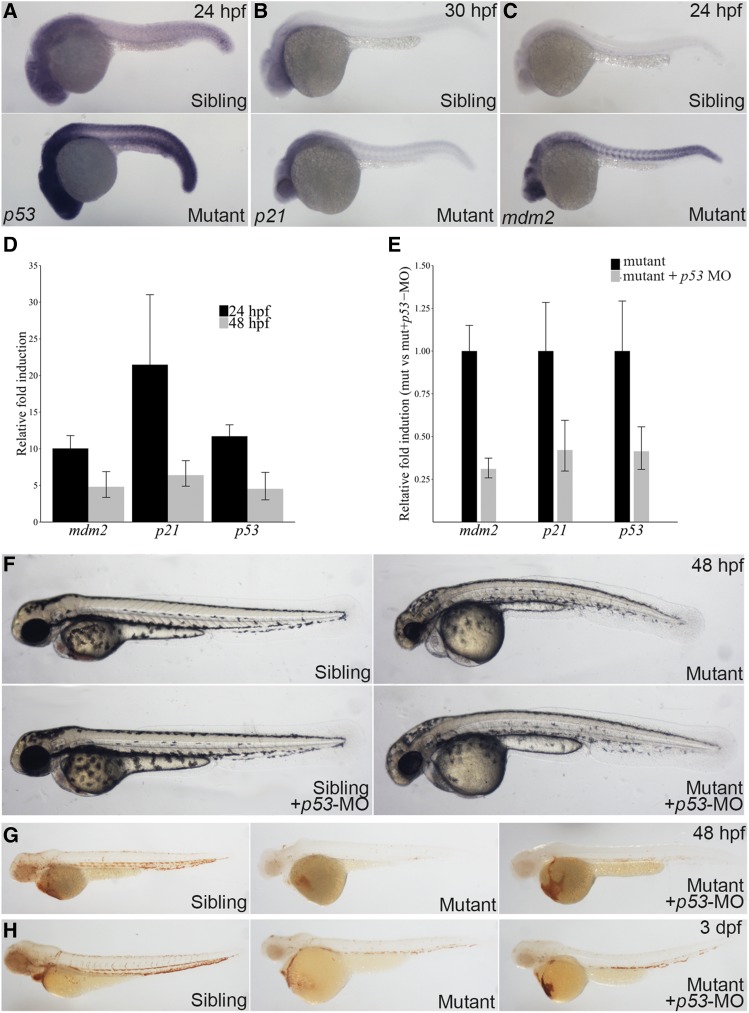
The connection between elevated apoptosis and p53 activation has been demonstrated in several ribosome protein mutants, and knockdown of p53 can alleviate the phenotype (Danilova et al. 2008; Ear et al. 2016). Consistent with increased apoptosis level, expression of p53 and p53-target genes was also increase, and this upregulation was observed throughout the entire embryo (Figure 5, A-D). By injecting morpholino (MO) targeting p53 in rps9 mutant, we analyzed whether down-regulating p53 could ameliorate both the morphological and erythroid defects. Notable morphological improvement was found in the mutant followed by the decrease of p53 and p53-target gene expression mediated by MO injection (Figure 5, E and F). Furthermore, erythropoiesis was also improved at 48 hpf and 3 dpf when detecting the hemoglobin level by o-dianisidine staining (Figure 5, G and H). Of note, no considerable changes to the blood level after p53 inhibition in wildtype sibling control embryos (Figure S7). This suggests that the anemia phenotype is largely dependent on the p53 pathway during both 48 hpf and 3 dpf in zebrafish rps9 mutants.
Figure 5.
Anemic phenotype is partially rescued by p53 konckdown in rps9 deficient embryos. (A-C) Whole mount in situ shows that p53, p21, and mdm2 are upregulated in mutants. (D) The expression of p53 and p53 target genes including mdm2 and p21 increases as measured by qPCR. The relative fold induction is represented as fold change over sibling control. (E) p53 inhibition by MO injection knockdown the gene expression of p53 and p53-target genes. (F) The morphological defects of rps9 mutant are largely rescued by p53 inhibition. (G-H) An increase of hemoglobin level appears at 48 hpf. (G) and 3 dpf (H) after p53 knockdown.
Rescue of anemia caused by rps9 deficiency using therapeutic agents
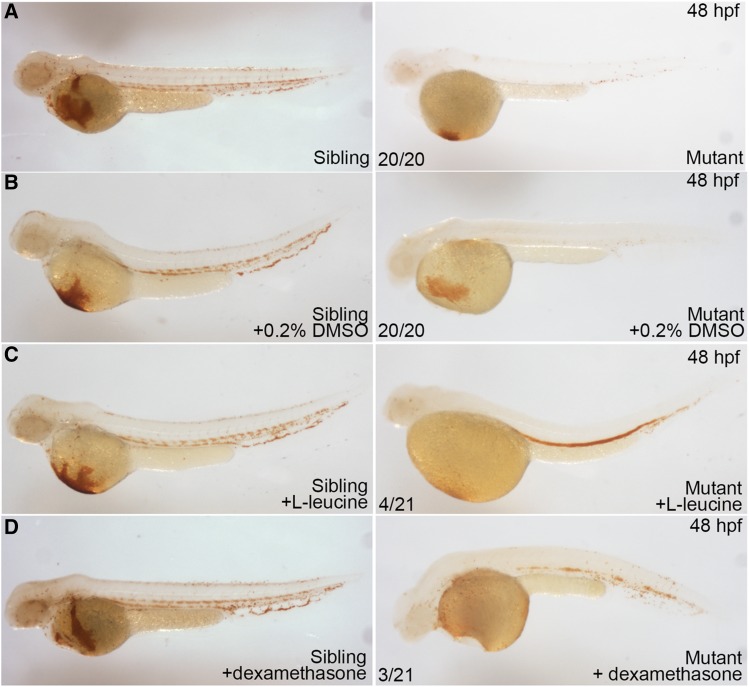
L-leucine, and dexamethasone have been previously shown to alleviate anemia in ribosomal deficiency models (Danilova et al. 2011; Payne et al. 2012; Narla et al. 2014; Ear et al. 2016). As shown in Figure 6, treatment of embryos with L-leucine or dexamethasone notably recovered hemoglobin level in the mutant embryos (Figure 6). This result suggests that these agents might be used as an option of treatment for anemia if rps9 mutation is identified in patients.
Figure 6.
Hemoglobin level is partially recovered by L-leucine, and dexamethasone treatment in rps9 deficient embryos. (A-B) Mutants without treatment and mutants treated with 0.2% DMSO were used as controls. (C) Mutants treated with L-leucine (200 mmol/L) show a considerable recovery in hemoglobin level. (D) Mutants treated with dexamethasone (250 μmol/L) display a less effective restoration in hemoglobin level.
Comparison of developmental defect of erythroid cells in four different ribosomal protein mutants
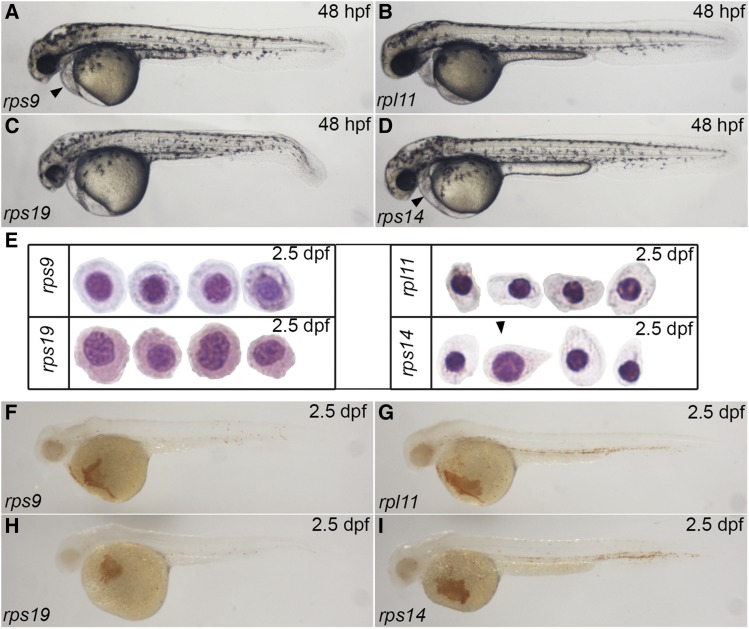
The developmental functions of ribosomal protein such as rps19, rpl11, and rps14 have been previously reported in zebrafish (Danilova et al. 2011; Zhang et al. 2014; Ear et al. 2016). We compared the phenotype of these mutants with rps9 mutant at the same developmental stage. At 48 hpf, rps19 mutant displayed the most severe morphological defects, while rpl11 mutant exhibited the mildest phenotype. The overall morphology of rps9 mutant and rps14 mutant appeared similar but rps9 had a slightly shorter trunk and a larger pericardium (Figure 7, A-D). By 2.5 dpf, rps9 mutant showed a less developed form of polychromatophilic erythroblast, whereas rpl11 mutant displayed a more mature form, with rps14 mutant being between them. At the same stage, erythroid cells in rps19 mutant arrested at the basophilic stage, suggesting the maturation of erythrocyte is most severely impaired in rps19 mutant (Figure 7E). Consistent with the cytomorphological change, the hemoglobin levels declined gradually in the order of rpl11, rps14, rps9 and rps19 mutants, implying that the status of erythrocyte maturation is correlated to anemic phenotype (Figure 7, F-I). In summary, this comparison illustrates that a variety of morphological abnormalities and anemic state are developed in different ribosomal protein mutants, thus requiring a detailed examination for each specific ribosomal protein mutants.
Figure 7.
The severity of phenotype of four ribosomal protein mutants increases in an order of rpl11, rps14, rps9, and rps19. (A-D) Morphological comparison of four mutants by 48 hpf. Arrowhead indicates pericadial anema. (E) Wright-Giemsa staining for four mutants at 2.5 dpf. Arrowhead indicates that an erythrocyte in rps14 mutants resembles the erythrocytes in rps9 mutants. (F-I) o-dianisidine staining of four mutants at 2.5 dpf.
Discussion
Through studying genetic mutation in zebrafish, we demonstrated that rps9 plays an important role in erythropoiesis in zebrafish. Although no rps9 mutation has been reported in any cases of anemia, this study suggests that rps9 can be a potential candidate for DBA or other human anemia diseases. A recent genome-wide association study has implicated rps9 as a candidate for Takayasu disease, which is a rare inflammatory disease that typically damages the aorta and its major branches (Renauer et al. 2015). To study whether rps9 is involved in maintaining vascular structure, we analyzed the blood vessel with flk and fli1, two common vascular markers (Figure S8, A and B). A moderate disruption of intersomitic vessel formation was observed at 24 hpf in the mutant, implicating that rps9 might be involved in the vessel formation in zebrafish. Similar to this observation, it was reported that rps29 mutation also caused the disruption of vessel formation (Burns et al. 2009). However, rpl11 mutation seemed not to cause any significant change of vessel morphology (Danilova et al. 2011). More experiments are needed to address the link between rps9 and Takayasu disease or ribosomal deficiency and vascular integrity in general.
Anemia in rps9 mutant is stronger than rps14 mutant. Erythroid failure of rps9 mutant appeared to be p53-dependent as early as 2 dpf. However, in rps14 mutant, erythroid failure was demonstrated to be p53-dependent at 3 dpf (Ear et al. 2016). This timing difference seems to be correlated with the severity of erythroid failure.
Comparison of four zebrafish ribosomal protein mutants revealed that severity of morphological and anemic phenotype ranks in the order of rpl11, rps14, rps9 and rps19. This raises the question why rpl11 deficiency develops defects later than the deficiency in the other three ribosomal proteins. A recent study suggests that ribosomal stress is mediated by free ribosomal proteins, in particular by free RPL11, to bind MDM2 and subsequently activate P53 (Zheng et al. 2015). As mentioned before, p53 activation induced by ribosomal protein disruption in zebrafish models has a great impact on the development of anemia, so it is possible that the other three mutants have more free RPL11 than rpl11 mutant, thus accelerating P53 activation and showing more severe phenotype. Further experiments are needed to test this hypothesis.
Acknowlegements
We thank Liuwen Huang and Linda Yuan for maintaining the fish used in this study. This work was supported by National Natural Science Foundation of China (31801209) and Science and Technology Program of Shenzhen (JCYJ20170818090542084).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.9948260.
Communicating editor: B. Andrews
Literature Cited
- Burns C. E., Galloway J. L., Smith A. C. H., Keefe M. D., Cashman T. J. et al. , 2009. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood 113: 5776–5782. 10.1182/blood-2008-12-193607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Zhu B., Li S. J., Yuan T., Yang Q. C.. et al. , 2017. Down-regulation of RPS9 inhibits osteosarcoma cell growth through inactivation of MAPK signaling pathway. J. Cancer 8: 2720–2728. 10.7150/jca.19130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N., Sakamoto K. M., and Lin S., 2011. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br. J. Haematol. 152: 217–228. 10.1111/j.1365-2141.2010.08396.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N., Sakamoto K. M., and Lin S., 2008. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112: 5228–5237. 10.1182/blood-2008-01-132290 [DOI] [PubMed] [Google Scholar]
- Ear J., Hsueh J., Nguyen M., Zhang Q. H., Sung V. et al. , 2016. A Zebrafish Model of 5q-Syndrome Using CRISPR/Cas9 Targeting RPS14 Reveals a p53-Independent and p53-Dependent Mechanism of Erythroid Failure. J. Genet. Genomics 43: 307–318. 10.1016/j.jgg.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Ear J., Huang H., Wilson T., Tehrani Z., Lindgren A. et al. , 2015. RAP-011 improves erythropoiesis in zebra fish model of Diamond-Blackfan anemia through antagonizing lefty1. Blood 126: 880–890. 10.1182/blood-2015-01-622522 [DOI] [PubMed] [Google Scholar]
- Jia Q., Zhang Q., Zhang Z., Wang Y., Zhang W. et al. , 2013. Transcriptome analysis of the zebrafish model of Diamond-Blackfan anemia from RPS19 deficiency via p53-dependent and -independent pathways. PLoS One 8 10.1371/annotation/54600aa1-0df0-4e0a-b2ab-9d6cdd46cade [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok F. O., Shin M., Ni C. W., Gupta A., Grosse A. S. et al. , 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32: 97–108. 10.1016/j.devcel.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström M. S., 2012. Elucidation of Motifs in Ribosomal Protein S9 That Mediate Its Nucleolar Localization and Binding to NPM1/Nucleophosmin. PLoS One 7: e52476 10.1371/journal.pone.0052476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström M. S., and Zhang Y., 2008. Ribosomal Protein S9 Is a Novel B23/NPM-binding Protein Required for Normal Cell Proliferation. J. Biol. Chem. 283: 15568–15576. 10.1074/jbc.M801151200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A., and Ebert B. L., 2010. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 115: 3196–3205. 10.1182/blood-2009-10-178129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A., Payne E. M., Abayasekara N., Hurst S. N., Raiser D. M. et al. , 2014. L–Leucine improves the anaemia in models of Diamond Blackfan anaemia and the 5q- syndrome in a TP53-independent way. Br. J. Haematol. 167: 524–528. 10.1111/bjh.13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne E. M., Narla A., Paw B. H., Look A. T., Virgilio M. et al. , 2012. L-leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood 120: 2214–2224. 10.1182/blood-2011-10-382986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost E., Wehner K. A., Zhong X., Ashar F., Nguyen E. et al. , 2012. Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development 139: 3232–3241. 10.1242/dev.077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian F., Zhen F., Xu J., Huang M., Li W. et al. , 2007. Distinct functions for different scl isoforms in zebrafish primitive and definitive hematopoiesis. PLoS Biol. 5: e132 10.1371/journal.pbio.0050132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauer P. A., Saruhan-Direskeneli G., Coit P., Adler A., Aksu K. et al. , 2015. Identification of susceptibility loci in IL6, RPS9/LILRB3, and an intergenic locus on chromosome 21q22 in takayasu arteritis in a genome-wide association study. Arthritis Rheumatol. 67: 1361–1368. 10.1002/art.39035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C. et al. , 2007. P53 Activation By Knockdown Technologies. PLoS Genet. 3: e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., and Thisse B., 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3: 59–69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Thomson E., Ferreira-Cerca S., and Hurt E., 2013. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126: 4815–4821. 10.1242/jcs.111948 [DOI] [PubMed] [Google Scholar]
- Torihara H., Uechi T., Chakraborty A., Shinya M., Sakai N. et al. , 2011. Erythropoiesis failure due to RPS19 deficiency is independent of an activated Tp53 response in a zebrafish model of Diamond-Blackfan anaemia. Br. J. Haematol. 152: 648–654. 10.1111/j.1365-2141.2010.08535.x [DOI] [PubMed] [Google Scholar]
- Uechi T., Nakajima Y., Chakraborty A., Torihara H., Higa S. et al. , 2008. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum. Mol. Genet. 17: 3204–3211. 10.1093/hmg/ddn216 [DOI] [PubMed] [Google Scholar]
- Uechi T., Nakajima Y., Nakao A., Torihara H., Chakraborty A. et al. , 2006. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS One 1: e37 10.1371/journal.pone.0000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao W. J., Liao J. M., Liao P., and Lu H., 2015. Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7: 92–104. 10.1093/jmcb/mjv014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ear J., Yang Z., Morimoto K., Zhang B. et al. , 2014. Defects of protein production in erythroid cells revealed in a zebrafish diamond-blackfan anemia model for mutation in RPS19. Cell Death Dis. 5: e1352 10.1038/cddis.2014.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Jia H., Zhang Q., Wan Y., Zhou Y. et al. , 2013. Assessment of hematopoietic failure due to Rpl11 deficiency in a zebrafish model of Diamond-Blackfan anemia by deep sequencing. BMC Genomics 14: 896 10.1186/1755-8794-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Lang Y., Zhang Q., Cui D., Sun H. et al. , 2015. Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation. Genes Dev. 29: 1524–1534. 10.1101/gad.261792.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files. Figure S1-S2 contains protein alignment and gene targeting illustration. Figure S3-S7 and Movie S1-2 contain blood related information. Figure S8 contains vessel related information. File S1 contains a full description of these 8 supplementary figures. Supplemental material available at figshare: https://doi.org/10.25387/g3.9948260.