Abstract
Litsea cubeba (Lour.) Pers. (mountain pepper, Lauraceae) is an important woody essential oil crop that produces fragrant oils in its fruits, especially in its peels. Identification of genes involved in the regulation of fruits and peel architecture is of economic significance for L. cubeba industry. It has been well known that the MADS-box genes are essential transcription factors that control flowers and fruits development. Here, we obtained 33 MADS-box genes first from the RNA-seq data in L. cubeba, and 27 of these genes were of the MIKC-type. LcMADS20, an AGAMOUS-like gene, was highly expressed in the developing stages of fruits, particularly at 85 days after full bloom. The ectopic expression of LcMADS20 in Arabidopsis resulted in not only curved leaves, early flowering and early full-opened inflorescences, but also shorter siliques and decreased percentage of peel thickness. Moreover, in the LcMADS20 transgenic Arabidopsis, the expression modes of several intrinsic ABC model class genes were influenced, among which the expression of FUL was significantly reduced and AP3, AG, and STK were significantly increased. This study systematically analyzed the MADS-box genes in L. cubeba at the transcriptional level and showed that LcMADS20 plays important roles in the regulation of fruit architecture.
Keywords: Lauraceae, mountain pepper, AGAMOUS-like genes, ectopic expression, fruit
Litsea cubeba, a dioecious species in the family Lauraceae, is regarded as an important industrial essential oil crop (Chen et al. 2013). L. cubeba essential oil, which is mainly extracted from the fresh fruits (especially from the peel), has been widely used for as botanical insecticides, natural additives, and ionone series flavors, in addition to other functions (Jiang et al. 2009; Agrawal et al. 2011; Wang et al. 2012; Su and Ho 2016). L. cubeba produces fragrant oils in its fruits, especially in its peels (Chen et al. 2013). Investigation of mechanisms involved in the regulation of fruits and peel architecture is specifically important for L. cubeba industry. However, studies on the growth and development of fruit and peel in L. cubeba remain rare. Generally, the majority fruit development begins after the ovary fertilization, and then the ovary wall differentiates into pericarp (endocarp, mesocarp, and exocarp) (Pabonmora et al. 2014). It has been well known that members of the MADS-box family genes encode for transcription factors essential in the control of flower and fruit development (Pinyopich et al. 2003; Lee and Lee 2010; Bowman et al. 2012). For example, the AG-like genes, which identity carpel, stamen and ovule, can affect fruit yield and quality (Pinyopich et al. 2003; Losa et al. 2010).
The MADS-box genes were initially identified from four founding proteins (Mcm1, Agamous, Deficiens A, Serum response element) from yeast (Saccharomyces cerevisiae), Arabidopsis thaliana, Antirrhinum majus, and mammals (human) respectively (Schwarz-Sommer et al. 1990). These proteins all have a contiguous conserved sequence of approximately 58 amino acids, which includes dimerization and a DNA binding domain (Schwarz-Sommer et al. 1990). In Arabidopsis, the MADS-box genes are classified into five groups named Mα, Mβ, Mγ, Mδ, and MIKC (Parenicová et al. 2003). Members of the MADS-MIKC group, which play important roles in flower development, include the M, I, K, and C domains (Causier et al. 2010). In general, the most conserved M domain is responsible for DNA binding and protein dimerization; the I intervening domain participates in dimerization specificity; the K domain is responsible for protein-protein interaction; and the C domain plays an essential role in specific functions (Causier et al. 2010; Kramer et al. 1998).
The MADS-box genes play variety functions in flower and fruit architecture. The flowering time regulators FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) can integrate numerous environmental and endogenous signals to regulate flower formation and development (Searle et al. 2006; Lee et al. 2007; Lee and Lee 2010). Moreover, the floral ABC model genes function in flower organ identity and formation (Theissen and Saedler 2001; Causier et al. 2010). The A class gene, APETALA1 (AP1), can control sepal identity and interacts with the B class genes PISTILLATA (PI) and APETALA3 (AP3) involved in the control petal identity. The C class gene AGAMOUS (AG) is a carpel identity gene and is necessary for stamen development, together with the B class genes (Causier et al. 2010; Smaczniak et al. 2012). The MADS-box transcription factors SEEDSTICK (STK), SHATTERPROOF1 (SHP1), and SHATTERPROOF2 (SHP2) belong to the D class genes and play a redundant role in controlling ovule integument identity (Pinyopich et al. 2003; Losa et al. 2010). The E class genes SEPALLATA1–4 (SEP1–4) are required for the specification of all four types of floral organs (Pelaz et al. 2000; Ditta et al. 2004).The MADS-box genes can also interact with the family genes and also with non-MADS transcriptional regulators or cofactors (Brambilla et al. 2007; Smaczniak et al. 2012).
AG is necessary for development of the reproductive organs (stamens and carpels). The loss of AG function caused homeotic conversion fromstamens and carpels to petals in Arabidopsis (Bowman et al. 1989), the overexpression of AG caused curled leaves, staminoid petals, carpelloid sepals, and inflorescence in Arabidopsis (Riechmann and Meyerowitz 1997). In rice (Oryza sativa L.), ectopic expression of OsMADS3, an AG-like gene, caused homeotic conversion of lodicules to stamens (Kyozuka and Shimamoto 2002). The redundant functions of AG and the SHP genes can promote carpel development, and there are redundant roles of AG, STK, and SHP genes in ovule identity (Pinyopich et al. 2003). Ectopic expression of either the STK or SHP gene is sufficient to induce the transformation of sepals into carpeloid organs bearing ovules (Favaro et al. 2003). To investigate the important roles of MADS-box transcription factors involved in the flower and fruit development in L. cubeba, we first identified the MADS-box genes in L. cubeba and then conducted a functional analysis on the AGAMOUS-like gene LcMADS20. This work was estimate to provide important information of genes involved in the fruit architecture for strategy of L. cubeba plant industry.
Materials and Methods
Plant materials
The plant material of L. cubeba used in this paper is the family of Anhui5#. L. cubeba is a dioecious plant, usual flowers in March, and full blooms at the middle of March. The pollen of the flower deteriorates in the female tree, while the ovary of the flower deteriorates in the male tree (Figure S1). After the flower fertilized in the female tree, the fruits begin to develop, and usually matured in August. To conduct the analysis on LcMADS-box genes and the expression mode during the three different developmental stages of flower bud, we used our previously published RNA-seq data for the three stages, which included the first stage (FD1, MD1) (the inflorescence primordium begins to differentiate), the second stage (FD2, MD2) (the flower primordium begins to differentiate) and the third stage (FD3, MD3) (the flower organ begins to differentiate) (He et al. 2018). The fruit samples for RT-qPCR picked on 25, 40, 55, 70, 85, 100, and 115 DAF (Days After Full bloom). All samples immediately frozen in liquid nitrogen and then stored at –80° until the RNA extraction. Arabidopsis of wide type (Columbia) used in this study.
Identification of MADS-box genes in L. cubeba
The HMM (Hidden Markov Models) profiles of the MADS domain (PF00319) were acquired from the PFAM database (Finn et al. 2016) and then searched against the RNA-seq translated coding sequence with HMMER software (3.1b2) (Eddy 1998). The candidate MADS-box proteins in this species subsequently verified through annotation in the databases of SMART (Simple Modular Architecture Research Tool), Pfam, and CDD(Conserved Domain Database) using InterProScan and then checked manually to confirm the reliability (Jones et al. 2014).
Phylogenetic construction
The encoded MADS-box sequences from L. cubeba and Arabidopsis (Huala et al. 2001) were aligned using Clustal W. Then, the alignment was refined and removed from the less conserved regions, leaving a conserved MADS domain of 58 characters. NJ (Neighbor-Joining) trees were generated using the MEGA6 program (Tamura et al. 2013).
Motif and gene expression analyses
MEME (Multiple Em for Motif Elicitation) was used to determine the conserved motif of the amino acid sequences in L. cubeba, and the parameters of the analysis established as follows: number of motifs at 10, motif width between 6 and 200 (Bailey et al. 2009). The identified motifs were annotated using SMART online (http://smart.embl-heidelberg.de/) (Letunic and Bork 2018). The expression of LcMADS-box genes in the flower bud developing stages was generated using Pheatmap in the R software (3.5.1) with data normalized of log2(FPKM+1).
RNA isolation and cDNA synthesis
The frozen flower organ samples were ground into powder in a mortar with liquid nitrogen, and then, the total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, China) following the manufacturer’s instructions. The cDNA synthesized from 1 µg of total RNA with a GoldenstarRT6 cDNA Synthesis Kit (Tsingke Biological Technology, China). The final cDNA products were diluted 1:10 with nuclease-free water for reverse-transcription quantitative real-time PCR (RT-qPCR).
Reverse-transcription quantitative real-time PCR
RT-qPCR amplification was performed in 20 µL mixtures consisting of 10 µL of 2× SYBR Premix Ex Taq II mix (Takara), 0.4 µL each of forward and reverse primers, 0.4 µL of Rox dye, 1 µL of cDNA template (50 ng/µL), and RNase-Free Water. The reaction mixture was incubated at 95° for 30 s; followed by 40 cycles of amplification (95° for 5 s, 60° for 34 s) in the PCR stage; and the melt curve stage was 95° for 15 s, 60° for 1 min, and 95° for 15 s, performed on an ABI QuantStudio 7 Flex Real-Time PCR System. Three technical replicates were run for every sample, and the data were analyzed using the 2−ΔΔCtmethods (Livak and Schmittgen 2001).
Transient transformation of LcMADS20 in L. cubeba leaves
To observe the subcellular localization of LcMADS20, we cloned the coding sequence of LcMADS20 and constructed the sequence into the pCambia 1300-GFP/C vector with the cauliflower mosaic virus (CaMV) 35 promoter. The fused plasmid was transformed into Agrobacterium tumefaciens strain GV3101 chemically competent cells, which were cultivated in Luria-Bertani medium with 50 mg/L kanamycin and 50 mg/L rifampicin (28°, 200 rpm, 12 h). The strain sediment was collected by centrifugation for 5 min at 5000 rpm at 4° and then suspended in 10 Mm 2-(N-Morpholino) ethanesulfonic acid and 20 µm acetosyringone at OD600 = 0.4 and further incubated for approximately 3 h. A needleless syringe was used to infiltrate the leaves of L. cubeba without penetration. Finally, the plantlet was maintained in the dark for approximately 2 days and then photographed using a ZEISS LSM 700 laser-scanning microscope at 488 nm to observe the subcellular localization.
Ectopic expression of LcMADS20 in Arabidopsis
The combined 35S::LcMADS20-GFP vectors and the empty vector (as control) were transformed into Arabidopsis via A. tumefaciens strain GV3101 using the flower dip method (Clough and Bent 1998).The expression vector contained hygromycin gene, which was useful in verification and screening. The positive transgenic Arabidopsislines were selected on 25 mg/L hygromycin Murashige and Skoog medium and verified using LcMADS20 specific primers. The seedlings were transplanted into soil cultivated in 7cm *7 cm pot with a 16 h light and 8 h dark photoperiod at 23° until the T3 generation.
Paraffin section detection of silique thickness
To observe the siliques architecture of the transgenetic Arabidopsis, we employed the paraffin section with reference the method of Ye (2012). The silique samples were first fixated in a FAA fixative and then dehydrated on the concentration of 30%, 50%, 70%, 80%, 90% and 100% of ethanol. Subsequently, the samples were embedded into paraffin blocks after treated with xylene, and then cut into thin paraplast sections (6-10μm) using the rotary microtome (ThermoHM325, USA). Additionally, the samples were deparaffinized by xylene and ethanol, and then stained with hematoxylin and eosin solution. Finally, the slices coated with neutral gum were observed using an Olympus BX53 microscope (Olympus, Tokyo, Japan), and the pericarp thickness in siliques were further measured.
Data availability
The MADS-box genes of L. cubeba were analyzed according to data from our previous published paper (He et al. 2018). The information of the MADS-box genes of L. cubeba was list on the Table S1. The sequences of MADS-box genes of Arabidopsis were downloaded from the database of The Arabidopsis Information Resource (https://www.arabidopsis.org/) with the accession number listed on the Table S2. Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.9978776.
Results
Identification and phylogenetic analysis of the MADS-box genes in L. cubeba
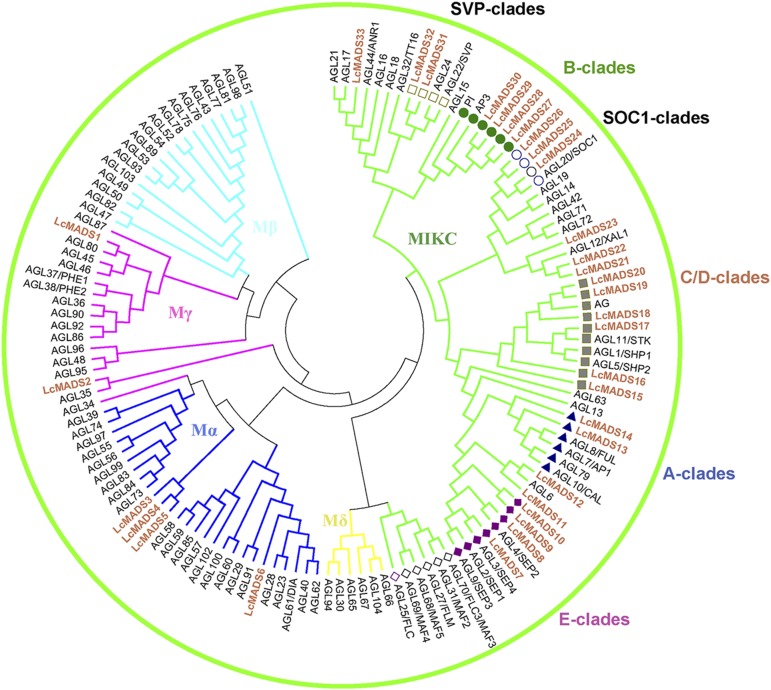
A total of 33 LcMADS domain sequences were obtained from RNA-seq data of male and female flower buds in three differentiation stages, respectively (Table S1), and the NJ tree was constructed utilizing the conserved domain with Arabidopsis MADS-box proteins (Table S2). Based on the Arabidopsis classified groups (Parenicová et al. 2003), 27 members were of MADS-MIKC-type, of which two, four, six, and four members belonged to the A, B, C/D, and E class clades, respectively. Three, two, two, and three members belonged to the SOC1, SVP, AGL6, and AGL12 clades, respectively (Figure 1).
Figure 1.
Phylogenetic analysis of MADS-box genes in L. cubeba and Arabidopsis. The 33 MADS-box genes in L. cubeba (light purple) and the MADS-box genes in Arabidopsis were used to construct the NJ tree based on the MADS conserved domain. The light green branches indicate the MIKC groups, the yellow branches the Mδ, the deep blue branches the Mα, the pink branches the Mγ, and the light blue branches the Mβ. Square indicates the SVP clade genes, circle labeled the SOCI branches, blue triangle labeled the A clades genes, green circle labeled the B clade genes, brown square labeled the C/D clade genes and purple diamond labeled the E clade genes.
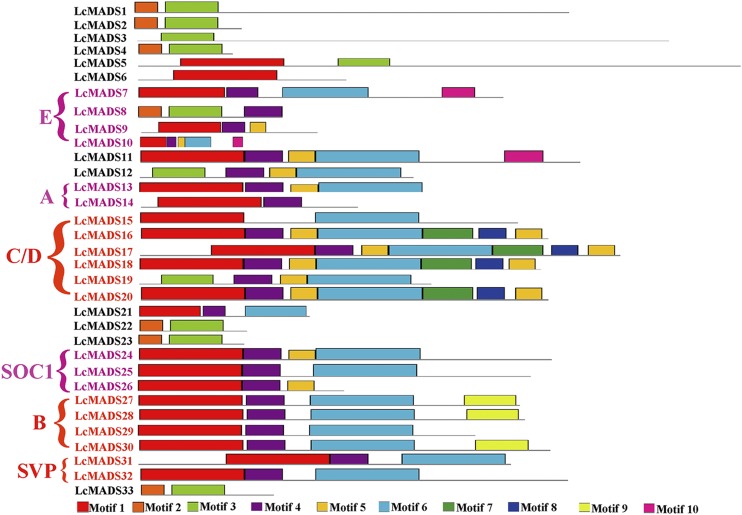
Conserved motif analysis of LcMADS-box genes
To identify whether the selected LcMADSs sequences contain the complete functional domains, we conduct the motif searching and clustering analysis for LcMADSs. A total of ten conserved motifs were detected in the 33 LcMADS-box genes (Table S3). Motif 1 represented the typical MADS domains, and motif 6 was the K-box domain that plays a crucial role in protein-protein interaction (Yang and Jack 2004; Kramer et al. 1998). The types Mα, Mβ, Mγ, and Mδ all lacked the K-box domain; however, most of the MADS-MIKC type genes had the complete K-box domain (Figure 2).
Figure 2.
The motif analysis of LcMADS-box genes in L. cubeba. Ten motifs were identified, which were filled with different colors. The ABC model genes were labeled. E means the E class genes colored with purple, C/D means the C and D class MADS-box genes colored with red, A means the A class genes colored with purple, B means the B class genes colored with red, SOC1 means the SOC1 clades MADS-box genes colored with purple, SVP means the SVP clades MADS-box genes colored with red.
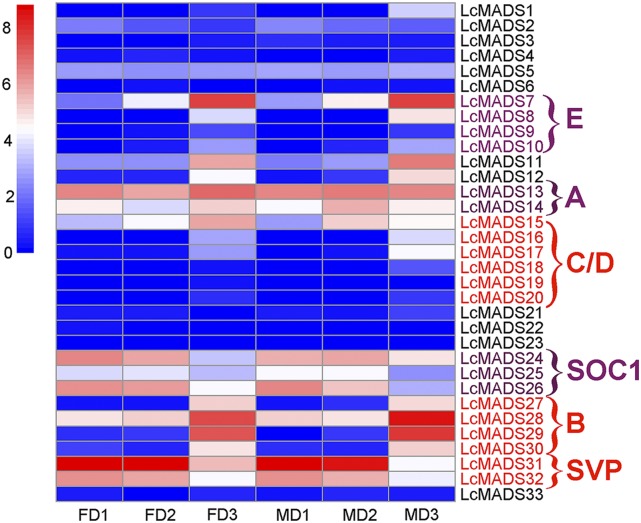
Analysis of LcMADS-box genes in flower bud development stages
Based on the expression patterns in the flower bud developmental stages of L. cubeba (Table S4), the A class genes, SOC1 and SVP clade genes all had high expression levels in all three flower bud differentiation stages. However, the expressions of SOC1 and SVP clade genes were relatively highly expressed in the first two stages, indicating that those genes might play a role in the differentiation of inflorescence and flower primordium. Furthermore, the B clade genes, including LcMADS27, LcMADS28, LcMADS29 and LcMADS30, all of them were highly expressed in the third stages, suggesting that those genes play an important role in flower organ differentiation. The majority of C/D clades genes were constantly lowly expressed in L. cubeba flower buds during the different developmental stages. LcMADS7, a member of SEP clade genes, had a complete M and K box domains and was expressed consistently with the floral bud differentiation period. LcMADS8 highly expressed in the third stage, but lower than LcMADS7 (Figure 3).
Figure 3.
Gene expression of LcMADS-box genes in different stages of flower bud development. The y-axis labels shows the LcMADS-box genes, while the x-axis, including FD1-3 and MD1-3 shows the three stages of female and male flower bud development, respectively. Red means high expression; blue means low expression. The ABC model genes were labeled. E means the E class genes colored with purple, C/D means the C and D class MADS-box genes colored with red, A means the A class genes colored with purple, B means the B class genes colored with red, SOC1 means the SOC1 clade MADS-box genes colored with purple, SVP means the SVP clade MADS-box genes colored with red. The expression data normalized with log2(FPKM+1).
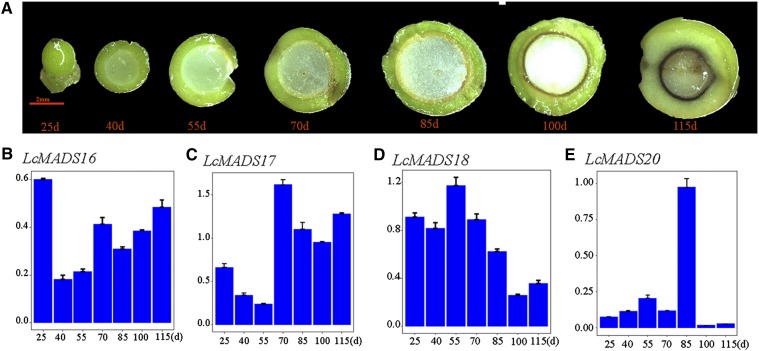
Expression of the C/D class genes in the fruit developmental stages
The C class gene AGAMOUS is characterized as a carpel identity gene, and plays a redundant role in controlling ovule identity with other D class MADS-box transcription factors STK, SHP1, and SHP2 (Coen and Meyerowitz 1991; Mizukami and Ma 1992; Losa et al. 2010). The expression of partial C/D class genes was detected in the developing stages of L. cubeba fruits (Figure 4A) using specific primers (Table S5) with the reference gene of ubiquitin-conjugating enzyme (UBC) (Lin et al. 2013). The expression of LcMADS16 was highest in the early fruit development stage (Figure 4B), whereas LcMADS17 was highly expressed in the middle and later fruit development stages (Figure 4C). The high expression of LcMADS18 was also likely in the early stages of fruit development (Figure 4D). However, LcMADS20 was specifically expressed in the middle stage of fruit development, particularly at 85 days after full bloom (Figure 4E) indicating LcMADS20 play an important role in special fruit development stages. Therefore, we choose LcMADS20 for further functional investigation.
Figure 4.
Expression of C/Dclass genes in seven stages of fruit development. (A) The fruit samples used for the RT-qPCR. (B-E) The relative expression ofLcMADS16, LcMADS17, LcMADS18, andLcMADS20, respectively. The y-axes show the relative gene expression, and the x-axes show the DAF of fruit samples were harvested.
Localization of LcMADS20
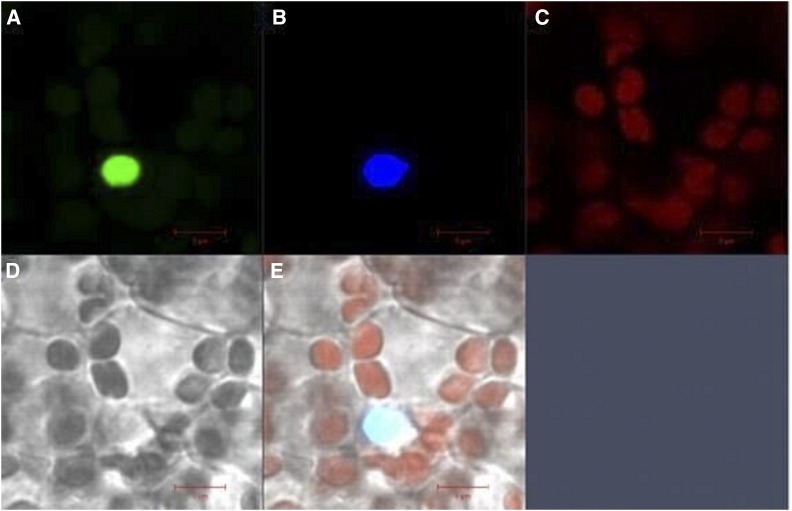
Plant transcription factors consist of a DNA-binding region, a transcription regulation domain and a nuclear localization signal (Liu et al. 1999). LcMADS20, an AG-like gene, encoded for a 223 amino acid protein with complete M and K domains, and was highly expressed in the fruit developmental stages, especially at 85 days after fertilization. The result suggested that this gene might play an important role in the essential oil production. Therefore, the 35S::LcMADS20-GFP vector was constructed to analyze the potential functions. The experiment of subcellular localization showed that the MADS-box transcription factor, LcMADS20, was located in the nucleus (Figure 5).
Figure 5.
Subcellular localization of 35S::LcMADS20-GFP in L. cubeba leaves.(A)The green signal is the GFP fluorescence. (B)The blue fluorescence is the nuclear localization dye of DAPI. (C) Red is the chloroplast fluorescence. (D) Bright field. (E) The light blue is the merged fluorescence.
Ectopic expression of LcMADS20
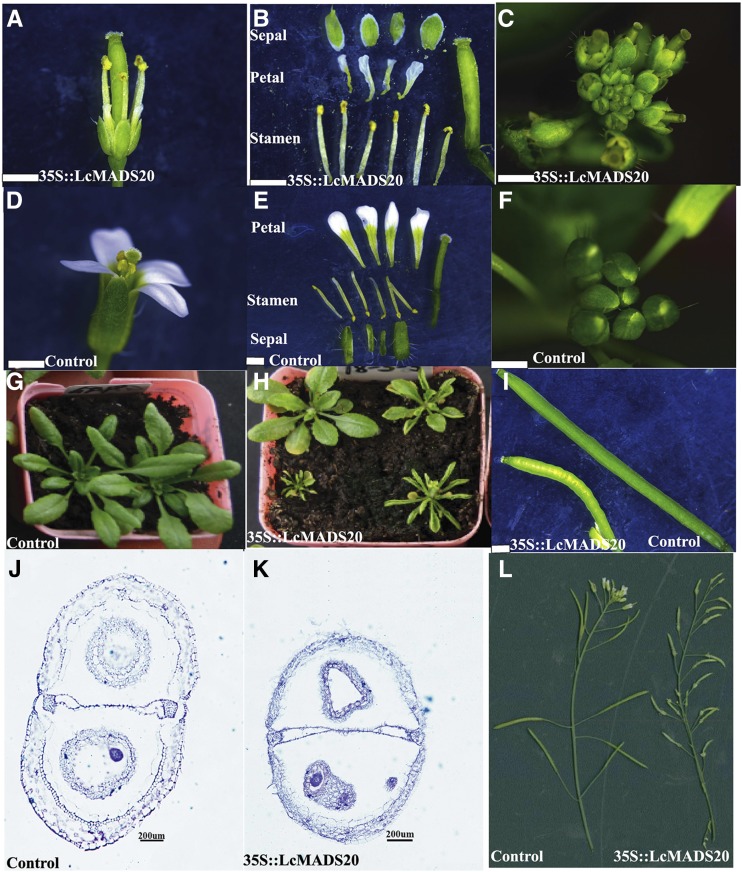
The combined 35S::LcMADS20-GFP vectors were transformed into Arabidopsis, and five positive transgenic Arabidopsis lines were acquired (Figure S2). The morphology changes appeared in T2 and T3 generation compared with the control at the same culture conditions. The severe phenotypes of the over expressed LcMADS20 Arabidopsis showed smaller plants, curly leaves and early flowering (Figure 6A-6C, 6H) compared with control (Figure 6D-6G). The curly leaves was similar to the phenotype of both 35S::AP3 and 35S::PI over expressed Arabidopsis (Krizek and Meyerowitz 1996). The ectopic expression of LcMADS20 showed abnormal flowers (Figure 6A) with shorter sepals and narrow petals (Figure 6B), and early opened inflorescence (Figure 6C), which were coincide with the phenotypes of the overexpression of AG and SHP in Arabidopsis (Mizukami and Ma 1992; Riechmann and Meyerowitz 1997; Pinyopich et al. 2003). Furthermore, the transgenetic plants showed twisted and shorter siliques compared with control (Figure 6I, 6L) and had similarity with the phenotype of siliques length caused by ful and stk mutant Arabidopsis (Pinyopich et al. 2003; Ripoll et al. 2015). More of interest, the percentage of pericarp thickness in silique was decreased in Arabidopsis overexpressed LcMADS20. In detail, the percentage of pericarp thickness in vertical silique was about 9.52% in transgenic Arabidopsis comparing with 21.05% in control, and the percentage of pericarp thickness in lateral silique was about 12.50% in transgenic Arabidopsis comparing with 37.50% in control (Figure 6J, K).
Figure 6.
The phenotypes of the 35S::LcMADS20-GFP overexpression Arabidopsis and Arabidopsis over expressed empty vector. (A) The flower in transgenic Arabidopsis. (B) The flower organs in transgenic Arabidopsis. (C) Flower inflorescence in transgenic Arabidopsis. (D) A single flower in control. (E) Flower organs in control. (F) Normal flower inflorescence in control. (G) Seedlings of control. (H) The seedlings of transgenic Arabidopsis. (I) The twisted silique of transgenic Arabidopsis and normal siliques from control. Bar = 1 mm. The control was Arabidopsis overexpressed the empty vector. (J) Paraffin section of siliques in control Arabidopsis. (K) Paraffin section of siliques in transgenic Arabidopsis. (L) The inflorescence from normal and transgenic Arabidopsis.
The expression of ABC model genes in transgenic Arabidopsis
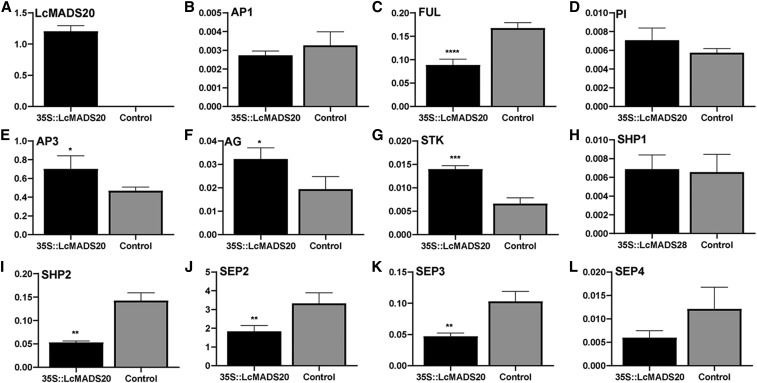
Overexpression of LcMADS20 in Arabidopsis showed severe phenotypes. However, the plants from the same line showed phenotype changing from slight to severe, even in the T3 generation indicating the expression of flower organ identity genes were affected. In order to verify this assumption, we detected the expression of endogenous ABC model genes using the siliques tissues with the reference gene of actin (Helliwell et al. 2006) in LcMADS20 over expressed Arabidopsis and control Arabidopsis both from the T3 generation (Figure 7A-L), which cultivated at same culture conditions. The results showed the exogenous gene LcMADS20 was especially highly expressed in the transgenic Arabidopsis (Figure 7A). The expression of FUL, SHP2, SEP2, and SEP3 was significantly decreased in the transgenic Arabidopsis (Figure 7C, I-K), while the expression of B class gene AP3, and C/D class genes AG and STK, were significantly increased (Figure 7E-G). The expression of AP1, PI, SHP1, and SEP4 were shown no obvious change in the Arabidopsis overexpressed exogenous LcMADS20 (Figure 7B, D, H, L). And all the primers used in this paper were shown on the supplementary table (Table S5).
Figure 7.
The expression of ABC model genes in siliques tissues in transgenic and control Arabidopsis. The y-axes show the gene relative expression level, the x-axes show the siliques materials from LcMADS20 overexpressed Arabidopsis and control Arabidopsis (The data statistic was analyzed by one-way analysis of variance, ANOVA).
Discussion
LcMADS7 was more important in flower bud development stages
The SEP3 clade genes in banana play important roles in peel and pulp development during normal fruit ripening (Elitzur et al. 2010). In strawberry, as non-climacteric fruits, the SEP-like gene FaMADS9 plays an important role in fruit development (Seymour et al. 2011). The SEP3-like gene LMADS3 in lily (Lilium longiflorum, a monocot species) ectopically expressed in Arabidopsis caused early flowering, reducing plant size, and the loss of floral determinacy, whereas another SEP3-like gene, LMADS4, does not cause any change in phenotype (Tzeng et al. 2003). In L. cubeba, the SEP clades gene LcMADS7 was highly expressed in the third stages (the flower organ begins to differentiate), suggestingLcMADS7 may play an important role in flower bud differentiation. Overall, SEP clades may be involved in flower bud differentiation and fruit development.
LcMADS20 was involved in the flowers and siliques development
Over expression of LcMADS20 in Arabidopsis caused early opened inflorescence, abnormal flowers, short sepals and narrow petals. It has been reported that the overexpression of AG and SHP can also cause early flowering, curly leaves and prematurely open flower bud (Mizukami and Ma 1992; Pinyopich et al. 2003). The results indicated LcMADS20 might have similar function with the AG and SHP genes. Correspondingly, the expression of AG was significantly increased in the LcMADS20 transgenic Arabidopsis. The phenotype of transgenic Arabidopsis showed smaller sepal and petal compare to the control. The sepal and petal identities are generally regulated by MADS-box A class gene (AP1) and B class genes (AP3, PI) in Arabidopsis (Bowman et al. 1989, Bowman et al. 1993; Jack et al. 1994; Riechmann and Meyerowitz 1997). However, the expression of the intrinsic AP1 and PI had no obvious change in Arabidopsis over expressed LcMADS20, only AP3 was increased compared with control. The phenotype of the short and twisted siliques caused by the overexpression of LcMADS20 was similar with the silique phenotype of the ful mutant (Gu et al. 1998; Ferrandiz et al. 2000), while FUL can activate the miR172 to activate valve growth through the MADS/ARF-miR172-AP2 regulatory module (Ripoll et al. 2015). The expression of FUL was found decreased in Arabidopsis overexpressed LcMADS20, indicating overexpression of LcMADS20 would inhibit the expression of FUL.
Overall, the regulation mechanism involved in pleiotropic phenotypes caused by overexpression of LcMADS20 may be complicated. The floral architecture seems to be not exclusively regulated by single gene or one certain MADS-box class genes. Each class of MADS-box genes is involved in the regulation of flower and fruit development through formation of homo- or hetero-dimer, or other kinds of complexes (Pelaz et al. 2000; Smaczniak et al. 2012). For example, the AP3/PIheterodimer can interact with SEP clades genes to function floral organ identity (Pelaz et al. 2000), the MADS-box products can form complexes to control floral architecture (Davies et al. 1996; Egea-Cortines et al. 1999). Moreover, the function of LcMADS20 should be further investigate in L. cubeba in the future. Based on the result, we considered LcMADS20 is a negative regulator involved in the fruits and peel development. The inhibition of LcMADS20 and its regulator will be consider in transgenic L. cubeba.
Conclusion
L. cubeba belongs to the Lauraceae family and is a dioecious shrub or small tree. The oil from L. cubeba is an aromatic essential oil extracted from the fresh fruits, especially from the peel. It is important to research the MADS-box genes, especially the carpel identity gene, the AGAMOUS-like genes. Thirty-three LcMADS-box genes were acquired in L. cubeba, and phylogenetic, motif, and expression pattern analyses were conducted. The A class genes, SVP clade, and SOC1 clade genes were all highly expressed in all the three stages of flower bud differentiation. LcMADS20 was highly expressed, particularly at 85 days after flowering, which indicated that LcMADS20 was important in fruit development. At the subcellular level, LcMADS20 was located in the nucleus. LcMADS20, a C class functional gene, can cause curved leaves, early flowering, early opened flower bud, and an abnormal silique morphology in Arabidopsis. In LcMADS20 transgenic Arabidopsis, the expression of intrinsic ABC model class genes was influenced, FUL was significantly reduced in the transgenic Arabidopsis, while AP3, AG, and STK were induced compared with control. The mechanism about how the transcription factor LcMADS20 and other endogenous MADS-box gene are involved in the regulation of flower and siliques development needs further investigation.
Acknowledgments
The work was financially supported by the Science and Technology Major Program on Agricultural New Variety Breeding of Zhejiang, China (no. 2016C02056); “Ten thousand people plan” Science and Technology Innovation Leading Talent of Zhejiang, China (no. 2018R52006); and a National Key R&D Program of China grant (no. 2017YFD0600704) awarded to Y.-C.C.
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.9978776.
Communicating editor: A. Doust
Literature Cited
- Agrawal N., Choudhary A. S., Sharma M. C., and Dobhal M. P., 2011. Chemical constituents of plants from the genus Litsea. Chem. Biodivers. 8: 223–243. 10.1002/cbdv.200900408 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M. C., Grant C. E. et al. , 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., and Meyerowitz E. M., 2012. The ABC model of flower development: then and now. Development 139: 4095–4098. 10.1242/dev.083972 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., and Meyerowitz E. M., 1989. Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52. 10.1105/tpc.1.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Alvarez J. N., Weigel D., Meyerowitz E. M., and Smyth D. R., 1993. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743. [Google Scholar]
- Brambilla V., Battaglia R., Colombo M., Masiero S., Bencivenga S. et al. , 2007. Genetic and molecular interactions between BELL1 and MADS Box factors support ovule development in Arabidopsis. Plant Cell 19: 2544–2556. 10.1105/tpc.107.051797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., and Bent A. F., 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Causier B., Schwarzsommer Z., and Davies B., 2010. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21: 73–79. 10.1016/j.semcdb.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang Y., Han X., Si L., Wu Q. et al. , 2013. Biology and chemistry of Litsea cubeba, a promising industrial tree in China. J. Essent. Oil Res. 25: 103–111. 10.1080/10412905.2012.751559 [DOI] [Google Scholar]
- Coen E., and Meyerowitz E. M., 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. 10.1038/353031a0 [DOI] [PubMed] [Google Scholar]
- Davies B., Egeacortines M., Silva E. D., Saedler H., and Sommer H., 1996. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15: 4330–4343. 10.1002/j.1460-2075.1996.tb00807.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G. S., Pinyopich A., Robles P., Pelaz S., and Yanofsky M. F., 2004. The SEP4 gene of Arabidopsisthaliana functions in floral organ and meristem identity. Curr. Biol. 14: 1935–1940. 10.1016/j.cub.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Eddy S. R., 1998. Profile hidden Markov models. Bioinformatics 14: 755–763. 10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., and Sommer H., 1999. Ternary complex formation between the MADS‐box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379. 10.1093/emboj/18.19.5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitzur T., Vrebalov J., Giovannoni J. J., Goldschmidt E. E., and Friedman H., 2010. The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J. Exp. Bot. 61: 1523–1535. 10.1093/jxb/erq017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R., Pinyopich A., Battaglia R., Kooiker M., Borghi L. et al. , 2003. MADS-Box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611. 10.1105/tpc.015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C., Liljegren S. J., and Yanofsky M. F., 2000. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289: 436–438. 10.1126/science.289.5478.436 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J. et al. , 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44: D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q., Ferrandiz C., Yanofsky M. F., and Martienssen R. A., 1998. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517. [DOI] [PubMed] [Google Scholar]
- He W., Chen Y., Gao M., Zhao Y., Xu Z.. et al, 2018. Transcriptome analysis of Litsea cubeba floral buds reveals the role of hormones and transcription factors in the differentiation process. G3 (Bethesda).8: 1103–1114. 10.1534/g3.117.300481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Wood C. C., Robertson M., Peacock W. J., and Dennis E. S., 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46: 183–192. 10.1111/j.1365-313X.2006.02686.x [DOI] [PubMed] [Google Scholar]
- Huala E., Dickerman A. W., Garciahernandez M., Weems D., and Reiser L. et al. , 2001. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29: 102–105. 10.1093/nar/29.1.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T., Fox G., and Meyerowitz E. M., 1994. Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716. 10.1016/0092-8674(94)90509-6 [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H., Fraser M., Li W. et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Akhtar Y., Bradbury R., Zhang X., and Isman M. B., 2009. Comparative toxicity of essential oils of litsea pungens and litsea cubeba and blends of their major constituents against the cabbage looper, trichoplusia ni. J. Agric. Food Chem. 57: 4833–4837. 10.1021/jf900274r [DOI] [PubMed] [Google Scholar]
- Kramer E. M., Dorit R. L., and Irish V. F., 1998. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149: 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A., and Meyerowitz E. M., 1996. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22. [DOI] [PubMed] [Google Scholar]
- Kyozuka J., and Shimamoto K., 2002. Ectopic expression of OsMADS3, a rice ortholog of AGAMOUS, caused a homeotic transformation of lodicules to stamens in transgenic rice plants. Plant Cell Physiol. 43: 130–135. 10.1093/pcp/pcf010 [DOI] [PubMed] [Google Scholar]
- Letunic I., and Bork P., 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46: D493–D496. 10.1093/nar/gkx922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Han X., Chen Y., Wu Q., and Wang Y., 2013. Identification of appropriate reference genes for normalizing transcript expression by quantitative real-time PCR in Litsea cubeba. Mol. Genet. Genomics 288: 727–737. 10.1007/s00438-013-0785-1 [DOI] [PubMed] [Google Scholar]
- Liu L., White M. J., and Macrae T. H., 1999. Transcription factors and their genes in higher plants Functional domains, evolution and regulation. FEBS J. 262: 247–257. 10.1046/j.1432-1327.1999.00349.x [DOI] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lee J. E., and Lee I., 2010. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61: 2247–2254. 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Yoo S. J., Park S., Hwang I., Lee J. S. et al. , 2007. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402. 10.1101/gad.1518407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losa A., Colombo M., Brambilla V., and Colombo L., 2010. Genetic interaction between AINTEGUMENTA (ANT) and the ovule identity genes SEEDSTICK (STK), SHATTERPROOF1 (SHP1) and SHATTERPROOF2 (SHP2). Sex. Plant Reprod. 23: 115–121. 10.1007/s00497-009-0130-3 [DOI] [PubMed] [Google Scholar]
- Mizukami Y., and Ma H., 1992. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131. 10.1016/0092-8674(92)90271-D [DOI] [PubMed] [Google Scholar]
- Pabon-Mora N., Wong G. K., and Ambrose B. A., 2014. Evolution of fruit development genes in flowering plants. Front. Plant Sci. 5: 300 10.3389/fpls.2014.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E., and Yanofsky M. F., 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- Parenicová L., De Folter S., Kieffer M., Horner D. S., Favalli C. et al. , 2003. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551. 10.1105/tpc.011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G. S., Savidge B., Liljegren S. J., Baumann E. et al. , 2003. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., and Meyerowitz E. M., 1997. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 8: 1243–1259. 10.1091/mbc.8.7.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll, J. J., L. J. Bailey, Q. Mai, S. L. Wu, C. T. Hon et al., 2015 microRNA regulation of fruit growth. Nat. Plants 1: 15036. 10.1038/nplants.2015.36 [DOI] [PubMed]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., and Sommer H., 1990. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250: 931–936. 10.1126/science.250.4983.931 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F. et al. , 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour G. B., Ryder C. D., Cevik V., Hammond J. P., Popovich A., et al. , 2011. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassaDuch.) fruit, a non-climacteric tissue. J. Exp. Bot. 62: 1179–1188. 10.1093/jxb/erq360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., and Ho C., 2016. Essential oil compositions and antimicrobial activities of various parts of Litsea cubeba from Taiwan. Nat. Prod. Commun. 11: 515–518. 10.1177/1934578x1601100425 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., Immink R. G., Muino J. M., Blanvillain R., Busscher M. et al. , 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565. 10.1073/pnas.1112871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D. S., Filipski A., and Kumar S., 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., and Saedler H., 2001. Plant biology: Floral quartets. Nature 409: 469–471. 10.1038/35054172 [DOI] [PubMed] [Google Scholar]
- Tzeng T., Hsiao C., Chi P., and Yang C., 2003. Two Lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol. 133: 1091–1101. 10.1104/pp.103.026997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiang Z., and Li R., 2012. Antioxidant activity, free radical scavenging potential and chemical composition of Litsea cubeba essential oil. J. Essent. Oil Bear. Pl. 15: 134–143. 10.1080/0972060X.2012.10644029 [DOI] [Google Scholar]
- Yang Y., and Jack T., 2004. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 55: 45–59. 10.1007/s11103-004-0416-7 [DOI] [PubMed] [Google Scholar]
- Ye C., 2012. Botany Experimental Guidance, Tsinghua University Press, Beijing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MADS-box genes of L. cubeba were analyzed according to data from our previous published paper (He et al. 2018). The information of the MADS-box genes of L. cubeba was list on the Table S1. The sequences of MADS-box genes of Arabidopsis were downloaded from the database of The Arabidopsis Information Resource (https://www.arabidopsis.org/) with the accession number listed on the Table S2. Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.9978776.