Abstract
Aspergillus flavus colonizes numerous oil seed crops such as maize, peanuts, treenuts and cottonseed worldwide, contaminating them with aflatoxins and other harmful toxins. Previously our lab characterized the gene rmtA, which encodes an arginine methyltransferase in A. flavus, and demonstrated its role governing the expression of regulators in the aflatoxin gene cluster and subsequent synthesis of toxin. Furthermore, our studies revealed that rmtA also controls conidial and sclerotial development implicating it as an epigenetic regulator in A. flavus. To confirm this, we performed a RNA sequencing analysis to ascertain the extent of rmtA’s influence on the transcriptome of A. flavus. In this analysis we identified over 2000 genes that were rmtA-dependent, including over 200 transcription factor genes, as well as an uncharacterized secondary metabolite gene cluster possibly responsible for the synthesis of an epidithiodiketopiperazine-like compound. Our results also revealed rmtA-dependent genes involved in multiple types of abiotic stress response in A. flavus. Importantly, hundreds of genes active during maize infection were also regulated by rmtA. In addition, in the animal infection model, rmtA was dispensable for virulence, however forced overexpression of rmtA increased mortality with respect to the wild type.
Keywords: Aspergillus flavus, Environmental stress, RmtA, Secondary metabolism, Transcriptome, Virulence
Aspergillus flavus is an opportunistic plant pathogen of great economic importance that infects oil seed crops such as maize, peanuts, cotton and certain treenuts, and in the course, produces potent mycotoxins (Hedayati et al. 2007), including the highly carcinogenic aflatoxins (Sarma et al. 2017). Ingestion of aflatoxin contaminated crops can result in jaundice, edema of the limbs, pain, vomiting, necrosis, potentially acute liver failure and in rare cases death (Lancaster et al. 1961; CDC, 2004; Fung and Clark 2004; Lewis et al. 2005). Chronic exposure can lead to suppression of the immune system, stunting of growth and wasting in children and several types of cancers such as those affecting the liver, lungs and gastrointestinal tract (CDC, 2004; Lewis et al. 2005; Marchese et al. 2018). In developed nations, legislation regulates levels of aflatoxins in food and feed commodities to prevent adulterated crops from entering the market place, however in most developing nations lacking such guidelines or restrictions, exposures becomes more prevalent (Ojiambo et al. 2018).
In the United States and other developed nations, the major impact of aflatoxin contamination of commodities is economic losses. It has been estimated that economic losses associated with aflatoxin contamination of maize can reach up to a billion dollars annually in the United States alone particularly in years with warm summers and drought (Mitchell et al. 2016).
In addition to its devastating effect on crops of economic importance, A. flavus is known to cause a deadly lung infection known as invasive aspergillosis. Although A. flavus is the second leading cause of IA after Aspergillus fumigatus, A. flavus is 100-fold more virulent than that of A. fumigatus (Ford and Friedman 1967; Mosquera et al. 2001; Kamai et al. 2002, Kaliamurthy et al. 2003 Hedayati et al. 2007).
Due to the adverse health and economic impacts associated with aflatoxin contamination and A. flavus, it is paramount to gain insight into its dispersal and survival mechanisms, as well as the regulatory pathways controlling its production of mycotoxins and its virulence. This knowledge could reveal novel genetic elements that could be used as possible targets to reduce the negative effects of this opportunistic pathogen of humans and plants.
Morphological development and secondary metabolism (SM) are genetically linked in A. flavus and other fungal species (i.e., Calvo et al. 2002; Calvo and Cary 2015). In A. flavus, one of those genetic links is rmtA, encoding an arginine methyltransferase that has been shown to regulate aflatoxin biosynthesis as well as development (Satterlee et al. 2016). Specifically, rmtA is a repressor of the production of conidia, air-borne asexual spores that constitute an efficient form of fungal dissemination, and a positive regulator of sclerotial production, resistant structures that can survive under adverse environmental conditions (Horn et al. 2014; Satterlee et al. 2016). Homologs of RmtA have been shown to be involved in transcriptional regulation, signal transduction, RNA processing and transport (Bedford and Clarke 2009). RmtA is known to have a role in methylation of histones, which in turns affects gene expression (Trojer et al. 2004; Tessarz & Kouzarides 2014).
As rmtA appeared to be functioning as a global regulator of secondary metabolism and development, our current study was performed to further assess its influence on the transcriptome of A. flavus. In this analysis we identified over two-thousand genes that are rmtA-dependent, some of those genes are associated with secondary metabolism, abiotic stress response and virulence of this agriculturally and medically important fungus.
Materials and Methods
Strains used and growth conditions
All strains used in this work are listed in Table 1. Strains were grown on potato dextrose agar (PDA) at pH 5.6 in the dark at 30°, unless otherwise stated. Stocks of each strain were maintained as conidia at -80° in 30% glycerol.
Table 1. Strains used in this study.
| Strain | Pertinent Genotype | Source |
|---|---|---|
| CA14-WT | Δku70 | USDA |
| CA14-ΔrmtA | ΔrmtA::pyrGA.fumigatus, niaDA.fumigatus, Δku70 | Satterlee et al. 2016 |
| CA14-comrmtA | ΔrmtA::pyrGA.fumigatus, rmtA::niaDA.fumigatus, Δku70 | Satterlee et al. 2016 |
| CA14-OErmtA | gpdA(p)::rmtA::trpC(t)::pyrGA.fumigatus, niaDA.fumigatus, Δku70 | Satterlee et al. 2016 |
| CA14 | pyrG-, niaD-, Δku70 | USDA |
| CA14- pyrG-1 | pyrG+, niaD-, Δku70 | USDA |
| CA14- ΔgliP | ΔgliP::pyrGA.fumigatus, niaD-, Δku70 | This Study |
| CA14- ΔrmtA | ΔrmtA::pyrGA.fumigatus, niaD-, Δku70 | Satterlee et al. 2016 |
Purification of RNA and sequencing
The wild type (WT), deletion rmtA (ΔrmtA), and overexpression rmtA (OE) strains were grown on potato dextrose top-agar in the dark at 30°. Spores (5x106) were inoculated into 5 ml of melted PDA top agar (0.5%), which was then placed onto 25 ml solid PDA medium. After 72 h of incubation, mycelia was collected, frozen in liquid nitrogen, and lyophilized. Total RNA was extracted from mycelia using an RNeasy Plant Mini Kit (Qiagen, Germantown, Maryland, USA) following the manufacture’s protocol. RNA was further purified using Dynabeads mRNA Purification Kit. RNA quality was assessed using an Agilent Bioanalyzer. Sequencing was performed as a HiSeq 2000 single read 1x100bp lane. The experiment was carried out with 2 biological replicates.
Analysis of RNA-sequencing data to identify differentially expressed genes
Read mapping:
The single-end reads of the WT, ΔrmtA, and OE samples were separately aligned to the A. flavus NRRL 3375 reference genome using HISAT2 (Kim et al. 2015) version 2.0.5. The command used was hisat2 -x reference_genome_index –U fastq_file -S output_file.sam. HISAT2 utilizes Bowtie2 (Langmead & Salzberg 2012) and was run using software version 2.3.HISAT2 utilizing Bowtie2 version 2.3.1. SAMtools version 1.3.1 was implemented to convert the SAM output file from TopHat into a BAM file for the next step.
Read counts:
The mapped reads in BAM format were then analyzed using the HTSeq.scripts.count command from the HTSeq python package version 0.9.1. This tool was employed to return a table of read counts for each gene. The command used was python –m HTSeq.scripts.count –i Parent gff_file. The GFF file downloaded from NCBI contains the pre-annotated gene models as well as their genomic locations.
Differential expressed genes (DEGs):
The table of read counts was used as input for the R limma package. This package was used to determine DEGs by comparing read counts between two conditions: WT vs. ΔrmtA and WT vs. OE. The two replicates of each condition were combined during this step of the analysis. The RPKM function in the R edgeR package determined the RPKM (reads per kilobase per million) values for all the genes. Bash and Perl scripts were developed to parse the DEGs data and RPKM data. An Excel file was created with the RPKM values for all genes across all conditions. FungiDB (Basenko et al. 2018) was used for functional enrichment of the data sets using Go Term (GO) annotations.
Selected groups of genes:
To gain more biological significance from the data sets, the differentially expressed genes were mapped to other databases. The list of SM gene clusters (SMGCs) information was extracted from Ehrlich and Mack (2014). A full list of transcription factors (TFs) in A. flavus was derived from the Fungal Transcription Factor Database (http://ftfd.snu.ac.kr/intro.php) (Park et al. 2008). Functional annotations of these transcription factors were obtained from NCBI. Genes related to environmental stress response were extracted from the database established by Miskei et al. 2009. The list of DEGs from the study performed by Dolezal and collaborators (Dolezal et al. 2013) was compared to this dataset to search for potentially rmtA–dependent virulence genes. R (R Core Team 2017) version 3.4.1, specifically the ggplot2 package (Wickham 2009), was used to make statistical figures.
Analysis of novel epidithiodiketopiperazine cluster
Generation of gliP deletion strain (ΔgliP):
To impair the function of gliP part of the gene was knocked out. First, the gliP deletion cassette was created by fusion PCR as described by Szewczyk et al. (Szewczyk et al. 2006). Primers AFLA_gliP_P1 and AFLA_gliP_P2 were used to PCR amplify the 5′ UTR of the gliP locus in the A. flavus genome, while AFLA_gliP_P3 and AFLA_gliP_P4 primers were used to amplify the 3′ end of the gliP coding region. The middle fragment containing the pyrG selection marker was PCR amplified from the genomic DNA of Aspergillus fumigatus using primers AFLA_gliP_P5 and AFLA_gliP_P6. The three fragments were then fused by PCR using primers AFLA_gliP_P7 and AFLA_gliP_P8. All primers used in this study are listed in Table 2. The fused PCR product was transformed into A. flavus CA14 host strain (pyrG-, niaD-) by a polyethylene glycol-mediated transformation as previously described (Cary et al. 2014). Transformants were selected on half-strength PDA without uracil. Potassium chloride (0.6 M) was used as an osmotic stabilizer in the regeneration medium. Transformants were confirmed by diagnostic PCR with primers AFLA_gliP_P1 and Afum_pyrG_R. A selected hbxA deletion transformant, TTRS6, was used in this study.
Table 2. Primers used in this study.
| Primers | Sequence 5′ -> 3′ |
|---|---|
| AFLA_gliP_P1 | TCGATTCAGCGAGCCAGATGG A |
| AFLA_gliP_P2 | GGAGGCTGCTTCAGGGTATAAGAGA |
| AFLA_gliP_P3 | CCGATGCTTACCTGTGCTTTACGTC |
| AFLA_gliP_P4 | GCCCAGTTGGAGGATATCACGGAA |
| AFLA_gliP_P5 | TCTCTTATACCCTGAAGCAGCCTCCACCGGTCGCCTCAAACAATGCTCT |
| AFLA_gliP_P6 | GACGTAAAGCACAGGTAAGCATCGGGTCTGAGAGGAGGCACTGATGCG |
| AFLA_gliP_P7 | GGTGGATAACGGCAAGTC ATCTCCT |
| AFLA_gliP_P8 | CGGCAATGAGATGGTTCCCCTG |
| Afum_pyrG_R | GAGCAGCGTAGATGCCTCGAC |
Chemical analysis of ΔgliP, ΔrmtA, and WT strains:
A. flavus ΔgliP, ΔrmtA, and WT strains were cultivated in 1 L zeolite medium (glycerol 30 g/L, glucose 10 g/L, peptone 5 g/L, NaCl 2 g/L, molecular sieve 0.5 nm 10 g/L, agar 1 g/L, pH 7.0, see Chankhamjon et al. 2014) in 2.8 L Fernbach flasks at 28° with orbital shaking (110 rpm) for 4 days. The culture broth and mycelia were extracted with ethyl acetate (500 mL). The organic extracts were filtered through miracloth and concentrated to dryness under reduced pressure. Extracts were analyzed as described previously (Lebar et al. 2018) on a Waters ACQUITY UPLC system using PDA UV and QDa nominal mass detection [column: BEH C18 1.7µm, 2.1 × 50 mm; gradient solvent system: (0.5 mL/min, solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile): 5% B (0-1.25 min), gradient to 25% B (1.25-1.5 min), gradient to 100% B (1.5-5.0 min), 100% B (5.0-7.5 min), then column equilibration 5% B (7.6-10.1 min)].
Environmental stress assay
To assess rmtA involvement in response of A. flavus to environmental stresses, fungal strains were assayed with osmotic stress inducers using temperatures above and below optimum growth conditions. For all assays, WT, ΔrmtA, OE and the genetically complemented ΔrmtA strain (COM) were point-inoculated onto PDA plates supplemented with osmotic agents (1 M sucrose, 1.2 M sorbitol, 0.6 M KCl, or 0.7 M NaCl) and incubated in the dark for 48 h. For temperatures assays, cultures were exposed to 25°, 28°, 30°, 37°, 40° and 42°.
Pathogenicity assay
Spores from WT, ΔrmtA, com, and OE strains were collected in a solution of 1x PBS with 0.1% tween and washed 5 times with additional equal volumes of 1x PBS. To reach the target concentration of 5x 103 spores per 10 μl-1, the spores suspensions were diluted further with 1x PBS. The infection procedure was carried out as previously described by Fuchs et al. (2010). Briefly, Galleria mellonella larvae (The Bug Company, Ham Lake, Minnesota) with a weight range between 275–300 mg and lacking gray markings were selected for the experiment. Groups of 30 larvae were infected with the WT, ΔrmtA, OE, and complementation (COM) strains. An additional two group of 30 larvae each were used as controls. One group received injections of 10 µl of 1x PBS while the other groups received no injections. Larvae were then placed in glass petri plates (90 mm × 15 mm) and wrapped in aluminum foil. Plates were placed in 37° in the dark. Larvae were check every 4 h following 16 h of incubation until one group of larvae experienced complete mortality.
Data availability
Table S1 contains calculated expression values comparing those of the wild-type strain to those of the deletion rmtA mutant and overexpression strain. Tables S2–S4 contain a subsets of Table S1 that includes DEGs corresponding to transcription factors, genes related to environmental stress, and genes that were shown to be DEG during maize infection. Figure S1 shows GO terms of DEG that are rmtA-dependent. Figure S2 details the strategy for the construction of the gliP mutant strain. Figures S3 and S4 show the rmtA-dependent effect of osmotic stress and temperature on A. flavus. The sequencing data are publicly available at NCBI’s SRA repository with the SRA Accession #: PRJNA573552. Supplemental material available at figshare: https://doi.org/10.25387/g3.9252530.
Results
rmtA-dependent transcriptome in A.flavus
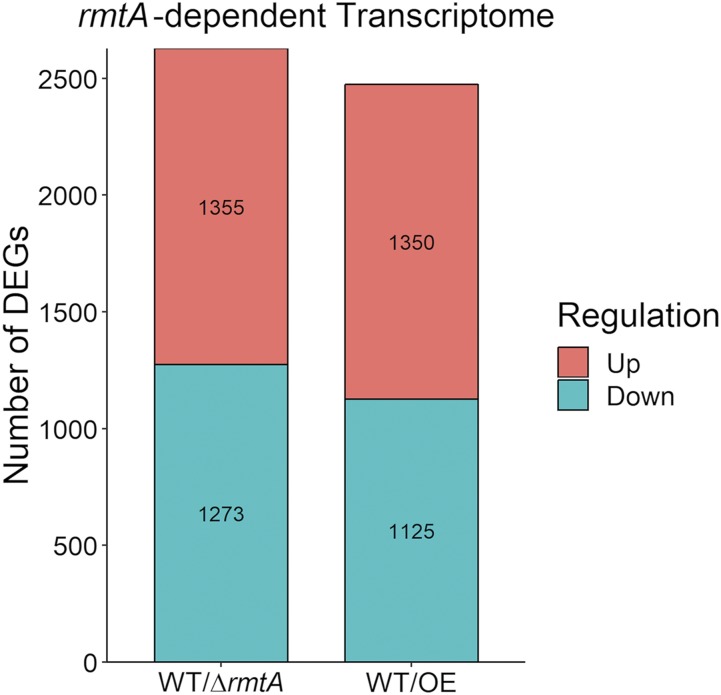
RNA-seq analysis of the influence of rmtA on the A. flavus transcriptome revealed that both deletion or over-expression of rmtA results in similar ratios of up or down differentially expressed genes (DEGs) with greater than twofold difference in expression compared to the wild type. This constituted more than 2000 rmtA-dependent DEGs, as shown in Figure 1. Absence of rmtA affected the expression of more genes than when rmtA was over-expressed. Only 27 DEGs showed opposite expression patterns, DEGs that are downregulated in the absence of rmtA while they are upregulated when rmtA is overexpressed and vice versa. Most of the DEGs require wild-type levels of rmtA expression to function properly, as both deletion and overexpression of rmtA cause alterations in their transcription. There were 719 genes with reduced expression when rmtA was either knocked out or over-expressed, and 632 genes with increased transcription when this occurs. Full analysis is located in Table S1. Enrichment analysis of the functional categories of the rmtA-dependent transcriptome did not indicate any particular areas of regulation that rmtA governs either by its absence or forced expression (Figure S1).
Figure 1.
Number of upregulated and downregulated genes when the expression of rmtA is altered by rmtA deletion or overexpression. Number of up-regulated (red) and down-regulated (green) DEGs in ΔrmtA/WT and OE/WT comparisons.
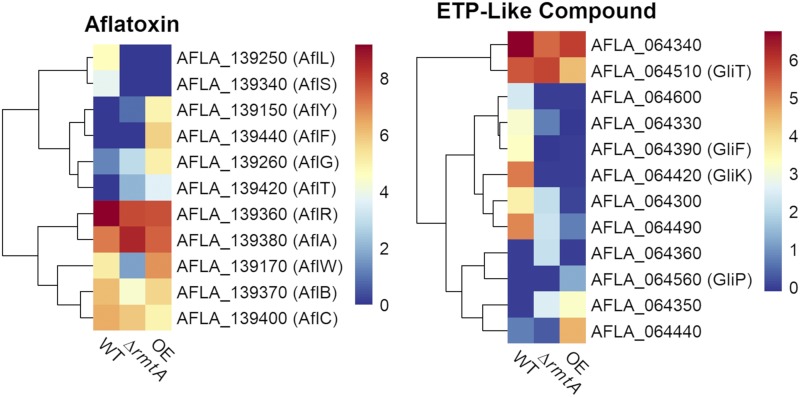
rmtA-Dependent Expression of SMGCs
Our results revealed that several genes within two secondary metabolite gene clusters (SMGCs) (defined in Georgianna et al. 2010) were rmtA-dependent (Figure 2). One cluster, denoted as #54, corresponds to the already characterized aflatoxin biosynthetic gene cluster. As shown previously, rmtA regulates production of aflatoxin (Satterlee et al. 2016). The other SMGC is the uncharacterized cluster #21 that contains several genes homologous to those associated with the gliotoxin gene cluster in A. fumigatus.
Figure 2.
Heat maps of DEGs in Aflatoxin and epidithiodiketopiperazine - like compound gene cluster in the rmtA-transcriptome. Heat map of RPKM values of genes that are differentially expressed in rmtA transcriptome and found in the secondary metabolite gene cluster #54 Aflatoxin (left), and cluster #21 an uncharacterized cluster that may produce an epidithiodiketopiperazine (ETP) compound similar to gliotoxin (right).
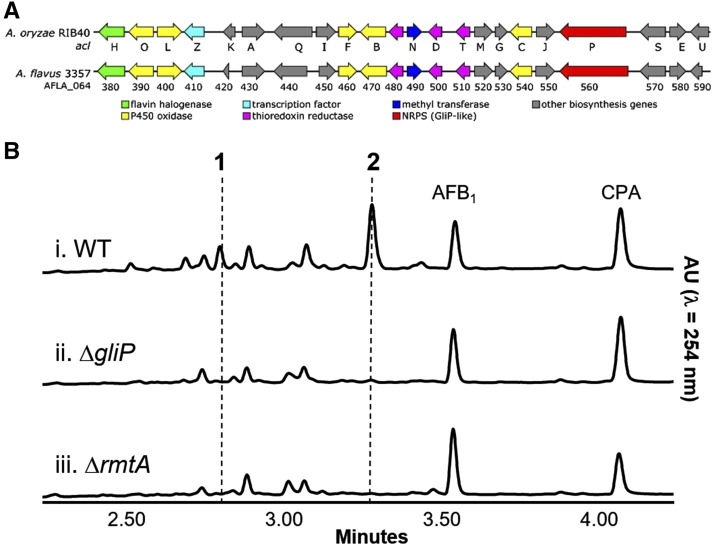
In order to determine what metabolite(s) is the product associated with cluster #21 an A. flavus strain with a deletion of the putative nonribosomal peptide synthetase gene (AFLA_064560), homologous to that of A. fumigatus gliP, was created (Figure S2). This strain was confirmed by PCR, yielding the expected 3.1 kb PCR product. Cluster 21 bears considerable resemblance to acl (Figure 3A), the cluster responsible for aspirochlorine biosynthesis in A. oryzae (Chankhamjon et al., 2014). However, initial chemical analysis of ΔgliP, ΔrmtA, and WT strains grown in PDA did not reveal any differences in secondary metabolite production, nor was aspirochlorine detected in any strains (data not shown). Chankhamjon et al. (2014) found that A. oryzae grown in optimized “zeolite” medium resulted in consistent aspirochlorine production. When A. flavus ΔgliP, ΔrmtA, and WT strains were cultured in zeolite medium, two peaks were observed in the WT extract (Figure 3B, i) that were absent in both ΔgliP (Figure 3Bii) and ΔrmtA extracts (Figure 3Biii). Neither of these peaks appear to be aspirochlorine, but may be biosynthetic intermediates or related epidithiodiketopiperazine analogs. The compounds ionized in negative mode (peak 1: [M-H]- =455 m/z; peak 2: [M-H]-=419 m/z) and have UV λmax = 230 nm.
Figure 3.
Chemical analysis of A. flavus strains. A) comparison of A. oryzae RIB40 aspirochlorine biosynthetic gene cluster acl and A. flavus 3357 biosynthetic gene cluster #21. B) UPLC chromatograms of A. flavus extracts: peaks 1 and 2 are present in WT (i), but not observed in ΔgliP (ii) or ΔrmtA (iii). CPA: cyclopiazonic acid, AFB1: aflatoxin B1.
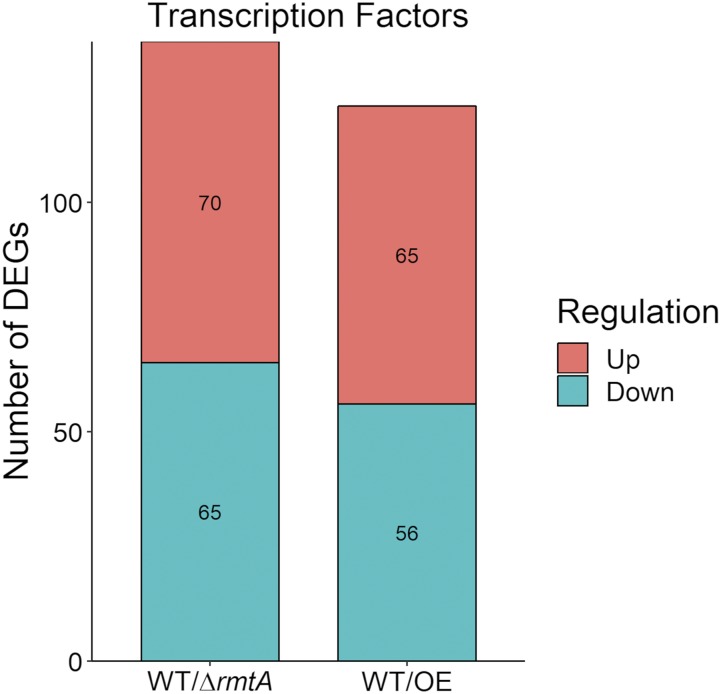
rmtA-dependent transcription factors
Based on our analysis, 251 out of the over 600 putative transcription factor genes in A. flavus were regulated by rmtA under the conditions used in the current study (Figure 4). Some of the transcription factors shown to be governed by rmtA are known to be involved in the regulation of development and metabolism, such as the aflatoxin transcription factor AflR (Woloshuk et al. 1994); MetR, a regulator of sulfur metabolism (Jain et al. 2018); and Rum1 which in A. flavus regulates both asexual development and metabolism (Hu et al. 2018). In addition, multiple transcription factors were previously shown to be associated with pathogenicity (Bultman et al. 2016; Issi et al. 2016). In other species genes such as con7, ctf1, metR and sreA were found to be connected with virulence, and their homologs are dependent on rmtA in A. flavus (Schrettl et al. 2008; Ramírez and Lorenz 2009; Ruiz-Roldán et al. 2015; Gai et al. 2019). Additionally, transcription factors attributed to different types of environmental stress response in Aspergillus were also found to be regulated by rmtA such as SrrA (oxidative and osmotic; Hagiwara et al. 2011), HacA (thermal; Zhou et al. 2016), AtfA (oxidative and osmotic; Balázs et al. 2010), and Seb1 (osmotic, oxidative, and thermal; Seidl et al., 2004). A complete list of rmtA-dependent transcription factors are located in Table S2.
Figure 4.
rmtA-dependent transcription factor genes. Graphical representation of rmtA-dependent transcription factor genes. The numbers of differentially expressed transcription factor genes whose expression is affected by either knock out or over-expression of rmtA. Red color indicates upregulated genes while green color indicates genes that are downregulated. A full list of transcription factors (TFs) in A. flavus was derived from the Fungal Transcription Factor Database (http://ftfd.snu.ac.kr/intro.php) (Park et al. 2008).
rmtA Acts as a Repressor of Conidiation even under Environmental Stress
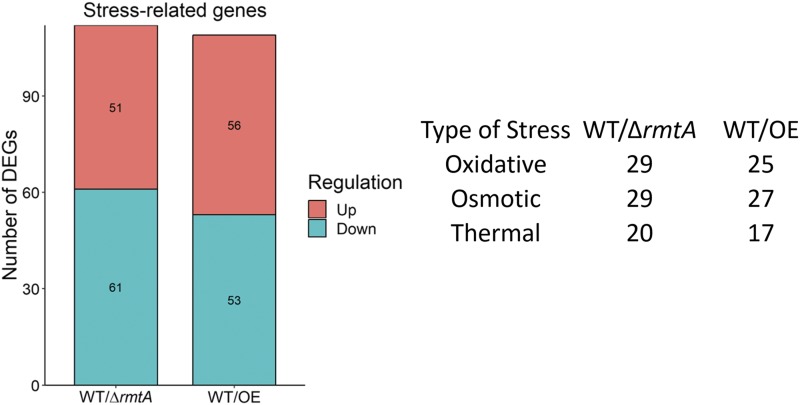
The identification, in the current study, of stress response transcription factors that are regulated by rmtA, led us to further investigate whether other genes involved in fungal stress response are also under rmtA control. A list of gene associated with stress response from Miskei et al. (2009) was used to parse our transcriptome data. The absence or over-expression of rmtA resulted in expression levels of approximately 100 genes significantly deviating from that of the wild type and were related to stress response (Figure 5). Analysis of the expression data indicated that rmtA does not regulate genes responding to a single type of stress but rather affects the expression of genes responding to multiple types of stress (Table S3).
Figure 5.
Impact of rmtA on the expression of stress-related genes in A. flavus. Graphical representation of the number of rmtA-regulated stress response genes based on the Miskei et al. (2009) database.
Whether rmtA plays a role in resistance to oxidative stress in A. flavus was previously assessed and it was found that when exposed to increasing concentrations of the oxidant menadione, alterations in rmtA expression (by deletion or overexpression of rmtA) improved resistance of A. flavus to this stress condition (Satterlee et al. 2016). However, the possible implications of rmtA on the effect of other environmental stresses have not been studied. Based on our findings that expression of several genes involved in osmotic and thermal stress response are influenced by rmtA, we examined whether rmtA is involved in resistance to those environmental stresses. Unlike under oxidative stress conditions, vegetative growth was slightly reduced in the absence of rmtA when cultures were exposed to high concentrations of NaCl (Figure S3A). However, colony growth was not affected when the strains were grown on high concentrations of sucrose, sorbitol or KCl. The hyperconidation phenotype of the deletion mutant was still detected even in the presence of high osmotic stress, although it was significantly attenuated.
In contrast to exposure to oxidative or osmotic stress, changes in expression of rmtA over a range of incubation temperatures did not cause any alterations in colony growth. While no change in growth was observed, the deletion mutant hyperconidiation phenotype persisted at all temperatures tested, except 42° (Figure S4).
rmtA regulates genes that are active during colonization of live plant tissue
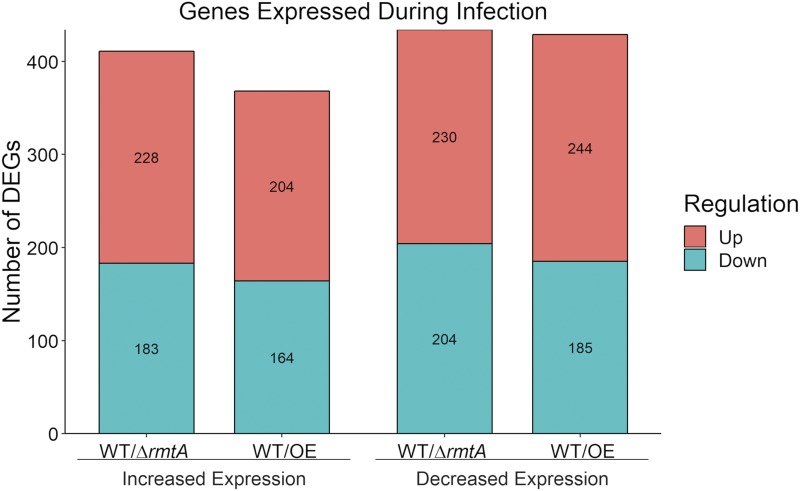
Dolezal et al. (2013) performed a transcriptome analysis of A. flavus during infection of maize that identified numerous DEGs during active infection vs. saprotrophic growth of this fungus using viable and nonviable maize kernels. The list of DEGs from the study performed by Dolezal was compared to our RNA-seq dataset to search for potential rmtA–dependent virulence genes. As shown in Figure 6, multiple genes that were differentially expressed during maize infection were also rmtA-dependent in the current experiment. Any modification of rmtA expression resulted in a decrease in the expression of 96 genes that were previously shown to be upregulated during maize seed infection. Conversely, we found 118 genes upregulated by changes in the rmtA locus that were suppressed during the seed infection study. A full list of genes that are regulated by rmtA and differentially expressed during plant infection are located in Table S4.
Figure 6.
Effect of rmtA on genes active during plant infection. Graphical representation of rmtA-dependent genes that are differentially expressed during maize infection. This graph represents transcription factor genes whose expression is affected by either loss of or forced expression of rmtA. Red color indicates upregulated genes while green color indicates genes that are downregulated. The two left columns represent genes with increased expression during infection of plant tissue, while the two columns on the right indicate genes with decreased expression under that condition.
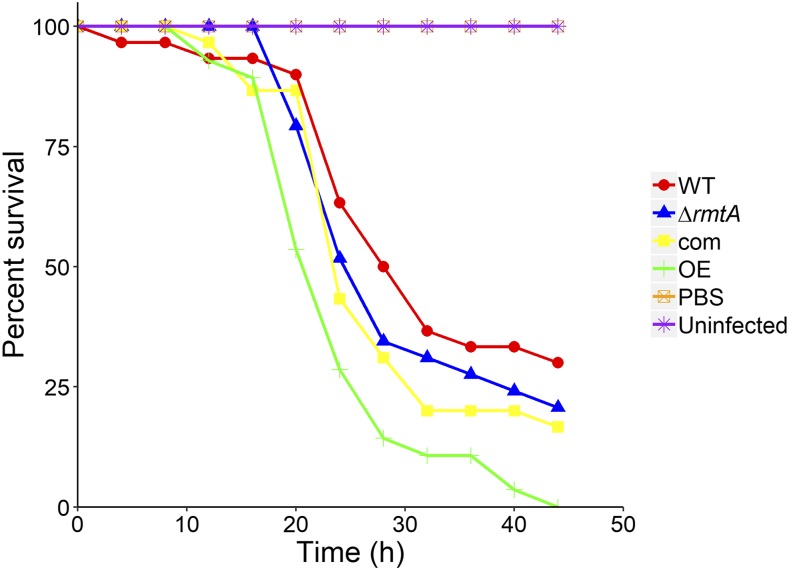
Overexpression of rmtA increases virulence in Galleria mellonella animal model
Aspergillus flavus is known to cause invasive aspergillosis in humans and animals. Since rmtA affects A. flavus infection in plants (Li et al. 2017), and our current study indicate that numerous genes active during plant infection are rmtA-dependent, we investigated whether rmtA also influences animal virulence. Galleria mellonella larvae were infected with WT, ΔrmtA, com, and OE strains to ascertain whether changes in rmtA expression affect virulence. While the deletion mutant showed a decrease in pathogenicity in maize and peanut infections, rmtA was dispensable for virulence in the animal model (Figure 7). We observed that overexpression of rmtA increased mortality in this model when compared to the control.
Figure 7.
Overexpression of rmtA positively regulates virulence of A. flavus in the G. mellonella model. Galleria mellonella larvae were infected with A. flavus wild type (WT), deletion (ΔrmtA), complementation (com), and overexpression (OE) strains as described in Material and Methods section. Two controls groups were used in this experiment, one received an injection of 1x PBS instead of fungal spores, and another received no injection. Statistical analysis of survival was carried out by a Kaplan-Meyer pairwise comparison using a long rank test.
Discussion
A comparative transcriptomic study of A. flavus control and rmtA mutants has provided further insights into the role of rmtA in the biology of A. flavus. Previously, it was shown that rmtA was a regulator of conidial and sclerotial production, as well as aflatoxin biosynthesis (Satterlee et al. 2016). Specifically, significant rmtA-dependent downregulation in expression of the conidiophore pathway developmental regulator brlA was shown to be the reason for reduction of conidiation in A. flavus rmtA knockout mutants. In terms of sclerotial production, knockout of rmtA halted production of these structures whereas increased levels of rmtA increased production compared to the wild type. RmtA was identified as positive regulator of aflatoxin production by directly affecting expression of aflatoxin cluster biosynthetic genes (Satterlee et al. 2016). Our present study revealed a broad regulatory scope for rmtA, where a significant portion of the A. flavus transcriptome is under its control. Hundreds of genes displayed altered expression upon comparison of expression levels in the wild type, ΔrmtA or overexpression rmtA strains; 719 genes in the A. flavus genome showed a reduction of their expression with this criterion, while 632 genes experienced an increase. In the model fungus A. nidulans, the rmtA homolog presented strong specificity for the methylation of histone H4 (Trojer et al. 2004). Epigenetic modifications of histone cores, such as histone methylation, affect nucleosome structures, leading to changes in the transcription of numerous genes (Tessarz and Kouzarides 2014). This agrees with the extensive effect of rmtA on the A. flavus transcriptome. In addition, based on our results, a balanced stoichiometry of RmtA with other partners seems to be required for its proper function.
As mentioned above, rmtA was found to be necessary for production of aflatoxin (Satterlee et al. 2016). Our transcriptome analysis revealed that out of 24 genes present in the aflatoxin SMGC (cluster #54 as in Georgianna et al. 2010), 11 genes were found to be rmtA-dependent. Outside of the aflatoxin cluster, only one other cluster, #21, demonstrated a large number of rmtA-dependent DEGs. In A. flavus cluster #21 has yet to be characterized. However, some genes in this cluster have homology to genes in the gliotoxin cluster in A. fumigatus (Dolan et al. 2015). Although there are similarities between these two clusters, the predicted cluster in A. flavus has nearly double the number of genes compared to that in the A. fumigatus cluster. Gliotoxin belongs to a class of metabolites known as epidithiodiketopiperazine. In Aspergillus oryzae, a cluster akin to the one described in A. flavus was characterized and found to produce another compound in this same family known as aspirochlorine (Chankhamjon et al. 2014). Production of this compound has been previously shown in A. flavus and documented to possess antifungal properties (Klausmeyer et al. 2005). Aspirochlorine was not detected in our A. flavus strain. However, chemical analysis of A. flavus WT and mutants did reveal two compounds present in WT that were not produced in SMGC#21 NRPS deletion mutant or the mutant lacking rmtA. Research on whether SMGC#21 is responsible for the synthesis of aspirochlorine or related epidithiodiketopiperazine compounds in A. flavus is ongoing. Surprisingly, levels of AF in ΔrmtA were similar to those in WT when the strains were grown in the zeolite medium, while production of this toxin was inhibited when the ΔrmtA strain was cultured on PDA (Satterlee et al., 2016), suggesting that the effect of rmtA on AF production is medium-dependent.
While methylation of histones by rmtA may directly regulate the expression of certain genes in the genome, it would also affect the transcription of others indirectly, including transcription factor genes. In our study we identified over 200 transcription factor genes with altered expression patterns caused by by either deletion or overexpression of the rmtA locus. While they are not all functionally characterized, some of these transcription factors are known to play crucial roles in fungal development, metabolism, response to environmental stresses, and virulence. A few examples of these genes investigated in A. flavus include aswA, a regulator of sclerotial production and related metabolism (Chang et al. 2017), and aflR which is the primary regulator of aflatoxin production in A. flavus (Masanga et al. 2015). Another example is rum1, a transcription factor that has a wide range of regulatory effects, controlling aflatoxin biosynthesis, and development of conidia and sclerotia in A. flavus (Hu et al. 2018). However, the majority are still uncharacterized or have been studied in other fungi such as the medusa transcription factor MedA which is shown to regulate conidiation in multiple fungi (Clutterbuck 1969; Chacko and Gold 2012; Gravelat et al. 2010).
Unexpectedly, the A. flavus ΔrmtA mutant is more resistant to sources of oxidative stress than the wild type (Satterlee et al. 2016). This was in contrast with the phenotype of the ΔrmtA mutant in A. nidulans (Trojer et al. 2004), suggesting a specialization in the regulatory output of rmtA in both fungi with respect to environmental stress resistance. In our transcriptome analysis several genes such as atfB (Sakamoto et al. 2008), fhdA (Malavazi et al. 2006), alb1 (Tsai et al. 1998), and pes1 (Reeves et al. 2006) were found to be upregulated in the absence rmtA. Expression of these genes has been shown to be indispensable for resistance to oxidative stress. It is possible that the effect of rmtA on environmental stress resistance could be mediated by its effect on the expression of these genes.
Furthermore, we identified additional rmtA-dependent genes involved in response of the fungus to other environmental insults, such as those involved in osmotic and thermal stress. Examples of these genes include members of the HOG pathway, nikA and shoA, a well-studied network that regulates osmotic stress in fungi (Furukawa et al. 2005; Hagiwara et al. 2013). Also, hacA and cypB are rmtA-dependent DEGs where HacA is a heat shock protein and cypB is expressed at high levels during heat shock conditions (Joseph et al. 1999; Zhou et al. 2016). Based on these transcriptome results, we also examined whether rmtA influences the growth of A. flavus colonies when challenged by osmotic or thermal stresses. In most cases no changes in vegetative growth were detected. Only high concentrations of NaCl resulted in a slight growth reduction compared to the wild type under the same experimental conditions, suggesting an effect of rmtA on sodium metabolism. Although some stress response genes were affected by alterations in rmtA expression, fungal colony growth was not notably changed, which suggests possible redundancies in a robust genetic system in A. flavus protecting it from environmental stresses. Interestingly, the hyperconidiation phenotype of ΔrmtA persisted in the presence of the stressors assayed, and it was only partially attenuated under osmotic stress, suggesting that even under exposure to environmental stress rmtA is still a required regulator of asexual development in A. flavus.
Li et al. (2017) reported that rmtA affects development and aflatoxin production during infection of peanuts seeds and maize kernels (Li et al. 2017). While this study did not examine whether removal of rmtA influenced fungal burden during infection, rmtA was found to regulate lipase and protease activity (Li et al. 2017). In our transcriptome analysis we investigated connections related to virulence in genes regulated by rmtA based on a study of Dolezal et al. (2013), which identified DEGs during A. flavus infection of maize. Our study revealed that several DEGs encoding classes of secretory enzymes such as lipases (PlaA & PLD), proteases (Pim1 & MEP1) and several putative hydrolases (AFLA_025360, AFLA_004540, & AFLA_062930) (Hong et al. 2005; Brown et al. 2007; Zhang et al. 2014; Ciesielski et al. 2016) were rmtA-dependent. As mentioned earlier, a regulator of conidiation in aspergilli, MedA (Clutterbuck 1969), is regulated by rmtA, but it is also important in virulence, as it was shown to be required for biofilm formation and normal adhesion in A. fumigatus (Gravelat et al. 2010). Additionally, its homolog in Ustilago maydis is also necessary for full virulence in maize (Chacko and Gold 2012). Although rmtA is relevant in the colonization of oil seeds, our results indicate that this gene is dispensable for virulence in the Galleria animal model. Furthermore, elevated expression of rmtA caused an increase in mortality rate. Our transcriptome analysis indicated that genes involved in iron metabolism such as sreA (Schrettl et al. 2008), and pes1, which in A. fumigatus was shown to be necessary for full virulence in G. mellonella (Reeves et al. 2006). Both of these genes were upregulated by increased expression of rmtA and may, at least in part, contribute to the increased mortality observed in the overexpression strain.
In conclusion, we have shown that the epigenetic regulator rmtA governs the expression of over 2000 genes, affecting multiple aspects of A. flavus biology, including development and virulence in plants. It also regulates some aspects of environmental stress response and secondary metabolism, including an uncharacterized biosynthetic gene cluster that may be responsible for the production of an epidithiodiketopiperazine-like compound. It is interesting that although some of these genes have been previously characterized, the function of most rmtA-dependent genes remains unknown, constituting a new avenue to be further explored in future research. Importantly, although RmtA is well conserved in eukaryotes (Satterlee et al. 2016), the similarly is low at the N-terminal and C-terminal regions of this protein. These regions could be potentially used as a target to develop a strategy to reduce the detrimental effects of this agriculturally important fungus.
Acknowledgments
This work was supported by USDA grant 58-6435-4-015 and the Department of Biological Sciences at Northern Illinois University. The authors would like to thank Dr. William Nierman for his constructive suggestions.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.9252530.
Communicating editor: A. Rokas
Literature Cited
- Balázs A., Pócsi I., Hamari Z., Leiter É., Emri T. et al. , 2010. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genomics 283: 289–303. 10.1007/s00438-010-0513-z [DOI] [PubMed] [Google Scholar]
- Basenko E., Pulman J., Shanmugasundram A., Harb O., Crouch K. et al. , 2018. FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J. Fungi (Basel) 4: 39 10.3390/jof4010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford M. T., and Clarke S. G., 2009. Protein arginine methylation in mammals: who, what and why. Mol. Cell 33: 1–13. 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. A., Henage L. G., Preininger A. M., Xiang Y., and Exton J. H., 2007. Biochemical analysis of phospholipase D. Methods Enzymol. 434: 49–87. 10.1016/S0076-6879(07)34004-4 [DOI] [PubMed] [Google Scholar]
- Bultman K. M., Kowalski C. H., and Cramer R. A., 2016. Aspergillus fumigatus virulence through the lens of transcription factors. Med. Mycol. 55: 24–38. 10.1093/mmy/myw120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A. M., and Cary J. W.. 2015. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol 6: 62 10.3389/fmicb.2015.00062. eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A. M., Wilson R. A., Bok J. W., and Keller N. P., 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66: 447–459. 10.1128/MMBR.66.3.447-459.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Harris-Coward P. Y., Ehrlich K. C., Mavungu J. D., Malysheva S. V. et al. , 2014. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet. Biol. 64: 25–35. 10.1016/j.fgb.2014.01.001 [DOI] [PubMed] [Google Scholar]
- CDC , 2004. (Center for Disease Control and Prevention), Outbreak of aflatoxin poisoning-eastern and central province, Kenya, January-July, 2004. MMWR 53: 790–792. [PubMed] [Google Scholar]
- Chang P., Scharfenstein L. L., Li R. W., Arroyo-Manzanares N., Saeger S. D. et al. , 2017. Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet. Biol. 104: 29–37. 10.1016/j.fgb.2017.04.006 [DOI] [PubMed] [Google Scholar]
- Chacko N., and Gold S., 2012. Deletion of the Ustilago maydis ortholog of the Aspergillus sporulation regulator medA affects mating and virulence through pheromone response. Fungal Genet. Biol. 49: 426–432. 10.1016/j.fgb.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Chankhamjon P., Boettger-Schmidt D., Scherlach K., Urbansky B., Lackner G. et al. , 2014. Biosynthesis of the halogenated mycotoxin aspirochlorine in koji mold involves a cryptic amino acid conversion. Angew. Chem. Int. Ed. 53: 13409–13413. 10.1002/anie.201407624 [DOI] [PubMed] [Google Scholar]
- Ciesielski S. J., Schilke B., Marszalek J., and Craig E. A., 2016. Protection of scaffold protein Isu from degradation by the Lon protease Pim1 as a component of Fe–S cluster biogenesis regulation. Mol. Biol. Cell 27: 1060–1068. 10.1091/mbc.E15-12-0815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck A. J., 1969. A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63: 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. K., O’Keeffe G., Jones G. W., and Doyle S., 2015. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 23: 419–428. 10.1016/j.tim.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Dolezal A. L., Obrian G. R., Nielsen D. M., Woloshuk C. P., Boston R. S. et al. , 2013. Localization, morphology and transcriptional profile of Aspergillus flavus during seed colonization. Mol. Plant Pathol. 14: 898–909. 10.1111/mpp.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich K., and Mack B., 2014. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins (Basel) 6: 1916–1928. 10.3390/toxins6061916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S., and Friedman L., 1967. Experimental study of the pathogenicity of Aspergilli for mice. J. Bacteriol. 94: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. B., O’Brien E., Khoury J. B., and Mylonakis E., 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1: 475–482. 10.4161/viru.1.6.12985 [DOI] [PubMed] [Google Scholar]
- Fung F., and Clark R. F., 2004. Health effects of mycotoxins: A toxicological overview. J. Toxicol. Clin. Toxicol. 42: 217–234. 10.1081/CLT-120030947 [DOI] [PubMed] [Google Scholar]
- Furukawa K., Hoshi Y., Maeda T., Nakajima T., and Abe K., 2005. Aspergillus nidulans HOG pathway is activated only by two-component signaling pathway in response to osmotic stress. Mol. Microbiol. 56: 1246–1261. 10.1111/j.1365-2958.2005.04605.x [DOI] [PubMed] [Google Scholar]
- Gai Y., Liu B., Ma H., Li L., Chen X. et al. , 2019. The methionine biosynthesis regulator AaMetR contributes to oxidative stress tolerance and virulence in Alternaria alternata. Microbiol. Res. 219: 94–109. 10.1016/j.micres.2018.11.007 [DOI] [PubMed] [Google Scholar]
- Georgianna D. R., Fedorova N. D., Burroughs J. L., Dolezal A. L., Bok J. W. et al. , 2010. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11: 213–226. 10.1111/j.1364-3703.2009.00594.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat F. N., Ejzykowicz D. E., Chiang L. Y., Chabot J. C., Urb M. et al. , 2010. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell. Microbiol. 12: 473–488. 10.1111/j.1462-5822.2009.01408.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Mizuno T., and Abe K., 2011. Characterization of the conserved phosphorylation site in the Aspergillus nidulans response regulator SrrA. Curr. Genet. 57: 223–224. 10.1007/s00294-011-0337-3 [DOI] [PubMed] [Google Scholar]
- Hagiwara D., Takahashi-Nakaguchi A., Toyotome T., Yoshimi A., Abe K. et al. , 2013. NikA/TcsC histidine kinase is involved in conidiation, hyphal morphology, and responses to osmotic stress and antifungal chemicals in Aspergillus fumigatus. PLoS One 8: e80881 10.1371/journal.pone.0080881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedayati M. T., Pasqualotto A. C., Warn P. A., Bowyer P., and Denning D. W., 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153: 1677–1692. 10.1099/mic.0.2007/007641-0 [DOI] [PubMed] [Google Scholar]
- Hong S., Horiuchi H., and Ohta A., 2005. Identification and molecular cloning of a gene encoding phospholipase A2 (plaA) from Aspergillus nidulans. Biochim. Biophys. Acta 1735: 222–229. 10.1016/j.bbalip.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Horn B. W., Sorensen R. B., Lamb M. C., Sobolev V. S., Olarte R. A. et al. , 2014. Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 104: 75–85. 10.1094/PHYTO-05-13-0129-R [DOI] [PubMed] [Google Scholar]
- Hu Y., Yang G., Zhang D., Liu Y., Li Y. et al. , 2018. The PHD transcription factor rum1 regulates morphogenesis and aflatoxin biosynthesis in Aspergillus flavus. Toxins (Basel) 10: 301 10.3390/toxins10070301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issi L., Farrer R. A., Pastor K., Landry B., Delorey T. et al. , 2016. Zinc cluster transcription factors alter virulence in Candida albicans. Genetics 205: 559–576. 10.1534/genetics.116.195024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Sekonyela R., Knox B. P., Palmer J. M., Huttenlocher A. et al. , 2018. Selenate sensitivity of a laeA mutant is restored by overexpression of the bZIP protein MetR in Aspergillus fumigatus. Fungal Genet Biol 117: 1–10. 10.1016/j.fgb.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J. D., Heitman J., and Means A. R., 1999. Molecular cloning and characterization of Aspergillus nidulans cyclophilin b. Fungal Genet. Biol. 27: 55–66. 10.1006/fgbi.1999.1131 [DOI] [PubMed] [Google Scholar]
- Kaliamurthy J., Geraldine J. P., and Thomas P. A., 2003. Disseminated aspergillosis due to Aspergillus flavus in an experimental model: efficacy of azole therapy. Mycoses 46: 174–182. 10.1046/j.1439-0507.2003.00865.x [DOI] [PubMed] [Google Scholar]
- Kamai Y., Harasaki T., Fukuoka T., Ohya S., Uchida K. et al. , 2002. In vitro and in vivo activities of CS-758 (R-120758), a new triazole antifungal agent. Antimicrob. Agents Chemother. 46: 367–370. 10.1128/AAC.46.2.367-370.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L., 2015. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausmeyer P., Mccloud T. G., Tucker K. D., Cardellina J. H., and Shoemaker R. H., 2005. Aspirochlorine class compounds from Aspergillus flavus inhibit azole-resistant Candida albicans. J. Nat. Prod. 68: 1300–1302. 10.1021/np050141k [DOI] [PubMed] [Google Scholar]
- Lancaster M. D., Jenkins F. P., and Phillip J. M., 1961. Toxicity associated with certain samples of groundnuts. Nature 192: 1095–1096. 10.1038/1921095a0 [DOI] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebar M. D., Cary J. W., Majumdar R., Carter-Wientjes C. H., Mack B. M. et al. , 2018. Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus. Fungal Genet. Biol. 116: 14–23. 10.1016/j.fgb.2018.04.009 [DOI] [PubMed] [Google Scholar]
- Lewis L., Onsong M., Njapau H., Schurz-Rogers H., Luber G. et al. , 2005. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicosis in Eastern and Central Kenya. Environ Health Perspect 113: 1763–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., He Y., Li X., Fasoyin O. E., Hu Y. et al. , 2017. Histone Methyltransferase aflrmtA gene is involved in the morphogenesis, mycotoxin biosynthesis, and pathogenicity of Aspergillus flavus. Toxicon 127: 112–121. Erratum: 134: 64. 10.1016/j.toxicon.2017.01.013 [DOI] [PubMed] [Google Scholar]
- Malavazi I., Savoldi M., Mauro S. M., Menck C. F., Harris S. D. et al. , 2006. Transcriptome analysis of Aspergillus nidulans exposed to camptothecin-induced dna damage. Eukaryot. Cell 5: 1688–1704. 10.1128/EC.00167-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese S., Polo A., Ariano A., Velotto S., Costantini S. et al. , 2018. Aflatoxin B1 and M1: biological properties and their involvement in cancer development. Toxins (Basel) 10: 214 10.3390/toxins10060214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masanga J. O., Matheka J. M., Omer R. A., Ommeh S. C., Monda E. O. et al. , 2015. Downregulation of transcription factor aflR in Aspergillus flavus confers reduction to aflatoxin accumulation in transgenic maize with alteration of host plant architecture. Plant Cell Rep. 34: 1379–1387. 10.1007/s00299-015-1794-9 [DOI] [PubMed] [Google Scholar]
- Miskei M., Karányi Z., and Pócsi I., 2009. Annotation of stress–response proteins in the Aspergilli. Fungal Genet. Biol. 46: S105–S120. 10.1016/j.fgb.2008.07.013 [DOI] [PubMed] [Google Scholar]
- Mitchell N. J., Bowers E., Hurburgh C., and Wu F., 2016. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit Contam: Part A 33: 540–550. 10.1080/19440049.2016.1138545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera J., Warn P. A., Morrissey J., Moore C. B., Gil-Lamaignere C. et al. , 2001. Susceptibility testing of Aspergillus flavus: inoculum dependence with itraconazole and lack of correlation between susceptibility to amphotericin b in vitro and outcome in vivo. Antimicrob. Agents Chemother. 45: 1456–1462. 10.1128/AAC.45.5.1456-1462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojiambo P. S., Battilani P., Cary J. W., Blum B. H., and Carbone I., 2018. Cultural and genetic approaches to manage aflatoxin contamination: recent insights provide opportunities for improved control. Phytopathology 108: 1024–1037. 10.1094/PHYTO-04-18-0134-RVW [DOI] [PubMed] [Google Scholar]
- Park J., Park J., Jang S., Kim S., Kong S. et al. , 2008. FTFD: An informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24: 1024–1025. 10.1093/bioinformatics/btn058 [DOI] [PubMed] [Google Scholar]
- R Core Team , 2017. R: a language and environment for statistical computing. https://www.Rproject.org/
- Ramírez M. A., and Lorenz M. C., 2009. The Transcription Factor Homolog CTF1 Regulates β-oxidation in Candida albicans. Eukaryot. Cell 8: 1604–1614. 10.1128/EC.00206-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves E. P., Reiber K., Neville C., Scheibner O., Kavanagh K. et al. , 2006. A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus. FEBS J. 273: 3038–3053. 10.1111/j.1742-4658.2006.05315.x [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán C., Pareja-Jaime Y., González-Reyes J. A., and Roncero M. I., 2015. The transcription factor Con7-1 Is a master regulator of morphogenesis and virulence in Fusarium oxysporum. Mol. Plant Microbe Interact. 28: 55–68. 10.1094/MPMI-07-14-0205-R [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Arima T., Iwashita K., Yamada O., Gomi K. et al. , 2008. Aspergillus oryzae atfB encodes a transcription factor required for stress tolerance in conidia. Fungal Genet. Biol. 45: 922–932. 10.1016/j.fgb.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Sarma U. P., Bhetaria P. J., Devi P., and Varma A., 2017. Aflatoxins: implications on health. Indian J. Clin. Biochem. 32: 124–133. 10.1007/s12291-017-0649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee T., Cary J. W., and Calvo A. M., 2016. RmtA, a Putative Arginine Methyltransferase, Regulates secondary metabolism and development in Aspergillus flavus. PLoS One 11: e0155575 10.1371/journal.pone.0155575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl V., Seiboth B., Karaffa L., and Kubicek C. P., 2004. The fungal STRE-element-binding protein Seb1 is involved but not essential for glycerol dehydrogenase (gld1) gene expression and glycerol accumulation in Trichoderma atroviride during osmotic stress. Fungal Genet. Biol. 41: 1132–1140. 10.1016/j.fgb.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C. et al. , 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70: 27–43. 10.1111/j.1365-2958.2008.06376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C., Edgerton H., Xiong Y. et al. , 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1: 3111–3120. Erratum in: 1: 31120. 10.1038/nprot.2006.405 [DOI] [PubMed] [Google Scholar]
- Tessarz P., and Kouzarides T., 2014. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 15: 703–708. 10.1038/nrm3890 [DOI] [PubMed] [Google Scholar]
- Trojer P., Dangl M., Bauer I., Graessle S., Loidl P. et al. , 2004. Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity. Biochemistry 43: 10834–10843. 10.1021/bi049626i [DOI] [PubMed] [Google Scholar]
- Tsai H., Chang Y. C., Washburn R. G., Wheeler M. H., and Kwon-Chung K. J., 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180: 3031–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2009. 2009 ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag, New York. [Google Scholar]
- Woloshuk C. P., Foutz K. R., Brewer J. F., Bhatnagar D., Cleveland T. E. et al. , 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60: 2408–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang Y., Chi W., Shi Y., Chen S. et al. , 2014. Metalloprotease genes of Trichophyton mentagrophytes are important for pathogenicity. Med. Mycol. 52: 36–45. 10.3109/13693786.2013.811552 [DOI] [PubMed] [Google Scholar]
- Zhou B., Xie J., Liu X., Wang B., and Pan L., 2016. Functional and transcriptomic analysis of the key unfolded protein response transcription factor HacA in Aspergillus oryzae. Gene 593: 143–153. 10.1016/j.gene.2016.08.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table S1 contains calculated expression values comparing those of the wild-type strain to those of the deletion rmtA mutant and overexpression strain. Tables S2–S4 contain a subsets of Table S1 that includes DEGs corresponding to transcription factors, genes related to environmental stress, and genes that were shown to be DEG during maize infection. Figure S1 shows GO terms of DEG that are rmtA-dependent. Figure S2 details the strategy for the construction of the gliP mutant strain. Figures S3 and S4 show the rmtA-dependent effect of osmotic stress and temperature on A. flavus. The sequencing data are publicly available at NCBI’s SRA repository with the SRA Accession #: PRJNA573552. Supplemental material available at figshare: https://doi.org/10.25387/g3.9252530.