Abstract
Drosophila sechellia is a dietary specialist fruit fly that evolved from a generalist ancestor to specialize on the toxic fruit of Morinda citrifolia. This species pair has been the subject of numerous studies where the goal has largely been to determine the genetic basis of adaptations associated with host specialization. Because one of the most striking features of M. citrifolia fruit is the production of toxic volatile compounds that kill insects, most genomic studies in D. sechellia to date have focused on gene expression responses to the toxic compounds in its food. In this study, we aim to identify new genes important for host specialization by profiling gene expression response to 3,4-dihydroxyphenylalanine (L-DOPA). Recent work found it to be highly abundant in M. citrifolia, critical for reproductive success of D. sechellia, and supplementation of diet with the downstream pathway product dopamine can influence toxin resistance phenotypes in related species. Here we used a combination of functional genetics and genomics techniques to identify new genes that are important for D. sechellia ecological adaptation to this new niche. We show that L-DOPA exposure can affect toxin resistance phenotypes, identify genes with plastic responses to L-DOPA exposure, and functionally test an identified candidate gene. We found that knock-down of Esterase 6 (Est6) in a heterologous species alters toxin resistance suggesting Est6 may play an important role in D. sechellia host specialization.
Keywords: L-DOPA, oogenesis, host specialization, adaptation, RNA-seq, esterase
In insects, the complex nature of plant-insect interactions, multi-trophic interactions involving predator and prey dynamics, and indirect effects between and among them contribute to adaptations. Most plant-feeding insects are dietary specialists that feed on a small number of closely-related plant species (Price et al. 1980; Jaenike 1990; Bernays and Chapman 1994). Often, this specialization is the result of host-specific adaptations due to variation in plant chemistry. However, determining the specific genetic changes that accompany specialization or host shifts remains challenging. Drosophila sechellia, a dietary specialist fruit fly endemic to the Seychelles Islands (Tsacas and Bachli 1981), is a well-suited model system for examining questions related to adaptive host specialization. D. sechellia has evolved to specialize its feeding, metabolism, oviposition, and development on the toxic fruit of Morinda citrifolia (Louis and David 1986; Matute and Ayroles 2014). Its close phylogenetic relationship with the model organism D. melanogaster (divergence time ∼3 MYA, Clark et al. 2007) and sister species D. simulans (divergence time <100 KYA, Schrider et al. 2018), both dietary generalists, gives D. sechellia exceptional power for dissecting the underlying genetics involved in host specialization and evolved resistance to plant defense compounds. Toxicity of M. citrifolia fruit is predominantly due to high levels of the medium chain fatty acid octanoic acid (OA) which D. sechellia has evolved resistance to and preference for (Legal et al. 1992, 1994; Farine et al. 1996). Many studies have investigated the genetic basis of toxin resistance in D. sechellia (R’Kha et al. 1991; Amlou et al. 1998a,b; Jones 1998, 2001; Hungate et al. 2013; Huang & Erezyilmaz 2015; Andrade López et al. 2017; Lanno et al. 2017; Peyser et al. 2017; Lanno et al. 2019), where most focus solely on the highly abundant and toxic compound OA. However, it is possible that an interaction or synergistic effect of other compounds found in the fruit contributes to both lethality and resistance.
In addition to being toxic to other insects, M. citrifolia fruit also contains high levels of 3,4-dihydroxyphenylalanine (L-DOPA), a chemical precursor of dopamine. Tyrosine Hydroxylase (TH), encoded by the pale (ple) locus, mediates the conversion of tyrosine to L-DOPA (Nagatsu et al. 1964; Budnik and White 1987; Neckameyer and White 1993). Homozygous null alleles of this gene as well as pharmacological inhibition of TH result in embryonic lethality (Neckameyer and White 1993; Neckameyer 1996; Pendleton et al. 1996). Alongside TH, a group of genes surrounding the Dopa Decarboxylase (Ddc) locus, which catalyzes the decarboxylation of L-DOPA to dopamine, also play an important role in catecholamine metabolism (Wright 1987; Stathakis et al. 1995). Of particular interest is Catecholamines up (Catsup), a gene within this group that encodes a protein that regulates TH activity that was discovered to contain loss of function mutations in D. sechellia, driving this species to become reliant on its obligate host, M. citrifolia (Stathakis et al. 1999; Lavista-Llanos et al. 2014). In contrast to other Drosophila species, D. sechellia has much lower levels of cellular L-DOPA while still maintaining high levels of dopamine. This is achieved by consuming M. citrifolia that produces large amounts of L-DOPA in its fruit. L-DOPA plays several roles in plants, including inhibiting growth of competing plant species, as well as acting as a secondary defense compound in some cases (Soares et al. 2014). Lavista-Llanos et al. (2014) found that supplementing food with dopamine increased D. melanogaster resistance to M. citrifolia fruit toxins (Lavista-Llanos et al. 2014). However, whether the high levels of L-DOPA naturally co-occurring in Morinda fruit (and not dopamine) contributes to OA resistance remains unknown.
Here we functionally test the role of L-DOPA in OA resistance by performing mortality assays in the presence of L-DOPA, perform RNA-sequencing on control vs. L-DOPA exposed flies to identify candidate genes that may be involved in L-DOPA mediated toxin resistance, and functionally test an identified candidate for effects on OA resistance.
Materials and Methods
Fly strains and culture
Drosophila sechellia (14021-0428.25), D. simulans (14021-0251.195), and D. melanogaster (14021-0231.36, BDSC:55927 (EST6 RNAi), GeneSwitch-GAL4 line (Tubulin-P[Switch]) flies were reared on cornmeal medium using a 16:8 light:dark cycle at 20°. Adult females of each species were collected at 0-3 days post-eclosion and exposed to either control food (0.75g Drosophila instant medium Equation 4-24, Carolina Biological Supply Company) or food containing L-DOPA (10mg/ml, concentration chosen from observations of dopamine supplementation described in Lavista-Llanos et al. 2014) for 24 hr. After exposure, flies were either used in mortality assays or for measurement of genome-wide gene expression.
Mortality assay
The mortality assays used in this work were performed according to methods described in prior studies (Andrade López et al. 2017; Peyser et al. 2017; Lanno & Coolon 2019; Lanno et al. 2019). Briefly, control and L-DOPA exposed flies of all three species were transferred into vials (10 per vial) containing 0.75g Drosophila medium supplemented with 1.2% octanoic acid (OA, Sigma) (Andrade López et al. 2017; Peyser et al. 2017; Lanno & Coolon 2019; Lanno et al. 2019). Flies used for mortality assays were all 1-4 day old females for each species and each sample type was collected in replicate six-eight times with 10 flies per replicate (N = 60 to 80 per sample type depending on the experiment, see below). OA resistance was measured by determining the number of flies ‘knocked down’ (a fly was determined to be knocked down when it was no longer able to walk or fly) every 5 min for a period of 60 min.
RNA interference
RNA interference (RNAi) was performed to knockdown the expression of Esterase 6 by first crossing the UAS-Est6-RNAi line that expresses a hairpin RNA under the control of UAS to GeneSwitch-GAL4 which expresses a modified chimeric GAL4 ubiquitously that will only become active in the presence of the synthetic antiprogestin mifepristone (RU486). Therefore, RNAi targeting Est6 will only occur in individuals that have both UAS-Est6-RNAi and GeneSwitch-GAL4 in the presence of RU486 (Osterwalder et al. 2001; Roman et al. 2001; Andrade López et al. 2017; Lanno et al. 2019). Adult female progeny from this cross were collected at 0-3 days post-eclosion and treated with either 10µg/ml RU486 (knockdown) or 10µl/ml EtOH (control) added directly to their media for 24 hr. After exposure, flies were used in mortality assays which also contained RU486 to maintain knockdown in those samples.
Cox proportional hazards regression analysis
A Cox proportional hazards statistical model was used to test the effect of L-DOPA exposure on OA associated mortality using the coxph command in the survival package in R (Cox 1972; Fox 2008; Therneau 2015; Andrade López et al. 2017; Peyser et al. 2017; R Core Development Team 2017; Lanno & Coolon 2019; Lanno et al. 2019). We report relative survival as the regression coefficient (-β) for each treatment group compared to its species-specific control group (with vs. without L-DOPA). Sample size for the effect of L-DOPA on OA resistance was N = 60 per treatment. Blocking by vial was included in the model and found to have no effect. In the RNAi experiment where we compare RNAi knockdown to uninduced controls (with vs. without knockdown) we used a separate Cox model to test for the effect of knockdown of Est6 on OA resistance and the sample size was N = 80 for each treatment. Blocking by vial was again included in the model and found to have no effect.
RNA extraction, library preparation and RNA- sequencing
After exposure to the control or L-DOPA food sources, flies of each species were flash frozen in liquid nitrogen and kept at -80° until RNA extraction. The Promega SV total RNA extraction system with modified protocol (Coolon et al. 2012) was used to extract RNA from a homogenate of 10 whole adult female flies per replicate per species per treatment. Three biological replicates were analyzed for each species and exposure environment for a total of 18 sequencing libraries (Table 1). Prior to library preparation, NanoDrop and subsequent gel electrophoresis were used to determine the quantity and quality of RNA extracted. All RNA samples were sent to the University of Michigan Medical School DNA Sequencing Core Facility for mRNA selected library preparation and sequencing. Bar-coded sequencing libraries were made using TruSeq library preparation kits and pooled for sequencing. Uniform library representation of each library was confirmed with qPCR prior to sequencing. The pooled barcoded libraries were sequenced on two lanes of an Illumina HiSeq-4000 generating single end sequence reads for subsequent analyses.
Table 1. Percent mapped reads for sequencing libraries.
| Sample | # Reads | # Mapped Reads | % Mapped |
|---|---|---|---|
| D. sechellia C1 | 19,222,060 | 18,496,450 | 96.23 |
| D. sechellia C2 | 20,704,811 | 19,440,620 | 93.89 |
| D. sechellia C3 | 17,696,868 | 17,123,579 | 96.76 |
| D. sechellia LD1 | 19,576,162 | 18,341,777 | 93.69 |
| D. sechellia LD2 | 14,508,205 | 12,988,684 | 89.53 |
| D. sechellia LD3 | 17,432,600 | 16,040,372 | 92.01 |
| D. simulans C1 | 28,056,123 | 26,210,691 | 93.42 |
| D. simulans C2 | 26,058,213 | 24,449,785 | 93.83 |
| D. simulans C3 | 24,095,284 | 22,589,715 | 93.75 |
| D. simulans LD1 | 17,841,650 | 16,739,731 | 93.82 |
| D. simulans LD2 | 14,608,378 | 13,562,252 | 92.84 |
| D. simulans LD3 | 17,628,201 | 16,471,452 | 93.44 |
| D. melanogaster C1 | 21,999,530 | 20,633,866 | 93.79 |
| D. melanogaster C2 | 20,950,464 | 19,779,953 | 94.41 |
| D. melanogaster C3 | 22,157,160 | 20,919,514 | 94.41 |
| D. melanogaster LD1 | 39,619,560 | 37,618,491 | 94.95 |
| D. melanogaster LD2 | 23,214,861 | 21,991,775 | 94.73 |
| D. melanogaster LD3 | 18,976,326 | 17,855,382 | 94.09 |
BIOL310 Genomics Analysis
The genomics analysis of RNA-seq data presented in this manuscript was performed by 18 undergraduate and 2 graduate students as part of a semester-long course at Wesleyan University called Genomics Analysis (BIOL310). This is the second such manuscript (see Lanno et al. 2017) made from this course where the aim is to provide undergraduate students with a course-based research experience where they actively participate in the process of scientific discovery. The students learn through engaging with never-before analyzed data where they learn how to use cutting edge genomics analysis techniques and bioinformatics tools through a discovery-based independent study. Each student in the course contributed to the analyses and write-up of the findings, providing their own unique interpretation of the results and text written by each and every student was combined into this manuscript.
After sequence reads were returned by the University of Michigan Sequencing Core (Table 1), an RNA-seq analysis pipeline was performed in the online Galaxy environment (https://usegalaxy.org/, Afgan et al. 2016). Sequencing output files for each sample were quality control checked using FASTQC (Andrews 2010) and identified overrepresented sequences were identified using NCBI BLAST (Altschul et al. 1990). Sequence reads were mapped to the corresponding species genome using Bowtie2 with default parameters (Langmead and Salzberg 2012). The most current genome files at the time of analysis were obtained from Ensembl (Yates et al. 2016) (D. sechellia: Drosophila_sechellia.dsec_caf1.dna.toplevel.fa, D. simulans: Drosophila_simulans.ASM75419v3.dna.toplevel.fa and D. melanogaster: Drosophila_melanogaster.BDGP6.dna.toplevel.fa). Quantification of gene expression and differential expression tests were performed with Cuffdiff (Trapnell et al. 2010, 2013) using the genome files described above and the gene annotation files available from Ensembl at the time of analysis (D. sechellia: Drosophila_sechellia.dsec_caf1.42.gff3, D. simulans: Drosophila_simulans.ASM75419v3.42.gff3 and D. melanogaster: Drosophila_melanogaster.BDGP6.95.gff3). Geometric normalization and gene length correction options in Cuffdiff were used to improve comparisons of gene expression. False discovery rate multiple testing correction (Benjamini and Hochberg 1995) was used to account for the multitude of simultaneously conducted tests. Data visualization and processing was performed in R (R Core Development Team 2017). In order to compare gene expression results across species, we obtained all 1:1:1 orthologs from D. sechellia, D. simulans and D. melanogaster from Flybase (Attrill et al. 2016). We performed Gene Ontology enrichment analysis with the Gene Ontology Consortium online tool (http://geneontology.org/, Ashburner et al. 2000; Blake et al. 2015) using the annotations from the D. melanogaster orthologs.
DNA coding and protein sequence analyses of Est6
DNA coding sequences (CDS) for Est6 were downloaded from FlyBase (Attrill et al. 2016) for D. melanogaster, D. sechellia, and D. simulans. Clustal Omega (Goujon et al. 2010; Sievers et al. 2011; McWilliam et al. 2013) was used to align DNA CDS and translated protein sequences in order to determine synonymous and nonsynonymous differences between these species. To investigate Est6 CDS variation in multiple D. sechellia genotypes, paired-end DNA sequencing files from 23 wild-caught D. sechellia genomes from the Seychelles islands were downloaded from NCBI’s Short Read Archive (BioProject number PRJNA395473) (Schrider et al. 2018). Each file was mapped to a fasta file containing the D. sechellia Est6 DNA CDS using Bowtie 2 (Langmead and Salzberg 2012). Aligned reads were then assessed for variation among D. sechellia lines for the Est6 allele using the Naïve Variant Caller (Blankenberg et al. 2014) in Galaxy using the D. sechellia Est6 DNA CDS downloaded from FlyBase as a reference (Attrill et al. 2016).
Data accessibility
All RNA-seq data generated in this manuscript have been submitted to the NCBI Gene Expression Omnibus under accession number GSE138119. Supplemental material available at figshare: https://doi.org/10.25387/g3.8938103.
Results
Testing species-specific L-DOPA mediated resistance to octanoic acid
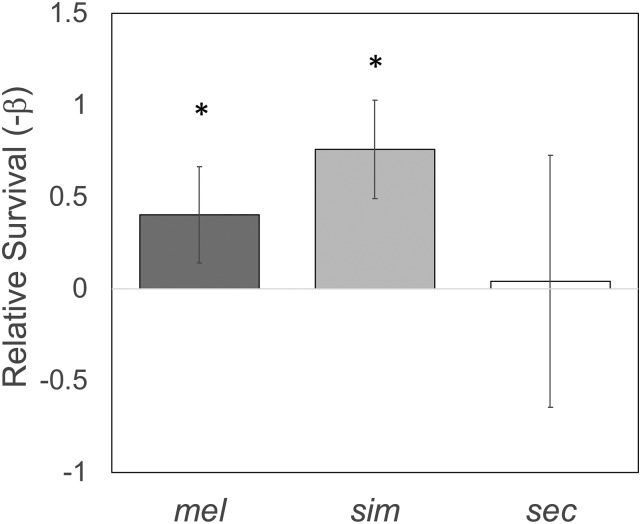
Prior work showed that D. melanogaster strains had increased resistance to a combination of octanoic and hexanoic acids when concurrently given dopamine in their media (Lavista-Llanos et al. 2014). However, M. citrifolia produces high levels of L-DOPA and not dopamine and the consequences of L-DOPA exposure on toxin resistance remain unknown. Furthermore, because D. sechellia was also not included in this test, it is unknown how D. sechellia responds to L-DOPA in their diet and possible effects on toxin resistance. To test for L-DOPA mediated toxin resistance, we performed octanoic acid (OA) resistance assays with and without L-DOPA supplementation using three closely-related species: D. sechellia, D. simulans and D. melanogaster. Flies were fed media containing 10mg/ml L-DOPA for 24 hr prior to measurement of OA resistance. We found that L-DOPA supplementation significantly increased both D. melanogaster (Cox Proportional Hazards Test, -β =1.06, P = 3.1 × 10−7) and D. simulans (Cox Proportional Hazards Test, -β =0.99, P = 3.1 × 10−7) resistance to OA, but there was no effect on D. sechellia (Cox Proportional Hazards Test, -β=0.63, P = 0.11) (Figure 1). Because D. sechellia is so highly resistant to OA (only 2/120 died in the experiment in total), our test using a concentration of OA consistent with the maximum biologically available OA in M. citrifolia fruit, representing a reasonable natural condition is not capable of identifying an increase (not statistically possible) in OA resistance for this species in response to L-DOPA. Therefore, it remains unknown whether L-DOPA influences OA resistance in D. sechellia.
Figure 1.
L-DOPA effect on OA resistance. After 24 hr of exposure to 10mg/ml L-DOPA, D. melanogaster, D. simulans and D. sechellia adult flies were tested for changes in OA resistance. Plotted are relative survival (-β) estimates comparing each species OA resistance with and without exposure to L-DOPA. Askterisk indicates significant effect of L-DOPA on OA resistance in that species (P < 0.05).
Investigating Drosophila gene expression responses to dietary L-DOPA
We next sought to identify gene expression responses of all three species to L-DOPA exposure with the goal of identifying candidate genes that may play a role in host specialization. Prior work has shown that genes whose expression is responsive to environmental conditions are important for fitness in those environments (Coolon et al. 2009; Lanno et al. 2017). Furthermore, Lavista-Llanos found that not only is toxin resistance altered by exposure to dopamine, but various aspects of egg production as well (Lavista-Llanos et al. 2014). In order to identify candidate genes that may play a role in OA resistance and egg production in the presence of L-DOPA exposure, we used RNA-seq to measure genome-wide gene expression in adult female D. melanogaster, D. simulans and D. sechellia with and without L-DOPA supplemented in their media (Figure 2).
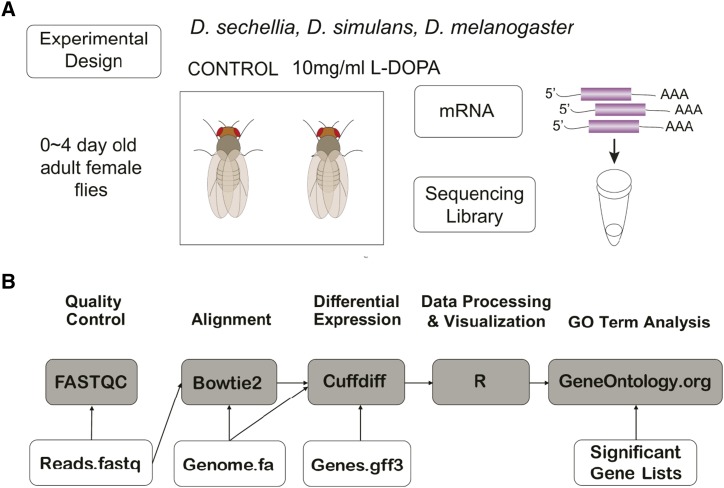
Figure 2.
RNA-seq experimental design. A) Adult female flies were fed either control food or food mixed with 10 mg/ml L-DOPA for 24 hr followed by RNA extraction, Illumina library preparation and sequencing on an Illumina Hiseq 4000. B) Data generated by the Illumina sequencing were analyzed using the bioinformatics pipeline implemented on the Galaxy platform (usegalaxy.org). The process included quality control with FASTQC, alignment with Bowtie2, normalization and differential expression testing with Cuffdiff, processing and visualization in R, and gene ontology (GO) term enrichment tests performed at GeneOntology.org (Ashburner et al. 2000; Blake et al. 2015).
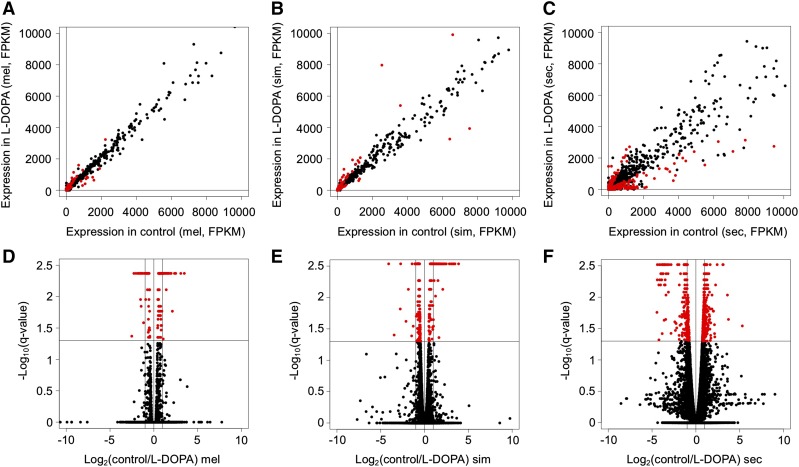
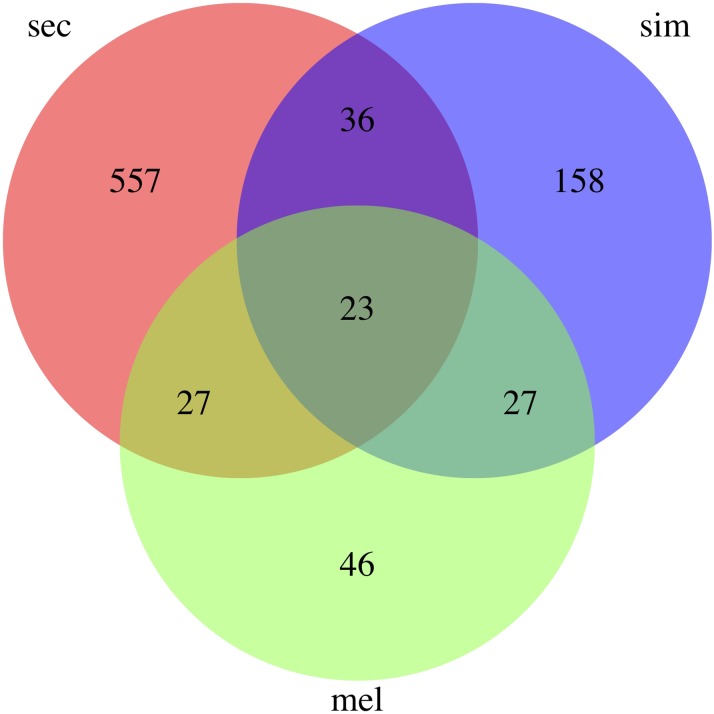
RNA-seq libraries were sequenced yielding a total of 384,346,456 sequence reads for the project and an average of 21 million per library (Table 1). FASTQC analysis (Andrews 2010) of the reads showed that they were high-quality and no trimming or sequence filtering was necessary prior to downstream analysis. We aligned the sequence reads to the corresponding genome with Bowtie2 (Langmead and Salzberg 2012) (see methods) and 94% of sequences on average aligned uniquely per library to the corresponding genome (Table 1). These alignments were then used for quantification of gene expression and differential expression testing with Cuffdiff (Trapnell et al. 2010, 2013). We found that 123 genes were significantly differentially expressed by D. melanogaster (Figure 3A,D, Supplemental Table 1), 244 by D. simulans (Figure 3B,E, Supplemental Table 2) and 643 by D. sechellia (Figure 3C,F, Supplemental Table 3) in response to dietary L-DOPA. Interestingly, the largest number was differentially expressed by D. sechellia, the only species that routinely has high concentrations of L-DOPA in its natural food source. There were 557 genes that were only plastic in response to L-DOPA by D. sechellia and not identified as differentially expressed by D. melanogaster or D. simulans (Figure 4, Supplemental Table 4), representing an interesting set of genes for further analysis because these genes may mediate L-DOPA associated phenotypic plasticity (e.g., increased egg production, Lavista-Llanos et al. 2014). Finally, there were 23 genes differentially expressed by all three species that are interesting for their conserved gene expression response across species (Supplemental Table 5).
Figure 3.
Genome-wide gene expression responses to L-DOPA. A-C) Scatterplots are shown for the comparison between flies fed control food (X-axis) and those fed food containing 10µg/ml L-DOPA (y-axis) with gene expression represented as F ragments P er K ilobase of sequence per M illion reads (FPKM) where each point is one gene with A) D. melanogaster, B) D. simulans and C) D. sechellia. D-F) Volcano plots are shown for the same comparison (control vs. 10µg/ml L-DOPA) for each species D) D. melanogaster, E) D. simulans and F) D. sechellia where expression response represented as Log(control FPKM/L-DOPA FPKM) on the x-axis and the significance of a statistical test for that gene represented as -Log10(q-value) where q is the false discovery rate corrected p-value on the y-axis.
Figure 4.
Venn diagram of L-DOPA responsive genes. The intersection of the genes identified by each of the three RNA-seq comparisons of flies fed control food vs. food supplemented with 10mg/ml L-DOPA are shown.
Gene ontology term enrichment analysis
In order to determine if there is enrichment for genes with particular functions among those differentially expressed in response to L-DOPA, we tested the sets of significantly upregulated and downregulated genes identified from each species for gene ontology (GO) term enrichment. We found that there were very few enriched terms for upregulated and downregulated genes from D. melanogaster or D. simulans (Supplemental Tables 6-9). There were no enriched GO terms for upregulated genes from D. melanogaster or D. simulans. There were only two enriched GO terms for in downregulated genes in D. melanogaster, chorion (GO:0042600) and external encapsulating structure (GO:0030312) (Supplemental Table 6). There were multiple enriched terms for D. simulans downregulated genes associated with cellular parts that were non informative (Supplemental Table 7) and there was enrichment for phosphoric ester hydrolase activity (GO:0042578). Finally, there were numerous functions enriched in both the up and down-regulated genes in D. sechellia with far more terms enriched in the upregulated gene set (Supplemental Table 8). The enriched functions from downregulated genes are predominantly associated with various classes of genes with peptidase activity (Supplemental Table 8). The numerous terms enriched for upregulated genes in D. sechellia include multiple terms associated with increased reproductive output including mitosis/meiosis, cell cycle, DNA replication, gamete generation (Supplemental Table 8). We next investigated GO term enrichment in the set of 23 genes found in all 3 comparisons and found that the majority of terms enriched had to do with egg production including aspects of vitelline membrane formation, egg coat formation, chorion and extracellular matrix formation (Supplemental Table 9).
Testing the function of a candidate gene in OA resistance
Many of the genes identified as responding to L-DOPA exposure are excellent candidates for various aspects of host specialization in D. sechellia including possible roles in reproduction (Lavista-Llanos et al. 2014), mate recognition/reproductive isolation (e.g., EloF, Supplemental Table 4, Combs et al. 2018), behavioral changes (Yamamoto and Seto 2014), cuticle formation (Lanno et al. 2017) and toxin resistance (Lavista-Llanos et al. 2014). For example, we found that D. sechellia response to L-DOPA includes marked increase in the expression of Esterase 6 (Est6, Supplemental Table 4). In our recent study, we found that one or more esterase genes are involved in D. sechellia derived resistance to OA through experiments with the chemical synergist tribufos (S,S,S-Tributyltrithiophosphate) that inhibits all the esterase genes simultaneously (Lanno and Coolon 2019). To determine if Est6 plays a role in evolved OA resistance we used RNAi in D. melanogaster to reduce the expression of Est6 and examined the effect on OA resistance. Using the ubiquitously expressed GeneSwitch-GAL4 crossed into UAS-EST6-RNAi line yields individuals that are wildtype unless they are exposed to the synthetic antiprogestin mifepristone (RU486), which induced RNAi knockdown of Est6. We compared genotypically identical siblings with and without RU486 exposure and found that knockdown of Est6 caused a significant decrease in resistance to OA (Cox Proportional Hazards Test, -β = -0.833, P = 1.7 × 10−7, Figure 5). To determine whether altered protein sequence could also contribute to Est6 functional differences in addition to the observed gene expression responses, we compared the protein sequences for Est6 from D. melanogaster, D. simulans and D. sechellia and identified 5 derived amino acids in D. sechellia. One of the derived residues (H187A) affects an amino acid that resides in the active site of the enzyme (Younus et al. 2017) that was previously shown to influence Est6 substrate specificity and enzyme kinetics (the specific amino acid substitution found in D. sechellia was not tested but other residue substitutions at this position had the stated effects (Myers et al. 1993). Using available genomes from several D. sechellia isolates (Schrider et al. 2018) we have confirmed that the mutations responsible for this amino acid substitution are present in all D. sechellia sequenced to date suggesting this a fixed sequence difference in this species.
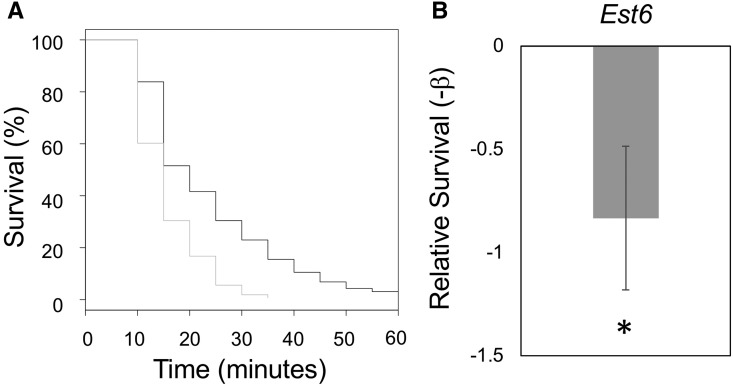
Figure 5.
Knockdown of Est6 causes reduction in OA resistance. After 24 hr of exposure to RU486, D. melanogaster GeneSwitch-GAL4 × UAS-EST6-RNAi adults female flies were tested for changes in OA resistance. Percent survival across time is plotted in A and relative survival (-β) estimates comparing individuals with and without exposure to RU486 and measuring OA resistance are shown in B. * indicates significant Est6 knockdown effect on OA (P < 0.05).
Discussion
Here we found that adult D. melanogaster and D. simulans exposed to L-DOPA have increased resistance to OA. We do not see this effect for D. sechellia because D. sechellia is so highly resistant to OA that our test could not test for increased resistance. In an effort to maintain ecological relevance of the results we have used a concentration of OA that best matches the maximum biologically available OA in M. citrifolia fruit. As such, it remains unknown whether L-DOPA influences OA resistance in D. sechellia. To identify genes that might contribute to host specialization downstream of L-DOPA exposure, we used RNA-seq, to determine the genes with expression plasticity in response to L-DOPA, with a specific interest in those that had derived plasticity in D. sechellia because they may represent good candidates for genes involved in D. sechellia specialization on M. citrifolia that contains high concentrations of L-DOPA in its fruit.
Identification of candidate genes
Included in the genes identified by the RNA-seq analysis were several candidates that may play a role in different aspects of D. sechellia host specialization. Specifically, we focused on genes with evolved transcriptional responses to L-DOPA in D. sechellia that were not identified as responsive to L-DOPA in D. melanogaster or D. simulans (Supplemental Table 4). Among these genes were several that have been identified in prior studies as possible contributors to different aspects of evolutionary changes in D. sechellia. For example, the fatty acid elongase eloF was shown to influence the abundance of different length fatty acids found on female flies with fewer longer cuticular hydrocarbon (CHC) species observed when the gene was knocked out (Combs et al. 2018). Interestingly, D. simulans flies are quite similar to the D. sechellia eloF knockout flies in CHC profiles suggesting that this may be a mechanism of species discrimination that influences mate choice. When eloF was knocked out in D. sechellia female flies, male D. simulans individuals courted them as if they were D. simulans females demonstrating the role eloF has played in reproductive isolation between these species (Combs et al. 2018). Here we found that D. sechellia significantly decreases expression of eloF in the presence of L-DOPA (Supplemental Table 4), something only observed in this species. This species-specific reduction in expression could suggest that in their natural context feeding on M. citrifolia that contains high levels of L-DOPA (Lavista-Llanos et al. 2014), D. sechellia flies may make CHC profiles more similar to D. simulans, lessening the reproductive isolation between these species. Because prior experiments examining aspects of interspecies courtship were conducted in the absence of L-DOPA, future experiments investigating the consequence of L-DOPA on courtship are warranted.
Another gene identified, Gasp, resides inside a fine-mapped QTL that explains the greatest amount of variation in octanoic acid resistance between D. simulans and D. sechellia (Hungate et al. 2013). However, this gene was knocked-down with RNAi and shown to not influence OA resistance in D. melanogaster functional tests (Andrade López et al. 2017) suggesting that it is not the gene that underlies this QTL. Despite the lack of effect on OA resistance, Gasp is involved in chitin metabolic processes which may influence other traits associated with host specialization. Interestingly, many other genes with functions in processes associated with the chitin-based cuticle have derived plastic expression responses to L-DOPA in D. sechellia, similar to that observed for gene expression responses to OA (Lanno et al. 2017) suggesting that D. sechellia is altering its cuticle in some way in response to its diet and this may be involved in specializing to eat toxic M. citrifolia fruit.
Genes associated with defense against microbial pathogens were also found to be responsive to L-DOPA exposure in this study. Similar to the finding for chitin-related genes, pathogen defense genes are also responsive to OA exposure, possibly suggesting that flies exposed to these chemicals have altered microbial resistance phenotypes. In response to both L-DOPA and OA we observe reduction in the expression of many bacterial defense genes including those involved in bacterial recognition (e.g., PGRPs, Supplemental Table 4), as well as many involved in other aspects of immune responses to bacteria (e.g., edin, Supplemental Table 4). Because the general trend is toward down-regulation of bacterial defense genes in response to chemicals associated with M. citrifolia fruit, our data suggest that flies eating this diet may be immune compromised and therefore more susceptible to bacterial infection when they are exposed to L-DOPA, OA or M. citrifolia fruit. Future studies will focus on how the Drosophila immune system is altered in these environments with the goal of testing the consequence of food chemical exposure on bacterial infection.
Knockdown of Esterase 6 alters OA resistance
Our work and that of other research groups have observed L-DOPA effects on various traits in D. sechellia (Lavista-Llanos et al. 2014) and one of the central aims of this study was to identify candidate genes potentially involved in these derived traits. To do this we focused on the genes that have derived plasticity only observed in D. sechellia and found that Est6 was included in this gene list (Supplementary Table 4). In response to L-DOPA exposure, we found that Est6 had a significant and marked increase in expression level. This observation was intriguing because our recent study using the chemical synergist tribufos to inactivate all the esterase enzymes simultaneously (Plapp et al. 1963; Snoeck et al. 2017) found that one or more esterase genes are involved in D. sechellia derived resistance to OA (Lanno and Coolon 2019). We therefore knocked down the expression of Est6 ubiquitously in D. melanogaster adults with RNAi and performed OA resistance assays. We found that knock-down of Est6 resulted in significant decrease in relative survival when individuals were exposed to OA (Figure 5). These experiments were performed in a heterologous host (D. melanogaster) based on availability of genetic tools, which is a common approach for tests of gene function, however, confirmation of Est6 function in OA resistance requires functional tests in D. sechellia in future studies. The combination of the OA resistance data and gene expression data collected in this study in conjunction with the prior data showing one or more members of the esterase gene family are involved in OA resistance (Lanno and Coolon 2019), and QTL mapping data showing Est6 is consistent with regions of the genome contributing to OA resistance all together strongly suggest that Est6 plays an important role in toxin resistance in D. sechellia. Altogether, identification of a derived amino acid substitution fixed in D. sechellia that alters a residue that resides in the enzymes active site and was shown to influence Est6 substrate-specificity and kinetics suggest that both protein coding as well as gene expression changes may contribute to evolutionary changes at this locus.
Acknowledgments
Research reported in this publication was supported by Wesleyan University (Startup to JDC, Department of Biology funds to JDC, College of the Environment funds to JDC), and the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number R15GM135901 (awarded to JDC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.8938103.
Communicating editor: J. Birchler
Literature Cited
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D. et al. , 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S., Gish W., Miller W., Myers E., and Lipman D., 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amlou M., Moreteau B., and David J. R., 1998a Genetic analysis of Drosophila sechellia specialization: Oviposition behavior toward the major aliphatic acids of its host plant. Behav. Genet. 28: 455–464. 10.1023/A:1021689312582 [DOI] [PubMed] [Google Scholar]
- Amlou M., Moreteau B., and David J. R., 1998b Larval tolerance in the Drosophila melanogaster species complex toward the two toxic acids of the D. sechellia host plant. Hereditas 129: 7–14. 10.1111/j.1601-5223.1998.00007.x [DOI] [PubMed] [Google Scholar]
- Andrade López J. M., Lanno S. M., Auerbach J. M., Moskowitz E. C., Sligar L. A. et al. , 2017. Genetic basis of octanoic acid resistance in Drosophila sechellia: functional analysis of a fine-mapped region. Mol. Ecol. 26: 1148–1160. 10.1111/mec.14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S., 2010 FastQC: A quality control tool for high throughput sequence data. Bioinformatics. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene Ontology: Tool for The Unification of Biology. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G. et al. , 2016. Flybase: Establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44: D786–D792. 10.1093/nar/gkv1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y., 1995. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bernays E. A., and Chapman R. F., 1994. Host-Plant Selection by Phytophagous Insects, Chapman and Hall, New York: 10.1007/b102508 [DOI] [Google Scholar]
- Blake J. A., Christie K. R., Dolan M. E., Drabkin H. J., Hill D. P. et al. , 2015. Gene ontology consortium: Going forward. Nucleic Acids Res. 43: D1049–D1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D., Von Kuster G., Bouvier E., Baker D., Afgan E. et al. , 2014. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 15: 403 10.1186/gb4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V., and White K., 1987. Genetic dissection of dopamine and serotonin synthesis in the nervous system of Drosophila melanogaster. J. Neurogenet. 4: 309–314. 10.3109/01677068709167191 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B. et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Combs P. A., Krupp J. J., Khosla N. M., Bua D., Petrov D. A. et al. , 2018. Tissue-Specific cis-Regulatory Divergence Implicates eloF in Inhibiting Interspecies Mating in Drosophila. Curr. Biol. 28: 3969–3975.e3. 10.1016/j.cub.2018.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., Jones K. L., Todd T. C., Carr B. C., and Herman M. A., 2009. Caenorhabditis elegans Genomic Response to Soil Bacteria Predicts Environment-Specific Genetic Effects on Life History Traits. PLoS Genet. 5: e1000503 10.1371/journal.pgen.1000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., Stevenson K. R., McManus C. J., Graveley B. R., and Wittkopp P. J., 2012. Genomic imprinting absent in Drosophila melanogaster adult females. Cell Reports 2: 69–75. 10.1016/j.celrep.2012.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. R., 1972. Regression Models and Life-Tables. J. R. Stat. Soc. B 34: 187–220. [Google Scholar]
- Farine, J-P., L. Legal, B. Moreteau, and J.-L. Quere, 1996 Volatile components of ripe fruits of Morinda citrifolia and their effects on Drosophila. 4: 433–438.
- Fox J., 2008. Cox Proportional-Hazards Regression for Survival Data The Cox Proportional-Hazards Model: Appendix to An R and S-PLUS Companion to Applied Regression, Sage Publications, Thousand Oaks, CA. [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S. et al. , 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38: W695–W699. 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., and Erezyilmaz D., 2015. The Genetics of Resistance to Morinda Fruit Toxin During the Postembryonic Stages in Drosophila sechellia. G3 (Bethesda) 5: 1973–1981. 10.1534/g3.114.015073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungate E. A., Earley E. J., Boussy I. A., Turissini D. A., Ting C.-T. et al. , 2013. A Locus in Drosophila sechellia Affecting Tolerance of a Host Plant Toxin. Genetics 195: 1063–1075. 10.1534/genetics.113.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J., 1990. Host Specialization in Phytophagous Insects. Annu. Rev. Ecol. Syst. 21: 243–273. 10.1146/annurev.es.21.110190.001331 [DOI] [Google Scholar]
- Jones C. D., 1998. The genetic basis of Drosophila sechellia’s resistance to a host plant toxin. Genetics 149: 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. D., 2001. The genetic basis of larval resistance to a host plant toxin in Drosophila sechellia. Genetics 78: 225–233. 10.1017/S0016672301005298 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanno S. M., and Coolon J. D., 2019. Derived esterase activity in Drosophila sechellia contributes to evolved octanoic acid resistance. Insect Mol. Biol. 10.1111/imb.12587 [DOI] [PubMed] [Google Scholar]
- Lanno S. M., Peyser R. D., Shimshak S. J., Linde S. C., and Coolon J. D., 2019. Investigating the role of Osiris genes in Drosophila sechellia larval resistance to a host plant toxin. Ecol. Evol. 9: 1922–1933. 10.1002/ece3.4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanno S. M., Gregory S. M., Shimshak S. J., Alverson M. K., Chiu K. et al. , 2017. Transcriptomic analysis of octanoic acid response in Drosophila sechellia using RNA-sequencing. G3 Genes, Genomes. Genet. 7: 3867–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavista-Llanos S., Svatos A., Kai M., Riemensperger T., Birman S. et al. , 2014. Dopamine drives Drosophila sechellia adaptation to its toxic host. eLife 3 10.7554/eLife.03785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legal L., Chappe B., and Jallon J. M., 1994. Molecular basis of Morinda citrifolia (L.): Toxicity on Drosophila. J. Chem. Ecol. 20: 1931–1943. 10.1007/BF02066234 [DOI] [PubMed] [Google Scholar]
- Legal L., David J. R., and Jallon J. M., 1992. Toxicity and attraction effects produced by Morinda citrifolia fruits on the Drosophila melanogaster complex of species. Chemoecology 3: 125–129. 10.1007/BF01370140 [DOI] [Google Scholar]
- Louis J., and David J., 1986. Ecological specialization in the Drosophila melanogaster species subgroup: A case study of D. sechellia. Acta Oecol. 7: 215–229. [Google Scholar]
- Matute D. R., and Ayroles J. F., 2014. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol. 27: 1057–1068. 10.1111/jeb.12391 [DOI] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M. et al. , 2013. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41: W597– W–600.. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M. A., Healy M. J., and Oakeshott J. G., 1993. Effects of the residue adjacent to the reactive serine on the substrate interactions of Drosophila esterase 6. Biochem. Genet. 31: 259–278. 10.1007/BF00553170 [DOI] [PubMed] [Google Scholar]
- Nagatsu T., Levitt M., and Udenfriend S., 1964. Tyrosine Hydroxylase The Initial Step in Norepinephrine Biosynthesis. J. Biol. Chem. 239: 2910–2917. [PubMed] [Google Scholar]
- Neckameyer W. S., 1996. Multiple roles for dopamine in Drosophila development. Dev. Biol. 176: 209–219. 10.1006/dbio.1996.0128 [DOI] [PubMed] [Google Scholar]
- Neckameyer W. S., and White K., 1993. Drosophila Tyrosine Hydroxylase is Encoded by the Pale Locus. J. Neurogenet. 8: 189–199. 10.3109/01677069309083448 [DOI] [PubMed] [Google Scholar]
- Osterwalder T., Yoon K. S., White B. H., and Keshishian H., 2001. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98: 12596–12601. 10.1073/pnas.221303298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton R. G., Robinson N., Roychowdhury R., Rasheed A., and Hillman R., 1996. Reproduction and development in Drosophila are dependent upon catecholamines. Life Sci. 59: 2083–2091. 10.1016/S0024-3205(96)00562-0 [DOI] [PubMed] [Google Scholar]
- Peyser R. D., Lanno S. M., Shimshak S. J., and Coolon J. D., 2017. Analysis of cytochrome P450 contribution to evolved plant toxin resistance in Drosophila sechellia. Insect Mol. Biol. 26: 715–720. 10.1111/imb.12329 [DOI] [PubMed] [Google Scholar]
- Plapp F. W., Bigley W. S., Chapman G. A., and Eddy G. W., 1963. Synergism of Malathion Against Resistant House Flies and Mosquitos. J. Econ. Entomol. 56: 643–649. 10.1093/jee/56.5.643 [DOI] [Google Scholar]
- Price P. W., Bouton C. E., Gross P., McPheron B., Thompson J. N. et al. , 1980. Interactions among three trophic levels: influence of plants on interaction between insect herbivores and natural enemies. Annu. Rev. Ecol. Syst. 11: 41–65. 10.1146/annurev.es.11.110180.000353 [DOI] [Google Scholar]
- R’Kha S., Capy P., and David J. R., 1991. Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc. Natl. Acad. Sci. USA 88: 1835–1839. 10.1073/pnas.88.5.1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G., Endo K., Zong L., and Davis R. L., 2001. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98: 12602–12607. 10.1073/pnas.221303998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Ayroles J., Matute D. R., and Kern A. D., 2018. Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14: e1007341 10.1371/journal.pgen.1007341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K. et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck S., Greenhalgh R., Tirry L., Clark R. M., Van Leeuwen T. et al. , 2017. The effect of insecticide synergist treatment on genome-wide gene expression in a polyphagous pest. Sci. Rep. 7: 13440 10.1038/s41598-017-13397-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares A. R., Marchiosi R., Siqueira-Soares Rde C., Barbosa de Lima R., and Dantas dos Santos W., 2014. The role of L-DOPA in plants. Plant Signal. Behav. 9: e28275 10.4161/psb.28275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis D. G., Burton D. Y., McIvor W. E., Krishnakumar S., Wright T. R. et al. , 1999. The catecholamines up (Catsup) protein of Drosophila melanogaster functions as a negative regulator of tyrosine hydroxylase activity. Genetics 153: 361–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis D. G., Pentz E. S., Freeman M. E., Kullman J., Hankins G. R. et al. , 1995. The genetic and molecular organization of the Dopa decarboxylase gene cluster of Drosophila melanogaster. Genetics 141: 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R. C., 2017. R: A language and environment for statistical computing, Scientific Research, Vienna, Austria. [Google Scholar]
- Therneau T. M., 2015. A Package for Survival Analysis in R.
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L. et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G. et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacas L., and Bachli G., 1981. Drosophila sechellia. n. sp., huitieme espece du sous-groupe melanogaster des iles Seychelles (Diptera, Drosophilidae). Rev. Fr. Entomol. 3: 146–150. [Google Scholar]
- Wright T. R. F., 1987. The Genetics Of Biogenic Amine Metabolism, Sclerotization, And Melanization In Drosophila melanogaster. Adv. Genet. 24: 127–222. 10.1016/S0065-2660(08)60008-5 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., and Seto E. S., 2014. Dopamine Dynamics and Signaling in Drosophila: An Overview of Genes, Drugs and Behavioral Paradigms. Exp. Anim. 63: 107–119. 10.1538/expanim.63.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Akanni W., Amode M. R., Barrell D., Billis K. et al. , 2016. Ensembl 2016. Nucleic Acids Res. 44: D710–D716. 10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younus F., Fraser N. J., Coppin C. W., Liu J. W., Correy G. J. et al. , 2017. Molecular basis for the behavioral effects of the odorant degrading enzyme Esterase 6 in Drosophila. Sci. Rep. 7: 46188 10.1038/srep46188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All RNA-seq data generated in this manuscript have been submitted to the NCBI Gene Expression Omnibus under accession number GSE138119. Supplemental material available at figshare: https://doi.org/10.25387/g3.8938103.